Abstract
Super-resolution microscopy has overcome a long-held resolution barrier – the diffraction limit – in light microscopy and enabled visualization of previously invisible molecular details in biological systems. Since their conception, super-resolution imaging methods have continually evolved, and can now be used to image cellular structures in three dimensions, multiple colors, and living systems with nanometer-scale resolution. These methods have been applied to answer questions involving the organization, interaction, stoichiometry, and dynamics of individual molecular building blocks and their integration into functional machineries in cells and tissues. In this review, we provide an overview of super-resolution methods, their state-of-the-art capabilities, and their constantly expanding applications to biology, with a focus on the latter. We will also describe the current technical challenges and future advances anticipated in super-resolution imaging.
Fluorescence microscopy has been central in shaping our understanding of the molecular organizations and interactions of biological systems. Its high molecular specificity and multi-color imaging capability allow direct visualization of interactions between specific molecular species, and its low invasiveness allows the study of living systems under physiological conditions. However, a main challenge in fluorescence microscopy was the limited spatial resolution set by the diffraction of light. This resolution limit, first described by Ernst Abbe in 1873, restricts the smallest objects that can be resolved using conventional light microscopes. As a result, objects separated by a distance smaller than approximately half of the wavelength of visible light, i.e. ~200–300 nm, are indistinguishable, making many molecular structures in cells unresolvable. The advent of super-resolution imaging methods has shattered this limit. In this review, we will provide an overview of the methods that surpass the diffraction limit in the far field, with emphasis on the new biological insights provided by these methods.
Overview of super-resolution imaging methods
The key to overcoming the diffraction limit lies in the ability to distinguish molecules that reside within the same diffraction-limited volume. This has been achieved by two main categories of approaches. The first category accomplishes this in a spatially coordinated manner by using patterned illumination to differentially modulate the fluorescence emission of molecules within the diffraction-limited volume and thereby achieve separate detection of these molecules. The pioneering method in this category is STimulated Emission Depletion (STED) microscopy (1, 2), subsequently generalized to REversible Saturable Optical Linear Fluorescence Transitions (RESOLFT) (3). STED and RESOLFT overcome the diffraction limit by accompanying a focused excitation beam with a spatially patterned “depletion” beam, typically in a donut shape, which serves to counteract excitation through either stimulated emission (STED) (1, 2) or other types of fluorescence transitions, such as photoswitching (RESOLFT) (3). As a result, only molecules at the very center of the donut-shaped beam (where the laser intensity is near zero) can emit light, thus creating a region of fluorescence emission that is much smaller than a typical focal spot of the light microscope. The reverse strategy is also possible, with the donut beam serving as patterned activation rather than depletion, limiting the emission-free region instead of emission region to the center of the beam (4). Scanning these beams across the sample then generates an image with a resolution much higher than the diffraction limit. Various other illumination patterns can also be used to increase the spatial frequency of the emission region and hence the image resolution (4). For example, in Structured Illumination Microscopy (SIM), the sample is excited by a series of standing waves with different orientations or phases to increase the spatial frequency detectable by the microscope (5). Because the standing-wave pattern is itself limited by diffraction, the linear form of SIM only extends the diffraction limit by a factor of two, whereas the nonlinear form of SIM (NL-SIM) overcomes the diffraction limit by using the nonlinear or saturated response of fluorophores to further increase the spatial frequency of the emission pattern (5), similar to STED and RESOLFT (4). Unlike STED and RESOLFT, which generate super-resolution images directly from the recorded raw data, SIM and NL-SIM require additional computational treatment to reconstruct final images (4, 5).
The second category of methods achieve the separation of molecules by stochastically turning on individual molecules within the diffraction-limited volume at different time points, including STochastic Optical Reconstruction Microscopy (STORM) (6) and (Fluorescence) Photoactivated Localization Microscopy ((F)PALM) (7, 8), and subsequent variations of these approaches (9, 10). When isolated in space, the positions of individual molecules can be determined to nanometer or even sub-nanometer precision by localizing the center positions of their images (11–13). However, molecules within the same diffraction-limited volume generate overlapping images, which is the fundamental cause of the diffraction limit in resolution. STORM and PALM overcome this limit by switching on only a stochastic subset of fluorescent molecules within a field of view at any given time such that their images do not substantially overlap, allowing their positions to be localized with high precision; these molecules are then switched off (or bleached) and a different subset of molecules are switched on and localized – iterating this process allows a super-resolution image to be constructed from numerous molecular localizations accumulated over time (6–8). Such stochastic activation of molecules is typically achieved by using photoswitchable dyes or fluorescent proteins (6–10). A variety of photoswitchable probes have been used for this approach, in some cases leading to the creation of different acronyms subsequently, but the imaging principles are the same as those for STORM and PALM. In addition to using photoswitchable probes, transient binding of fluorescent probes can also be used to stochastically “turn on” fluorescent signals in space and time, as in Point Accumulation for Imaging in Nanoscale Topography (PAINT) (14).
Recently, a new super-resolution imaging method named MINFLUX has been developed that combines strengths from both categories of approaches, using stochastic switching of individual molecules to enable the separate detection of nearby molecules, along with patterned illumination, such as a donut-shaped beam, to achieve ultrahigh-precision localization of individual molecules by detecting local emission minima (15).
In addition to the above methods which directly overcome the diffraction limit optically, a different form of super-resolution microscopy, Expansion Microscopy (ExM), has been recently developed, which increases the image resolution effectively through physical expansion of samples (16). In ExM, the specimen is embedded in a gel with the labeling probes attached to the gel. The sample is then digested to leave only the labeling probes attached to the gel followed by gel expansion to increase the probe separation, allowing super-resolution images to be taken with diffraction-limited microscopes.
Super-resolution technologies are constantly expanding, including both variations of the above approaches and other distinct methods, such as fluctuation-based methods and computer-vision-based methods. Due to the limited space of this short review and its focus on biological applications, we cannot describe all methods here but refer interested readers to other reviews (4, 9, 10) for additional coverage on super-resolution technologies.
Imaging capabilities of super-resolution microscopy
Three-dimensional (3D) imaging
The 3D nature of biological structures calls for super-resolution in all three dimensions. For methods based on stochastic activation of single molecules, such as STORM and PALM, achieving 3D super-resolution imaging requires high-precision localization not only in the xy plane, but also in the z direction along the optical axis. This was first achieved by astigmatism imaging (using a cylindrical lens to create z-dependent point-spread-functions (PSF)) (17), followed by various other approaches including bi-focal plane imaging (18), PSF engineering (19), and interferometry (20), among others (9, 10, 21). In STED and RESOLFT, isotropic 3D super-resolution imaging was achieved by generating a depletion illumination pattern to counteract excitation in all directions surrounding the focal point, for example, by using donut-shaped STED beam in conjunction with two opposing objectives (4, 22).
Image resolution
Both the methods based on patterned illumination, like STED, RESOLFT, and NL-SIM, and the methods based on single-molecule switching and localization, like PALM and STORM, are diffraction-unlimited, and thus do not have a theoretical resolution limit. In practice, however, many factors can influence the achievable resolution, including the excitation and detection schemes, the photo-physical properties and size of fluorescent probes, as well as the labeling and sampling density of these probes. In biological applications, resolutions achieved by these methods are typically in the range of 10–70 nm, with sub-10 nm resolution achieved in some cases (9, 10).
For the patterned-illumination-based methods, the spatial frequency (or sharpness) of the final emission pattern determines the image resolution. For example, in STED and RESOLFT, the donut-shaped depletion beam limits the fluorescence emission zone to the very center of the donut beam. The stronger the depletion light, the narrower this emission zone and the higher the achievable image resolution (4). However, strong illumination can lead to significant photobleaching, phototoxicity, and enhanced background noise. Hence the typically achieved resolutions are tens of nanometers, though resolution as high as a few nanometers has also been demonstrated using probes with ultra-high photostability, such as diamond nitrogen-vacancy centers (4). With isoSTED, isotropic 3D resolution of ~30 nm has been demonstrated (4, 22). Combining patterned illumination with photoswitchable probes, RESOLFT (3, 4) has also achieved ~30 nm isotropic 3D resolution (23). Similarly, combining sinusoidal patterned illumination and photoswitchable probes, and using additional computational image reconstruction, NL-SIM has demonstrated ~45–60 nm resolution in 2D using saturated depletion (SD NL-SIM) or patterned activation (PA NL-SIM) (24, 25). PA NL-SIM has been extended to 3D with the help of lattice light sheet microscopy (26), providing a resolution of ~120–230 nm in 3D (25).
For single-molecule-switching-based methods, like STORM and PALM, the resolution depends on the photophysical properties of the fluorophores. Although many fluorophores exhibit blinking or switching behavior, only those with sufficient brightness and proper on-off switching kinetics yield high-quality images (27). The achievable image resolution depends on the number of photons detected from individual molecules, known as the photon budget. Typical experiments with bright photoswitchable dyes provide ~20–30 nm xy resolution, whereas the resolution is worse for fluorescent proteins due to lower photon budget. The resolution is often worse in the z direction, but the use of interferometry (20, 28, 29) or specially engineered PSF (30) can improve the z resolution to become equal to or even better than the xy resolution. For example, interferometry can provide <10 nm z-resolution though a more complicated imaging setup is needed (20, 28, 29). In general, the resolution in both xy and z can be increased by improving the photon budget of the fluorophores. For example, the development of ultra-bright photo-activatable dyes allowed resolution as high as a few nanometers to be achieved on biological structures using STORM (31). More recently, using stochastic binding of dye-labeled DNA probes, DNA-PAINT also achieved similar image resolution on DNA-origami nanostructures (32). However, the time required to detect such a large number of photons for each molecule substantially increased the acquisition time per image. The recently developed MINFLUX thus represents an important advance in that it uses patterned excitation to drastically increase the localization precision (or reduce the number of photons required to reach a set precision), achieving an impressive localization precision of ~1 nm with orders of magnitude lower photon budget (15). In ExM, the resolution depends on the number of rounds of sample expansion and the expansion factor per round, and resolution of ~25 nm has been demonstrated with two rounds of expansion (33).
Other factors can also limit the final image resolution, such as the size of the fluorescent probes and the labeling density, which affect all super-resolution methods. For single-molecule-based approaches, methods that increase the number of times each target is sampled can also increase the final resolution if the resolution is limited by sampling density. This includes using fluorophores that undergo many on-off switching cycles (27), diffusible probes that can sample multiple locations of the target (34), and PAINT approaches that sample the target numerous times using reversible probe binding (35).
Finally, thick samples pose additional challenges, including reduced localization precision for out-of-focus molecules, optical aberration and light scattering, as well as increase in background noise, all of which can lower image resolution. Various PSF engineering methods have been developed to allow high localization precision over substantially longer focal depths (19, 21, 30). Adaptive optics have been used to correct for aberrations in super-resolution imaging of thick samples (29), and light-sheet illumination provides an effective optical-sectioning approach to reduce background in thick-sample super-resolution imaging (36). Tissue clearing methods can reduce not only aberration but also scattering, and are particularly powerful for thick sample imaging (37). Alternatively, serial physical sectioning has also been used to reconstruct super-resolution images over large volumes of tissue (38).
Live-cell imaging, temporal resolution and photo-toxicity
Several super-resolution methods have demonstrated live-cell imaging. As scanning-based techniques, STED and RESOLFT can image a relatively small field of view (FOV) with very high temporal resolution, and thus have the impressive capability of probing millisecond dynamics of cellular structures at the spatial resolution of tens of nanometers (39). Although the time resolution decreases with increasing FOVs, highly parallelized RESOLFT with 100,000 intensity minima, effectively akin to 100,000 tightly spaced donut patterns, allows sub-second time resolution for large FOVs (40).
As wide-field imaging methods, the time resolutions of STORM and PALM do not change as rapidly with the FOV size. Sub-second time resolution at ~20–30 nm spatial resolution has been achieved for large FOVs in live-cell imaging by STORM using fast-switching dyes (41) and fast scientific-CMOS cameras (42). Several recently developed algorithms to localize a high density of molecules with overlapping images (43) can further increase the time resolution of these methods. In addition, the single-particle-tracking mode of PALM, STORM and PAINT (44, 45) allows movement of individual molecules to be tracked with time resolutions of milliseconds to tens of milliseconds at high molecular concentrations. The ability of MINFLUX to achieve high localization precision with a minimal photon budget has led to a drastic increase in the tracking time resolution of molecules in live cells to sub-millisecond (~100 μs) coupled with a corresponding increase in the number of snapshots possible for each molecule before photobleaching (15).
For live cell imaging, in addition to achieving high spatiotemporal resolution, it is also important to reduce photobleaching and phototoxicity in order to prolong the overall duration of imaging and to minimize perturbations to the biological systems. Because of the trade-offs between spatial and temporal resolutions and between spatiotemporal resolution and phototoxicity/imaging duration, it is possible to reduce the spatial resolution of both the patterned-illumination-based methods, such as STED and RESOLFT, and the single-molecule-switching-based methods, such as STORM and PALM, to trade for higher time resolution, or lower phototoxicity and longer imaging duration. In addition, adaptively changing the intensity of the STED beam based on the presence/absence of fluorophores, as in DyMIN, results in a substantial reduction in photobleaching and phototoxicity (46). By using photoswitching instead of stimulated emission, RESOLFT requires a much lower light intensity than STED, and thus drastically reduces phototoxicity in live-cell super-resolution imaging (4). When the spatial resolution requirement is not particularly high (~100 nm), SIM is a popularly used live-cell imaging method due to its ability for high-speed wide-field imaging with low phototoxicity. The recently reported PA NL-SIM demonstrated live-cell imaging with ~60 nm spatial resolution and sub-second time resolution over large FOVs and tens of time points (25). In general, using light-sheet illumination for optical sectioning can also reduce phototoxicity in imaging (36). The recently developed lattice light sheet microscopy (26) further decreases phototoxicity and improved optical sectioning (to ~300 nm) compared to previous light-sheet schemes, and has been used in conjunction with super-resolution approaches to improve their volumetric live-cell imaging capability (25, 26, 35).
Quantitative biological insights offered by super-resolution imaging
Super-resolution imaging has transformed our understanding of biological systems and the applications are rapidly expanding, prohibiting comprehensive descriptions in a short review. Instead, we will highlight in this section the types of quantitative insights that can be obtained by super-resolution imaging, with representative examples illustrating each case (Fig. 1). In the next section, we will provide more detailed descriptions of a few examples to further illustrate the power of super-resolution imaging (Figs. 2–4).
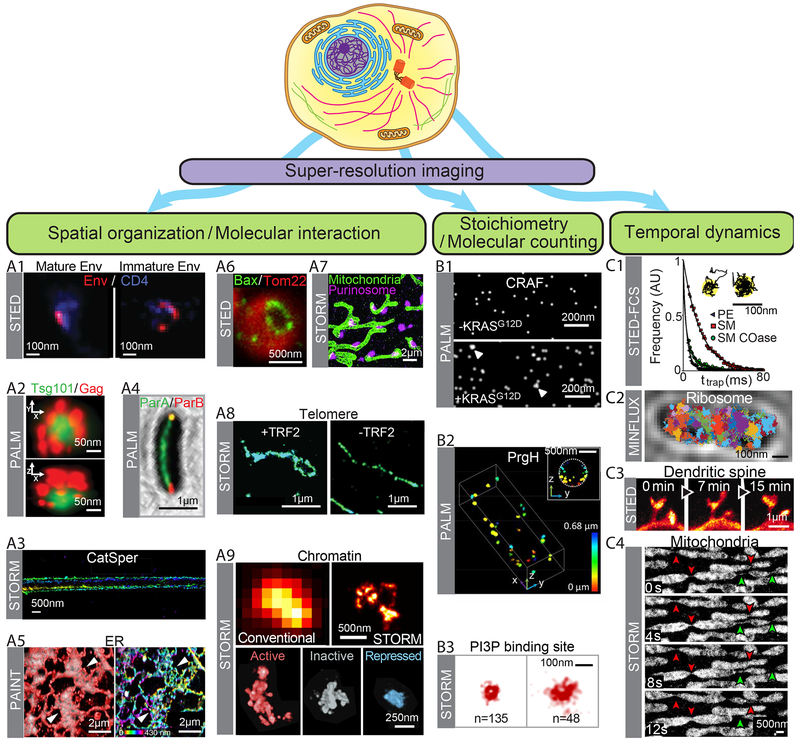
Fig. 1. Quantitative biological insights offered by super-resolution imaging.
Schematic of a cell together with three major application directions for super-resolution imaging. (A1–A9) Spatial organization and molecular interactions of cellular structures. (A1) STED images showing distinct distribution patterns of the envelope protein Env (red) in mature (left) and immature (right) HIV-1 particles attached to the cell, overlaid with the cell surface HIV-1 receptor CD4 (blue). Scale bar, 100 nm. Modified with permission from Ref. (49). (A2) PALM images showing the organization of ESCRT-I subunit Tsg101 (green) in a HIV assembly site marked by HIV Gag poteins (red) in lateral (top) and axial (bottom) views. Scale bar, 250 nm. Modified with permission from Ref. (50). (A3) 3D STORM images of a sperm-specific calcium channel (CatSper1) showing four linear domains along the sperm flagella. The z-position information is color-coded. Scale bars, 500 nm. Modified with permission from Ref. (51). (A4) PALM image on a bacterial cell showing the distribution of the ParA ATPase (green) with the ParB DNA binding protein (red) localized to the cell poles, for the coordination of chromosome segregation and cell division. Scale bar, 1 μm. Modified with permission from Ref. (52). (A5) Left: Overlay of PAINT (red) and diffraction-limited (gray) images of the ER obtained using lattice light-sheet microscopy. Right: PAINT image from the left panel, but color-coded by the z-position information. White arrowheads indicate areas that appear as sheets in diffraction-limited images but are resolved as connected tubular structures in super-resolution images. Scale bars, 2 μm. Modified with permission from Ref. (35). (A6) STED image of the pro-apoptotic cell-death mediator Bax (green) showing ring structures in apoptotic mitochondria marked by Tom22 (red). Scale bar, 500 nm. Modified from with permission Ref. (54). (A7) STORM image showing interactions between mitochondria (green) and purinosomes marked by the core protein FGAMS (magenta). Scale bar, 2 μm. Modified from with permission Ref. (56). (A8) Comparison of STORM images of telomeric DNA in mouse embryo fibroblasts in the presence (left) and absence (right) of the shelterin protein TRF2 that is required for t-loop formation. Scale bar, 1 μm. Modified with permission from Ref. (59). (A9) Top: Comparison of diffraction-limited (left) and 3D STORM (right) images for DNA in a chromatin domain in the nucleus of drosophila Kc167 cells. Scale bar, 500 nm. Bottom: Differential DNA compaction of transcriptionally active (red), inactive (gray) or polycomb-repressed (blue) epigenetic domains visualized using STORM. Scale bar, 250 nm. Modified with permission from Ref. (60). (B1–B3) Molecular counting and stoichiometric characterization. (B1) PALM images of proto-oncogene cRAF clusters on the cell plasma membrane, with (bottom) and without (top) coexpression of KRASG12D which induces cRAF clustering. Scale bar, 200 nm. Modified with permission from Ref. (66). (B2) 3D PALM image of molecular clusters with various sizes formed by a secretion system protein PrgH near the membrane of a bacterial cell. Scale bar, 500 nm. Modified with permission from Ref. (67). (B3) STORM images of endocytic vesicles displaying distinct vesicle size and PI3P content. The number of PI3P binding sites on each vesicle (n) is indicated. Scale bar, 100 nm. Modified with permission from Ref. (64). (C1–C4) Temporal dynamics. (C1) Durations (ttrap) for three lipid types, Phospho-ethanolamine (PE, grey), sphingomyelin (SM, red), sphingomyelin after cholesterol depletion (SM Coase, green), that are differentially trapped in ~20 nm nanodomains at the plasma membrane, which are detected and distingused by STED-FCS and confocal single-molecule tracking. Scale bar: 100 nm. Modified with permission from Ref. (69). (C2) Single particle tracking of a 30S ribosomal subunit protein in a bacterial cell using MINFLUX. Trajectories of individual molecules are shown in different colors. Scale bar, 500 nm. Modified with permission from Ref. (15). (C3) Time-lapsed STED images of a region of the somatosensory cortex of a living mouse with EYFP-labeled neurons showing dynamics of dendritic spines. Scale bar,1 μm. Modified with permission from Ref. (71). (C4) Time-lapsed STORM images showing fission (green arrowheads) and fusion (red arrowheads) events of mitochondria, with thin tubular structures connecting neighboring mitochondria as fission and fusion intermediates. Scale bar, 500nm. Modified with permission from Ref. (34).
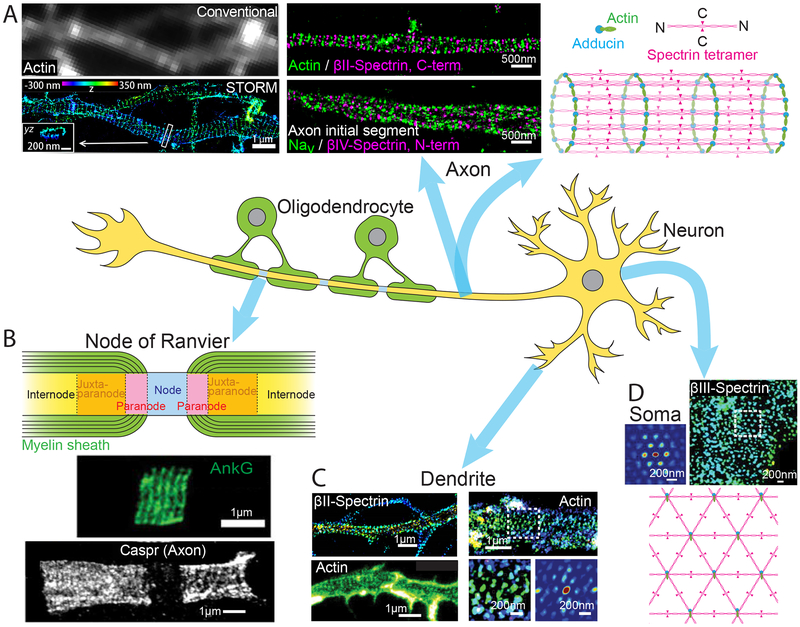
Fig. 2. The membrane-associated periodic skeleton (MPS) in neurons discovered by super-resolution imaging.
(A) Quasi one-dimensional (1D) periodic MPS observed in axons using STORM. Left: Comparison of diffraction-limited (top) and 3D-STORM (bottom) images of actin in axons. STORM image shows the periodic distribution of actin rings along the axon that is obscured by diffraction-limited imaging. Scale bar, 1 μm. Middle: Two color STORM images showing the periodic distributions of and spatial relationship between actin, spectrins (βII- and βIV-spectrin), and voltage gated sodium channels (Nav). Scale bar, 500 nm. Right: Schematic of the 1D MPS structure showing the organization of actin, spectrin tetramers, and adducin. Modified with permission from Ref. (47). (B) Top: Schematic of a node of Ranvier. Center: STED image showing the periodic distribution ankyrin-G (AnkG) on the 1D MPS structure at a node of Ranvier. Modified with permission from Ref. (74). Bottom: STED image showing the periodic distribution of the adhesion molecule Caspr on the 1D MPS structure observed flanking a node of Ranvier. Modified with permission from Ref. (76). Scale bars, 1 μm. (C) MPS structures observed in dendrites: Top-left: 1D MPS in a dendritic region observed by STORM imaging of βII-spectrin. Modified with permission from (72). Bottom-left: 1D MPS observed in a dendritic region by STED imaging of Actin. Modified with permission from Ref. (73). Right (top): 2D polygonal lattice-like arrangement of MPS components observed in a dendritic region by STORM imaging of actin. Right (bottom): A magnified region of the STORM image (left) and the corresponding 2D autocorrelation analysis (right) are shown. Modified with permission from Ref. (77). Scale bars, 1 μm. (D) 2D MPS observed on the soma of neuron by STORM imaging of βIII-spectrin (top-right) along with a 2D autocorrelation analysis of the boxed region (top-left). Bottom: Schematic of the 2D MPS structure. Scale bars, 200 nm. Modified with permission from Ref. (77).
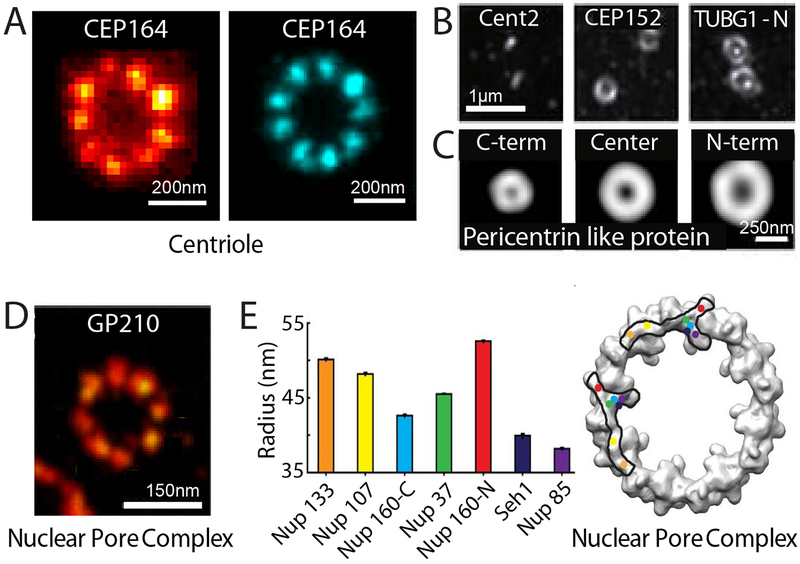
Fig. 4. Super-resolution visualizing molecular complexes: Centriole-containing complexes and the nuclear pore complex.
(A) Single STED image (left) and particle-averaged STORM image (right) of the centriolar protein (CEP164), showing a radial 9-fold symmetry. Scale bars, 200 nm. Modified with permission from Ref. (89, 90). (B) Three centriolar and peri-centriolar proteins, Cent2, CEP152 and γ-Tubulin (TUBG1), imaged using SIM showing a concentric organization of the pericentriolar matrix. Scale bar, 1 μm. Modified with permission from Ref. (93). (C) Average distribution of C-terminus, central domain, and N-terminus of the pericentrin-like protein (PLP) demonstrating a radial, spoke-like orientation for PLP through the pericentriolar matrix as determined by SIM. Scale bar, 250 nm. Modified with permission from Ref. (94). (D) Single STORM image of the nucleoporin protein GP210 showing an 8-fold symmetry within the nuclear pore complex (NPC) of Xenopus oocytes. Scale bar, 150 nm. Modified with permission from Ref. (95). (E) Left: Radial distribution of several nucleoporins including Nup133, Nup107, Nup160 (C-terminus), Nup37, Nup160 (N-terminus), Seh1, and Nup85, that comprise the Y-shaped Nup107–160 complex determined using STORM and particle averaging. Right: A projection of the electron density of the cytoplasmic ring of the NPC determined by EM is overlaid with two possible arrangements of the Nup107–160 complex, determined by super-resolution imaging. Each protein is represented by a colored dot corresponding to the color and radius in the graph (left). Modified with permission from Ref. (96).
Spatial organization and molecular interaction of cellular structures
The nanometer-scale resolution afforded by super-resolution imaging has substantially advanced our ability to interrogate the spatial organization of molecular structures in cells (9, 10). In addition, multi-color super-resolution imaging has allowed molecular interactions to be examined at unprecedented resolution (9, 10). With these abilities, super-resolution imaging has provided new insights into numerous cellular structures, and even led to discoveries of previously unknown cellular structures, such as the membrane-associated periodic skeleton (MPS) in neurons (47) detailed in the next section.
At the cell surface, membrane proteins such as receptors, channels, vesicle scission proteins, and viral fusion proteins have been investigated by various super-resolution approaches and are often found to assume functionally important spatial organizations. For example, it was shown that the SNARE complex component syntaxin-1 is densely packed within discrete clusters that are regulated by the lipid composition (48). The HIV envelope protein (Env) was observed to reorganize upon maturation, which is important for viral entry (49) (Fig. 1A1), while the ESCRT complex is localized to the virus budding site and plays an important role in HIV budding (50) (Fig. 1A2). The calcium channel CatSper was shown to adopt a linear-domain organization along the sperm tail together with other signaling and scaffolding molecules, playing an important role in calcium signaling and sperm activity (51) (Fig. 1A3). In the cytoplasm, super-resolution imaging has provided new insights into the organizations of cytoskeleton structures and membrane organelles, as well as other molecular assemblies. In addition to the discovery of the MPS in neurons (47), as will be detailed in the next section, novel organization has also been observed for other cytoskeletal structures, such as the ParA/ParB system in bacteria (52) (Fig. 1A4) and focal adhesions connecting the cytoskeleton to the plasma membrane (53). For membrane organelles, super-resolution imaging has revealed, for example, densely packed and dynamic tubular structures in ER sheets (35) (Fig. 1A5), ring structures of Bax on apoptotic mitochondria (54, 55) (Fig. 1A6), and synergistic interactions between mitochondria and purinosomes (56) (Fig. 1A7). In addition to protein structures, super-resolution imaging has also provided new insights into RNA distributions and interactions in cells (57, 58). In the cell nucleus, super-resolution imaging has revealed interesting organizations of DNA and DNA-interacting proteins, such as the TRF2-dependent telomere loop (t-loop) formation important for DNA end protection (59) (Fig. 1A8), distinct chromatin organization and compaction in different epigenetic states (60) (Fig. 1A9), and cell-type dependent organizations of nucleosomes (61).
Stoichiometry of molecular complexes
Although measuring stoichiometry by spatially resolving individual subunits within molecular complexes is still challenging, the abilities to activate and localize individual molecules by PALM and STORM have triggered growing interest in stoichiometric characterizations within intact cells. However, it is important to note that the number of measured single-molecule localizations is not equivalent to the number of molecules due to two complications. The first arises from imperfect labeling. New labeling approaches, such as using gene editing to label endogenous proteins with rapidly maturing, monomeric fluorescent proteins or with protein/peptide tags that can be conjugated to dyes with near 100% efficiency can help mitigate this challenge. The second complication arises from complex fluorophore switching: no dye or fluorescent protein has the ability to give precisely one localization per molecule because fluorophores tend to blink (multiple times) and most fluorophores also have an inactivatable fraction. Multiple methods have been developed to combat this problem, including calibrations of fluorophores using standards of known stoichiometry or quantification and modeling of blinking properties (62–64). STED has also been used to quantify the number of molecules based on coincident photon detection (65). These methods have been applied to quantifying, for example, the number of proteins in flagellar motors (62), receptor complexes (63), kinase complexes (66) (Fig. 1B1), secretion machinery (67) (Fig. 1B2), as well as lipid binding sites in endocytic vesicles (64) (Fig. 1B3).
Temporal dynamics of cellular structures
Super-resolution imaging has enhanced our ability to extract dynamic information of cellular structures, allowing the mobility of biomolecules and the shape or structural dynamics of molecular complexes and organelles to be tracked with higher accuracy. For example, STED imaging has been used in combination with fluorescence correlation spectroscopy (FCS) to study the diffusion properties of molecules on the membrane. The drastic reduction in the region of fluorescence emission by the STED beam has allowed the detection of membrane nanodomains of < 20 nm in size, within which different lipid molecules show distinct diffusion properties (68, 69) (Fig. 1C1). Super-resolution imaging has also enhanced our ability to perform single-particle tracking (SPT) in live cells. Conventional SPT experiments requires a low labeling density for the molecule of interest to avoid signal overlap between molecules. Stochastically turning on only a subset of labeled molecules at a given time, as in PALM, STORM, and PAINT, allows SPT at much higher molecular concentrations at the endogenous expression level (34, 44, 45), facilitating the studies of gene expression, protein-nucleic acid interaction, and dynamic processes on cell membranes. With its unique capabilities, MINFLUX has allowed the tracking of ribosomes in bacterial cells with unprecedented spatiotemporal resolution, achieving a localization precision of <50 nm with a time resolution of ~100μs (15) (Fig. 1C2).
In addition, various super-resolution microscopy methods have been used to measure structural and shape dynamics of molecular assemblies, organelles and small cellular compartments, such as the dynamics of neuronal processes and dendritic spines in tissue (70) and even in live animals (Fig. 1C3) (71), fission and fusion dynamics of mitochondria (34) (Fig. 1C4), and structural dynamics of ER (35).
Super-resolution studies of specific molecular assemblies
The membrane-associated periodic skeleton in neurons
Super-resolution imaging enabled the discovery of the membrane-associated periodic skeleton (MPS) in neurons, which was initially observed in the axons by STORM imaging (47). In the MPS, short actin filaments, capped by actin-capping proteins, such as adducin, are organized into repetitive, ring-like structures that wrap around the circumference of axon; adjacent actin rings are connected by spectrin tetramers, forming a long-range quasi-one-dimensional (1D) periodic structure with a periodicity of ~180–190 nm underneath the axonal membrane (47) (Fig. 2A).
The MPS spans the entire axon shaft, in both myelinated and unmyelinated axonal segments, including the axon initial segments (AIS) and nodes of Ranvier where action potentials are generated and amplified, respectively (47, 72–76) (Fig. 2A, B). This structure was observed in all neuronal types examined, including excitatory and inhibitory neurons in both central and peripheral nervous systems (74, 75), and is evolutionarily conserved across diverse animal species (75). Subsequent to its discovery in axons, this 1D periodic structure was also observed in dendrites by both STORM and STED (72, 73) (Fig. 2C), but the formation propensity and development rate of MPS appear to be lower in dendrites than in axons (77). In addition, a 2D polygonal lattice structure formed by MPS components was observed in the soma and dendrites (Fig. 2D), resembling the membrane skeleton structure observed in erythrocytes (77).
This highly ordered sub-membrane skeletal structure can play diverse functional roles in neurons. It provides flexible mechanical support for axons that is likely critical for axon stability under mechanical stress (47); indeed, axons tend to break in spectrin-deficient animals under movement induced stress (78). The MPS was also implicated in mechanosensation (79). Moreover, the MPS organizes membrane proteins, such as ion channels and adhesion molecules, into periodic distributions along axons (47, 72, 73, 76), potentially influencing the generation and propagation of action potentials, and other signaling pathways in axons. The MPS also influences axon and dendrite morphology (80), and is important for the formation of the AIS and nodes of Ranvier (72, 76, 80), and may also act as a diffusion barrier at the AIS (81). Disruption of the MPS causes widespread neurodegeneration and a range of neurological impairments (80), and mutations of MPS components are implicated in various neurodegenerative diseases. The discovery of the MPS, which escaped detection by previous imaging methods, demonstrates the power of super-resolution imaging for uncovering new cellular structures.
Molecular organization in synapses
Neuronal synapses are typically only several hundred nanometers in size but contain elaborate protein machineries to orchestrate neurotransmitter-mediated signal transmission, hence the structural interrogation of synapses requires high spatial resolution and has benefited from extensive super-resolution imaging efforts. For example, STED has revealed the spatial organization of several important components within the Drosophila neuromuscular junction, including the clustered organization of Ca2+ channels, as well as the organization of scaffolding proteins required for both Ca2+ channel clustering and synaptic vesicle tethering at the presynaptic active zone (82, 83) (Fig. 3A). STORM imaging has mapped the spatial organization of many proteins in the pre- and post-synaptic terminals, which show oriented organization of presynaptic scaffolding proteins, laminar organization of postsynaptic density proteins, and synapse-to-synapse variability in the lateral distributions of neurotransmitter receptors (84) (Fig. 3B).
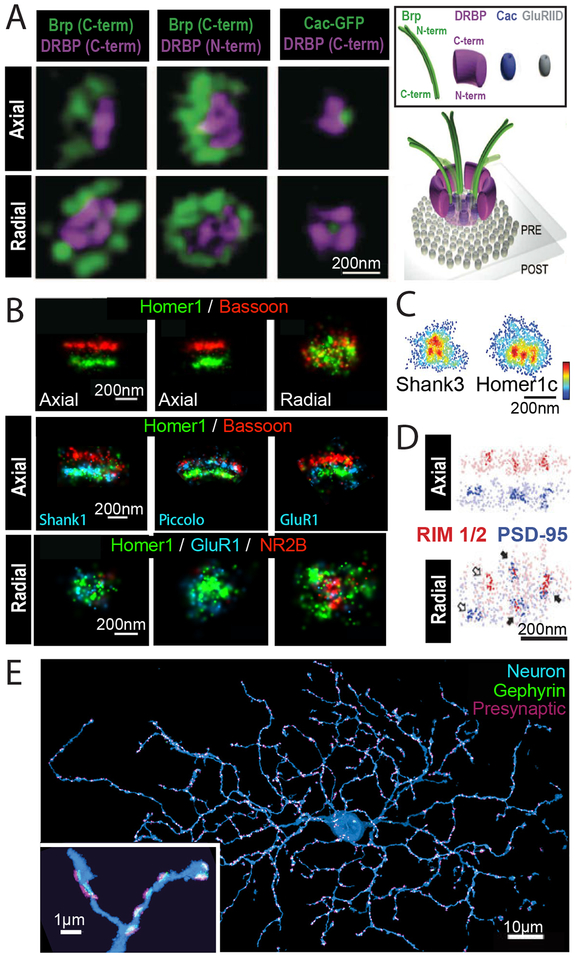
Fig. 3. Super-resolution imaging of synaptic structures.
(A) Left: 3D STED images of the presynaptic active zone including Bruchpilot (Brp) and Drosophila RIM binding protein (DRBP) as well as the voltage gated calcium channel Cacophony (Cac) at Drosophila neuromuscular junction synapses. Both axial (top) and radial (bottom) projections are shown. Scale bar, 200 nm. Right: Schematic of the active zone showing positions and orientations of components of the active zone cytomatrix including Brp, DRBP, and Cac in relation to the post-synaptic glutamate receptor (GluRIID) determined using STED. Modified with permission from Ref. (83). (B) Top: 3D STORM images of presynaptic protein Bassoon (red) and postsynaptic protein Homer (green). Two orthogonal axial views (left and middle) and the radial view (right) are shown. Scale bars, 200 nm. Center: Axial views of three synapses. In addition to Bassoon (red) and Homer1 (green), a third color (blue) was used to map the positions of additional postsynaptic (Shank1, left; GluR1, right) and presynaptic (Piccolo, middle) components at synapses. Bottom: Radial views of three example synapses showing differential abundance and spatial distribution of neurotransmitter receptors, NR2B and GluR1. Scale bars, 200 nm. Modified with permission from Ref. (84). (C) Radial projections of PALM images showing the clustered organization of postsynaptic proteins Shank3 (left) and Homer1c (right). Scale bar, 200 nm. Modified with permission from Ref. (85). (D) STORM and PALM images show that areas of higher protein density (darker colors) of both presynaptic (RIM1/2, red) and postsynaptic (PSD-95, blue) components are often trans-synaptically aligned to form “nanocolumns” (indicated by filled arrows). Both axial (top) and radial (bottom) projections are shown. Scale bar, 200 nm. Modified with permission from Ref. (87). (E) STORM maximum intensity projection of a retinal ganglion cell (blue) with associated synapses marked by postsynaptic scaffolding protein gephyrin (green) and presynaptic proteins (Bassoon, Piccolo, Munc13–1, and ELKS)(magenta), reconstructed from ultra-thin serial sections. Inset shows a zoomed in view of a region of dendrite. Scale bar, 10 μm, inset, 1 μm. Modified with permission from Ref. (38).
In addition, recent super-resolution studies revealed that the neurotransmitter receptors and post-synaptic scaffolding proteins adopt activity-dependent clustered organization (85, 86) (Fig. 3C). Such clustered organization also extends across the synaptic cleft, giving rise to “nanocolumns” formed by spatially aligned presynaptic vesicle fusion sites and postsynaptic receptor clusters (87) (Fig. 3D). This nanocolumn organization provides a mechanism for the coordination of synaptic vesicle release and neurotransmitter receptor response.
Super-resolution studies of synapses have been recently extended to both proteomic-scale analysis of synaptic structures and circuit-scale analysis of synapse distributions. For example, STED has been used to image numerous protein components in the presynaptic terminals, creating a model of an “average” synaptic bouton (88). A volumetric STORM platform has been developed to determine the entire synaptic fields of neurons (38) (Fig. 3E), providing synaptic connectivity at the neural circuit scale.
Protein complexes with structural symmetry
On the shorter length scale of individual protein complexes, it is possible to obtain higher resolution reconstructions from many super-resolution images through particle averaging in a way similar to electron microscopy (EM) reconstruction, especially for structures with well-defined symmetry. Two notable examples are centrioles and nuclear pore complexes (NPCs).
STED and STORM, the latter in combination with particle averaging, have been used to visualize the 9-fold symmetry of centrioles (89, 90) (Fig. 4A). In addition to resolving this symmetric arrangement, super-resolution imaging has also been used to map the 3D organization of several centriolar proteins and determine the order of protein recruitment during centriole formation (91, 92). At centrosomes, centrioles are surrounded by the less structured pericentriolar material (PCM), and the radial distribution of proteins within the centrosome and PCM have also been mapped by STORM and SIM (93, 94) (Fig 4B, C).
Similarly, STORM imaging showed the 8-fold radial symmetry of NPCs (95) (Fig. 4D). In combination with particle averaging, STORM allowed the positions of seven nucleoporin components to be determined with ~1 nm precision, which in turn allowed the orientation of the Nup107–160 subcomplex within the pore to be determined (96). These super-resolution pictures allowed discrimination between contradictory models of the structural organization of the NPC scaffold (96) (Fig. 4E).
Outlook
Super-resolution fluorescence microscopy has transformed understanding of the structure and function of many biological systems. However, challenges are still present, and to maximize the impact of super-resolution microscopy, further technological advancements are still needed.
The spatial resolution achieved by super-resolution microscopy in biological systems typically ranges from 10–70 nm, larger than most biomolecules. Achieving true molecular-scale resolution (~1 nm) would allow molecular interactions and conformations to be directly probed inside cells, but remains a challenging task. In principle, the two main categories of optical approaches to overcome the diffraction limit, including the patterned-illumination-based methods represented by STED, RESOLFT, and NL-SIM and the single-molecule-switching-based methods represented by STORM, PALM, and PAINT, can both achieve unlimited high resolution. However, practical factors, such as the requirement of increasing illumination intensity (in the former category) and increasing fluorophore photon budget (in the latter category) for higher resolution, limit the resolution that can be achieved. Re-inspection of the fundamental principles of super-resolution methods can lead to powerful new innovations and concepts, as demonstrated by MINFLUX, which combines strengths from both approaches and achieves ultrahigh, previously inaccessible resolutions. In addition, these methods can be combined with ExM, an orthogonal approach that achieves resolution increase through physical sample expansion, potentially leading to a direct multiplication in the fold-increase in resolution that are separately achievable by individual methods. However, it is worth noting that the final image resolution is also limited by probe size and labeling density. Thus, in order to ultimately benefit from the ultra-high resolution, parallel development in probes and labeling methods are needed to allow molecules in cells to be labeled with small molecule probes with high efficiency.
Furthermore, although super-resolution imaging has demonstrated sub-second and even millisecond time resolution in some cases, due to the trade-off between spatial and temporal resolutions, the limited photon budget of the fluorophores, and phototoxicity to samples, live cell imaging with high spatiotemporal resolution for a long period of time remains difficult and an active area of development. In addition, in vivo super-resolution imaging deep inside tissues remains challenging, notwithstanding considerable efforts combating tissue-induced background, aberration and light scattering.
Another challenge, but also an exciting new direction, is to increase the number of molecular species that can be simultaneously imaged. Cells contain thousands of distinct genes and other molecules that act collectively to give rise to behavior and function, yet multicolor imaging usually only allows simultaneous visualization of a few different molecular species. Recent advances have broken new ground in this direction and genomic-scale imaging is now within reach. For example, single-cell transcriptome-imaging methods have allowed RNAs of 1000 or more genes to be simultaneously imaged in individual cells using multiplexed FISH (97, 98) or in situ sequencing (99, 100). A similar level of multiplexity may be achievable for DNA and proteins in the future. Combination of these approaches with super-resolution microscopy could potentially allow genomic-scale super-resolution imaging. Technologically, a major challenge in genomic-scale imaging is molecular crowding, which can prevent resolution of neighboring molecules by conventional imaging, and super-resolution microscopy provides a promising solution. Biologically, the ability to image all molecules in a complex molecular machinery or in a whole signaling pathway, and ultimately at the whole genome scale, will provide a comprehensive picture of the molecular basis of cellular behavior and function. It is exhilarating to imagine how such a picture of a cell, with all molecules imaged at a resolution that allows direct inference of molecular interactions, would open new opportunities for understanding life at the molecular level.
Acknowledgments
Funding: This work is in part supported by the National Institutes of Health. X.Z. is a Howard Hughes Medical Institute Investigator.
Footnotes
Competing interests: None declared.
References and Notes
- 1.Hell SW, Wichmann J, Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Optics letters 19, 780–782 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Klar TA, Hell SW, Subdiffraction resolution in far-field fluorescence microscopy. Optics letters 24, 954–956 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Hofmann M, Eggeling C, Jakobs S, Hell SW, Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proceedings of the National Academy of Sciences of the United States of America 102, 17565–17569 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eggeling C, Willig KI, Sahl SJ, Hell SW, Lens-based fluorescence nanoscopy. Quarterly reviews of biophysics 48, 178–243 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Heintzmann R, Gustafsson MGL, Subdiffraction resolution in continuous samples. Nature Photonics 3, 362–364 (2009). [Google Scholar]
- 6.Rust MJ, Bates M, Zhuang X, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nature Methods 3, 793–795 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betzig E et al. , Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Hess ST, Girirajan TP, Mason MD, Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophysical journal 91, 4258–4272 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang B, Babcock H, Zhuang X, Breaking the diffraction barrier: super-resolution imaging of cells. Cell 143, 1047–1058 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahl SJ, Hell SW, Jakobs S, Fluorescence nanoscopy in cell biology. Nature Reviews Molecular Cell Biology 18, 685–701 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Moerner WE, Orrit M, Illuminating single molecules in condensed matter. Science 283, 1670–1676 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Yildiz A et al. , Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Pertsinidis A, Zhang Y, Chu S, Subnanometre single-molecule localization, registration and distance measurements. Nature 466, 647–651 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Sharonov A, Hochstrasser RM, Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proceedings of the National Academy of Sciences of the United States of America 103, 18911–18916 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balzarotti F et al. , Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 355, 606–612 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Chen F, Tillberg PW, Boyden ES, Optical imaging. Expansion microscopy. Science 347, 543–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang B, Wang W, Bates M, Zhuang X, Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juette MF et al. , Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nature Methods 5, 527–529 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Pavani SR et al. , Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proceedings of the National Academy of Sciences of the United States of America 106, 2995–2999 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shtengel G et al. , Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proceedings of the National Academy of Sciences of the United States of America 106, 3125–3130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Diezmann A, Shechtman Y, Moerner WE, Three-Dimensional Localization of Single Molecules for Super-Resolution Imaging and Single-Particle Tracking. Chemical Reviews 117, 7244–7275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt R et al. , Spherical nanosized focal spot unravels the interior of cells. Nature Methods 5, 539–544 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Böhm U, Hell SW, Schmidt R, 4Pi-RESOLFT nanoscopy. Nature Communications 7, 10504 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rego EH et al. , Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proceedings of the National Academy of Sciences of the United States of America 109, E135–143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D et al. , ADVANCED IMAGING. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 349, aab3500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen BC et al. , Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X, Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nature Methods 8, 1027–1036 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aquino D et al. , Two-color nanoscopy of three-dimensional volumes by 4Pi detection of stochastically switched fluorophores. Nature Methods 8, 353–359 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Huang F et al. , Ultra-High Resolution 3D Imaging of Whole Cells. Cell 166, 1028–1040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia S, Vaughan JC, Zhuang X, Isotropic three-dimensional super-resolution imaging with a self-bending point spread function. Nature Photonics 8, 302–306 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaughan JC, Jia S, Zhuang X, Ultrabright photoactivatable fluorophores created by reductive caging. Nature Methods 9, 1181–1184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai M, Jungmann R, Yin P, Optical imaging of individual biomolecules in densely packed clusters. Nature Nanotechnology 11, 798–807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang JB et al. , Iterative expansion microscopy. Nature Methods 14, 593–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shim SH et al. , Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proceedings of the National Academy of Sciences of the United States of America 109, 13978–13983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nixon-Abell J et al. , Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 354, aaf3928 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gustavsson AK, Petrov PN, Moerner WE, Light sheet approaches for improved precision in 3D localization-based super-resolution imaging in mammalian cells [Invited]. Optics express 26, 13122–13147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gradinaru V, Treweek J, Overton K, Deisseroth K, Hydrogel-tissue chemistry: Principles and applications. Annual review of biophysics 47, 355–376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sigal YM, Speer CM, Babcock HP, Zhuang X, Mapping Synaptic Input Fields of Neurons with Super-Resolution Imaging. Cell 163, 493–505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider J et al. , Ultrafast, temporally stochastic STED nanoscopy of millisecond dynamics. Nature Methods 12, 827–830 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Chmyrov A et al. , Nanoscopy with more than 100,000 ‘doughnuts’. Nature Methods 10, 737–740 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Jones SA, Shim SH, He J, Zhuang X, Fast, three-dimensional super-resolution imaging of live cells. Nature Methods 8, 499–508 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang F et al. , Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nature Methods 10, 653–658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sage D et al. , Quantitative evaluation of software packages for single-molecule localization microscopy. Nature Methods 12, 717 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Manley S et al. , High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nature Methods 5, 155–157 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Giannone G et al. , Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophysical journal 99, 1303–1310 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heine J et al. , Adaptive-illumination STED nanoscopy. Proceedings of the National Academy of Sciences of the United States of America 114, 9797–9802 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu K, Zhong G, Zhuang X, Actin, Spectrin, and Associated Proteins Form a Periodic Cytoskeletal Structure in Axons. Science 339, 452–456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Bogaart G et al. , Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chojnacki J et al. , Maturation-Dependent HIV-1 Surface Protein Redistribution Revealed by Fluorescence Nanoscopy. Science 338, 524–528 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Van Engelenburg SB et al. , Distribution of ESCRT Machinery at HIV Assembly Sites Reveals Virus Scaffolding of ESCRT Subunits. Science 343, 653–656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung J-J et al. , Structurally Distinct Ca2+ Signaling Domains of Sperm Flagella Orchestrate Tyrosine Phosphorylation and Motility. Cell 157, 808–822 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ptacin JL et al. , A spindle-like apparatus guides bacterial chromosome segregation. Nature Cell Biology 12, 791–798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanchanawong P et al. , Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Große L et al. , Bax assembles into large ring‐like structures remodeling the mitochondrial outer membrane in apoptosis. The EMBO journal 35, 402–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salvador‐Gallego R et al. , Bax assembly into rings and arcs in apoptotic mitochondria is linked to membrane pores. The EMBO journal 35, 389–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.French JB et al. , Spatial colocalization and functional link of purinosomes with mitochondria. Science 351, 733–737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fei J et al. , RNA biochemistry. Determination of in vivo target search kinetics of regulatory noncoding RNA. Science 347, 1371–1374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moffitt JR, Pandey S, Boettiger AN, Wang S, Zhuang X, Spatial organization shapes the turnover of a bacterial transcriptome. eLife 5, e13065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doksani Y, Wu John Y., de Lange T, Zhuang X, Super-Resolution Fluorescence Imaging of Telomeres Reveals TRF2-Dependent T-loop Formation. Cell 155, 345–356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boettiger AN et al. , Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ricci Maria A., Manzo C, García-Parajo MF, Lakadamyali M, Cosma Maria P., Chromatin Fibers Are Formed by Heterogeneous Groups of Nucleosomes In Vivo. Cell 160, 1145–1158 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Lee S-H, Shin JY, Lee A, Bustamante C, Counting single photoactivatable fluorescent molecules by photoactivated localization microscopy (PALM). Proceedings of the National Academy of Sciences of the United States of America 109, 17436–17441 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Renz M, Daniels BR, Vamosi G, Arias IM, Lippincott-Schwartz J, Plasticity of the asialoglycoprotein receptor deciphered by ensemble FRET imaging and single-molecule counting PALM imaging. Proceedings of the National Academy of Sciences of the United States of America 109, E2989–2997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Puchner EM, Walter JM, Kasper R, Huang B, Lim WA, Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory. Proceedings of the National Academy of Sciences of the United States of America 110, 16015–16020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ta H et al. , Mapping molecules in scanning far-field fluorescence nanoscopy. Nature communications 6, 7977 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nan X et al. , Single-molecule superresolution imaging allows quantitative analysis of RAF multimer formation and signaling. Proceedings of the National Academy of Sciences of the United States of America 110, 18519–18524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Lara-Tejero M, Bewersdorf J, Galán JE, Visualization and characterization of individual type III protein secretion machines in live bacteria. Proceedings of the National Academy of Sciences of the United States of America 114, 6098–6103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eggeling C et al. , Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1162 (2008). [DOI] [PubMed] [Google Scholar]
- 69.Sahl SJ, Leutenegger M, Hilbert M, Hell SW, Eggeling C, Fast molecular tracking maps nanoscale dynamics of plasma membrane lipids. Proceedings of the National Academy of Sciences of the United States of America 107, 6829–6834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tonnesen J, Inavalli V, Nagerl UV, Super-Resolution Imaging of the Extracellular Space in Living Brain Tissue. Cell 172, 1108–1121 e1115 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Berning S, Willig KI, Steffens H, Dibaj P, Hell SW, Nanoscopy in a living mouse brain. Science 335, 551 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Zhong G et al. , Developmental mechanism of the periodic membrane skeleton in axons. eLife 3, e04581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D’Este E, Kamin D, Gottfert F, El-Hady A, Hell SW, STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Reports 10, 1246–1251 (2015). [DOI] [PubMed] [Google Scholar]
- 74.D’Este E et al. , Subcortical cytoskeleton periodicity throughout the nervous system. Scientific reports 6, 22741 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He J et al. , Prevalent presence of periodic actin-spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proceedings of the National Academy of Sciences of the United States of America 113, 6029–6034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.D’Este E, Kamin D, Balzarotti F, Hell SW, Ultrastructural anatomy of nodes of Ranvier in the peripheral nervous system as revealed by STED microscopy. Proceedings of the National Academy of Sciences of the United States of America 114, E191–E199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han B, Zhou R, Xia C, Zhuang X, Structural organization of the actin-spectrin-based membrane skeleton in dendrites and soma of neurons. Proceedings of the National Academy of Sciences of the United States of America 114, E6678–E6685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hammarlund M, Jorgensen EM, Bastiani MJ, Axons break in animals lacking beta-spectrin. Journal of Cell Biology 176, 269–275 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krieg M, Dunn AR, Goodman MB, Mechanical control of the sense of touch by beta-spectrin. Nature Cell Biology 16, 224–233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang CY, Zhang C, Zollinger DR, Leterrier C, Rasband MN, An alphaII Spectrin-Based Cytoskeleton Protects Large-Diameter Myelinated Axons from Degeneration. The Journal of Neuroscience 37, 11323–11334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Albrecht D et al. , Nanoscopic compartmentalization of membrane protein motion at the axon initial segment. Journal of Cell Biology 215, 37–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kittel RJ et al. , Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312, 1051–1054 (2006). [DOI] [PubMed] [Google Scholar]
- 83.Liu KSY et al. , RIM-Binding Protein, a Central Part of the Active Zone, Is Essential for Neurotransmitter Release. Science 334, 1565–1569 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Dani A, Huang B, Bergan J, Dulac C, Zhuang XW, Superresolution Imaging of Chemical Synapses in the Brain. Neuron 68, 843–856 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.MacGillavry HD, Song Y, Raghavachari S, Blanpied TA, Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron 78, 615–622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nair D et al. , Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. The Journal of Neuroscience 33, 13204–13224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang AH et al. , A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536, 210–214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilhelm BG et al. , Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Lau L, Lee Yin L., Sahl Steffen J., Stearns T, Moerner WE, STED Microscopy with Optimized Labeling Density Reveals 9-Fold Arrangement of a Centriole Protein. Biophysical journal 102, 2926–2935 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi X et al. , Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nature Cell Biology 19, 1178–1188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gartenmann L et al. , A combined 3D-SIM/SMLM approach allows centriole proteins to be localized with a precision of ~4–5 nm. Current Biology 27, R1054–R1055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hamel V et al. , Identification of Chlamydomonas Central Core Centriolar Proteins Reveals a Role for Human WDR90 in Ciliogenesis. Current Biology 27, 2486–2498.e2486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lawo S, Hasegan M, Gupta GD, Pelletier L, Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nature Cell Biology 14, 1148–1158 (2012). [DOI] [PubMed] [Google Scholar]
- 94.Mennella V et al. , Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nature Cell Biology 14, 1159–1168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loschberger A et al. , Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. Journal of Cell Science 125, 570–575 (2012). [DOI] [PubMed] [Google Scholar]
- 96.Szymborska A et al. , Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341, 655–658 (2013). [DOI] [PubMed] [Google Scholar]
- 97.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X, RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shah S et al. , Dynamics and Spatial Genomics of the Nascent Transcriptome by Intron seqFISH. Cell 174, 363–376.e316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang X et al. , Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science, aat5691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee JH et al. , Highly Multiplexed Subcellular RNA Sequencing in Situ. Science 343, 1360–1363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]