Abstract
G protein–coupled receptors (GPCRs) mediate the majority of cellular responses to external stimuli. Upon activation by a ligand, the receptor binds to a partner heterotrimeric G protein and promotes exchange of GTP for GDP, leading to dissociation of the G protein into α and βγ subunits that mediate downstream signals. GPCRs can also activate distinct signaling pathways through arrestins. Active states of GPCRs form by small rearrangements of the ligand-binding, or orthosteric, site that are amplified into larger conformational changes. Molecular understanding of the allosteric coupling between ligand binding and G protein or arrestin interaction is emerging from structures of several GPCRs crystallized in inactive and active states, spectroscopic data, and computer simulations. The coupling is loose, rather than concerted, and agonist binding does not fully stabilize the receptor in an active conformation. Distinct intermediates whose populations are shifted by ligands of different efficacies underlie the complex pharmacology of GPCRs.
Keywords: 7-TM receptors, G protein–coupled receptor, GPCR, β2-adrenergic receptor, allostery, energy landscape
INTRODUCTION: GPCRs AS ALLOSTERIC PROTEINS
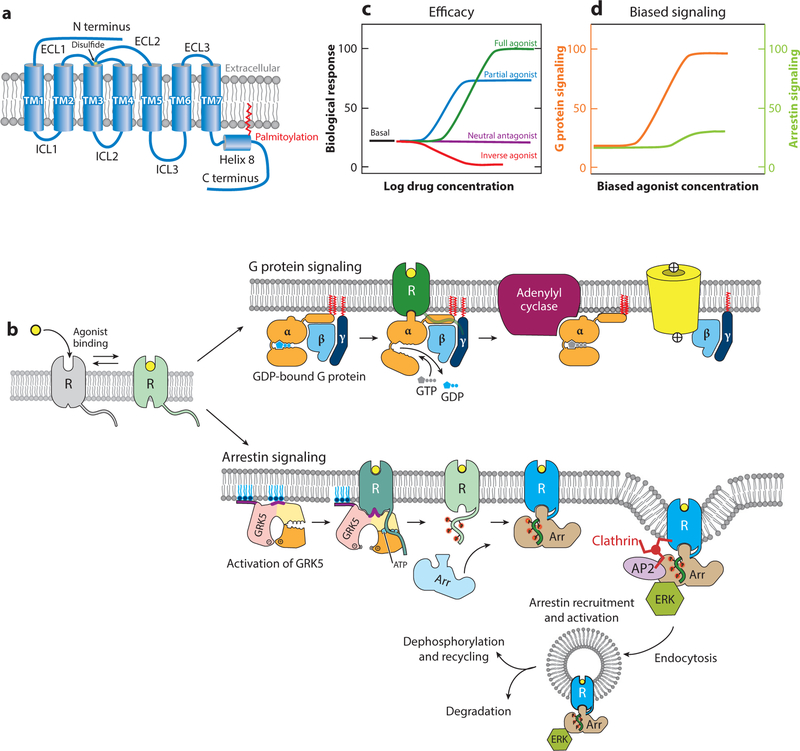
G protein–coupled receptors (GPCRs) mediate the majority of cellular responses to external stimuli, including light, odors, hormones, and growth factors. GPCRs are integral membrane proteins that contain seven transmembrane (TM) α-helices (Figure 1a). Activating ligands, or agonists, stabilize a GPCR conformation that can interact with a heterotrimeric G protein to promote exchange of GTP for GDP from the Gα subunit. GTP-bound Gα dissociates from Gβγ, and Gα and Gβγ separately mediate downstream signaling activities (Figure 1b). In addition to signaling through G proteins, GPCRs can signal through arrestins (Figure 1b). Arrestins were first described as proteins that turn off G protein signaling: Phosphorylation of the receptor C-terminal tail by a G protein–linked kinase leads to recruitment of arrestin, which prevents interaction with G proteins and promotes receptor internalization. However, certain GPCR ligands can activate arrestin binding directly or possibly by promoting interaction with kinases that phosphorylate the receptor to enable arrestin binding, thereby activating downstream signaling pathways distinct from those mediated by G proteins.
Figure 1.
(a) General architecture of a family A G protein–coupled receptor (GPCR). The seven transmembrane (TM) helices, the connecting intracellular loops (ICLs) and extracellular loops (ECLs), and a conserved disulfide bond are indicated. (b) Outline of GPCR activity upon binding of an agonist to a receptor (R). Top, the classical G protein pathway. Exchange of GDP for GTP in the G protein α subunit leads to dissociation and interaction with downstream effectors such as Gαs stimulation of adenylyl cyclase and Gβγ activation of ion channels. Bottom, activated GPCRs can also signal through arrestins. Phosphorylation of the receptor C-terminal tail by a G protein–coupled receptor kinase (GRK) promotes arrestin (Arr) recruitment and activation, including endocytosis through interactions with the clathrin adaptor protein 2 (AP2) complex and activation of extracellular signal-regulated kinase (ERK). (c) Efficacy of ligands. (d) Biased agonists stimulate one pathway preferentially over another. Figure modified with permission from Reference 72.
In the simplest scheme, agonist binding stabilizes a receptor in an active conformation that interacts with its cytoplasmic partner (Figure 1b). However, GPCRs are not simple on/off switches: Most exhibit a basal level of GTP exchange activity in the absence of a ligand, indicating that there is an equilibrium population of the receptor in an active conformation. The notion of a preexisting equilibrium between inactive and active conformations is essential for understanding the basic pharmacology of GPCR ligands. The efficacy of a ligand describes the extent to which it can activate a particular signaling pathway (Figure 1c). Agonists promote exchange activity above the basal level and therefore favor an active conformation. Inverse agonists lower activity below that of the unliganded, basal state and thus favor an inactive conformation. Neutral antagonists do not affect the basal activity (i.e., the inactive–active equilibrium) but compete with inverse agonists or agonists for the same ligand-binding site. Partial agonists produce weaker maximal activity at saturation than do full agonists. Some ligands can stimulate both G protein and arrestin pathways, or multiple G proteins, and the greater efficacy toward one or the other is known as ligand bias (Figure 1d). The ability of some ligands to stimulate both pathways may be responsible for many of the undesired effects of drugs targeted to GPCRs.
GPCRs are classic allosteric proteins: Agonist binding at one site that is accessed from the extracellular space, termed the orthosteric site, promotes binding to another partner (e.g., a heterotrimeric G protein) at the cytoplasmic side. The coupled equilibrium of agonist and G protein binding is well known: Not only does agonist binding promote binding of the receptor to a G protein, but the affinity of the agonist for the receptor is increased by the G protein (1, 2).
Conformational selection is widely believed to underlie allosteric behavior (3, 4). All molecules exist in a thermal equilibrium in which they sample multiple conformations; some states may have relatively high free energies and therefore be sparsely populated. Ligands bind to a subset of conformations, which results in a population shift that establishes a new equilibrium. For example, an agonist shifts the conformational distribution such that more molecules are able to bind the partner G protein, whereas an inverse agonist depopulates such states. The relative efficacies of ligands reflect the free energy differences of their complexes with receptors, which alters the distribution of functional states.
Functionally distinct states of proteins exhibiting allosteric behaviors are typically related by conformational differences that can involve relative motions of whole domains, subdomains, secondary structure elements, and/or loops and are ultimately mediated by alterations in hydrogen bonding and side chain packing. These states can also differ in the extent of motion (fluctuations) around these local conformations, which contributes an entropic component to their free energy differences. Conformational changes occur on timescales that depend on the energy barrier between states and range from nanoseconds (e.g., side chain rotamer changes) to milliseconds or longer (e.g., whole domain movements) (5). Molecular structures obtained from X-ray crystallography or electron microscopy are conformational snapshots in a local energy minimum.
Approximately 800 GPCRs are encoded in the human genome (6). The majority belong to the so-called family A or rhodopsin-like receptors, which is the most well-studied group. Structural and spectroscopic data indicate that conserved structural elements confer basic similarities in the functional conformations of family A GPCRs. Sequence differences tune the relative energies of these conformations and the barriers between them, which gives rise to differences in basal activity and in the strength of coupling between ligand and G protein or arrestin binding. Recent structural data suggest that many of the general principles uncovered in family A receptors apply to other GPCR families.
In this review, we describe the molecular basis of GPCR activation from the perspective of conformational selection, focusing on family A GPCRs that bind diffusible ligands, in particular the β2-adrenergic receptor (β2AR), which has been studied extensively by a range of biophysical and structural methods. The ongoing challenge is to define the GPCR conformational states structurally, the dynamics of the conformational ensemble including how the different binding partners shift populations, and the relationship of these states to biochemical activity. The combined approaches of X-ray crystallography, NMR, fluorescence and electron paramagnetic resonance spectroscopies, and molecular dynamics (MD) simulations have made significant progress toward these goals.
To facilitate comparisons of different receptors, we employ the Ballesteros–Weinstein nomenclature (7) for family A GPCRs: The most conserved residue of each TM helix is assigned position 50, and other residues within the helix are numbered relative to this position. A particular residue is then indicated with a superscript x.yy, where x is the TM helix number and yy is the position in the helix. For example, Trp286 in the β2AR lies two residues N terminal to Pro2886.50, the most conserved position in TM6, and is designated Trp2866.48. Residues on extracellular or in-tracellular loops are denoted with a superscript ECLx/ICLx, where E or I indicates extracellular or intracellular and x is the loop. For example, a residue in the intracellular loop connecting TM1 and TM2 would have the superscript ICL1 (Figure 1a).
THREE-DIMENSIONAL STRUCTURES OF GPCR FUNCTIONAL STATES
Molecular understanding of GPCR activation was founded upon crystal structures in both inactive and active states. To date, crystal structures of approximately 40 different GPCRs are available in at least one functional state (see http://gpcrdb.org). Most are from family A, but at least one from each of the other families is known. The first crystal structure of a GPCR was that of dark rhodopsin, the receptor for light (8). Rhodopsin is an unusual GPCR because visual physiology requires that it has essentially no basal activity. Its covalently bound ligand, 11-cis retinal, undergoes a photoisomerization that activates the receptor, and the large energy barrier to retinal isomerization in the absence of light maintains the inactive structure. In contrast, most other GPCRs are activated by diffusible ligands, and the energy difference between ligand-free and agonist-bound states is smaller than in rhodopsin, which enables many ligand-free receptors to sample the activated conformation and therefore display basal activity.
The conformational flexibility of GPCRs, as well as the unique challenges posed by membrane proteins, has required several approaches to their crystallization. First, high affinity, low off-rate ligands are typically used, because once the ligand dissociates conformational equilibrium reduces the fraction of molecules in the desired state. Second, GPCRs are not particularly stable in detergents used for crystallization. Two principal approaches have been used to overcome this problem. Crystallization in lipids, using lipid bicelles (9, 10) and more commonly with lipidic mesophases (11), has been employed for the majority of GPCR crystal structures to date. An alternative approach, pioneered by Tate and colleagues (12), is to introduce thermostabilizing mutations, which has enabled crystallization in detergents. Third, membrane protein crystals typically form through contacts between the extramembranous (water soluble) portions of the molecule. Many GPCRs have only small extramembranous regions, and these elements are often flexible. To overcome this problem, particularly in lipid-based crystallization, chimeric proteins in which a small, rigid protein is fused to the N terminus (13) or inserted into an intracellular loop (typically ICL3) (14, 15) have been employed, and flexible termini are truncated. The inserted protein mediates contacts with other molecules in the crystal lattice. These modified proteins display similar ligand-binding properties to those of the parent protein, indicating that conformations observed in crystal structures are functionally relevant. Indeed, in cases where the same receptor–ligand complex was crystallized with or without the fusion, no major differences in conformation were observed (e.g., 13, 16–18). Spectroscopic studies, however, have shown that the presence of a fusion protein in ICL3 can alter conformational equilibria (19).
Inactive State Structures
The inactive state is the more stable conformation of GPCRs, and the first structures were of this state. Dark rhodopsin, purified directly from cow retinas, was crystallized in detergent (8). This is considered the reference inactive structure as it displays no basal activity. The first structure of GPCR bound to a diffusible ligand was β2AR, which was crystallized bound to a high-affinity inverse agonist, carazolol, in lipid bicelles bound to a Fab fragment and in lipidic cubic phase as a chimeric protein in which T4 lysozyme (T4L) replaced most of ICL3 (10, 14, 16). Although carazolol is classified as an inverse agonist, it reduces, but does not completely abolish, basal activity (10, 14, 16), making the differences between carazolol-bound β2AR and dark rhodopsin informative.
Both of these structures revealed the basic architecture of GPCRs (Figure 2a). Each contains the expected seven TM helices, followed by a short helix (H8) that lies parallel to the cytoplasmic surface of the membrane. In most family A receptors, a conserved disulfide bond stabilizes extra-cellular loop 2 (ECL2). The orthosteric ligand–binding site is a pocket in the extracellular side of the receptor TM region formed principally by TM3, TM5, TM6, and TM7. Residues from the extracellular loops also contact the bound ligand. The extracellular loops adopt different structures in different GPCRs, and to varying degrees they can occlude the orthosteric ligand–binding site from the extracellular milieu and thereby affect the kinetics of ligand binding (20, 21).
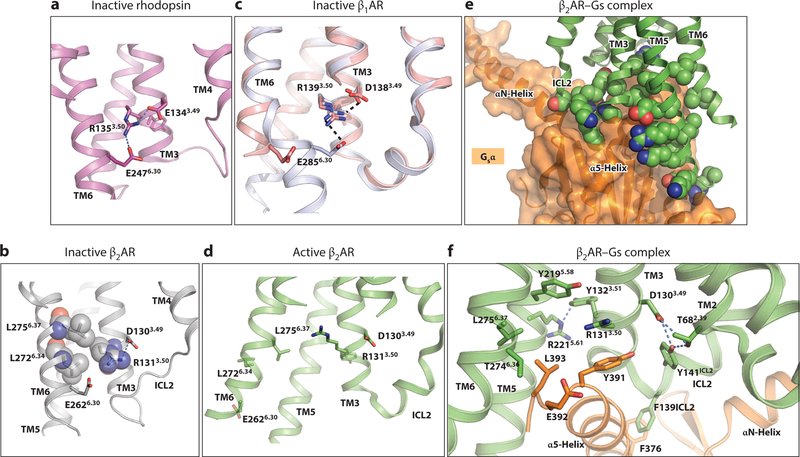
Figure 2.
Overall GPCR structures. (a) Inactive state of β2AR, bound to carazolol (PDB 2RH1). (b) Active state of β2AR bound to adrenaline (left and center, PDB 3SN6, with adrenaline coordinates from PDB 4LDO; right, PDB 4LDO). Ligands are shown in space-filling representation, with carbon atoms of carazolol in purple and of epinephrine in yellow. The conserved prolines P2115.50, P2886.50, and P3237.50 are shown as sticks. The center panels show the receptor with its extracellular face up and cytoplasmic face down. The left and right panels show views from the intracellular and extracellular surfaces, respectively. The labeling sites used for NMR probes (C265) and DEER and FRET probes (N148C and L266C) are shown in the left panel, and the opening of the G protein pocket upon receptor activation is indicated by the change in distance between the α carbons at N148 and L266. Abbreviations: β2AR, β2-adrenergic receptor; DEER, double electron-electron resonance; FRET, fluorescence resonance energy transfer; GPCR, G protein–coupled receptor; PDB, Protein Data Bank.
A key feature of GPCR architecture is the presence of kinks in TM5, TM6, and TM7 caused by interruptions in the intrahelical hydrogen bonding at conserved proline residues (β2AR residues P2115.50, P2886.50, and P3237.50). Kinks and helical bulges are found at nonconserved proline or glycine residues in other helices as well. These interruptions are essential for coupling ligand binding to global changes in conformation needed to achieve the active state, as they lower the energy barrier to main-chain conformational changes associated with the formation of different functional states. In contrast, TM3 is straight and forms the core of the protein, where it interacts with many neighboring TMs (22).
Active State Crystal Structures
For most GPCRs, the active state is not sufficiently stable to crystallize even in the presence of a high-affinity or covalently bound agonist (23); the G protein is needed to stabilize a state of lower free energy—in other words, to shift the equilibrium to make the active conformation the dominant one in solution. In only one case, the β2AR, has the complex with a partner heterotrimeric G protein, Gs, been solved by crystallography, and we use this as a reference active state structure. This structure served as a basis to engineer a mini-Gs comprising the Ras-like domain of the Gsα subunit without the N-terminal α-helix or the α-helical subdomain, which was then complexed with agonist-bound adenosine 2a receptor (A2aR) (24). Rhodopsin is unusual in that its active state is sufficiently stable and it does not require stabilization by its cognate Gα protein, transducin: The active state can be achieved using retinal-free opsin and low pH, and the structures of this state, as well as a complex with the C-terminal helix of transducin, have been obtained (25–27).
Active state crystal structures of several GPCRs have been obtained using nanobodies, the variable portion of single-chain camelid antibodies (28). These reagents serve two roles. First, they can bind to the extramembranous receptor loops and thereby provide more surface area for lattice contacts. Second, and more importantly, they can stabilize particular states of a receptor, most notably mimicking a G protein by binding to the G protein–binding site and enhancing the affinity of agonists to the same degree as a G protein (29). Nanobodies are generated by immunizing animals with a receptor or receptor complex bound to a high-affinity ligand. A phage display library derived from the B cells is used to select nanobodies that bind preferentially to the agonist-bound receptor (30). After expression of selected nanobodies in bacteria, they are tested for their ability to increase agonist-binding affinity equivalently to G protein and thereby create a pharmacological mimic of the active state. A related approach employs yeast display, in which a framework nanobody with randomized complementarity determining regions is displayed on the surface of yeast cells, which are incubated with fluorescently labeled receptor protein and subjected to fluorescence-activated cell sorting to identify clones expressing nanobodies with the desired properties. Yeast display has been used to optimize initial animal-derived nanobodies for higher affinity (31–33). Nanobodies have been used to crystallize active states of the β2AR (29, 31), the μ-opioid receptor (34), the M2 muscarinic receptor (32), and a virally encoded GPCR related to family A receptors, US28 (33).
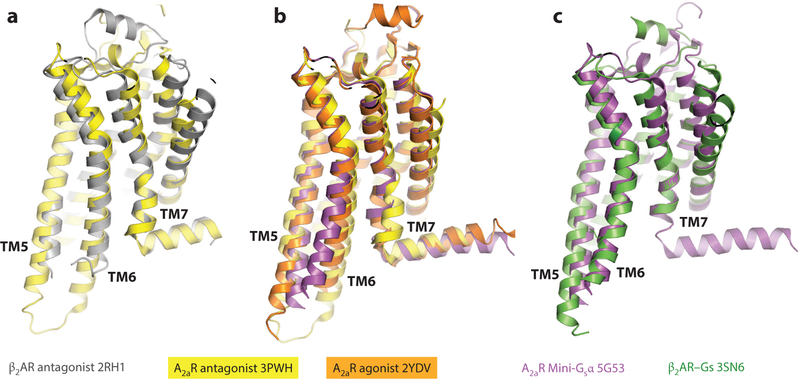
Figure 2 compares the inactive (inverse agonist–bound) and active (agonist- and Gs-bound) structures of the β2AR. The largest difference occurs near the cytoplasmic side of the receptor, which opens to create a binding pocket for the C-terminal helix of a cognate G protein α subunit. This pocket, lined by residues from TM3, TM5, and TM6, forms by changes in the positions of TM5, TM6, and TM7 relative to TM3. The largest change occurs in TM6, which moves outward from the central TM3 by 14Å, measured at the Cα of Glu2686.30. Residues in TM3 and TM5 that interact with TM6 in the inactive state form direct contacts with the G protein in the active state. Similar results were found for activated opsin bound to the C-terminal peptide of the transducin α subunit (26, 27) and the A2aR–mini-Gsα complex (24). The global changes observed in these structures were also seen in nanobody-stabilized active states of the muscarinic M2 and μ-opioid receptors (32, 34).
Recently, the structures of the calcitonin receptor and the Glp-1R receptor, both class B GPCRs bound to the Gs heterotrimer, were determined by cryo–electron microscopy (35, 36). The changes in architecture described for class A receptors, notably the outward movement of TM6 and the inward movement of TM7, are similar in the calcitonin and Glp-1R receptors, although there are notable differences in the extramembranous helix 8. Thus, despite significant differences in sequence, the overall architecture of GPCRs and their changes upon activation appear to be preserved throughout the GPCR superfamily.
ROLE OF CONSERVED SEQUENCE/STRUCTURAL MOTIFS IN COUPLING AGONIST AND G PROTEIN BINDING
The key mechanistic problem in GPCR signaling is how agonists favor the G protein–interacting conformation. Comparison of five GPCR structures solved in both inactive and active states reveals a diversity of residue contact changes throughout the receptor (37). Nonetheless, there are common features in the coupling between orthosteric and G protein–binding sites (37), and we describe the changes in the β2AR as an example. The transitions between different functional states involve sequence motifs strongly conserved in family A GPCRs.
The G Protein–Binding Site
Near the cytoplasmic end of TM3, the sequence D(E)3.49-R3.50-Y3.51 lies adjacent to TM5 and TM6 (Figure 3). In inactive GPCR structures, the position of R3.50 is fixed by an intrahelical salt bridge with D(E)3.49, and the aliphatic portion of R3.50 packs against L6.34 and L6.37 of TM6 (Figure 3a,b). Of interest, the mutation L2726.34A in the β2AR leads to increased basal activity and biochemical instability, consistent with the role of L2726.34 in stabilizing the inactive state (38). In dark rhodopsin, a salt bridge, termed the ionic lock, is made between R3.50 and E6.30 (Figure 3a). Notably, this salt bridge is absent in the inactive, carazolol-bound β2AR (Figure 3b), where TM6 is moved outward and E2686.30 points to the cytoplasmic space. In some crystal structures of the related β1AR (thermostabilized) bound to inverse agonists, Arg3.50 interacts with Glu6.30 (although the two side chains are too far apart to form a formal salt bridge), and in others, TM6 straightens and further separates these two residues (39) (Figure 3c). Long timescale MD simulations of the β2AR indicate that the ionic lock may form only transiently (40, 41). Collectively, these observations suggest that the salt bridge stabilizes the inactive state of family A GPCRs but forms transiently in receptors that display basal activity. Consistent with this notion, the mutations D1303.49A or E2686.30A in β2AR increase basal activity but do not change agonist-induced activity (42, 43). In contrast, the β2AR R1313.50A mutation does not alter basal or agonist-induced activity. As described below, R1313.50 forms part of the G protein–binding pocket, so its roles in both states may negate the effect of this mutation (43). The importance of the salt bridge in stabilizing the inactive state is further illustrated by the virally encoded US28 chemokine receptor, a constitutively active GPCR, in which E3.46 interacts with an arginine in ICL2, preventing the latter from stabilizing the inactive DRY conformation (33).
Figure 3.
The conserved D(E)RY motif in the G protein–binding site. (a) The ionic lock in dark rhodopsin formed by R1353.50 and E2476.30. (b) The ionic lock is not present in the crystal structure of carazolol-bound β2AR. The packing of R3.50 with L2726.34 and L2756.37 is shown with space-filling models of these side chains. (c) Two conformations of the ionic lock region are observed in inverse-agonist bound structures of the β1AR. Bending near the cytoplasmic end of TM6 results in an electrostatic interaction between R1393.50 and E2856.30 ( gray; PDB 2YCX). The alternative straight conformation of TM6 (salmon; PDB 2VT4) moves these two residues apart. (d) Movements of TM6 and TM7 in the β2AR active state prevent ionic lock formation, and the intrahelical salt bridge between D1303.49 and R1313.50 is broken. (e) Packing of β2AR ( green, with side chains in space-filling representation) and Gsα (orange, shown as a transparent surface). ( f) Interactions of β2AR with the C-terminal region of bound Gsα. Gsα is shown in orange. Polar interactions are shown with dashed lines. Abbreviations: β2AR, β2-adrenergic receptor; G sα, Gsβ, stimulatory heterotrimeric G protein α and β subunits; PDB, Protein Data Bank; TM, transmembrane.
The D(E)-R3.50-Y motif has critical roles in forming the G protein–binding site, as it interacts directly with the bound Gsα and also stabilizes the binding pocket (Figure 3d–f). In the active structure of β2AR, the portions of TM3 and TM5 on the intracellular side of the receptor form extensive contacts with Gsα-helix 5, and TM6 interacts with the nonhelical C-terminal loop of Gsα (Figure 3e). In Gs-bound β2AR, R1313.50 extends into the Gsα-binding pocket and packs against Y391 of Gsα and Y2195.58 (Figure 3f). In active rhodopsin and the μ-opioid receptor structures, R3.50 also forms a hydrogen bond with the Y5.58 phenolic hydroxyl group. The interactions of Y5.58 with R3.50 appear to be critical for forming the active state, as the β2AR Y2195.58A mutant displays no constitutive activity and does not activate Gs (43).
The conserved D3.49 and Y3.51 residues of the DRY motif stabilize the active state conformation but do not interact directly with Gs. D1303.49 hydrogen bonds to Y141ICL2, and both D1303.49 and Y141ICL2 form hydrogen bonds with T68 of TM2, which in the inactive structure is linked to R1313.50. The interaction of D(E)3.49 with ICL2 as well as the end of TM2 is seen in several other active structures (32, 34). These interactions stabilize a helical conformation of ICL2, which positions F139ICL2 to pack against conserved residues in Gsα (Figure 3f). Y1323.51 maintains packing with TM5 but in addition forms a hydrogen bond with R2215.61 of TM5 (Figure 3f). The changes in the DRY motif also indirectly enable other contacts with the bound G protein. For example, L2756.37, which packs against R3.50 in the inactive structure, now packs against Gs L393, a highly conserved residue in Gα proteins. Gsα residues L393 and E392 (not conserved) also pack against β2AR T2746.36 (Figure 3f).
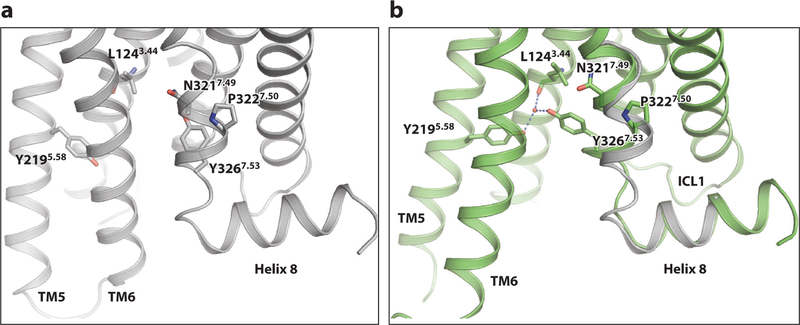
A second conserved motif, NP7.50xxY in TM7, also participates in forming the G protein–binding site. This motif does not interact directly with the bound G protein but is essential for forming the active conformation. Owing to the break in hydrogen bonding, TM7 can rotate at P3237.50 (Figure 4). This moves Y3267.53 toward the position that was occupied by TM6 in the inactive structure, where it packs against L1243.44 and I1273.47 and forms a water-mediated hydrogen bond with Y2195.58. Also, the last turn of the helix in TM7 in the inactive structure unravels as part of this transition.
Figure 4.
The NP7.50xxY motif. (a) The NPxxY motif in the inactive state of β2AR. (b) Superposition of the NPxxY region in the inactive (gray; PDB 2RH1) and active ( green; PDB 3SN6) β2AR structures. Abbreviations: β2AR, β2-adrenergic receptor; PDB, Protein Data Bank.
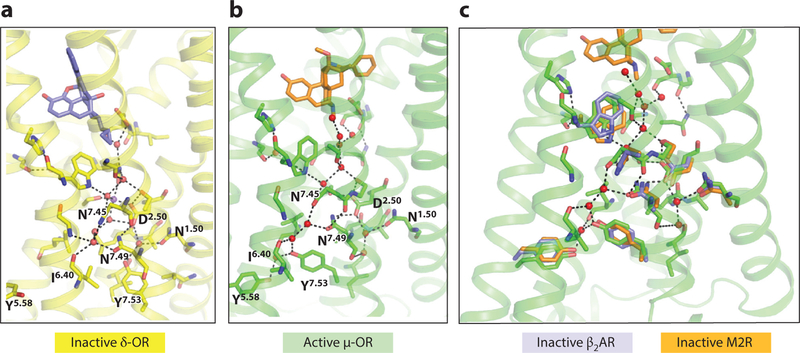
Water-mediated networks that connect side chains of conserved polar residues as well as the backbone, most prominently on the cytoplasmic halves of TM2, TM3, TM6, and TM7, have been observed in a number of GPCR crystal structures (34) (Figure 5). These networks rearrange in the transition between the inactive and active states (Figure 5a,b). For example, in the inactive state, water molecules link the conserved N7.49 in TM7 to N1.50 in TM1, D2.50 in TM2, I6.40 in TM6, and N7.45 and Y7.53 in TM7 (Figure 5). In the active conformation, N7.49 repositions and directly hydrogen bonds to D2.50, and Y7.53 is now linked to Y5.58 and the backbone at L3.43, thereby stabilizing the positions of the TM3 and TM5 residues for Gα binding. Closer to the orthosteric site and connector that links the orthosteric and G protein sites (see below), N3.35 in the inactive state forms a network of polar interactions that link it to D2.50, G7.22, N7.45, S7.46, and W6.48; in the active state, the movement of TM6 and TM7 breaks several of these interactions. Although the resolution of many GPCR crystal structures does not allow visualization of water molecules, the conservation of polar residues in the interior of the protein strongly suggests that these water-mediated networks are common to family A GPCRs and must rearrange as part of forming the active state. The barrier to rearranging hydrogen-bonded water networks is likely lower than that involving extensive repacking of hydrophobic groups.
Figure 5.
Water networks linking TM2, TM5, TM6, and TM7. Polar side chains conserved in the family A receptors are shown in stick representation and are labeled with their Ballesteros–Weinstein numbers. (a,b) Comparison of the inactive δ-OR (PDB 4N6H) and active μ-OR (PDB 5C1M) structures. These two receptors are highly homologous, and these structures are at sufficiently high resolution (1.8 A and 2.1 A, respectively) to visualize extensive water networks. (c) Overlay of inactive β2AR (PDB 2RH1) and the muscarinic M2R structures (PDB 3UON) reveals similar polar side chain and water positions as those in the δ-OR. Abbreviations: β2AR, β2-adrenergic receptor; M2R, M2 muscarinic receptor; OR, opioid receptor; PDB, Protein Data Bank; TM, transmembrane.
The Orthosteric Site
In accord with the diverse ligands with which they interact, the detailed geometry and chemical nature of the ligand-binding sites vary among different GPCRs, and the changes in the sites upon activation also vary. A general feature of agonist binding relative to inverse agonists is a small compaction of the orthosteric ligand–binding site arising from inward movements of one or more surrounding helices as well as changes in side chain rotamers.
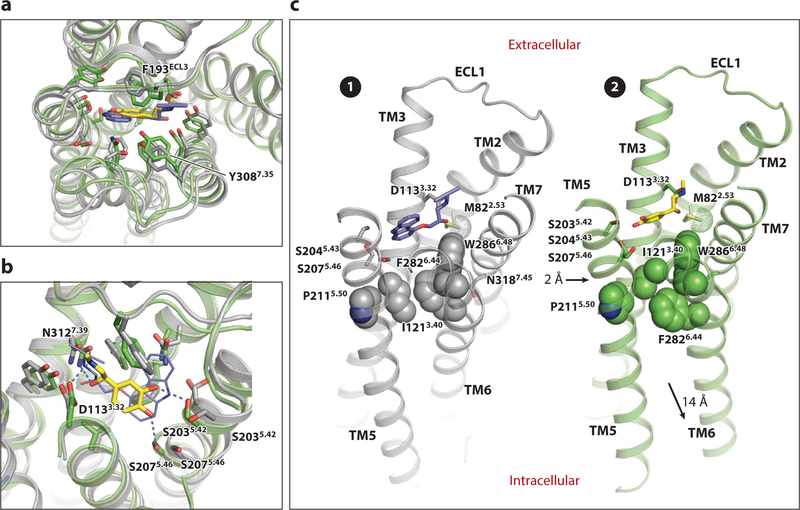
The differences between the inactive and active structures in the orthosteric site are remarkably subtle (Figure 6a,b). β-adrenergic receptor ligands have in common an ethanolamine moiety featuring a secondary amine and a β-OH group. The amine interacts with β2AR D1133.32 and N3127.39, and the β-OH group with N3127.39. In agonists, the ethanolamine moiety is linked to a catechol ring or structures of similar size bearing hydrogen-bonding groups with equivalent spacings; inverse agonists typically contain a larger ring such as the carbazole ring system of carazolol and lack one of these polar groups. Structures bound to the high-affinity BI-167107 agonist as well as to the natural agonist adrenaline (epinephrine) reveal that agonists form a hydrogen bond with S2075.46 (29, 31, 44; Figure 6b). This interaction arises from an inward bulge of the TM5 helix enabled by the break in hydrogen bonding at the conserved Pro2115.50, which moves S207 2 A inward. Antagonists lack this hydrogen-bonding group and are typically bulkier in this region; therefore, they require a more expanded binding site. The positions of F193ECL2 and Y3087.35, located near the extracellular space, move to cover the bound agonist (Figure 6a), and K3057.32, which is salt bridged to D192ECL2 in the inactive structure, now forms a hydrogen bond with the backbone at F193ECL2, thereby helping to stabilize this closed site. Binding of the agonist also slightly changes the position of W2866.48 at the base of the orthosteric ligand–binding site, a highly conserved residue in family A GPCRs.
Figure 6.
Changes in the orthosteric site and connector of the β2AR upon activation. (a,b) Top (a) and side (b) views of superimposed carazolol-bound inactive ( gray) and super adrenaline-bound active ( green) structures, highlighting changes in the orthosteric site. Carazolol is shown in blue sticks, and adrenaline in yellow sticks. In panel b, key hydrogen bonds formed with adrenaline are shown with dashed lines. (c) The connector region in the inactive (❶) and active (❷) states. The conserved nonpolar residues P2115.50, I1213.40, and F282 6.44 are shown in space-filling representation to highlight changes in their packing. The Cε methyl group of M822.53 is indicated with a dotted surface. The inward bulge of TM5 near P2115.50 in the active state activation and the outward movement of TM6 are indicated by the black arrows in state ❷. Abbreviations: β2AR, β2-adrenergic receptor; TM, transmembrane.
The Connector
The inward bulge of TM5 in the agonist-bound β2AR orthosteric site moves P2115.50 inward toward the core of the protein, which requires the I1213.40 side chain to adopt a different rotamer to prevent steric clash (Figure 6b,c). This in turn requires a shift in the position of F2826.44 to maintain close packing and an inward movement of TM7 at N3187.45, part of the helical turn associated with the rotation of TM7 at P3237.50. In addition, W2866.48 moves in concert with these residues. The shifts needed to repack W2866.48 and F2826.44 arise from a small rotation of TM6 in the turn preceding P2886.50, again facilitated by the absence of intrahelical hydrogen bonding in this region. These changes swing the lower half of TM6 outward, as described above. Similarly, the changes at N3187.45 are associated with a rotation in the turn preceding P3237.50 in the NPxxY motif, which moves TM7 (Figure 4). Thus, the small changes in the ligand-binding site that form the agonist-bound conformation favor repacking of neighboring residues, which is coupled to the rotations in TM6 and TM7 that produce the G protein–binding conformation. The rigidity of the helices surrounding the kinks provides a lever arm that amplifies these small changes into the large movements that create the G protein–binding pocket.
HOW DO LIGANDS ALTER THE CONFORMATIONAL LANDSCAPE OF GPCRs?
The inactive- and active-state crystal structures of the β2AR described above serve as a basis for examining conformational fluctuations that underlie allosteric coupling of the orthosteric and G protein sites. Data from crystallography, spectroscopy, and MD simulations have provided evidence for stable intermediate states that may link the fully inactive and fully active conformations.
Structural data from several GPCRs strongly suggest that ligand binding to the orthosteric site and the large changes observed in the cytoplasmic portion of the receptor are not strictly coupled. In the crystal structure of the β2AR bound to a covalently attached (i.e., irreversibly bound) agonist, S2075.42 forms a hydrogen bond with the ligand like that observed in the active state structures, but this interaction arises from a change in the side chain rotamer without the accompanying inward movement of TM3 (23). Moreover, no other changes relative to the inactive structure occurred, such as the repacking of the P5.50/F6.44/I3.40/W6.48 connector or movements of TM6 and TM7 (23). Similar results were reported for β1AR bound to various partial and full agonists (45). In contrast, agonist-bound structures of the A2aR in the absence of a G protein revealed the inward movement and contraction of the orthosteric site and also the altered packing of P5.50/F6.44/I 3.40/W6.48 associated with the G protein–bound conformation (17, 18) (Figure 7). In these A2aR structures, there are also changes in the cytoplasmic side of the TM region, but they are not as extensive as those observed in the fully activated state: TM5 shifts slightly toward TM6, TM6 moves outward slightly, and TM7 moves inward as much as 4–5 Å (17, 18, 24) (Figure 7). These structural data imply that the relative energies of the different intermediates differ in different receptors.
Figure 7.
Likely activation intermediate observed in the A2aR. (a) Overlay of A2aR ( yellow; PDB 3PWH) and β2AR inactive ( gray; PDB 2RH1) states. (b) A2aR bound to agonist (orange; PDB 2YDV) superimposed on the antagonist-bound structure (PDB 3PWH) and the active structure bound to the mini-Gsα (magenta; PDB 5G53). (c) Overlay of A2aR (PDB 5G53) and β2AR ( green; PDB 3SN6) active states. Abbreviations: A2aR, adenosine 2a receptor; β2AR, β2-adrenergic receptor; Gsα, heterotrimeric G protein α subunit; PDB, Protein Data Bank.
Long timescale (μs) MD calculations initiated from the active, agonist- and nanobody-bound β2AR showed that the open G protein–binding site in the cytoplasmic side of agonist-bound β2AR relaxes back to its inactive conformation in the absence of a G protein or a nanobody surrogate of a G protein (23). An intermediate that resembles the agonist-bound intermediate of the A2aR is seen in these simulations (46). Moreover, changes in the orthosteric site (bulging of TM5 at S2075.46), the G protein–binding region (changes in TM3–TM6 distance and movement of the TM7 NPxxY region), and the intervening connector (I1213.40, F2826.44) frequently occurred independently, indicating that ligand and G protein binding are not strictly coupled. The simulations also suggested that the G protein–binding site can leave its inactive conformation before the connector assumes the active conformation (46). This is consistent with observations that binding of a nucleotide-free G protein or nanobody mimic to ligand-free β2AR impedes binding of agonists and antagonists to the receptor, implying that it can stabilize the contracted or closed conformation of the orthosteric site observed in the agonist-bound active structures (21). Moreover, the G protein or nanobody slows dissociation of the agonist from the orthosteric site (21). As described above, F193ECL2 and Y3087.35 move toward one another to occlude the ligand-binding site (Figure 6a). Mutation of Y3087.35 to alanine significantly diminished the effect of the nanobody on antagonist and agonist binding (21). These observations demonstrate that the G protein can stabilize the high-affinity, agonist-binding conformation of the receptor.
Analysis of correlated movements defines allosteric pipelines that couple different regions of the receptor (47) and confirms that the intermediate and active states have fewer correlated movements than the inactive states and are more conformationally heterogeneous. Overall, the biochemical, structural, and simulation data indicate the formation of stable intermediates in the presence of agonists, an idea that is consistent with the notion that the G protein lowers the free energy of the agonist-bound state and thereby shifts the conformational equilibrium such that the active conformation is significantly populated.
MAPPING THE ENERGY LANDSCAPE OF GPCRs
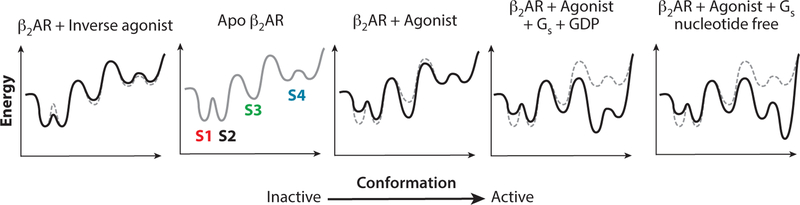
Energy landscapes describe the relative energies of conformational states and the barriers to their interconversion (48) (Figure 8). Ligand binding changes the free energy of states and thereby changes their relative populations at equilibrium, so defining the energy landscape that governs the equilibrium among functional GPCR states is essential for understanding GPCR pharmacology. In the absence of ligands, the inactive β2AR states appear to be more stable (i.e., of lower free energy) than the active states, and agonists and G protein change the landscape to lower the free energy of the active state (Figure 8) (49, 50). Spectroscopic measurements of equilibrium populations and their rates of interconversion have been used to map the energy landscape of the β2AR and other GPCRs. When combined with structural data, spectroscopically distinct states can be related to particular conformations and thereby deepen our understanding of how ligands activate GPCRs.
Figure 8.
Schematic energy landscape of β2AR. The gray lines (solid and dashed) indicate the energy landscape of the ligand-free (basal) state. The solid black lines indicate the effect of the indicated ligand and G protein on the energy landscape. The two inactive states detected by spectroscopy are denoted S1 (intact ionic lock) and S2 (broken ionic lock). S3 is the intermediate detected in the presence of an agonist without G protein, and S4 states are active states in the presence of an agonist and a G protein. Single-molecule FRET analysis provides evidence for distinct active states in the presence of an agonist that are dependent on the nucleotide state of the G protein. Abbreviations: β2AR, β2-adrenergic receptor; FRET, fluorescence resonance energy transfer; Gs, stimulatory heterotrimeric G protein.
NMR spectroscopy using labels at different positions has been extremely powerful for constructing GPCR energy landscapes. For an NMR label at a given site, the observation of peaks at more than one chemical shift value provides evidence for multiple structural states, and measurements of exchange rates between these states provide information about the energy barrier between them. Spectra measured in the presence of ligands of different efficacies allow assignment of these peaks to a functional conformation. Differences in the relative peak intensities in the presence of different ligands without changes in chemical shift values provide strong evidence for ligands being able to shift populations among different preexisting conformations. However, the appearance of peaks at distinct chemical shifts that depend on the ligand is more challenging to interpret. This can arise either from populating conformations that are rarely sampled in the absence of the ligand or from changes in exchange rate between preexisting populations.
Double electron-electron resonance (DEER) spectroscopy, which measures the interaction between spin probe labels, provides information complementary to NMR. DEER provides distance distributions (i.e., a probability plot of interprobe distances), so when two or more distinct conformations are present, the relative populations (free energies) of these states are obtained. Changes in the distance distributions in the presence of different ligands give a readout of the effect of a ligand on the relative populations of the conformations. DEER spectroscopy was used to demonstrate movements of TM6 in light-activated rhodopsin (51).
Fluorescence resonance energy transfer (FRET), which is sensitive to changes in distances between probes, can also be used to monitor conformational changes. Because spin centers in the probes used for DEER are more localized than the dipoles of fluorophores, DEER distance measurements are more accurate than FRET. DEER probes are also smaller than typical fluorophores, making it less likely that they will perturb the conformational equilibrium (52). Nonetheless, single-molecule FRET has provided important insights into the dynamics of the equilibrium (see below).
Dynamics of the G Protein–Binding Site
The outward movement of TM6 associated with forming the G protein–binding site is the largest change upon receptor activation and has proven experimentally accessible with different probes. Early studies employing fluorescent labels at a native cysteine near the cytoplasmic region of TM6 (C2656.27) of β2AR (Figure 2) showed that agonists led to a change in solvent exposure of the fluorophore, consistent with a major conformational change in this region (53, 54). These experiments also confirmed that receptors sample multiple conformational states in the ligand-free or neutral antagonist–bound (basal) state as well as when bound to ligands of different efficacies.
NMR experiments with 19F probes attached to β2AR C2656.27 confirmed that this region of β2AR undergoes a major change in its environment upon activation. Some experiments were performed on dodecyl maltoside (DDM)–solubilized β2AR labeled on residues C2656.27 with 19F-trifluoroethanethiol (TET) in the presence of ligands ranging from a strong inverse agonist to a full agonist (55, 56). Two peaks were observed, even in the presence of the strong inverse agonist carazolol. Crucially, the positions and line widths of the NMR peaks were essentially unchanged in the presence of different ligands; only their intensities differed. These observations demonstrated that ligands promote population shifts between preexisting conformations rather than stabilizing unique conformations. Single-molecule fluorescence studies of the β2AR labeled at C2656.27 also revealed population shifts associated with binding to different ligands (57). Conformational se-lection has also been observed directly in NMR studies of the A2aR labeled at an equivalent site (58).
As noted above, the ionic lock between R3.50 and E6.30 observed in dark rhodopsin is absent in β2AR structures but appears to form transiently in MD simulations. Evidence that β2AR conformations with an intact or broken lock are significantly populated in the inactive state has been obtained from DEER and NMR experiments (59). DEER measurements using spin labels at positions N148C and L266C (located respectively at the cytoplasmic ends of TM4 and TM6; Figure 2) in the presence of the strong inverse agonist carazolol revealed a bimodal distance distribution that corresponds approximately to the distances expected for the bent and straight TM6 conformations observed in the inactive β1AR structures (39) (Figure 3c), as well as in MD simulations (59). In the presence of carazolol, NMR measurements of affinity-purified β2AR labeled at C2656.27 labeled with 19F-BTFA (trifluoroacetanilide) revealed a broad peak that could be modeled as two interconverting states, which likely correspond to the intact and broken ionic lock conformations of TM6 observed by DEER spectroscopy (59). These spectroscopically defined states, designated S1 and S2 (Figure 8), were equally populated (i.e., equal free energy) and had a 325-μs lifetime. In the ligand-free receptor, the populations were also equal but the lifetimes increased and the exchange rate dropped approximately two-fold, indicating that the energy of these states is the same but that the inverse agonist lowers the barrier between them (Figure 8). Multiple states in the presence of carazolol or unliganded β2AR were also found using a smaller 19F-trifluoromethyl probe attached to C2656.27 (50). [Distinct states in the presence of carazolol were not detected in the earlier study (56), which may reflect slower exchange between these two conformations in the detergent lauryl maltose neopentyl glycol (LMNG) versus DDM (60), use of non-affinity-purified material, or the use of a different probe.]
In the presence of full agonists, NMR analysis of LMNG–solubilized β2AR labeled with 19F-BTFA at C2656.27 revealed a third state, S3, and a concomitant drop in the populations of the two inactive states (59). When a nanobody that mimics the G protein was added to the agonist-bound receptor, a fourth state (S4) with a distinct chemical shift was observed. This state is relatively homogeneous, consistent with a single, sharp distance distribution observed in the DEER experiments corresponding to the β2AR–Gs or β2AR–nanobody crystal structures. These observations suggest that S3 is an activation intermediate favored by agonist binding but is not the fully active conformation, and they also suggest that the nanobody shifts the equilibrium strongly to the activated conformation (Figure 8).
Both DEER and NMR measurements of β2AR (59) showed that in the presence of saturating amounts of the agonist isoproterenol, a relative of adrenaline that binds with 760-nM affinity and displays rapid association and dissociation kinetics, only 15–20% of the receptors are in the fully active S4 conformation. Isoproterenol also increases the fraction of receptors in the broken ionic lock state S2 relative to the intact ionic lock state S1 (Figure 8). Even when the high affinity, low off-rate agonist BI-167107 is used, approximately 40–60% of the receptors occupy the two inactive conformations. In the presence of a high-affinity agonist, the inactive and active intermediate conformations exchange slowly, in the millisecond–second regime, indicating a high energy barrier between these functional states (Figure 8). The relatively high barrier is likely important for conferring an approximately on/off response to the receptor.
A recent single-molecule FRET analysis of the β2AR purified in DDM and labeled with fluorophores at N148C and L266C (Figure 2), the same sites spin labeled in the DEER analysis, extended the observations made by ensemble DEER and NMR methods (61). In the presence of inverse agonists or neutral antagonists, the receptor was in a high FRET state corresponding to the distances predicted from the inactive structure. In the presence of partial or full agonists, lower FRET states were observed, indicating relative separation of the fluorophores resulting from an outward displacement of TM6. When nucleotide-free Gs was added, a further lowering of FRET was observed, and the low FRET value was consistent with the distance predicted from the β2AR–Gs crystal structure. In agreement with the NMR and DEER studies, agonists alone did not stabilize the β2AR in a fully active conformation. The extent to which agonists and partial agonists reduced FRET correlated with ligand efficacy.
The single-molecule FRET study also provided evidence for distinct receptor conformational states associated with the nucleotide state of bound Gs (61). Nucleotide-free Gs dissociated very slowly (lifetimes of minutes) from receptors bound to partial or full agonists. The dissociation rate increased 20-to 100-fold in the presence of physiological concentrations of GTP or GDP. Thus, in the presence of guanine nucleotides, Gs remains associated with the receptor for multiple seconds. Upon rapid addition of GDP to the nucleotide-free β2AR–Gs complex, a transient intermediate low-FRET state was observed, suggesting the existence of a GDP-bound β2AR–Gs complex that is structurally distinct from the nucleotide-free complex (Figure 8). The stability of the GDP-bound β2AR–Gs complex correlated with agonist efficacy. Thus, when bound to a full agonist, the GDP-bound β2AR–Gs complex is more likely to release GDP and bind GTP, while GDP-bound Gs is more likely to dissociate from the β2AR bound to a partial agonist before GDP release. These data imply that the receptor accesses conformations that can bind GDP-loaded Gs prior to nucleotide exchange, and the post-exchange GTP-bound Gs may associate with a distinct receptor conformation. The structural differences among these states is not known, however.
Different GPCRs display differences in their energy landscapes. As noted above, 19F-BTFA– labeled β2AR in the ligand-free state did not show evidence for the activation intermediate or the fully active states (59). In contrast, the A2aR appears to more readily form an active state, as NMR analysis of the A2aR labeled with 19F-BTFA at a position near the cytoplasmic end of TM6 revealed two activated states, as well as two inactive states, in the absence of a ligand (58). Addition of a partial or full agonist shifted the populations to favor these active states, supporting the conformational selection mechanism. Notably, the inactive and active states exchange slowly, in the 1–3 s time frame, and one of the active states is long lived. Moreover, the C-terminal Gα peptide could bind to the ligand-free receptor and shift the equilibrium to the two active states. Importantly, one active state was stabilized by a partial agonist, whereas the other was stabilized by a full agonist, suggesting that these agonists form distinct conformations even in the presence of a G protein. The state stabilized by the full agonist was shifted upfield relative to that associated with the partial agonist, indicating that the label at the end of TM6 in the full agonist form is solvent exposed. These data support a model in which partial agonists form a unique, less efficacious state than full agonists, rather than simply not being able to shift the equilibrium as far toward a single active structure.
Dynamics of Agonist–G Protein Coupling
The spectroscopic studies summarized above focused on the large conformational changes associated with formation of the G protein–binding site. To understand how these changes are coupled to engagement of orthosteric ligands, isotopically labeled side chains have been used for NMR measurements of conformational dynamics of sites in the β2AR that undergo smaller changes. These regions are unlikely to be accessible to labeling by large probes, which also would likely perturb the local structure. Labeling the ε-methyl group of native or introduced methionine residues with 13C (62, 63) and examining the effects of different ligands gives a similar picture of the β2AR energy landscape derived from the 19F studies. The native M822.53, which lies near the orthosteric site (Figure 6c,d), showed two peaks in millisecond exchange in apo form or in the presence of an inverse agonist. This timescale is too slow to be attributed to side chain rotamer changes and suggests that the two peaks reflect larger conformational differences. In the inactive structure, Met822.53 packs on Y316 and S319, residues in TM7 that shift upon activation, and also is close to W2866.48, part of the connector that shifts as part of forming the agonist-bound state (Figure 6c,d). Thus, it appears that in the inactive state the structure in this region samples at least two conformations, one of which may be an intermediate to the formation of the active state. Upon addition of the agonist BI-167107 and a nanobody mimic of the G protein, most of the methionine peaks shift, indicating a distinct conformation (63). With BI-167107 alone, M822.53 appears to be in an environment very similar to that observed in the presence of the nanobody, whereas other methionine residues closer to the G protein–binding site display distinct resonances associated with a different conformation. Again, this is consistent with loose coupling of agonist and G protein–binding described from MD simulations. These data also show that with an agonist alone, the receptor does not simply populate a mixture of the inactive and active crystallographic conformations but adopts new, intermediate conformations, particularly in the vicinity of M2155.54 and M2796.34, near the cytoplasmic ends of TM5 and TM6, respectively. MD simulations suggest that in this state, TM7 is in an inactive conformation but TM5 and TM6 differ, with their intracellular ends having substantial mobility (63). It is important to note that the intermediates detected spectroscopically likely depend on the probe, so it cannot be rigorously established that different probes are detecting the same intermediate even in the presence of a particular ligand.
The receptor environment has a critical role in determining the dynamics of the receptor. For example, the exchange between conformations of the β2AR probed by a 19F label at C265 was found to be slower in MNG than in DDM (60). Likewise, Kofuku et al. (64) examined changes in labeled methionine residues in β2AR reconstituted into lipid nanodiscs. Transitions between the two inactive conformations and between them and the active conformation observed at Met822.53 were considerably slower than in detergent. Moreover, in the presence of a partial agonist, the population of the active conformation was higher than in detergents. Specific lipids also influence the energy landscape. MD simulations in the presence of different lipid mixtures showed different conformations of β2AR (65). β2AR in mixed lipid-detergent micelles had a higher affinity for agonists and higher basal coupling to a G protein mimetic nanobody in micelles containing the synthetic lipids DOPG or DOPS than in micelles containing the lipids DOPE or DOPC (66), suggesting that lipid head groups could alter both the energy of an active intermediate as well as the energy barrier between inactive and active states.
Isogai et al. (67) examined changes in the β1AR backbone using 15N-labeled valine. Individual valine peaks were assigned by mutational analysis and examined in the presence of an inverse agonist, no ligand, or an agonist. Changes in the orthosteric site correlated well with contacts made by different classes of ligands. This study employed a thermostabilized β1AR variant that did not activate G protein even in the presence of an agonist. Nonetheless, changes in the back-bone at several key positions in TM5, TM6, and TM7 observed between inactive and activated GPCR crystal structures were detected as chemical shifts, suggesting that agonists can promote an intermediate conformation on the pathway to full activation. The stabilized protein included the mutations Y5.58A and Y3437.53L (the latter part of the NP7.50xxY motif). These positions are critical for forming the active state (Figure 4), and when the native tyrosines were restored, the protein could now activate G protein. In the presence of a nanobody mimic of a G protein, the protein displayed the expected conformational changes. In addition to highlighting the importance of the conserved tyrosines, these results support the notions that the active state is less stable and that its relative instability is essential for agonist-mediated activation.
Isogai et al. (67) also examined changes in chemical shifts at the various valine backbone positions in the β1AR as a function of ligand efficacy toward G protein activation when key residues I3.40, Y5.58, and Y7.53 were mutated, thereby mapping which conformational changes were altered by the mutation. They found that the changes that link TM2 and TM3 to TM7 are only weakly connected to the network that connects TM3, TM5, and TM6, consistent with the idea of a loose coupling between ligand binding and G protein activation.
The loose coupling between agonist and G protein binding sites is not unique to adrenergic receptors. NMR analysis of the μ-opioid receptor with 13C-methylated lysine residues revealed significant spectral changes of lysines on TM5 and TM6 in the presence of an agonist and a G protein nanobody mimic but not in the presence of the agonist alone (68). In contrast, lysine residues in ICL1 and helix 8 changed in the presence of an agonist alone, leading to the suggestion that these regions may first engage the G protein before formation of the stable ternary complex.
FUTURE CHALLENGES
Partial Versus Full Agonism
The mechanism of partial agonism, that is, how certain ligands at saturation activate activity to a level below that of full agonists, is not established. In an NMR study employing 13C-Met β2AR, partial agonists appeared to alter the inactive–active equilibrium rather than stabilizing a unique state (62). This could mean that, in the presence of a partial agonist, a G protein cannot shift the population as far toward the fully active conformation as it does when the receptor is bound to a full agonist. Conversely, the A2aR study employing 19F label at the base of TM6 suggested distinct conformations for the partial and fully active states (58). This difference could be due to the different positions probed in these experiments. Alternatively, although the partial agonist-bound state observed by NMR might be an intermediate similar to that on the pathway to full activation, it is possible that in the presence of a G protein the partial agonist-bound receptor adopts a unique conformation that is less effective in promoting nucleotide exchange. A recent study using a nanobody that stabilizes the active state and another that stabilizes the inactive state (69) supports the notion that different partial agonists stabilize distinct conformations of the β2AR. Gregorio et al. (61) found that complexes with Gs bound to β2AR in the presence of partial agonists have higher affinity for GDP than those in the presence of full agonists, implying that more efficacious ligands increase probability of GDP release needed for exchange. Overall, it appears that the receptor can adopt even more conformations than those observed using conformational probes at just a few sites and that these conformations probably relate to the ultimate level of efficacy of particular ligands.
Biased Agonism
We have focused on activation of GPCR guanine nucleotide exchange activity. Much less is known about coupling to the arrestin pathway (Figure 1b). Recently, the crystal structure of rhodopsin bound to visual arrestin was determined (70, 71). The receptor displays similar rearrangements on the cytoplasmic side of the TM region that occur as part of G protein binding. However, the principal contacts made by arrestin are with TM7; in contrast, the G protein does not interact directly with this part of the receptor. In the Liu et al. (56) NMR study of the β2AR, G protein–biased ligands produced little change in a 19F probe at C3277.54, which is not involved in contacts with the G protein, whereas, β-arrestin–biased ligands produced larger changes at C3277.54, consistent with the rhodopsin/arrestin complex structure. Interestingly, C3277.54 showed two populations even in the presence of the inverse agonist carazolol, consistent with conformational heterogeneity at the end of TM7 in the inactive state. Thus, while the pharmacology implies distinct coupling mechanisms for agonists that target arrestin versus those that activate G proteins, these agonists likely share allosteric transmission mechanisms. Low barriers between different activation intermediate states may enable signaling through different pathways (47) by a given ligand. Higher resolution structures of arrestin complexes with GPCRs that bind to diffusible ligands, as well as spectroscopic characterization, are needed to address these mechanistic problems.
GPCRs:
G protein–coupled receptors
Agonists:
ligands that stimulate receptor activity over basal activity
Efficacy:
the extent to which a ligand can activate a particular signaling pathway
Inverse agonists:
ligands that suppress receptor activity below basal level
Neutral antagonists:
ligands that compete with other ligands for binding to the orthosteric site but that do not alter basal activity
Partial agonists:
ligands that produce weaker maximal activity than full agonists
Orthosteric site:
the site occupied by the cognate hormone or neurotransmitter
β2AR:
β2-adrenergic receptor
Gs:
stimulatory heterotrimeric G protein
A2aR:
adenosine 2a receptor
ACKNOWLEDGMENTS
This work was supported by a grant from the Mathers Foundation (W.I.W. and B.K.K.) and grants from the National Institutes of Health R01NS028471 and R01GM083118 (B.K.K). B.K.K. is supported by the Chan Zuckerberg Biohub.
Footnotes
DISCLOSURE STATEMENT
Brian Kobilka is co-founder of ConfometRx, Inc.
LITERATURE CITED
- 1.Maguire ME, Van Arsdale PM, Glilman AG. 1976. An agonist-specific effect of guanine nucleotides on binding to the beta adrenergic receptor. Mol. Pharmacol 12:332–39 [PubMed] [Google Scholar]
- 2.De Lean A, Stadel JM, Lefkowitz RJ. 1980. A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled β-adrenergic receptor. J. Biol. Chem 255:7108–17 [PubMed] [Google Scholar]
- 3.Cui Q, Karplus M. 2008. Allostery and cooperativity revisited. Protein Sci 17:1295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nussinov R, Ma B, Tsai CJ. 2014. Multiple conformational selection and induced fit events take place in allosteric propagation. Biophys. Chem 186:22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henzler-Wildman K, Kern D. 2007. Dynamic personalities of proteins. Nature 450:964–72 [DOI] [PubMed] [Google Scholar]
- 6.Fredriksson R, Langerstrom MC, Lundin LG, Schioth HB. 2003. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol 63:1256–72 [DOI] [PubMed] [Google Scholar]
- 7.Ballesteros JA, Weinstein H. 1995. Integrated methods for the construction of three-dimensional models and computational probing of structure function relations in G protein-coupled receptors. Methods Neurosci 25:366–428 [Google Scholar]
- 8.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, et al. 2000. Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289(5480):739–45 [DOI] [PubMed] [Google Scholar]
- 9.Faham S, Bowie JU. 2002. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J. Mol. Biol 316(1):1–6 [DOI] [PubMed] [Google Scholar]
- 10.Rasmussen SGF, Choi H-J, Rosenbaum DM, Kobilka TS, Thian FS, et al. 2007. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature 450(7168):383–87 [DOI] [PubMed] [Google Scholar]
- 11.Caffrey M 2009. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Annu. Rev. Biophys 38:29–51 [DOI] [PubMed] [Google Scholar]
- 12.Magnani F, Serrano-Vega MJ, Shibata Y, Abdul-Hussein S, Lebon G, et al. 2016. A mutagenesis and screening strategy to generate optimally thermostabilized membrane proteins for structural studies. Nat. Protoc 11:1554–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou Y, Weis WI, Kobilka BK. 2012. N-terminal T4 lysozyme fusion facilitates crystallization of a G protein coupled receptor. PLOS ONE 7(10):e46039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, et al. 2007. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science 318(5854):1266–73 [DOI] [PubMed] [Google Scholar]
- 15.Chun E, Thompson AA, Lu W, Roth CB, Griffith MT, et al. 2012. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure 20:967–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, et al. 2007. High-resolution crystal structure of an engineered human β2-adrenergic G protein–coupled receptor. Science 318(5854):1258–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lebon G, Warne T, Edwards PC, Bennett K, Langmead CJ, et al. 2011. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 474:521–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu F, Wu H, Katritch V, Han GW, Jacobson KA, et al. 2011. Structure of an agonist-bound human A2A adenosine receptor. Science 332:322–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddy MT, Didenko T, Stevens RC, Wuthrich K. 2016. β2-Adrenergic receptor conformational response to fusion protein in the third intracellular loop. Structure 24:2190–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, et al. 2012. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature 482(7386):552–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeVree BT, Mahoney JP, Vélez-Ruiz GA, Rasmussen SGF, Kuszak AJ, et al. 2016. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature 535:182–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler Babu MM. 2013. Molecular signatures of G-protein-coupled receptors. Nature 494:185–88 [DOI] [PubMed] [Google Scholar]
- 23.Rosenbaum DM, Zhang C, Lyons J, Holl R, Aragao D, et al. 2011. Structure and function of an irreversible agonist-β2 adrenoceptor complex. Nature 469(7329):236–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter B, Nehme R, Warne T, Leslie AGW, Tate CG. 2016. Structure of the adenosine A2A receptor bound to an engineered G protein. Nature 536:104–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, et al. 2008. Crystal structure of opsin in its G-protein-interacting conformation. Nature 455:497–502 [DOI] [PubMed] [Google Scholar]
- 26.Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, et al. 2011. Crystal structure of metarhodopsin II. Nature 471:651–55 [DOI] [PubMed] [Google Scholar]
- 27.Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, et al. 2011. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature 471:656–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manglik A, Kobilka BK, Steyaert J. 2017. Nanobodies to study G protein–coupled receptor structure and function. Annu. Rev. Pharmacol. Toxicol 57:19–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen SG, Choi H-J, Fung JJ, Pardon E, Casarosa P, et al. 2011. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469(7329):175–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steyaert J, Kobilka BK. 2011. Nanobody stabilization of G protein-coupled receptor conformational states. Curr. Opin. Struct. Biol 21:567–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ring AM, Manglik A, Kruse AC, Enos MD, Weis WI, et al. 2013. Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature 502(7472):575–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruse AC, Ring AM, Manglik A, Hu J, Hu K, et al. 2013. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504(7478):101–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burg JS, Ingram JR, Venkatakrishnan AJ, Jude KM, Dukkipati A, et al. 2015. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science 347:1113–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W, Maglik A, Venkatakrishnan AJ, Laeremans T, Feinberg EN, et al. 2015. Structural insights into μ-opioid receptor activation. Nature 524:315–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang YL, Khoushouei M, Radjainia M, Zhang Y, Glukhova A, et al. 2017. Phase-plate cryo-EM structure of a class B GPCR–G-protein complex. Nature 546:118–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Sun B, Feng D, Hu H, Chu M, et al. 2017. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546:248–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkatakrishnan AJ, Deupi X, Lebon G, Heydenreich FM, Flock T, et al. 2016. Diverse activation pathways in class A GPCRs converge near the G protein-coupling region. Nature 536:484–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gether U, Seifert R, Ballesteros JA, Sanders-Bush E, Weinstein H, Kobilka BK. 1997. Structural instability of a constitutively active G protein-coupled receptor. J. Biol. Chem 272:2587–90 [DOI] [PubMed] [Google Scholar]
- 39.Moukhametzianov R, Warne T, Edwards PC, Serrano-Vega MJ, Leslie AGW, et al. 2011. Two distinct conformations of helix 6 observed in antagonist-bound structures of a β1-adrenergic receptor. PNAS 108:8228–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dror RO, Arlow DH, Borhani DW, Jensen MØ, Piana S, Shaw DE. 2009. Identification of two distinct inactive conformations of the β2-adrenergic receptor reconciles structural and biochemical observations. PNAS 106:4689–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romo TD, Grossfield A, Pitman MC. 2010. Concerted interconversion between ionic lock substates of the β2 adrenergic receptor revealed by microsecond timescale molecular dynamics. Biophys. J 98:76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ballesteros JA, Jensen AD, Liapakis G, Rasmussen SGF, Shi L, et al. 2001. Activation of the β2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J. Biol. Chem 276(31):29171–77 [DOI] [PubMed] [Google Scholar]
- 43.Valentin-Hensen L, Groenen M, Nygaard R, Frimurer TM, Holliday ND, Schwartz TW. 2012. The arginine of the DRY motif in transmembrane segment III functions as a balancing micro-switch in the activation of the β2-adrenergic receptor. J. Biol. Chem 287:31973–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, et al. 2011. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477(7366):549–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warne T, Moukhametzianov R, Baker JG, Néhme R, Edwards PC, et al. 2011. The structural basis for agonist and partial agonist action on a β1-adrenergic receptor. Nature 469:241–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dror RO, Arlow DH, Maragakis P, Mildorf TJ, Pan AC, et al. 2011. Activation mechanism of the β2-adrenergic receptor. PNAS 108:18684–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhattacharya S, Vaidehi N. 2014. Differences in allosteric communication pipelines in the inactive and active states of a GPCR. Biophys. J 107:422–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frauenfelder H, Sligar SG, Wolynes PG. 1991. The energy landscapes and motions of proteins. Science 254:1598–603 [DOI] [PubMed] [Google Scholar]
- 49.West GM, Chien EYT, Katrich V, Gatchalian J, Chalmers MJ, et al. 2011. Ligand-dependent perturbation of the conformational ensemble for the GPCR β2 adrenergic receptor revealed by HDX. Structure 19:1424–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim TH, Chung KY, Manglik A, Hansen AL, Dror RO, et al. 2013. The role of ligands on the equilibria between functional states of a G protein-coupled receptor. J. Am. Chem. Soc 135:9465–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. 2008. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. PNAS 105(21):7439–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klare JP. 2013. Site-directed spin labeling EPR spectroscopy in protein research. Biol. Chem 394:1281–300 [DOI] [PubMed] [Google Scholar]
- 53.Ghanouni P, Steenhuis JJ, Farrens DL, Kobilka BK. 2001. Agonist-induced conformational changes in the G-protein-coupling domain of the β2 adrenergic receptor. PNAS 98(11):5997–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao X, Parnot C, Deupi X, Ratnala VRP, Swaminath G, et al. 2006. Coupling ligand structure to specific conformational switches in the β2-adrenoceptor. Nat. Chem. Biol 2(8):417–22 [DOI] [PubMed] [Google Scholar]
- 55.Horst R, Liu JJ, Stevens RC, Wuthrich K. 2013. β2-Adrenergic receptor activation by agonists studied with 19F NMR spectroscopy. Angew. Chem 52(41):10762–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu JJ, Horst R, Katrich V, Stevens RC, Wuthrich K. 2012. Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335:1106–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lamichhane R, Liu JJ, Pljevaljcic G, White KL, van der Schans E, et al. 2015. Single-molecule view of basal activity and activation mechanism of the G protein-coupled receptor β2AR. PNAS 112:14254–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye L, Van Eps N, Zimmer M, Ernst OP, Prosser RS. 2016. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature 533:265–68 [DOI] [PubMed] [Google Scholar]
- 59.Manglik A, Kim TH, Masureel M, Altenbach C, Yang Z, et al. 2015. Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell 161:1101–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chung KY, Kim TH, Manglik A, Alvares R, Kobilka BK, Prosser RS. 2012. Role of detergents in conformational exchange of a G protein-coupled receptor. J. Biol. Chem 287:36305–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gregorio GG, Masureel M, Hilger D, Terry DS, Juette M, et al. 2017. Single-molecule analysis of ligand efficacy in β2AR–G-protein activation. Nature 547:68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kofuku Y, Ueda T, Okude J, Shiraishi Y, Kondo K, et al. 2012. Efficacy of the β2-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat. Commun 3:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nygaard R, Zhou Y, Dror RO, Mildorf TJ, Arlow DH, et al. 2013. The dynamic process of β2-adrenergic receptor activation. Cell 152:532–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kofuku Y, Ueda T, Okude J, Shiraishi Y, Kondo K, et al. 2014. Functional dynamics of deuterated β2-adrenergic receptor in lipid bilayers revealed by NMR spectroscopy. Angew. Chem 53(49):13376–79 [DOI] [PubMed] [Google Scholar]
- 65.Mahmood I, Liu X, Neya S, Hoshino T. 2013. Influence of lipid composition on the structural stability of g-protein coupled receptor. Chem. Pharm. Bull 61:426–37 [DOI] [PubMed] [Google Scholar]
- 66.Dawaliby R, Trubbia C, Delporte C, Masureel M, Van Antwerpen P, et al. 2016. Allosteric regulation of G protein-coupled receptor activity by phospholipids. Nat. Chem. Biol 12:35–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Isogai S, Deupi X, Opitz C, Heydenreich FM, Tsai C-J, et al. 2016. Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nature 530:237–41 [DOI] [PubMed] [Google Scholar]
- 68.Sounier R, Mas C, Steyaert J, Laeremans T, Manglik A, et al. 2015. Propagation of conformational changes during μ-opioid receptor activation. Nature 524:375–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staus DP, Strachan RT, Manglik A, Biswaranjan P, Kahsai AW, et al. 2016. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature 535:448–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kang Y, Zhou XE, Gao X, He Y, Liu W, et al. 2015. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523(7562):561–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou XE, He Y, de Waal PW, Gao X, Kang Y, et al. 2017. Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell 170:457–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Komolov KE, Du Y, Duc NM, Betz RM, Rodrigues JPGLM, et al. 2017. Structural and functional analysis of a β2-adrenergic receptor complex with GRK5. Cell 169:407–421 [DOI] [PMC free article] [PubMed] [Google Scholar]