Abstract
The circadian clock is a complex cellular mechanism that, through the control of diverse metabolic and gene expression pathways, governs a large array of cyclic physiological processes. Epidemiological and clinical data reveal a connection between the disruption of circadian rhythms and cancer that is supported by recent preclinical data. In addition, the use of animal models and molecular studies indicate emerging links between cancer metabolism and the circadian clock. This has implications for therapeutic approaches and we discuss the possible design of chrono-pharmacological strategies.
The circadian clock sustains self-perpetuating oscillations with a 24-hour periodicity, while also being synchronized by external environmental cues such as light, temperature and food intake, the so-called zeitgebers or time-givers that maintain proper timekeeping (Figure 1). Disruptions in biological rhythms result in numerous physiological disorders in organismal homeostasis, the consequences of which have been linked to several pathologies, including cancer1,2. Specifically, epidemiological and laboratory evidence has long suggested that a relationship exists between the circadian clock and cancer3, yet the precise molecular mechanisms of this connection are not fully elucidated. Interestingly, epidemiological evidence shows a link between hormone-dependent cancers and environmental disruption of the circadian clock by shift-work and light exposure at night4–6 which is supported by preclinical data, although the precise mechanisms of clock disruption related to cancer initiation versus progression remain unknown.
Figure 1. The mammalian circadian clock.
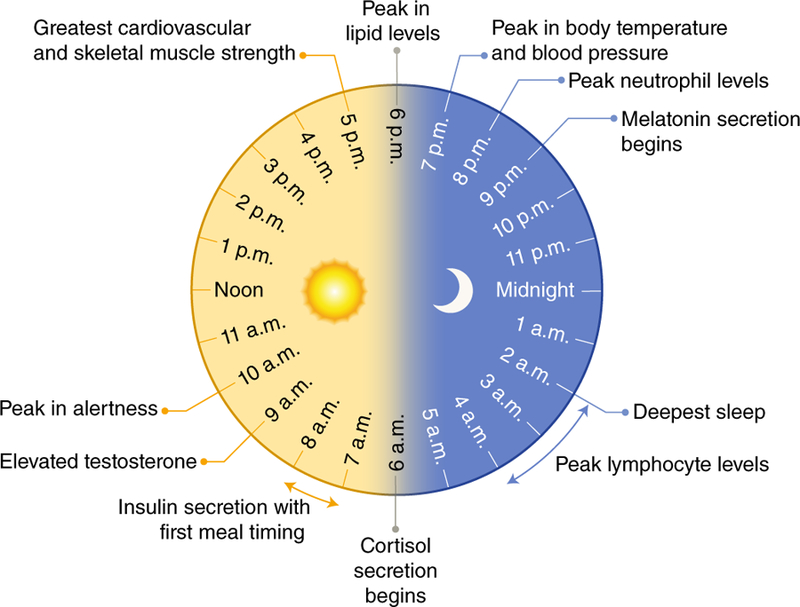
An overview of rhythmic functions which are critically controlled by the human circadian pacemaker are outlined. Time of day is indicated for peak endocrine functions, deepest sleep, metabolic control, immune responses, alertness, and cardiovascular parameters over the 24-hour cycle. Information was adapted from several references96–99.
Therefore, this Perspective serves to summarize the current state of knowledge regarding the links between the circadian clock (see Box 1, Figure 2) and cancer in an effort to highlight new avenues for therapeutic intervention. Specifically, we discuss recent advances in teasing out how the clock is implicated in regulating cancer-initiating cells versus utilization of genetic mouse models of cancer where circadian disruption alters disease progression. Additionally, we focus on several facets of cancer metabolism that can be rewired in response to circadian disruption. We intend to point to new directions where further research emphasis is required to fully understand how clock disruption and cancer converge, in addition to new avenues of pharmacological intervention and treatment of cancer.
Box 1: Mammalian Circadian Clocks.
Physiological and metabolic variations follow cycles linked to the time of the day. These circadian (from the Latin words, circa diem, about a day) rhythms include sleep-wake cycles, feeding behavior, body temperature and hormonal oscillations (Figure 1). All cells harbor molecular clocks which operate in concert to control circadian rhythms98,109. The architecture of the circadian system is based on a hierarchical structure whereby the central oscillator receives environmental cues or zeitgebers (time givers) that can adjust the otherwise self-sustained rhythms driven by the clock110,111. From an anatomical standpoint, the circadian system is based on a central clock located in the suprachiasmatic nucleus (SCN) within the hypothalamus112,113. Neurons within the SCN function autonomously and are reset in response to light to coordinate the timekeeping of peripheral clocks located in all other tissues and cells in the body111,114,115. Therefore, proper synchronization and coordination between central and peripheral clocks is thought to be of upmost importance for systemic homeostasis.
The molecular organization of the core circadian clock has been unveiled during the past couple of decades (Figure 2). The molecular machinery that constitutes the circadian clock is comprised of two DNA-binding transcription factors, CLOCK and BMAL1, that heterodimerize and direct transcriptional activation of core clock genes and additional clock-controlled genes (CCGs)104,105, by binding to E-box sites on their promoters30. Among these CCGs, CLOCK and BMAL1 direct transcription of their own repressors, period (PER) and cryptochrome (CRY) family members, creating a tightly self-regulated transcriptional/translational feedback loop110,113,116. During the day, transcription of Per and Cry is high, resulting in translation of the circadian repressors, and subsequent formation of the inhibitory complex with CLOCK and BMAL1 that abolishes transcription of CCGs at night106. Degradation of the circadian repressors, PER and CRY, alleviates transcriptional repression and restores CLOCK:BMAL1-mediated transcription, establishing an oscillatory rhythm in circadian gene expression that operates within a precise 24-hour period. An additional level of circadian regulation exists with the orphan nuclear receptors RORa and REV-ERBa which activate and repress transcription of the Bmal1 gene, respectively107,108,110,113.
Transcriptome studies have shown that the clock directs the expression of a large number of genes in different tissues, illustrating that a significant fraction of the genome is under clock control102–104. This notion implicates cyclic chromatin transitions to occur on a genome-wide scale and a number of chromatin remodelers have been found to display circadian activity. For example, the core regulator CLOCK displays acetyltransferase activity on non-histone proteins 117,118 and on H3 at K9 and K14119, both marks associated to a chromatin state permissive for transcription. The CLOCK:BMAL1 heterodimer appears to interact also with CBP (CREB binding protein), p300 and with the CBP-associated factor PCAF120,121, suggesting that a number of HATs may contribute to circadian epigenome . Additionally, various histone deacetylases (HDACs) have been implicated, such as HDAC1106 and HDAC3122,123, in addition to the nicotinamide adenine dinucleotide (NAD+)-dependent sirtuins, SIRT173,74,124 and SIRT675 which are regulated by cyclic availability of NAD+ through the salvage pathway77,78. In addition to acetylation, histone methylation has been also implicated in circadian chromatin remodeling through histone methyltransferase (myeloid/lymphoid or mixed-lineage leukemia 1) MLL1125 and MLL3126.
Specific molecular components of the circadian clock machinery are listed below:
CLOCK [Circadian Locomotor Output Cycles Kaput], Core Transcription Factor
BMAL1 [Aryl Hydrocarbon Receptor Nuclear Translocator-like protein 1 (ARNTL or BMAL1)], Core Transcription Factor
PER 1–3 [Period 1–3], Transcriptional Repressors
CRY 1–2 [Cryptochrome 1–2], Transcriptional Repressors
REV-ERBα [Nuclear Receptor Subfamily 1, group D, member 1 (NR1D1)], Transcriptional Repressor
RORα [Retinoid-Related Orphan Receptor alpha], Transcriptional Activator
Figure 2. The Molecular Components of the Mammalian Circadian Clock.
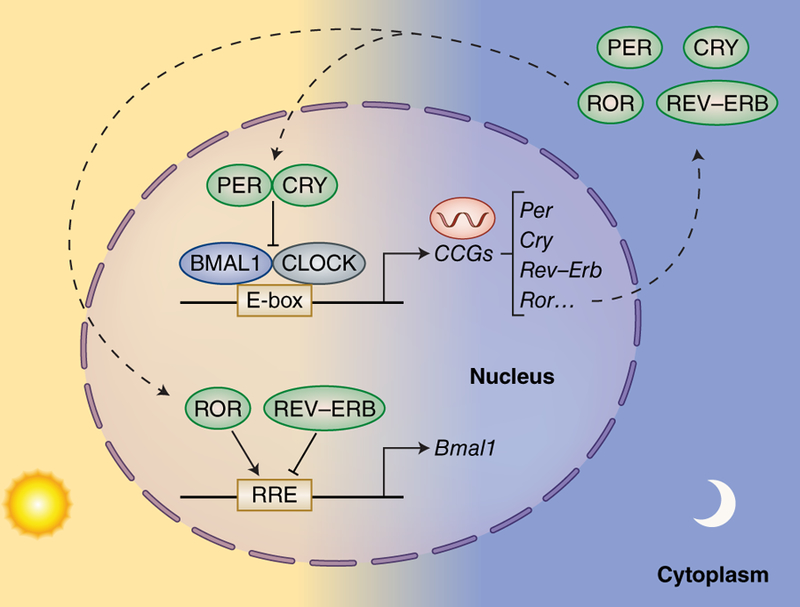
The circadian transcriptional/translational feedback loop occurs within a period of 24-hours. The core circadian transcriptional machinery consists of the bHLH DNA-binding transcription factors, CLOCK and BMAL130,100, which bind E-Box sequences to control the rhythmic expression of ~1015% of genes98,101–103. CLOCK:BMAL1-dependent transcription of core clock and clock-controlled genes (CCGs) peaks during the day, while transcription is inhibited by the circadian repressors, Period (PER) and Cryptochrome (CRY), at night104–106. An additional level of circadian regulation exists with the nuclear receptors RORa and REV-ERBa that activate and repress transcription of the Bmal1 gene, respectively107,108.
Links between Circadian Disruption and Cancer
Epidemiological Links between the Clock and Cancer
Circadian disruption has been associated with increased incidence of specific cancers in various epidemiological studies, though the causes and factors linked with this disruption remain somewhat unclear. Yet, in 2007, the International Agency for Research on Cancer (IARC) listed “shift work leading to a disruption in circadian rhythm” as a probable human carcinogen, classified as Group 2A7. The most convincing epidemiological evidence relates to studies on shift workers, and more recently meal-timing. Accumulating data show correlation between shift work and breast and prostate cancer4–6. For example, according to the Nurses’ Health Study and case-controlled studies conducted with Norwegian nurses, women that worked night shifts for less than 30 years had a moderately increased risk for breast cancer and risk was further increased upon working 30 or more years of rotating shift work [relative risk (RR) = 1.36]4,6,8. Similarly, reports suggest that the risk of prostate cancer is increased in night-shift workers, and this risk is augmented with longer duration of shift work, particularly in high grade cancers5,9. Moreover, light at night exposure may not be the only risk factor influencing circadian disruption and tumorigenesis. A prospective study of 41,398 day-working adults in the French NutriNet-Sante cohort identified that ‘late-eaters’ (those that exhibited eating episodes after 9:30PM) have an increased risk of breast and prostate cancer, with Hazard ratios (HR) of 1.48 and 2.20, respectively10. Based on these epidemiological studies, the effect of environmental disruption of the clock on cancer is further discussed below.
Linking the Circadian Clock and Cancer in animal models
The epidemiological evidence summarized above broadly linking circadian disruption with cancer, although limited, has triggered a significant number of studies using genetic mouse models (Figure 3). While drawing parallels between epidemiological information and results from mouse experimental models is always difficult, significant evidence of a link between circadian clock disruption and increased cancer risk has accumulated. Early landmark studies have shown that mice carrying mutations in individual clock genes (mutation of Per2m/m; heterozygous ablation of Bmal1+/−; and double null Cry1/2-/ ) are more susceptible to lymphoma, and when irradiated, these mutant mice have increased rates of lymphoma and hepatocellular carcinoma11,12. Recent evidence demonstrates that crossing Per2m/m mice with a KrasLSL-G12D/+;p53fl/fl genetically engineered mouse model (GEMM) of lung adenocarcinoma resulted in increased tumor burden, more aggressive Grade 3 and 4 lung tumors, and subsequent decreased overall survival13. Altogether these studies provide compelling information that genetic disruption of key components of the clock mechanism increases tumorigenesis, it is still unclear whether there is any specificity for distinct types of cancer.
Figure 3. Circadian regulation of tumor initiation and progression.
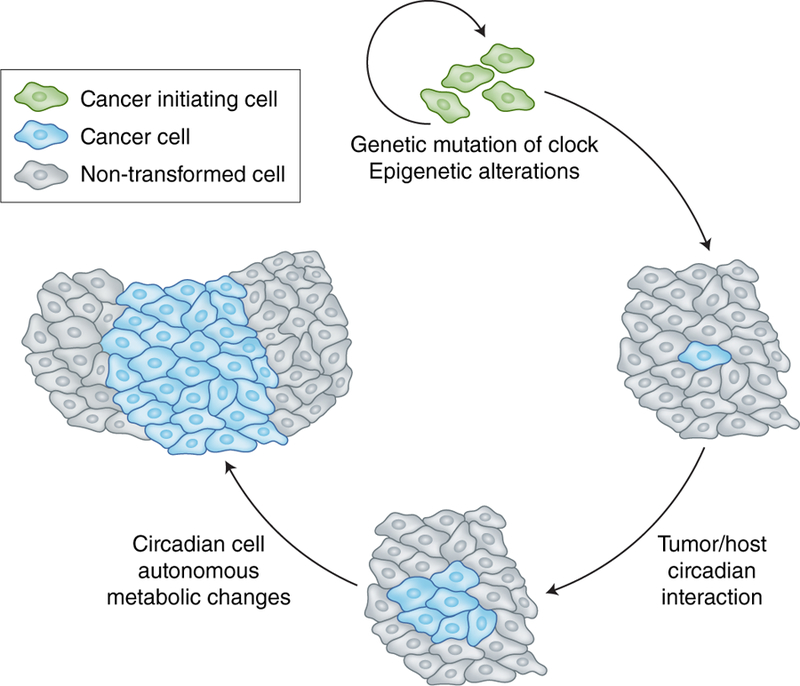
During tumorigenesis, several aspects of altered circadian control have been described at the stages of initiation and progression. These include genetic disruption of the canonical circadian transcriptional machinery and changes in epigenetic control mechanisms that regulate circadian gene expression, steps which are likely more implicated in tumor initiation. Subsequent deregulation of metabolism could further drive tumor progression, both in a cell autonomous manner and circadian metabolic changes that influence tumor/host interactions.
Notably, mouse studies indicate that tumorigenesis may be linked to anatomical disruption of the central circadian pacemaker housed in the suprachiasmatic nucleus (SCN), though the effects on peripheral clocks remains unresolved. Bilateral electrolytic lesions of the SCN enhanced tumor growth of implanted Glasgow osteosarcoma and pancreatic ductal adenocarcinoma (PDAC) versus sham operated mice14. Yet, the molecular mechanisms by which disruption of the central pacemaker results in enhanced tumorigenesis is unknown and likely involves changes in the synchrony between the SCN and peripheral clocks.
Environmental Disruption of the Circadian Clock and Cancer
Epidemiological studies indicate that disruption of circadian rhythms in humans is associated to increased cancer risk (see above). In addition, pilot data now suggests that shift work can also regulate the epigenetic landscape, which could likely be a mechanism by which circadian gene expression is altered15–17. For instance, DNA methylation studies have found genome-wide alterations to cancer-specific and circadian loci in female shift workers from a prospective cohort in Denmark, including estrogen receptor a (Esr1) and circadian genes, Clock and Cry217. On a genome-wide scale, 5,409 CpG sites were found to be differentially methylated in day-time versus night-time shift workers, and a remarkable 66% of these loci were hypermethylated17. Similarly, CpG methylation of the Per1, Per2 and Per3 promoters was found to correlate with changes in protein expression in 50% of breast tumors versus normal tissue taken from the same patient15. Moreover, in 126 cases of several types of hematologic malignancies, Bmal1 gene silencing due to CpG promoter methylation was found in 19.7% of diffuse large B-cell lymphomas, 33.3% of acute lymphocytic leukemia, and 19.2% of acute myeloid leukemias16.
These findings are intriguing and suggest an active mechanism by which epigenetic modifications may alter genome-wide gene expression programs under shift work conditions. Yet, several questions remain: are these shift-work induced changes to the epigenome a direct result of night-time light exposure, how does this work mechanistically, how quickly do these events occur, and are they reversible? These questions remain fully unresolved but addressing them would shed light on an active epigenetic mechanism(s) that is environmentally controlled by day/night rhythms to impinge on tumorigenesis.
Importantly, environmental disruption of the central/peripheral circadian axis by chronic jet lag has been modeled in mice. For instance, wild-type (WT) mice undergoing repeated jet lag manipulation display disrupted gene expression of the circadian repressors Per2 and Rev-Erba, resulting in increased growth of Glasgow osteosarcoma18 as well as enhanced incidence of lymphoma and hepatocellular carcinoma (HCC)12. While it is unclear why circadian disruption impacts specific tumors, similar experiments have been performed with a variety of mice with mutations in clock genes, specifically Cry1/2−/−, Per2−/−, or Per1−/−;Per2m/m. These mutant mice display a heightened incidence of lymphoma, osteosarcoma and HCC when subjected to severe chronic jet lag versus WT mice12. While these findings provide experimental evidence that genetic disruption of the clock leads to enhanced tumorigenesis, to date there is no direct counterpart of these studies in human cancers.
Recent laboratory data has shed light on the molecular mechanisms of circadian disruption through jetlag and its link with tumorigenesis. Long-term jetlag initiates a program of non-alcoholic fatty liver disease (NAFLD) that progresses to steatohepatitis, fibrosis and eventually HCC in mice19. Jetlag operates by disrupting both circadian gene expression programs as well as circadian metabolism in the liver, and central to this rewiring is an induction of hepatic cholesterol and bile acid levels that activate the oncogenic program of the nuclear receptor constitutive androstane receptor (CAR), and downstream activation of β-catenin19. Collectively these findings implicate a critical signaling axis that coordinates the central pacemaker with peripheral circadian transcription and metabolism, though the implications of these findings in human remains unresolved.
Circadian Control of Cancer Metabolism
Crosstalk between Myc and the Circadian Clock
Given that the clock is intimately involved in regulating metabolism in peripheral tissues20,21, and the majority of metabolites in liver and serum are controlled in a cyclic manner22–24, the intersection of cancer metabolism and its control by the circadian clock is an area of active investigation. A study reported the role of the c-Myc oncogene in regulating rhythmic metabolism in cultured U2OS human osteosarcoma cells25. The MYC protein has oncogenic potential due to its capacity to activate gene expression programs related to survival and proliferation26–28. Using an inducible system, ectopic expression of MYC was found to disrupt the expression of circadian genes. MYC was found to activate the negative transcriptional arm of the clock through REV-ERBa (see Figure 2 and Box 1), and stimulate metabolic sensing pathways such as AMPK (a kinase whose enzymatic activity depends on cellular metabolic state) ultimately leading to increased consumption of glucose and glutamine25. This specific case illustrates another scenario, where an oncoprotein disrupts circadian function with subsequent effects on cellular metabolism. It is tempting to speculate that altered metabolism could feedback on cellular growth and clock function, contributing to the unbalanced state characteristic of tumor cells.
These findings in cells cultured in vitro have been further validated in vivo by using genetically engineered mice. One study took advantage of the KrasLSL-G12D/+;p53fl/fl mouse model. These mutant mice are prone to cancer because they carry an activated KRAS oncogene and an inactivation of the tumor suppressor p5313,29. The authors combined the ablation of the clock genes, Per2 or Bmal1, with the KrasLSL-G12D/+;p53fl/fl mice and identified that genetic disruption of the circadian clock resulted in elevated consumption of glucose and glutamine and increased excretion of lactate as compared to the KrasLSL-G12D/+;p53fl/fl mice13. These studies suggest that disruption of the clock caused by the ablation of the Per2 of Bmal1 genes exacerbates the effect of the KrasLSL-G12D/+;p53fl/fl mutations, thereby illustrating that a functional clock is required to maintain proper rhythms of cancer cell metabolism in vivo.
CLOCK, BMAL1 and MYC are all transcription factors that share a highly similar basic helix-loop-helix protein domain. This allows these proteins to recognize the same promoter element, the E-box sequence30,31, in the regulatory regions of target genes. It is thus tempting to speculate that a rewiring could be taking place during tumorigenesis whereby the balance of clock-controlled transcription can be lost and consequently compensated for by oncogenic MYC signaling. In further support of this idea, the circadian repressor CRY2 has been reported to promote MYC degradation through the FBXL3-containing E3 ligase, and Cry2 deletion resulted in enhanced Myc-driven lymphomas in mice32. Also, it has been shown that MYC and its binding partner MIZ1 are responsible for forming a repressive complex which down-regulates core clock gene expression33. Further confirming these findings, the expression of BMAL1 was found to be inversely correlated with MYC in 102 human lymphoma samples33. These data suggest a counterbalance may exist between the transcriptional networks of the circadian clock and MYC. Yet, what remains to be fully elucidated is the functional significance of this potential transcriptional switch from canonical CLOCK:BMAL1 control to MYC-dependent signaling, and therefore the extent of the prospective shared gene expression network in normal and transformed cells that regulates metabolism.
Although several studies described above have utilized human cells cultured in vitro or genetically engineered mouse models, the relevance of these studies clinically is hard to assess. Yet, it could be speculated that alteration in human metabolism brought by, for example, nutritional challenges, could disrupt circadian homeostasis in ways that would parallel some mouse models of clock disruption. Indeed, the effects of nutritional challenge on circadian reprogramming and alterations in rhythmic homeostasis are reported23,24,34–37. Studies in mouse models as well as in human subjects have shown the remarkable effect of specific dietary regimes such as high-fat diet23,24,38 as well as the effect of time-restricted feeding24,34,39–41 on the circadian clock. Also, metabolomics analysis in human serum/plasma and skeletal muscle has identified major populations of rhythmic metabolites over the circadian cycle42,43. Therefore, further investigation is required to determine the role of nutritional inputs on circadian metabolism and tumorigenesis.
The Circadian Clock and Oxidative Stress in Cancer
Oxygen is critical for cellular respiration and hypoxia has been shown to play a regulatory role in tumorigenesis linked to metabolism and angiogenesis26,27. Moreover, a transcriptional crosstalk exists between the clock and the transcription factor hypoxia-inducible factor (HIF), a potential scenario that may be extended to tumorigenesis. The promoter element recognized by HIF, the hypoxia-response element (HRE), is an E-Box like sequence that contributes to transcription under low oxygen conditions. The HRE is recognized by HIF heterodimers consisting of two highly similar proteins, HIF1a and HIF1b. Recent reports indicate that the hypoxic response is gated by the circadian clock as the Hif1a promoter is directly controlled by CLOCK:BMAL144. Similarly, blood oxygen levels were found to exhibit daily rhythms which influences expression of core clock genes in a HIF1a-dependent manner in kidney, brain and hepatocytes45. This transcriptional crosstalk was further supported by the fact that genetic disruption of Bmal1 in C2C12 myotubes results in reduced anaerobic glycolysis, the gene targets of which are HIF1a-dependent46. These studies establish an intriguing link between the transcriptional networks of the clock system with HIF1a, and potentially suggest that this crosstalk could be involved in the hypoxic response during tumorigenesis, an area that requires further investigation. In addition, it remains to be determined if glycolytic metabolism of cancer cells could depend on a HIF1a axis that would interplay with the clock machinery.
Interestingly, several lines of epidemiological evidence connecting circadian disruption by shift-work with cancer has largely focused on hormone-dependent diseases such as breast and prostate cancer, which raises the possibility that additional clock-controlled endocrine factors may be at play. One such link has been made with melatonin, a hormone produced by the pineal gland in a circadian manner to regulate sleep47,48. Melatonin has been linked to regulation of oxidative stress mostly in the mitochondria49, where melatonin is reported to stimulate the activity of glutathione peroxidase (GPx) and glutathione reductase (GRd), two enzymes involved in the regulation of GSH:GSSG glutathione ratio50,51. Interestingly, melatonin regulates the mitochondrial respiratory complexes I and IV, and thereby modulates ATP production50,52. Therefore, melatonin may antagonize the glycolytic dependency of cancer cell metabolism in a time-dependent manner by targeting mitochondrial function53. Notably, dampened melatonin secretion caused by sleep disruption may increase ROS levels and reactive nitrogen species (RNS) production as suggested by studies in night-shift workers54.
Tumor macroenvironment and circadian metabolism
The tumor environment can be extended beyond the microenvironment to a systems level approach looking at tumor/host interactions that are especially relevant to the circadian metabolic clock (Figure 4). The tumor ‘macroenvironment’ consists of metabolites and other tumor-secreted factors (such as cytokines and chemokines) that circulate in the blood55. Accumulating evidence illustrates the significant role that the tumor macroenvironment may play in connecting systemic metabolism and cell proliferation56,57. In this respect, specific components of the circadian clock machinery may be highly susceptible to factors secreted by the tumor, given that the clock is especially vulnerable to metabolic fluctuations such as those caused by different types of nutritional regimes23,24,34,37. Using the KrasLSL-G12D/+;p53fl/fl mouse model, lung adenocarcinoma was found to distally rewire the circadian transcriptome and metabolome in the liver58. This finding illustrates a communication system from the lung to the liver mediated by the tumor macroenvironment, as revealed by changes in the metabolite composition in the serum. Tumor-dependent inflammation through the IL-6 pathway dampened insulin/glucose sensitivity and altered hepatic circadian lipid metabolism58. Similarly, using a mouse model of triple negative breast cancer, rewiring of circadian gene expression was distally observed in the liver, resulting in increased oxidative stress59. These findings raise the possibility that the tumor-host interaction may influence cancer cell viability, and specifically circadian oscillations of metabolism may be highly susceptible to systemic cues that reorganize physiological homeostasis. To date, rewiring of circadian metabolism in response to tumorigenesis has been reported within the context of lung and breast cancer in mouse models, but other types of cancers that function similarly as well as the extent of this rewiring in all peripheral tissues systemically remain unknown.
Figure 4. Tumor/host communication involving circadian metabolic tissues.
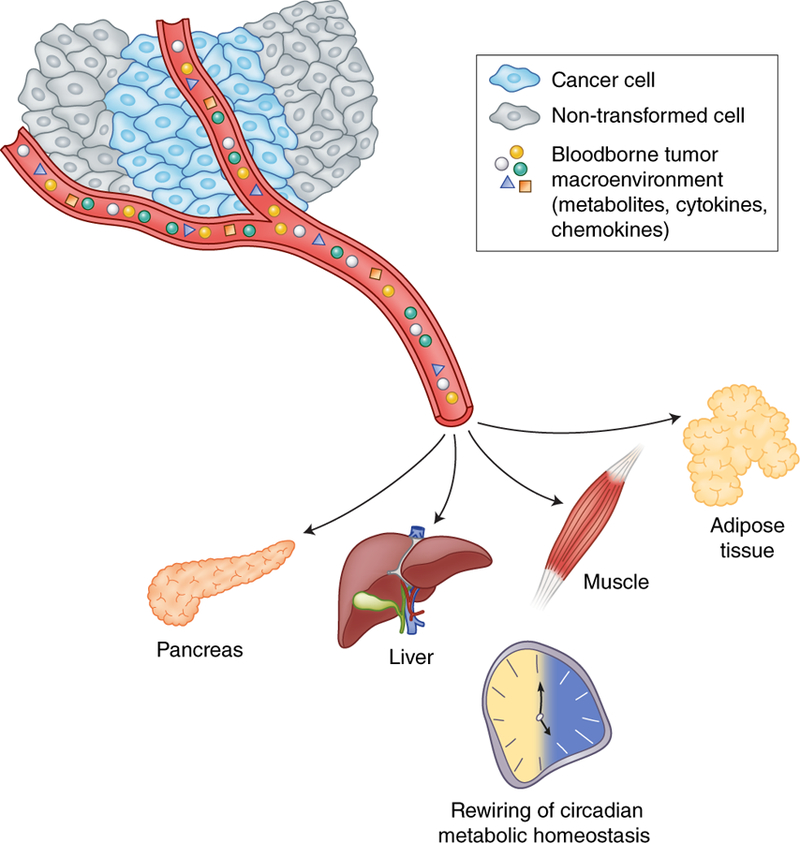
The tumor macroenvironment, comprised of inflammatory cytokines, chemokines, glycolytic byproducts, and other tumor-derived waste is secreted into the blood. Emerging evidence now suggests that metabolic waste byproducts, such as lactate, can potentially be utilized as carbon sources to satisfy the demand of rapidly proliferating cells61. The circulating tumor macroenvironment has been described to rewire circadian metabolism at a distance58,59. Target metabolic tissues include the liver, but likely extend to the pancreas, adipose tissue and skeletal muscle as shown. In addition, the role of peripheral tissues in further driving tumorigenesis, by potentially supplying metabolic fuel, emphasizes the significance of the tumor/host interaction.
Yet, can the circadian clock feedback and regulate properties of tumorigenesis? If so, what are these circadian cues that modulate cancer progression systemically? Interestingly, BMAL2 has been reported to regulate lung adenocarcinoma distal metastasis through a unique tumor-dependent ‘secretome.’ Using the KrasLSL-G12D/+;p53fl/fl;R26LSL-Tom lung adenocarcinoma model that utilizes a tomato reporter to track sites of metastases, BMAL2 was found to be highly expressed in primary metastatic tumors60. Also, BMAL2 was found to be required for metastasis by regulating the expression of secreted modular calcium-binding protein 2 (Smoc2), which is critical for anchorage-independent growth and metastatic seeding in vivo60. BMAL2 expression has been associated with lung adenocarcinoma metastases in humans while being silenced within the tumor. Parallel experiments in cell culture and in the mouse have shown that BMAL2 contributes to the clonogenic ability of lung adenocarcinoma cells and it directs the activation of genes encoding secreted factors. These in turn are involved in pro-metastatic function, indeed revealing that the clock protein BMAL2 is involved in regulating a metastatic ‘secretome’ consisting of Smoc2, Wnt5a and Ccl760. These findings suggest a crosstalk between the circadian clock and cancer cells through the tumor-derived macroenvironment and describe the ability of canonical and non-canonical clock components in regulating a metastatic ‘secretome’. Further investigation is needed to better understand how the clock controls metastasis in different cancer types, and how this tumor-dependent ‘secretome’ may differ based on disease profile.
Importantly, evidence also suggests that tumor-derived waste can be repurposed and subsequently utilized as fuel for tumors. Clinical and laboratory studies have provided evidence that non-small cell lung cancer (NSCLC) exhibit metabolic heterogeneity in fuel utilization and, while glucose oxidation is metabolically imperative, lactate can also be used as a carbon source61–63. A similar example exists with nitrogen repurposing of ammonia waste that was reported to be recycled into central amino acid metabolism through the enzymatic activity of glutamate dehydrogenase (GDH) in breast cancer cells64. These studies suggest a mechanism by which the tumor-derived macroenvironment can be recycled as alternative fuel sources for rapidly proliferating cells. Given the fundamental role of the circadian clock in regulating metabolism, including pathways involved in carbohydrate, amino acid, fatty acid/lipid metabolism20,65, it is plausible that the circadian clock may be involved in regulating these processes in the context of cancer. Also, it is possible that tumor-dependent metabolic waste may be secreted in a temporal manner, suggesting peak times of day whereby these metabolic pathways could be targeted pharmacologically.
Clinical implications: relevance of ‘chronotherapy’ for cancer
The multiple connections discussed here between clock disruption and cancer beg the question of whether circadian therapeutic intervention (such as time of day) should be considered. This so-called ‘chronotherapy’ has been recently reviewed in depth66, and strongly supports the idea that metabolic and xenobiotic detoxification enzymes exhibit temporal peaks in activity and thus should be pharmacologically targeted based on optimal time of day.
Several pathways relevant to the circadian clock are currently being targeted for cancer therapy, and we highlight several potential avenues whereby circadian intervention approaches (such as time of day) could be considered. For instance, inhibition of acetyl-CoA carboxylase (ACC) and subsequent decreased fatty acid synthesis in mouse models of lung adenocarcinoma effectively dampened tumor growth67. Similarly, acetyl-CoA synthetase (ACSS2) has been reported to supply tumors with acetate-dependent acetyl-CoA that is used for de novo lipogenesis and histone acetylation68,69. These studies implicate pathways that have been previously reported to be clock-controlled. For example, the enzymatic activity of ACSS2 is modulated by NAD+-dependent SIRT1 and a functional clock to control acetyl-CoA production and fatty acid synthesis70. Therefore, this raises the possibility that changes in clock-controlled ACSS2 activity can potentially influence tumorigenesis and suggest that a time of day intervention approach should be considered. Recent evidence also suggests that pharmacological agonists of REV-ERB are selectively lethal to cancer cells and impair growth of glioblastoma in vivo71. These findings require further examination to determine if this type of approach would be effective in other cancer types.
Metformin has been reported to impair respiration by inhibiting mitochondrial complex I and altering the NAD+/NADH ratio72. This treatment strategy for cancer is especially appealing from a circadian perspective given that the clock machinery is subject to NAD+-dependent control through the mammalian sirtuins73–76. Also, the circadian clock is reported to regulate the cyclic availability of NAD+ by controlling the rhythmic activity of nicotinamide phosphoribosyltransferase (NAMPT)77,78, a critical enzyme in the NAD+-salvage pathway. Therefore, future pharmacological intervention may need to consider possible strategies to restore clock function, especially at late stages of cancer progression when a decline of circadian output is believed to occur. Indeed, rescue of carcinogen-dependent decline of the NAD+/NADH ratio in mammary epithelial cells restored SIRT1 activity and rhythmic expression of the circadian gene Per279. Conversely, an alternative treatment strategy involves suppression of NAD+ levels to inhibit tumor growth, given the elevated metabolic demand of rapidly proliferating cells. For instance, pharmacological suppression of NAMPT and the subsequent depletion of intracellular NAD+ levels has been demonstrated to induce apoptosis in several leukemic cell lines and abrogate tumor growth in mouse models of AML80,81. This NAD+-starvation strategy requires further experimental validation as it remains unclear what is the resulting effect on circadian function. Yet, given that the clock is reported to be required for AML development 82, suppression of NAD+ levels in hematologic tumors may differ from solid tumors in treatment efficacy.
Additionally, the circadian hormone melatonin has been used in combination with cancer therapy to minimize toxicity or enhance chemotherapeutic viability in clinical and laboratory settings. For example, melatonin protects against cisplatin-induced ROS production and mitochondrial damage through glutathione in mouse ovaries to minimize reproductive toxicity83. Melatonin has also been used in combination with 5-Fluorouracil (5-FU) to enhance the inhibitory effect on colon cancer cell proliferation by suppression of the PI3K/AKT survival pathway and NFkB-dependent activation of inducible nitric oxide synthase (iNOS) signaling pathway84. Lastly, melatonin has been reported to inhibit the epithelial to mesenchymal transition (EMT) by increasing E-cadherin expression and decreasing the migratory/invasive capacity of breast cancer cells in culture85 and inhibiting EMT through a GSK3b-dependent mechanism in vivo86. Clinical evidence similarly suggests an improvement in the 5-year survival rate of metastatic NSCLC patients treated with melatonin in combination with cisplatin and etoposide87. Yet, conflicting clinical evidence exists regarding the use and efficacy of melatonin and its potential benefits towards quality of life88, therefore further investigations are required to fully understand the potential beneficial effects of melatonin action.
Finally, circadian analyses of the tumor macroenvironment could have unique value clinically. Indeed, considering the complexity of the tumor-host relationship and the potential for this communication to be constantly changing, the tumor-dependent ‘secretome’ has value as a non-invasive prognostic tool. In this respect it is noteworthy that an increase in plasma branched-chain amino acids (BCAA) is associated with an increased risk for diagnosis of pancreatic cancer89. Also, the use of blood-borne tumor-secreted metabolites has been documented with colorectal cancer90. Similar strategies have been used in patients with advanced breast cancer91 or diagnosed with pancreatic cancer92. Interestingly, profiling the metabolome can also be utilized to gauge treatment efficacy. Indeed, serum metabolome from breast cancer patients treated with neo-adjuvant chemotherapy identified metabolites responsive to treatment, including threonine, isoleucine, glutamine and linoleic acid93. Thus far, these studies have not been conducted over the day/night cycle to determine if availability of these metabolites is found to be rhythmic. While these studies could have a clinical value, we would argue that the time of day could be an important factor to consider given that metabolites such as amino acids and lipids display circadian profiles94,95.
Concluding Remarks
The complex network of communication between the circadian clock and tumorigenesis is only beginning to be unraveled (Figure 3). At the foundation is strong epidemiological evidence that implicates circadian disruption with cancer. Several lines of genetic evidence from laboratory studies connect disruption of the circadian molecular machinery with lymphoma, HCC, lung cancer, and other tumor types. Intriguingly, preliminary evidence supports a circadian connection with cancer metabolism, in an oncogene-driven cell autonomous manner, such as with MYC. Given that cancer cells utilize nutrients at a high metabolic rate, it can be envisioned that several pathways may be controlled by the circadian clock. However, it remains to be determined how the clock may impinge on such pathways, including amino acid metabolism and availability, the pentose phosphate pathway, and other intracellular energy generating mechanisms that can potentially be hijacked by cancer cells. Several metabolic pathways are dynamically circadian and hence the time at which therapeutic targeting of these pathways occurs may be critical. Furthermore, several lines of evidence discussed in this review point to systemic nutrient repurposing as a means of circumventing typical energy requirements in order to sustain heightened cell proliferation in cancer. These ideas focusing on the circadian clock should be considered when developing pharmacological approaches to target tumor metabolism to dictate survival and proliferation.
Box 2: Cancer-Initiating Cells and the Circadian Clock.
Studies using genetic mouse models have revealed that the circadian clock is implicated in determining unique properties of tissue-specific stem cell populations127–129. As these stem cells are thought to share important features with cancer initiating cells, a question is raised: does a stem cell clock differ from other clocks and, do cancer-initiating cells have a distinct circadian transcriptional program? By extension, it could be speculated that common changes in clock function could be causal to determine both ‘stemness’ and cancer phenotypes. A number of studies support the notion that elements of the circadian clock regulate stem cell functions. For example, rhythmic oscillations of key secreted factors in the skin such as bone morphogenetic protein (BMP) regulate stem cells involved in hair regeneration130 and BMAL1 transcriptionally controls stem cell regulatory genes to specify epidermal stem cell heterogeneity127. Also, a critical chemokine for hematopoietic stem cell (HSC) migration, CXCL12, displays clock-controlled expression131. Recent evidence in aged epidermal and muscle stem cells versus hepatocytes demonstrates very different tissue-specific transcriptional control pathways that are unique to stem cell populations128,129.
Yet, can the circadian clock alter tumor initiation potential through cancer-initiating cells? Mice carrying an epidermal deletion of Bmal1 crossed with oncogenic Sos (which activates the Ras pathway) displayed fewer squamous tumors in the tail versus control keratin5 expressing Sos mice which display 100% penetrance of neoplastic lesions127. However, using a similar keratinocyte-specific mouse model, it was also found that Bmal1 ablation increased cell proliferation and elevated susceptibility to UV-induced DNA damage132. Therefore, how Bmal1 is involved in tumorigenesis through cancer-initiating cells remains unresolved. Moreover, disruption of the canonical circadian molecular machinery depletes leukemic stem cells and Clock and Bmal1 were reported to be required for growth of acute myeloid leukemia (AML)82. These studies suggest that clock function could act in a dual manner in regulating tumorigenesis at the levels of initiation and disease progression. Based on these differing reports, the role of the circadian clock in cancer-initiating cells requires further investigation.
This concept of a dual functioning circadian clock can be further extended, and interestingly, there are conflicting reports of clock disruption in specific cancer types. For instance, hypermethylation at the Clock promoter was found to reduce risk of breast cancer and reduced levels of Clock expression was found in healthy controls relative to breast cancer tumors133. Yet, breast cancer tissue was also found to exhibit hypermethylation of the Cry2 promoter relative to normal controls in ER/PR-negative breast cancers, but not in ER/PRpositive tumors 134. These studies point to possible differences in circadian function and transcriptional output based on cell of origin. Therefore, this prompts further investigation into how the circadian machinery in stem cells versus differentiated cell types differs. Also, further dissection of the changes in circadian gene expression programs within cancer cell subtypes is needed in addition to analysis of how the clock machinery is altered over the course of transformation, from adenomas to adenocarcinomas.
Acknowledgements
The Masri laboratory is supported by a K22 Transition Career Development Award through the National Cancer Institute (NCI), the Concern Foundation, and the V Foundation for Cancer Research. Work in the Sassone-Corsi laboratory is supported by NIH (National Institutes of Health), INSERM (Institut National de la Sante et la Recherche Medicale, France) and KAUST (King Abdullah University of Science and Technology, Saudi Arabia).
Footnotes
The authors declare no conflicts of interest.
References:
- 1.Fu L & Lee CC The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer 3, 350–361 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Sahar S & Sassone-Corsi P Metabolism and cancer: the circadian clock connection. Nat Rev Cancer 9, 886–896 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Masri S, Kinouchi K & Sassone-Corsi P Circadian clocks, epigenetics, and cancer. Curr Opin Oncol 27, 50–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lie JA, Roessink J & Kjaerheim K Breast cancer and night work among Norwegian nurses. Cancer Causes Control 17, 39–44 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Papantoniou K, et al. Night shift work, chronotype and prostate cancer risk in the MCCSpain case-control study. Int J Cancer 137, 1147–1157 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Schernhammer ES, et al. Rotating night shifts and risk of breast cancer in women participating in the nurses’ health study. J Natl Cancer Inst 93, 1563–1568 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Straif K, et al. Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol 8, 1065–1066 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Knutsson A, et al. Breast cancer among shift workers: results of the WOLF longitudinal cohort study. Scand J Work Environ Health 39, 170–177 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Kakizaki M, et al. Sleep duration and the risk of prostate cancer: the Ohsaki Cohort Study. Br J Cancer 99, 176–178 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srour B, et al. Circadian nutritional behaviours and cancer risk: New insights from the NutriNet-sante prospective cohort study: Disclaimers. Int J Cancer (2018). [DOI] [PubMed] [Google Scholar]
- 11.Fu L, Pelicano H, Liu J, Huang P & Lee C The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111, 41–50 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Donehower LA, Herron AJ, Moore DD & Fu L Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One 5, e10995 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papagiannakopoulos T, et al. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab 24, 324–331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filipski E, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst 94, 690–697 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Chen ST, et al. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis 26, 1241–1246 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi H, et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res 69, 8447–8454 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Zhu Y, et al. Epigenetic impact of long-term shiftwork: pilot evidence from circadian genes and whole-genome methylation analysis. Chronobiol Int 28, 852–861 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filipski E, et al. Effects of chronic jet lag on tumor progression in mice. Cancer Res 64, 7879–7885 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Kettner NM, et al. Circadian Homeostasis of Liver Metabolism Suppresses Hepatocarcinogenesis. Cancer Cell 30, 909–924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asher G & Sassone-Corsi P Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Marcheva B, et al. Circadian clocks and metabolism. Handb Exp Pharmacol, 127–155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbondante S, Eckel-Mahan KL, Ceglia NJ, Baldi P & Sassone-Corsi P Comparative Circadian Metabolomics Reveal Differential Effects of Nutritional Challenge in the Serum and Liver. J Biol Chem 291, 2812–2828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckel-Mahan KL, et al. Reprogramming of the Circadian Clock by Nutritional Challenge. Cell 155, 1464–1478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatori M, et al. Time-Restricted Feeding without Reducing Caloric Intake Prevents Metabolic Diseases in Mice Fed a High-Fat Diet. Cell Metab 15, 848–860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altman BJ, et al. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metab 22, 1009–1019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646674 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Hanahan D & Weinberg RA The hallmarks of cancer. Cell 100, 57–70 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Gaucher J, Montellier E & Sassone-Corsi P Molecular Cogs: Interplay between Circadian Clock and Cell Cycle. Trends Cell Biol 28, 368–379 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15, 3243–3248 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ripperger JA & Schibler U Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet 38, 369–374 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Walhout AJ, Gubbels JM, Bernards R, van der Vliet PC & Timmers HT cMyc/Max heterodimers bind cooperatively to the E-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene. Nucleic Acids Res 25, 1493–1501 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber AL, et al. CRY2 and FBXL3 Cooperatively Degrade c-MYC. Mol Cell 64, 774–789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shostak A, et al. MYC/MIZ1-dependent gene repression inversely coordinates the circadian clock with cell cycle and proliferation. Nat Commun 7, 11807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaix A, Zarrinpar A, Miu P & Panda S Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab 20, 991–1005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tognini P, et al. Distinct Circadian Signatures in Liver and Gut Clocks Revealed by Ketogenic Diet. Cell Metab 26, 523–538 e525 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Oishi K, Uchida D & Itoh N Low-carbohydrate, high-protein diet affects rhythmic expression of gluconeogenic regulatory and circadian clock genes in mouse peripheral tissues. Chronobiol Int 29, 799–809 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Dyar KA, et al. Atlas of Circadian Metabolism Reveals System-wide Coordination and Communication between Clocks. Cell 174, 1571–1585 e1511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A 106, 21453–21458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14, 2950–2961 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stokkan KA, Yamazaki S, Tei H, Sakaki Y & Menaker M Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Dallmann R, Viola AU, Tarokh L, Cajochen C & Brown SA The human circadian metabolome. Proc Natl Acad Sci U S A 109, 2625–2629 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato S, Parr EB, Devlin BL, Hawley JA & Sassone-Corsi P Human metabolomics reveal daily variations under nutritional challenges specific to serum and skeletal muscle. Molecular Metabolism 16(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Y, et al. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab 25, 73–85 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Adamovich Y, Ladeuix B, Golik M, Koeners MP & Asher G Rhythmic Oxygen Levels Reset Circadian Clocks through HIF1alpha. Cell Metab 25, 93–101 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Peek CB, et al. Circadian Clock Interaction with HIF1alpha Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle. Cell Metab 25, 86–92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arendt J, Bojkowski C, Franey C, Wright J & Marks V Immunoassay of 6hydroxymelatonin sulfate in human plasma and urine: abolition of the urinary 24-hour rhythm with atenolol. J Clin Endocrinol Metab 60, 1166–1173 (1985). [DOI] [PubMed] [Google Scholar]
- 48.Zawilska JB, Skene DJ & Arendt J Physiology and pharmacology of melatonin in relation to biological rhythms. Pharmacol Rep 61, 383–410 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Reiter RJ, Tan DX, Manchester LC & Qi W Biochemical reactivity of melatonin with reactive oxygen and nitrogen species: a review of the evidence. Cell Biochem Biophys 34, 237–256 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Reiter RJ, Tan DX, Manchester LC & El-Sawi MR Melatonin reduces oxidant damage and promotes mitochondrial respiration: implications for aging. Ann N Y Acad Sci 959, 238–250 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Leon J, Acuna-Castroviejo D, Escames G, Tan DX & Reiter RJ Melatonin mitigates mitochondrial malfunction. J Pineal Res 38, 1–9 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Martin M, et al. Melatonin-induced increased activity of the respiratory chain complexes I and IV can prevent mitochondrial damage induced by ruthenium red in vivo. J Pineal Res 28, 242–248 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Proietti S, Cucina A, Minini M & Bizzarri M Melatonin, mitochondria, and the cancer cell. Cell Mol Life Sci (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhatti P, et al. Oxidative DNA damage during night shift work. Occup Environ Med (2017). [DOI] [PubMed] [Google Scholar]
- 55.Al-Zoughbi W, et al. Tumor macroenvironment and metabolism. Semin Oncol 41, 281295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee YM, Chang WC & Ma WL Hypothesis: solid tumours behave as systemic metabolic dictators. J Cell Mol Med 20, 1076–1085 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rutkowski MR, Svoronos N, Perales-Puchalt A & Conejo-Garcia JR The Tumor Macroenvironment: Cancer-Promoting Networks Beyond Tumor Beds. Adv Cancer Res 128, 235–262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masri S, et al. Lung Adenocarcinoma Distally Rewires Hepatic Circadian Homeostasis. Cell 165, 896–909 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hojo H, et al. Remote reprogramming of hepatic circadian transcriptome by breast cancer. Oncotarget 8, 34128–34140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brady JJ, et al. An Arntl2-Driven Secretome Enables Lung Adenocarcinoma Metastatic Self-Sufficiency. Cancer Cell 29, 697–710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hensley CT, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell 164, 681–694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Faubert B, et al. Lactate Metabolism in Human Lung Tumors. Cell 171, 358–371 e359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hui S, et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spinelli JB, et al. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bass J & Takahashi JS Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dallmann R, Okyar A & Levi F Dosing-Time Makes the Poison: Circadian Regulation and Pharmacotherapy. Trends Mol Med 22, 430–445 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Svensson RU, et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med 22, 11081119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Comerford SA, et al. Acetate dependence of tumors. Cell 159, 1591–1602 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schug ZT, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 27, 57–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sahar S, et al. Circadian Control of Fatty Acid Elongation by SIRT1-mediated Deacetylation of Acetyl-CoA Synthetase 1. J Biol Chem (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sulli G, et al. Pharmacological activation of REV-ERBs is lethal in cancer and oncogeneinduced senescence. Nature 553, 351–355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gui DY, et al. Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab 24, 716–727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Masri S, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aguilar-Arnal L, Katada S, Orozco-Solis R & Sassone-Corsi P NAD(+)-SIRT1 control of H3K4 trimethylation through circadian deacetylation of MLL1. Nat Struct Mol Biol 22, 312–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakahata Y, Sahar S, Astarita G, Kaluzova M & Sassone-Corsi P Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang M, Guo WR, Park Y, Kang HG & Zarbl H Enhancement of NAD(+)-dependent SIRT1 deacetylase activity by methylselenocysteine resets the circadian clock in carcinogen-treated mammary epithelial cells. Oncotarget 6, 42879–42891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nahimana A, et al. The NAD biosynthesis inhibitor APO866 has potent antitumor activity against hematologic malignancies. Blood 113, 3276–3286 (2009). [DOI] [PubMed] [Google Scholar]
- 81.Thakur BK, et al. Involvement of p53 in the cytotoxic activity of the NAMPT inhibitor FK866 in myeloid leukemic cells. Int J Cancer 132, 766–774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Puram RV, et al. Core Circadian Clock Genes Regulate Leukemia Stem Cells in AML. Cell 165, 303–316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barberino RS, et al. Melatonin protects against cisplatin-induced ovarian damage in mice via the MT1 receptor and antioxidant activity. Biol Reprod 96, 1244–1255 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Gao Y, et al. Melatonin synergizes the chemotherapeutic effect of 5-fluorouracil in colon cancer by suppressing PI3K/AKT and NF-kappaB/iNOS signaling pathways. J Pineal Res 62(2017). [DOI] [PubMed] [Google Scholar]
- 85.Goncalves Ndo N, et al. Effect of Melatonin in Epithelial Mesenchymal Transition Markers and Invasive Properties of Breast Cancer Stem Cells of Canine and Human Cell Lines. PLoS One 11, e0150407 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mao L, et al. Circadian gating of epithelial-to-mesenchymal transition in breast cancer cells via melatonin-regulation of GSK3beta. Mol Endocrinol 26, 1808–1820 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lissoni P, Chilelli M, Villa S, Cerizza L & Tancini G Five years survival in metastatic non-small cell lung cancer patients treated with chemotherapy alone or chemotherapy and melatonin: a randomized trial. J Pineal Res 35, 12–15 (2003). [DOI] [PubMed] [Google Scholar]
- 88.Del Fabbro E, Dev R, Hui D, Palmer L & Bruera E Effects of melatonin on appetite and other symptoms in patients with advanced cancer and cachexia: a double-blind placebo-controlled trial. J Clin Oncol 31, 1271–1276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mayers JR, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med 20, 1193–1198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nishiumi S, et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS One 7, e40459 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jobard E, et al. A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett 343, 33–41 (2014). [DOI] [PubMed] [Google Scholar]
- 92.Kobayashi T, et al. A novel serum metabolomics-based diagnostic approach to pancreatic cancer. Cancer Epidemiol Biomarkers Prev 22, 571–579 (2013). [DOI] [PubMed] [Google Scholar]
- 93.Wei S, et al. Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol Oncol 7, 297–307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aviram R, et al. Lipidomics Analyses Reveal Temporal and Spatial Lipid Organization and Uncover Daily Oscillations in Intracellular Organelles. Mol Cell 62, 636–648 (2016). [DOI] [PubMed] [Google Scholar]
- 95.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A 109, 5541–5546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haus E, Lakatua DJ, Swoyer J & Sackett-Lundeen L Chronobiology in hematology and immunology. Am J Anat 168, 467–517 (1983). [DOI] [PubMed] [Google Scholar]
- 97.Scheiermann C, Kunisaki Y & Frenette PS Circadian control of the immune system. Nat Rev Immunol 13, 190–198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Partch CL, Green CB & Takahashi JS Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24, 90–99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bass J Circadian topology of metabolism. Nature 491, 348–356 (2012). [DOI] [PubMed] [Google Scholar]
- 100.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 (1998). [DOI] [PubMed] [Google Scholar]
- 101.Masri S & Sassone-Corsi P Plasticity and specificity of the circadian epigenome. Nat Neurosci 13, 1324–1329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 (2002). [DOI] [PubMed] [Google Scholar]
- 103.Akhtar RA, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12, 540–550 (2002). [DOI] [PubMed] [Google Scholar]
- 104.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9, e1000595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Duong HA, Robles MS, Knutti D & Weitz CJ A molecular mechanism for circadian clock negative feedback. Science 332, 1436–1439 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 (2002). [DOI] [PubMed] [Google Scholar]
- 108.Sato TK, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43, 527–537 (2004). [DOI] [PubMed] [Google Scholar]
- 109.Masri S & Sassone-Corsi P The circadian clock: a framework linking metabolism, epigenetics and neuronal function. Nat Rev Neurosci 14, 69–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Welsh DK, Takahashi JS & Kay SA Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol 72, 551–577 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685 (2000). [DOI] [PubMed] [Google Scholar]
- 112.Welsh DK, Logothetis DE, Meister M & Reppert SM Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706 (1995). [DOI] [PubMed] [Google Scholar]
- 113.Reppert SM & Weaver DR Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 63, 647–676 (2001). [DOI] [PubMed] [Google Scholar]
- 114.Welsh DK, Yoo SH, Liu AC, Takahashi JS & Kay SA Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14, 2289–2295 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pando MP, Morse D, Cermakian N & Sassone-Corsi P Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell 110, 107–117 (2002). [DOI] [PubMed] [Google Scholar]
- 116.Thresher RJ, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282, 1490–1494 (1998). [DOI] [PubMed] [Google Scholar]
- 117.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450, 1086–1090 (2007). [DOI] [PubMed] [Google Scholar]
- 118.Nader N, Chrousos GP & Kino T Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: potential physiological implications. FASEB J 23, 1572–1583 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Doi M, Hirayama J & Sassone-Corsi P Circadian regulator CLOCK is a histone acetyltransferase. Cell 125, 497–508 (2006). [DOI] [PubMed] [Google Scholar]
- 120.Hung HC, Maurer C, Kay SA & Weber F Circadian transcription depends on limiting amounts of the transcription co-activator nejire/CBP. J Biol Chem 282, 31349–31357 (2007). [DOI] [PubMed] [Google Scholar]
- 121.Hosoda H, et al. CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol Brain 2, 34 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Alenghat T, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456, 997–1000 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang HC & Guarente L SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 153, 1448–1460 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Katada S & Sassone-Corsi P The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol 17, 1414–1421 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Valekunja UK, et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci U S A 110, 1554–1559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Janich P, et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature 480, 209–214 (2011). [DOI] [PubMed] [Google Scholar]
- 128.Sato S, et al. Circadian Reprogramming in the Liver Identifies Metabolic Pathways of Aging. Cell 170, 664–677 e611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Solanas G, et al. Aged Stem Cells Reprogram Their Daily Rhythmic Functions to Adapt to Stress. Cell 170, 678–692 e620 (2017). [DOI] [PubMed] [Google Scholar]
- 130.Plikus MV, et al. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature 451, 340–344 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mendez-Ferrer S, Lucas D, Battista M & Frenette PS Haematopoietic stem cell release is regulated by circadian oscillations. Nature 452, 442–447 (2008). [DOI] [PubMed] [Google Scholar]
- 132.Geyfman M, et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci U S A 109, 11758–11763 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hoffman AE, et al. CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses. Cancer Res 70, 1459–1468 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hoffman AE, et al. The core circadian gene Cryptochrome 2 influences breast cancer risk, possibly by mediating hormone signaling. Cancer Prev Res (Phila) 3, 539–548 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]


