Abstract
Background: MicroRNAs (miRNAs) play a key role in the development of gastric cancer (GC). MiRNA arrays showed that lymph node metastasis in GC is correlated with the expression of miR-1284. Although its function and mechanisms in GC have not been fully described, the regulation of EIF4A1 by miR-1284 and its role in drug-resistant GC has been reported in our previous studies.
Methods: qRT-PCR was used to study the level of miR-1284 expression in GC cell lines and tissues. Subsequently, the CCK-8 assay was used to detect cell proliferation, while transwell assay was used to detect invasion and migration of the GC cells. Flow cytometry was used to detect the effect of miR-1284 on GC cells in vivo by building subcutaneous GC nude mice transplantation tumor model. In addition, the influence of miR-1284 gene expression profile in SGC-7901 cells was detected by total gene expression chip, and the target gene of miR-1284 was detected by luciferase reporter assay, qRT-PCR, and western blotting.
Results: The miR-1284 level was down-regulated in GC tssues and cell lines. MiR-1284 was significantly associated with tumor size, degree of differentiation and patients’ distant metastasis. MiR-1284 inhibited invasion, migration, and proliferation of GC cells. During the G1/S phase, miR-1284 arrested the cycle of GC cells in vitro. MiR-1284 also suppressed tumor from growing and metastasizing in xenograft models as well as influenced the gene expression profile in SGC-7901 cells. Also, EIF4A1 was the direct target gene for miR-1284. Further, an inverse correlation between the miR-1284 expression and EIF4A1 was found in GC tissues. Over-expressed miR-1284 decreased c-Myc, MMP12, JUN expression, while increased CDH1 expression.
Conclusion: These data suggested that miR-1284 acts as a tumor suppressor, and directly blocked EIF4A1 in GC.
Keywords: MiR-1284, gastric cancer, EIF4A1, metastasis, proliferation
Introduction
Gastric cancer (GC) is the most common cancer around the world. MicroRNA (MiRNA) has been widely used in the treatment of GC, for example, metastasis, invasion, and carcinogenesis.1 MiRNAs suppress the protein-coding genes from target gene expression through posttranscription. In mammals, the length of miRNAs is about 22 nucleotides, most of the recognition sites of these are located in the 3′-untranslated region (UTR) of transcripts.2 Chen et al3 initially found out that miR-1284 was lowly expressed in lymph node metastases positive tissue when compared to lymph node metastases negative tissue in the chip expression of miRNA of patients with GC. The regulatory mechanisms of elongation initiation factor 4A1 (EIF4A1) by miR-1284 and its role in drug-resistant GC; and its inhibition in GC proliferation and induction of apoptosis have been reported by our previous studies.4,5 These findings suggested that miR-1284 play a vital role in the treatment of GC by suppressing metastasis.
The linked human EIF4A1 is considered as an initiation factor in the gene map of chromosome 17p13, and is also known as EIF4A.6 The mRNA structure was unwinded by EIF4A1 using the energy provided by ATP hydrolysis, and with the conjunct action of other translational factors.7 According to the latest study, mRNAs and EIF4A1 are adjacently associated to the structure of cap, and reach the downstream of 52 bases from the cap.8 EIF4A1 has been identified to be the target of miR-US25-2-3p, and it also inhibited human cytomegalovirus from replicating, and overexpression of human miR-US25-2-3p downregulated EIF4A1.7 Furthermore, EIF4A1 is correlated with malignant phenotype of cancer cells, tumor-specific survival and sensitivity of therapeutic drug.9–11 These indicate that EIF4A1 is closely involved in tumors and can be regulated by miRNAs. Interestingly, EIF4A1 was reported to be a crucial downstream target gene of miR-1284 using bioinformatics tools (TargetScan: https://www.targetscan.org, miRDB: http://www.mirdb.org, microRNA: http://www.microrna.org). Thus, we postulated that EIF4A1 might be directly regulated by miR-1284.
Hence, in this study, the differential expression of miR-1284 and its clinicopathological implication in GC, the function of miR-1284 both in vivo and in vitro in GC, whether miR-1284 directly targets EIF4A1 gene and the relationship between the expression of EIF4A1 and miR-1284 in GC tissues were evaluated. Also, our study investigated the mechanism of miR-1284 and EIF4A1 in GC. The results of this study showed that miR-1284 suppressed GC migration and invasion through targeting EIF4A1 gene, contributing to the understanding of how miR-1284 works and its possible application as a therapeutic target in GC.
Material and methods
Cell lines and tissue samples
The First Affiliated Hospital of Guangxi Medical University, in Nanning, China, provided 74 sets of GC tissues and their corresponding nonmalignant tissues. Written informed consent forms were obtained from patients before collecting the tissues. All the tissues were taken from patients by undergoing a gastrectomy. All the samples are cut into two parts, where some specimens are quickly immersed into liquid nitrogen and stored at a temperature of −80°C for future use, while few others are stored in 10% formalin and underwent analysis by hematoxylin-eosin (HE) staining and immunohistochemical staining. Subjects were informed about the purpose of the study according to the declaration of Helsinki. The Ethics Committee of the First Affiliated Hospital of Guangxi Medical University approved this study in 2015 (KY-E-003). The pathological diagnoses were re-confirmed according to 2007 WHO classification of gastric carcinoma.
AGS, GES-1, HGC-27, MGC-803, and SGC-7901 GC cell lines were obtained from Shanghai Institute of Cell Biology Cell Bank, Chinese Academy of Sciences, Shanghai, China. The cells were stored in RPMI 1640 medium at 37°C in 5% CO2 (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum from Thermo Fisher Scientific, USA, 100 µg/mL streptomycin, and 100 units/mL penicillin.
Quantitative real time
The expression of miRNA and mRNA was analyzed by polymerase chain reaction (qRT-PCR). RNA was extracted from the cells and fresh tissues using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription was performed using the RNA PCR kit (AMV) (TaKaRa). Quantification of miRNAs and mRNA was done using qRT-PCR with SYBR Premix Ex TaqTM (TaKaRa, Otsu, Japan) according to the manufacturer’s instructions. U6 was employed for the expression of miR-1284, and used as an internal control. The primers (Table 1) for gene detection of qRT-PCR were synthesized by TaKaRa. The 2−△△Ct method was used to calculate the fold changes in expression.
Table 1.
Base sequence of primers used in this work
| Name | Base sequences |
|---|---|
| miR-1284 forward primer | 5′-UCUAUACAGACCCUGGCUUUUC-3′ |
| miR-1284 reverse primer | 5′-CGTCTATACAGACCCTGGCTTTTC-3′ |
| U6 forward primer | 5′-TTATGGGTCCTAGCCTGAC-3′ |
| U6 forward primer | 5′-CACTATTGCGGGTCTGC-3′ |
| EIF4A1 forward primer | 5′-ATCCCAGAGGCTCTCCTCAC-3′ |
| EIF4A1 reverse primer | 5′-CTACCATTTTCTCTCCCCTGCTT-3′ |
| GAPDH forward primer | 5′-GCACCGTCAAGGCTGAGAAC-3′ |
| GAPDH reverse primer | 5′-TGGTGAAGACGCCAGTGGA-3′ |
| Pri-miR-1284-S | 5′-GAGGATCCCCGGGTACCGGTTTTCACAGAGGAAAATGTTC-3′ |
| Pri-miR-1284-A | 5′-CACACATTCCACAGGCTAGGGACTCAAAAGTCAACACTC-3′ |
Migration and invasion assays
For cell migration assay, the matrigel did not cover the transwell chambers on the top (the chamber is a 24-well insert, with 8 um pore size, BD) and for cell invasion assay, 0.5 mg/mL matrigel covered the top of the chambers (BD Bioscience, San Jose, USA). SGC-7901, MGC-803, HGC-27, and AGS were maintained in a serum-free RPMI 1640 medium, and the cell lines were transferred onto the top of the transwell chambers (1×105 cells/pore). For the lower level chambers, RPMI 1640 medium containing 5% FBS was added, and used as an inducible factor of a cell. The cells were cultured for 16 hrs, and the un-penetrated cells were removed using a cotton ball from the top of the wells. The cells present on the membrane of the lower surface of the transwell chambers were fixed in a solution of 0.2% crystal violet and 4% formaldehyde. Eight fields were chosen with the aim of counting the invading cells under a microscope, as well as photographs were taken. The cells with stronger abilities of migration and invasion of cell lines were selected for subsequent experiments.
Establishment of stable miR-1284 expressing cell lines
Lentivirus expressing hsa-miR-1284 and control vector were structured and provided by Genechem (Shanghai, China). A Primer was used to amplify the sequence of pri-miR-1284. The transfection of SGC-7901 and MGC-803 cells was done by using Lipofectamine 2000 reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. One day after transfection, the culture medium was replenished. At 48 hrs of post-transfection, 0.4 mg/mL puromycin (Sigma, MO, USA) was added into the medium, and puromycin-resistant colonies were selected and separately expanded. After the cells became confluent, they were harvested for subsequent experiments.
In vitro assays of invasion and migration
With the aim to analyze the effects of miR-1284 on cell invasion and migration, the transfected cells were detected by the assays of migration and invasion as described in the experiment section.
Cell proliferation assay
To measure the effect of miR-1284 on the rate of cell proliferation, the cells were incubated in 10% CCK-8 (DOJINDO, Japan), and then were diluted in a normal culture medium at 37°C till a change of color was observed by naked eyes. From 0 hrs to an interval of 24 hrs, the rate of proliferation was studied after transfection for five times and the absorbance was measured at a wavelength of 450 nm using a microplate reader.
In vitro cell cycle assays
To analyze the effects of miR-1284 on cell cycle, the transfected cells were detached and stored for 24 hrs in 70% cold ethanol containing 1 mL PBS. The cells were washed once in 3 mL PBS and then were resuspended in a mixture of 1 mg/mL propidium iodide (Biosciences, NJ, USA) and 0.5 mg/mL RNaseA (Keygen Biotech, Nanjing, China) in PBS for 0.5 hrs at 37°C. Approximately, 1.5×105 cells were studied by CyFlow® flow cytometer (BD).
Microarray hybridization of whole genome expression profile
TRIzol reagent (Thermo Fisher Scientific, Carlsbad, USA) was used to extract RNA from the cells and fresh tissues. After RNA extraction, all quantitation and microarray of subsequent experiments were carried out at the Phalanx Biotech Group Laboratory (Taiwan, China). Concentration and quality of RNA were analyzed by NanoDropTM (Thermo Scientific, China) and Agilent Bio-analyzer2100 system (Agilent Technologies, Palo Alto CA) according to the manufacturer’s instructions. The arrays were washed and stained with streptavidin phycoerythrin in a black chip box and scanned using an Agilent Microarray Scanner (G2505C). The primary microarray imaging data were captured by GenePix™, and the data were saved as gpr files. All the files were integrated into a gpr file. The gene expression spectrum detection data of the chip were analyzed by the Rosetta Resolver® System (Rosetta Biosoftware, Kirkland, WA). The differentially expressed genes were selected at |log2(Ratio)| ≥1 and P-value (Differentially expressed) <0.05. The principal component analysis and clustering analysis were analyzed by Heartbeat 3.0.3.
MiRNA target prediction
Previously, the target genes of MiR-1284 have been described using TargetScan (https://www.targetscan.org), miRNA (http://www.microrna.org) and miRDB (http://www.mirdb.org) algorithms.
Dual luciferase reporter assay
Generally, UTR is the 3′-UTR of EIF4A1 mRNA, and has a concentration of miR-1284 with side binding, with PCR amplification. Mutant EIF4A1 3′-UTR and wild-type with putative binding regions of miR-1284 content have been cloned and constructed into the vector of pmiR-REPORT (Ambion). Site-directed Gene Mutagenesis Kit (Roche, Basle, Switzerland) was used in producing the target site in the miR-1284 mutations. After 48 hrs of co-transfection, luciferase activities were studied using the dual luciferase reporter (promega, Madison, WI) assay system. The vector containing Renilla luciferase was used as the reference control for co-transfection. All the experiments were performed in triplicate.
Western blot analysis
Cell Signaling Technology (Beverly, MA, USA) and Abcam (Cambridge, USA) provided the anti-human antibodies of GAPDH, CDH1, JUN, MMP12, c-Myc, and EIF4A1 from rabbit. Li-Cor Biosciences (NE, USA) provided the anti-rabbit antibodies and goat antibodies IRDye 800 as infrared-labeled secondary antibodies. The cell was washed twice in PBS and allowed to lyse in the special buffer for western blotting and immunoprecipitation (Beyotime, Beijing, China) and protease inhibitor phenylmethanesulfonyl fluoride (Beyotime, Beijing, China). Equal amounts of protein were warmed up using the Laemmli sample buffer at 100°C for 300 s, and 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate the protein samples. The cells were then transferred onto polyvinylidene fluoride membranes. The primary antibodies at a ratio of 1:1,000 were used to incubate the membranes at a temperature of 4°C for 12 hrs. The membranes were then washed with PBST thrice. After washing, the peroxidase-conjugated secondary antibodies at a ratio of 1:10,000 were incubated with the membranes for 60 mins at 25°C and the membranes were washed thrice using PBST. The membranes were scanned and quantified the intensities of the bands by using the Odyssey Software Version 3.0 system (Li-Cor Biosciences Lincoln, NE, USA). The GAPDH protein was used as the reference, and the expression of the target proteins was calculated.
Tumor xenograft experiment
The Ethics Committee of Guangxi Medical University approved all animal experiments. The Chapter Cheng of Guangxi Medical University Experimental Animal Welfare and Ethics Committee rules governing the process of animal testing were used. Male, 4–5 weeks old, athymic nude BALBC/c mice were provided by Guangxi Animal Center. The mice were maintained in an isolated temperature controlled and pathogen-free environment. Sterilized food and autoclaved water were provided according to the experimental animal guidelines.
The mice were subcutaneously injected with inoculation dosage of 1×107 cells in 200 μL of PBS per mouse. Group 1 was transfected with negative control vectors MGC-803-LV-miR-NC cells (MGC-803-NC), Group 2 was transfected with MGC-803-LV-hsa-miR-1284 cells stably expressing miR-1284 (MGC-803-miR-1284), Group 3 was transfected with negative control vectors transfected SGC-7901 – LV-miR-NC cells (SGC-7901-NC), and Group 4 was transfected with SGC-7901-LV-hsa-miR-1284 cells stably expressing miR-1284 (SGC-7901-miR-1284). V= tumor size was calculated at 3 days internal using a digital caliper. The formula used was as follows: V (mm3)=1/2× a (length) × b(width)2. The treatment duration was 21 days, and then the mice were killed to access the tumor. The liver tissues, lung tissues, and the formed tumors were removed, fixed, and embedded in paraffin. HE staining was performed on the paraffin-embedded sections.
Tissue immunohistochemistry
The patient specimens were sectioned using a 4-microtome. IHC was used to test EIF4A1. The primary rabbit anti-EIF4A1 antibodies were used in this study (Abcam, England). The secondary antibodies of goat anti-rabbit immunoglobulin were used based on the SP-9000 SPlink Detection Kits (ZSGB-BIO, Beijing, China). The slides were then examined under ×200 magnification, and then the ratio of positively stained cell number vs the total cell number was calculated.
Statistical analysis
The data were analyzed by SPSS 17.0. The data were presented as mean ± SD. For statistical analysis, Student’s t test and Chi-squared test were used. For each experiment, P<0.05 was regarded as statistically significant.
Results
miR-1284 was down-regulated in GC
qRT-PCR was used to examine the miR-1284 expression level in GC. Initially, the expression level of miR-1284 in normal gastric tissues and adjacent tissues of GC were examined. The results showed that 48/74 cases had reduced levels of miR-1284 in GC tissues than in normal tissues, which was about 64.86%. Comparatively, 26/74 (35.14%) had increased miR-1284 levels in the tissues of GC than those in adjacent normal tissues (P<0.05, Figure 1A).
Figure 1.

MiR-1284 is lowly expressed in GC tissues and cell lines. (A) The level of MiR-1284 was detected by quantitative real-time polymerase chain reaction in 74 GC tissues and normal tissues. (B) MiR-593-5p expression was detected by quantitative real-time polymerase chain reaction in GC cells (HGC-27, AGS, SGC-7901, MGC-803) and normal gastric epithelial cells (GES). U6 was used as a reference baseline gene. *P<0.05 compared to GES-1. **P<0.01 compared to GES-1.
Consequently, the mature expression of miR-1284 was examined in four different GC cell lines, namely, SGC-7901, MGC-803, HGC-27, and AGS, as well as GES-1 cell line of human gastric epithelium. The expression levels of miR-1284 in GES-1 were initially higher and then lower in the four GC cell lines. Among them, MGC-803 cells had the lowest (Figure 1B). Our results suggested that the miR-1284 was greatly attenuated in tumor tissues as compared to adjacent normal tissues. Therefore, miR-1284 may function as a tumor suppressor in GC.
The low expression of miR-1284 was influenced by metastasis, degree of differentiation, and tumor size of GC
The correlation of miR-1284 expression with clinical and pathological features was analyzed. The results demonstrated that the prognosis of gastric patients’ affected the correlation. Furthermore, expression of miR-1284 was significantly affected by distant metastasis, degree of differentiation, and the size of tumor (P<0.05). However, the expression of miR-1284 in GC patients was not affected by T classification, gender, or age (P>0.05, Table 2).
Table 2.
Clinicopathological variables and the expression of miR-1284 in gastric cancer patients
| Characteristics | Cases | miR-1284 expression level | P-value | |
|---|---|---|---|---|
| No. of high expression | No. of low expression | |||
| Age (years) | ||||
| ≥60 | 30 | 11 | 19 | 0.979 |
| <60 | 44 | 16 | 28 | |
| Gender | ||||
| Male | 52 | 17 | 35 | 0.297 |
| Woman | 22 | 10 | 12 | |
| Tumor size (cm) | ||||
| >3 | 59 | 18 | 41 | 0.034* |
| ≤3 | 15 | 9 | 6 | |
| TNM stage | ||||
| I–II | 27 | 10 | 17 | 0.941 |
| III–IV | 47 | 17 | 30 | |
| Lymph node metastasis | ||||
| Yes | 49 | 16 | 33 | 0.338 |
| No | 25 | 11 | 14 | |
| Distant metastasis | ||||
| Yes | 22 | 4 | 18 | 0.033* |
| No | 52 | 23 | 29 | |
| Differentiation | ||||
| Low | 44 | 9 | 35 | 0.001* |
| Well | 30 | 18 | 12 | |
Note: *Statistically significant.
Up-regulation of miR-1284 can inhibit GC cell invasion and migration
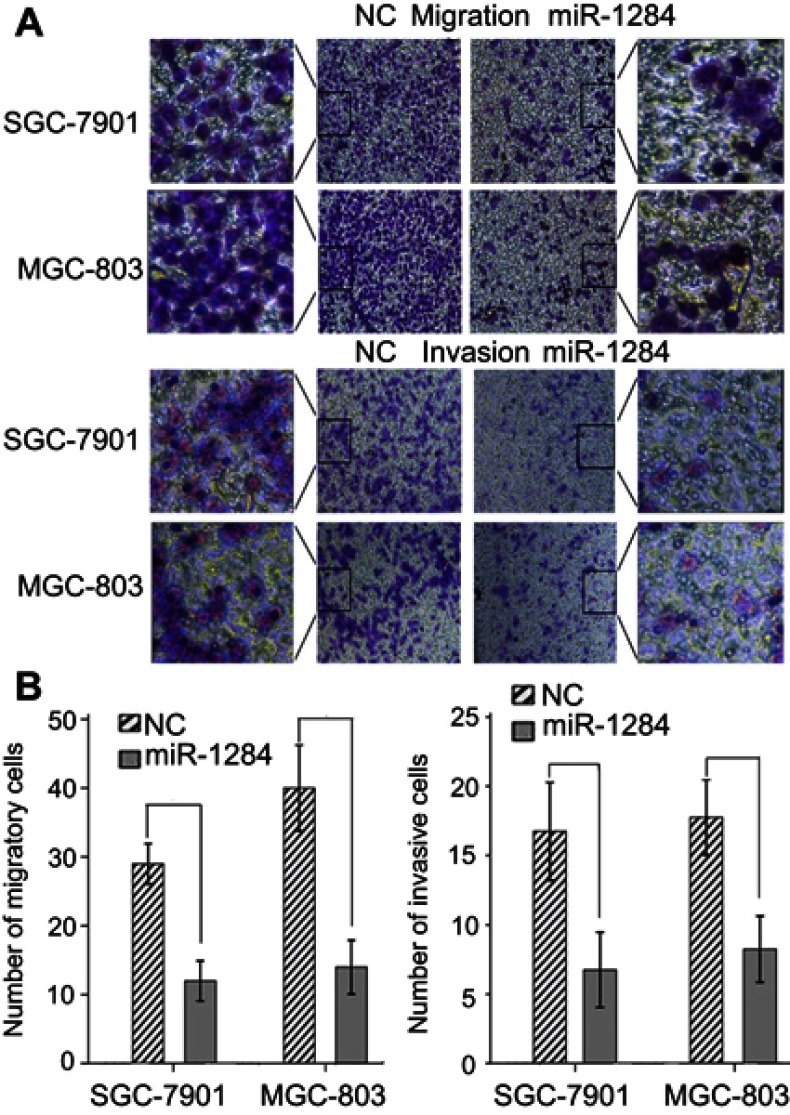
The effects of miR-1284 on migration and invasion of GC cell lines (HGC-27, MGC-803, AGS, and SGC-7901) were investigated. The results showed that the abilities of migration and invasion of MGC-803 and SGC-7901 were stronger than those of the others (Figure 2A and B). Subsequently, SGC-7901 and MGC-803 cell lines were selected for the transfection by lentiviral vectors that contained the over-expression of miR-1284 sequences. qPCR confirmed the overexpression of miR-1284 (Figure 2C). Further, the effects of miR-1284 on the migration and invasion of MGC-803 and SGC-7901 cell lines were investigated by transwell methods. The untreated cells and scramble control were compared, and according to the results of transwell assay the overexpression of miR-1284 greatly decreased the migration and invasion of two GC cell lines (Figure 3A and B). In summary, miR-1284 was considered as important in the regulation of cell invasion and migration in GC.
Figure 2.
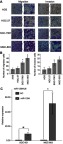
Comparison of migratory cells and invasive cells in each GC cell line, and miR-1284 transfection efficiency after transfection with miR-1284. (A) Images of cells were assessed by transwell assay in (AGS, HGC-27, SGC-7901, and MGC-803) cells (×200). (B) Data are represented in means ± SD of three duplication number of migratory cells and aggressive cells in each group. The number of migratory cells is more in MGC-803 and SGC-7901 than that in other cell lines. *P<0.05 compared to AGS and HGC-27 cells. (C) MiR-1284 transfection efficiency after transfection with miR-1284 lentivirus was detected by Quantitative real-time polymerase chain reaction in SGC-7901and MGC-803 cells. U6 was used as a reference baseline gene. The expression of miR-1284 is more in the miR-1284 treatment group than in the NC group. *P<0.05 compared to NC group.
Figure 3.
Effect of miR-1284 on the migration and invasion of GC cells. (A) Images of cell migration and invasion after transfection with miR-1284 lentivirus were assessed by transwell assay in SGC-7901and MGC-803 cells (×200). (B) Data are presented as means ± SD of three duplication amount of aggressive cells of each group. *P<0.05 compared to NC group.
miR-1284 regulates cell cycle and cell proliferation in vitro
Flow cytometry and CCK-8 assay were performed to examine the cell cycle and proliferation to understand the regulatory mechanism of miR-1284 on cell cycle and tumor cell growth. CCK-8 assay results revealed that both SGC-790 and MGC-803 cells demonstrated that the exogenous expression of miR-1284 inhibited cell proliferation (Figure 4A). On the other hand, flow cytometry showed similar results that exogenous expression of miR-1284 inhibited transition of cells in C1 phase (Figure 4B and C). These findings revealed that the cell cycle G1/S transition and cell proliferation were inhibited by miR-1284.
Figure 4.

Proliferation and cell cycle distribution of GC cells in different groups. (A) The hyperplastic GC cells after transfection with miR-1284 lentivirus were assessed by CCK-8 assay in SGC-7901and MGC-803 cells. (B) Images of cells in a cell cycle after transfection with miR-1284 lentivirus was assessed by Flow cytometry in SGC-7901and MGC-803 cells. (C) Data are presented as means ± SD of three duplications in the respective phase of cell cycle rate in each group. *P<0.05 compared to NC group. **P<0.01compared to NC group.
miR-1284 directly targets EIF4A1 in GC
Further investigation of the mechanism showed that miR-1284 acts as a tumor suppressor in GC. The total gene expression chip of homo-sapiens was used to find significant differential expression of genes in MGC-803 cells transfected with lentivirus containing over-expression of miR-1284. The results showed that 97 cells were up-regulated, while 46 cells were down-regulated in stably expressing miR-1284 cells (MGC-803-1284), as compared with the negative control transfected MGC-803-LV-miR-NC cells (MGC-803-NC). The ten most differentially down-regulated genes were: ZFAND4, EIF4A1, ATF3, CHAC1, EGR1, DUSP5, PHLDA1, JUN, CTGF, and ETV4. The ten most differentially up-regulated genes were: GRM3, SEMG1, ACTR2, OXNAD1, MVB12B, DIP2C, LRRTM3, FTCD, F7, and KCNQ4. In addition, miR-1824 targets can be stimulated by using algorithms such as miRDB (http://www.mirdb.org), TargetScan (https://www.targetscan.org), and miRNA (http://www.microrna.org). Bioinformatics website software predicted 3,334 target genes of SGC-7901miR-1284. The intersection genes were taken as the results between gene expression chip (candidate genes) and the bioinformatics website software predicted target genes. EIF4A1 is the intersection gene, and was a candidate of the target gene for miR-1284. To verify whether EIF4A1 gene was the target of miR-1284, luciferase activity assay was performed and the results showed that miR-1284 suppressed the reporter gene EIF4A1 activity (Figure 5A).
Figure 5.

MiR-1284 negatively regulates EIFA4A1 and its relative signaling pathway directly. (A) The base sequence of the interaction of miR-1284 and nominee gene EIF4A1 wild-type 3′-UTR and continuous missing style of 3′UTR reporters. Data are presented as means ± SD (n=3) of proportional expression in each group by Reporter Gene Assays, and each group was infected separately by miR-1284 and IRREPORT-EIF4A1 plasmid (miR-NC and miR-1284 with EIF4A1 WT 3′UTR, miR-NC and miR-1284 with EIF4A1 MUT 3′UTR) after 24 h. *P>0.05 compared to controls. (B) GO gene biological function indicated that JUN and c-Myc genes were involved in the MAPK signaling pathway.
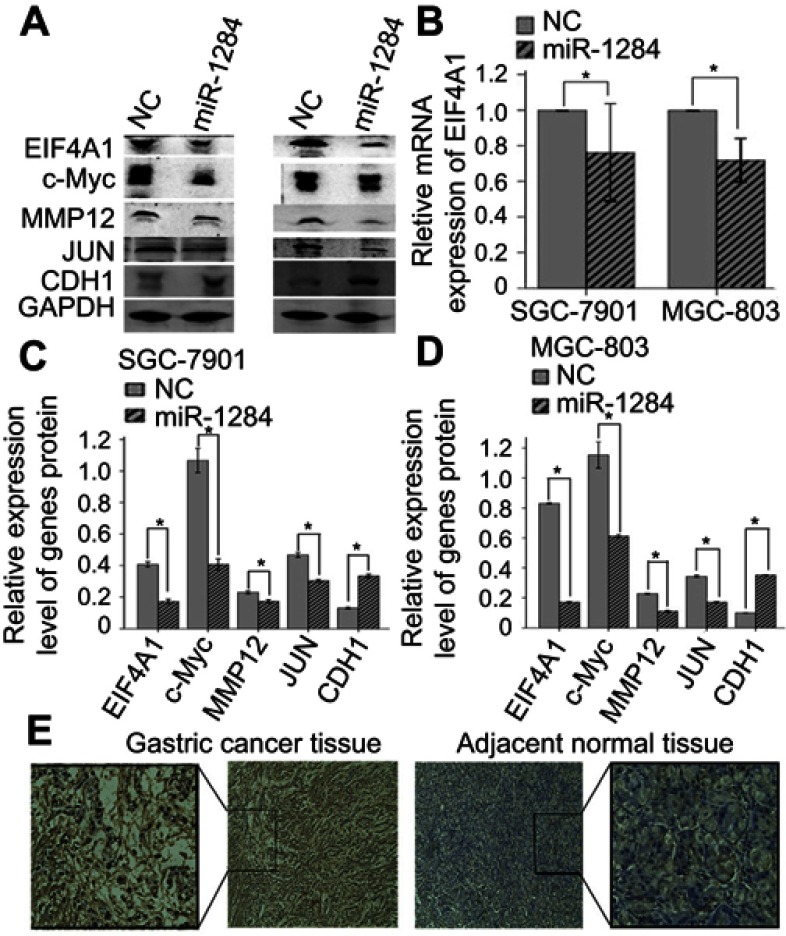
Western blotting and qRT-PCR were used to further investigate whether the expression of EIF4A1 could be regulated in SGC-7901 and MGC-803 cells by miR-1284 transfected with scramble lentivirus and miR-1284. The results revealed that the protein levels and mRNA of EIF4A1 greatly reduced when transfected by miR-1284 as compared to transfection by scramble lentivirus (Figure 6A and B). From these results, it was concluded that EIF4A1-UTR can be directly targeted by miR-1284 in vitro in cells of GC.
Figure 6.
MiR-1284 regulates EIF4A1 relative signal pathway. (A) Images of genes and protein expression after transfection with miR-1284 lentivirus by western blotting in SGC-7901 and MGC-803 cells. (B) Data are presented as means ± SD of three duplications of mRNA expression of target gene EIF4A1 by qRT-PCR in each group, and GAPDH was used as an internal control. (C, D) Data are presented as means ± SD of relative protein expression of genes in each group. GAPDH was used as a reference baseline gene. *P<0.05 compared to NC group. (E) The expression of EIF4A1 in GC tissues and the adjacent normal tissues was evaluated by immunohistochemical staining (×200), and the expression of EIF4A1 is more in cancer tissues than in adjacent normal tissues.
Over-expression of miR-1284 decreases expression of c-Myc, MMP12, and JUN, while increases CDH1 expression
To probe how miR-1284 over-expression inhibits cell migration and invasion in GC, western blotting was performed to detect the metastatic-related genes, such as c-Myc, MMP12, JUN, and CDH1 (Figure 6A). Compared with vector control cells group, the results showed that the protein levels of MMP12, c-Myc, and JUN were lowered in the miR-1284 group, but were increased for CDH1 in the miR-1284 group (P<0.05) (Figure 6C and D). These results suggested that over-expression of miR-1284 effectively decreased the expression of c-Myc, MMP12, JUN and increased the expression of CDH1 in MGC-803 and SGC-7901cell lines in vitro.
EIF4A1 expression and miR-1284 expression have an inverse correlation in GC
To investigate the correlation of the expression levels of EIF4A1 and miR-1284 in GC tissues, the immunohistochemical method was used to study the expression of protein levels of EIF4A1 in 74 clinical samples. Compared with the results of miR-1284 expression by qRT-PCR, EIF4A1 was up-regulated, but combined with miR-1284 showed down-regulation in GC tissues. EIF4A1 showed down-regulation, but combined with miR-1284 showed up-regulation in GC tissues (Figure 6E and Table 3). These results suggested that the expression of EIF4A1 can be suppressed by miR-1284.
Table 3.
Correlation between miR-1284 and EIF4A1 expression
| Parameter | No. of cases | MiR-1284 expression | P-value | |
|---|---|---|---|---|
| N=74 | Low (%) | High (%) | ||
| EIF4A1 expression | ||||
| Negative | 31 | 15 (48.39) | 16 (51.61) | 0.022* |
| Positive | 43 | 32 (74.42) | 11 (25.58) | |
Note: *Statistically significant.
miR-1284 metastasis suppressed tumorigenesis in a xenograft model
To evaluate the role of miR-1284 in the growth and formation of tumor in vivo, SGC-7901 cells and cells from the xenograft model of human GC were adopted from nude mice. MGC-803 and SGC-7901 cells transfected with scramble lentivirus or miR-1284 were injected into the flank of nude mice subcutaneously. After injection, the tumor size was observed at an interval of 3 days, and the observation was recorded to plot the tumor growth curves. The mice were then killed to collect the xenografts. The mean size of tumors generated from the miR-1284 overexpression group was much lowered than that of the control group (Figure 7A and B). The livers and lungs were dissected to study the in vivo effect of miR-1284 toward tumor metastasis. HE staining was used to evaluate the tissue morphology (Figure 7C). The results showed that the macroscopic liver metastases’ number was low when miR-1284 was used to transfect SGC-803 cells as compared with the control group (Table 4). All these results suggested a high possibility that miR-1284 represses metastasis and proliferation of GC.
Figure 7.
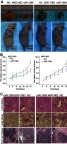
The effect of miR-1284 in GC growth in vivo. (A) The whole specimens of transfected human GC cells with subcutaneous transplanted tumors in each group, and the images of subcutaneous transplantation tumor for each cell line group after 21 days. (B) Tumor growth curve after injecting the SGC-7901 or MGC-803 cells into nude mice. The length and width of tumor were measured by calipers. *P<0.05 compared to NC group. (C) The tumor, lung, and liver tissues were assessed by H&E staining in each group (×100), HE staining was used to observe the morphological changes in tumors, and the tissues referred by the black line represent metastatic tumors.
Abbreviations: NC, Negative control; GC, Gastric cancer.
Table 4.
The metastasis cases of liver and lung tissues in cells transplanted with tumor of nude mice in each group
| Groups | Cases | Liver metastasis | P-value | Lung metastasis | P-value | ||
|---|---|---|---|---|---|---|---|
| Positive cases | Positive rate (%) | Positive cases | Positive rate (%) | ||||
| MGC-803-miR-1284 | 10 | 1 | 10.00 | 0.020* | 1 | 10.00 | 0.303 |
| MGC-803-NC | 10 | 7 | 70.00 | 4 | 40.00 | ||
| SGC-7901-miR-1284 | 10 | 3 | 30.00 | 0.650 | 1 | 10.00 | 0.582 |
| SGC-7901-NC | 10 | 5 | 50.00 | 3 | 30.00 | ||
Note: *Statistically significant.
Discussion
The effects of micro RNAs on GC have been widely studied previously and miRNA dysregulation led to carcinogenesis.12,13 Therefore, aberrant expression of miRNAs in GC might be associated with the occurrence of diseases. Chen et al, had initially reported that the expression of miR-1284 was not normal in patients with GC,3 indicating that the abnormal expression of miR-1284 might have a great effect in the development of GC. Huang et al, have described that over-expression of miR-1284 reduced SGC-7901 cell proliferation and improved cell apoptosis, and might be related to the up-regulation of p27, Bax, procaspase-3, and active caspase-3.4 However, reports on clinicopathologic characteristics and function of miR-1284 are not concrete. To further explore the role of miR-1284 in GC, we focused on the role of miR-1284 both in vitro and in vivo.
In this study, the expression analysis was expanded by examining the expression in 74 pairs of GC tissues and adjacent normal gastric tissues, GC cell lines, and human GC epithelial cell lines. The results demonstrated that miR-1284 greatly downregulated GC in both GC cell lines and tissues (Figure 1). In the metastatic tissues, the expression level of miR-1284 was comparatively lower than that in non-distant metastatic tissues (Table 2). These results suggest that miR-1284 may be a suppressor of tumor genes. Lentiviral expressing miRNAs were transfected into cancer cells to observe their function, and this has been done widely in the previous studies.14,15 To assess the influence of miR-1284 on GC metastasis, transwell assay was used to detect the abilities of cell invasion and metastasis. The GC cell lines MGC-803 and SGC-7901 showed stronger invasion and metastasis abilities (Figure 2), and the two cell lines were transfected with miR-1284 precursor to obtain stable miR-1284 expressing cell lines. The results showed that miR-1284 overexpression inhibited the invasion and migration of GC cells in vitro (Figure 3). Similarly, the in vivo overexpression of miR-1284 in MGC-803 and SGC-790 cells caused slow growth when compared with control xenografts of the vector (Figure 7). Tumor liver metastatic rate was lower in the mouse xenograft model that overexpress miR-1284 relative to the vector control group (Table 4). Collectively, these results revealed that miR-1284 inhibited GC growth and metastasis.
Several research studies have confirmed that the functions of miRNAs depend on their main downstream target genes, and miRNAs are used to bind to 3′UTRs of transcripts and may also target the open reading frames.16,17 However, the direct target gene of miR-1284 has not yet been reported. Recently, bioinformatics websites such as miRNA (http://www.microrna.org), miRDB (http://www.mirdb.org), and TargetScan (https://www.targetscan.org) have predicted miRNAs as the target genes. However, the bioinformatics websites predicted hundreds of miRNAs as target genes. To make an informed choice, a total gene expression chip method was used to establish the genes that are mostly affected by overexpression of miR-1284 in GC. Intersection genes between the results of gene expression chip (candidate genes) method and the results of bioinformatics website software predicted the target genes. The intersection gene EIF4A1 was confirmed as the candidate target gene of miR-1284 and was verified by luciferase activity assay. The results showed that miR-1284 directly targeted EIF4A1 according to dual luciferase reporter assay, as well as by western blotting and qRT-PCR (Figure 6). Additionally, EIF4A1 and miR-1284 expression are inversely correlated in GC (Figure 6). Therefore, the metastasis and growth of GC cells are inhibited by miR-1284 by targeting EIF4A1 gene, by adjusting the levels of c-Myc, MMP12, JUN, and CDH (Figure 6). Furthermore, suppression of EIF4A1 could repress the invasion and migration of GC cells in vitro (Figure 3). Additionally, the cell gene chip results suggested that miR-1284 was associated with MAPK signaling pathway (Figure 5B), because JUN was down-regulated and CDH1 was up-regulated when miR-1284 was overexpressed (Figure 6). Loss of E-cadherin impaired cell–cell adhesion and inhibition of E-cadherin inhibited cell migration.18 In carcinoma, abrogation of E-cadherin caused accumulation of c-Jun protein dramatically,19 cell–cell adhesion is activated by E-cadherin-mediated accumulation of JUN,20, and the activity of JUN and the expression of protein are decreased by re-expression of E-cadherin.21 In addition, suppression of JUN can reduce the expression of MMP protein,22 which in turn is positively correlated with the invasive ability of cancer,23,24 and down-regulated MMP12 can inhibit the migration and invasion of GC cells.25 Thus, MAPK signaling pathway is suppressed by target gene EIF4A1 of miR-1284 in GC.
Conclusion
The effect of miR-1284 has been down-regulated in GC. The in vivo and in vitro data showed that up-regulation of miR-1284 acts as a tumor suppressor in GC, and inhibits cell metastasis through binding to 3′UTR of EIF4A1. Therefore, miR-1284 is expected to play an important role in the development of therapeutic strategies for GC.
Acknowledgments
This study was supported by National Natural Science Foundation of China (NO. 81660511 and NO. 81060201), the Natural Science Foundation of Guangxi (NO. 2018GXNSFBA138016 and NO. 2015GXNSFDA227001) and the Guangxi Medical University Training Program for Distinguished Young Scholars.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Slezak-Prochazka I, Durmus S, Kroesen B-J, van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA. 2010;16(6):1087–1095. doi: 10.1261/rna.1804410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Svoboda P, Flemr M. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Rep. 2010;11(8):590–597. doi: 10.1038/embor.2010.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Tang Z, Sun Y, et al. miRNA expression profile in primary gastric cancers and paired lymph node metastases indicates that miR-10a plays a role in metastasis from primary gastric cancer to lymph nodes. Exp Ther Med. 2012;3(2):351–356. doi: 10.3892/etm.2011.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang M, Wu L, Luo S, et al. MicroRNA-1284 inhibits proliferation and induces apoptosis in SGC-7901 human gastric cancer cells. Biotechnol Lett. 2017;39(1):33–38. doi: 10.1007/s10529-016-2213-1 [DOI] [PubMed] [Google Scholar]
- 5.Cao W, Wei W, Zhan Z, Xie Y, Xiao Q. MiR-1284 modulates multidrug resistance of gastric cancer cells by targeting EIF4A1. Oncol Rep. 2016;35(5):2583–2591. doi: 10.3892/or.2016.4643 [DOI] [PubMed] [Google Scholar]
- 6.Jones E, Quinn CM, See CG, et al. The linked human elongation initiation factor 4A1 (EIF4A1) and CD68 genes map to chromosome 17p13. Genomics. 1998;53(2):248–250. doi: 10.1006/geno.1998.5515 [DOI] [PubMed] [Google Scholar]
- 7.Qi M, Qi Y, Ma Y, et al. Over-expression of human cytomegalovirus miR-US25-2-3p downregulates eIF4A1 and inhibits HCMV replication. FEBS Lett. 2013;587(14):2266–2271. doi: 10.1016/j.febslet.2013.05.057 [DOI] [PubMed] [Google Scholar]
- 8.Lindqvist L, Oberer M, Reibarkh M, et al. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS One. 2008;3(2):e1583. doi: 10.1371/journal.pone.0001583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modelska A, Turro E, Russell R, et al. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015;6:e1603. doi: 10.1038/cddis.2014.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu C-Z, Shi R-J, Chen D, et al. Potential biomarkers for paclitaxel sensitivity in hypopharynx cancer cell. Int J Clin Exp Pathol. 2013;6(12):2745–2756. [PMC free article] [PubMed] [Google Scholar]
- 11.Liang S, Zhou Y, Chen Y, Ke G, Wen H, Wu X. Decreased expression of EIF4A1 after preoperative brachytherapy predicts better tumor-specific survival in cervical cancer. Int J Gynecol Cancer. 2014;24(5):908–915. doi: 10.1097/IGC.0000000000000152 [DOI] [PubMed] [Google Scholar]
- 12.Wu WK, Lee CW, Cho CH, et al. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29(43):5761–5771. doi: 10.1038/onc.2010.352 [DOI] [PubMed] [Google Scholar]
- 13.Pan H-W, Li S-C, Tsai K-W. MicroRNA dysregulation in gastric cancer. Curr Pharm Des. 2013;19(7):1273–1284. [DOI] [PubMed] [Google Scholar]
- 14.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. doi: 10.1038/nature09284 [DOI] [PubMed] [Google Scholar]
- 15.Varambally S, Cao Q, Mani R-S, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322(5908):1695–1699. doi: 10.1126/science.1165395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- 18.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28(1–2):151–166. doi: 10.1007/s10555-008-9179-y [DOI] [PubMed] [Google Scholar]
- 19.Knirsh R, Ben-Dror I, Spangler B, et al. Loss of E-cadherin-mediated cell-cell contacts activates a novel mechanism for up-regulation of the proto-oncogene c-Jun. Mol Biol Cell. 2009;20(7):2121–2129. doi: 10.1091/mbc.e08-12-1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S-J, Jung YH, Oh SY, Yong MS, Ryu JM, Han HJ. Netrin-1 induces MMP-12-dependent E-cadherin degradation via the distinct activation of PKCα and FAK/Fyn in promoting mesenchymal stem cell motility. Stem Cells Dev. 2014;23(16):1870–1882. doi: 10.1089/scd.2013.0632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spangler B, Vardimon L, Bosserhoff AK, Kuphal S. Post-transcriptional regulation controlled by E-cadherin is important for c-Jun activity in melanoma. Pigment Cell Melanoma Res. 2011;24(1):148–164. doi: 10.1111/j.1755-148X.2010.00787.x [DOI] [PubMed] [Google Scholar]
- 22.Nishitani K, Ito H, Hiramitsu T, et al. PGE2 inhibits MMP expression by suppressing MKK4-JNK MAP kinase-c-JUN pathway via EP4 in human articular chondrocytes. J Cell Biochem. 2010;109(2):425–433. doi: 10.1002/jcb.22421 [DOI] [PubMed] [Google Scholar]
- 23.Bradbury PA, Zhai R, Hopkins J, et al. Matrix metalloproteinase 1, 3 and 12 polymorphisms and esophageal adenocarcinoma risk and prognosis. Carcinogenesis. 2009;30(5):793–798. doi: 10.1093/carcin/bgp065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu D, Ye T, Xiang Y, et al. Quercetin inhibits epithelial-mesenchymal transition, decreases invasiveness and metastasis, and reverses IL-6 induced epithelial-mesenchymal transition, expression of MMP by inhibiting STAT3 signaling in pancreatic cancer cells. Onco Targets Ther. 2017;10:4719–4729. doi: 10.2147/OTT.S136840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon CH, Moon HJ, Park HJ, Choi JH, Park DY. S100A8 and S100A9 promotes invasion and migration through p38 mitogen-activated protein kinase-dependent NF-κB activation in gastric cancer cells. Mol Cells. 2013;35(3):226–234. doi: 10.1007/s10059-013-2269-x [DOI] [PMC free article] [PubMed] [Google Scholar]