Abstract
Purpose: Esophageal cancer is a major cause of cancer-related mortality worldwide. The long noncoding RNA LINC00152 has been confirmed to play an oncogenic role in many cancers. However, the expression pattern and function of LINC00152 in human esophageal squamous cell carcinoma (ESCC) remain unclear.
Materials and methods: We evaluated LINC00152 expression in ESCC by qPCR and in situ hybridization. Proliferation, apoptosis, cell cycle, migration and invasion were examined in ESCC cells knocked down for LINC00152 knockdown by siRNA. Furthermore, an mRNA microarray was performed in ESCC cells with LINC00152 knockdown.
Results: LINC00152 was significantly upregulated in human ESCC clinical samples (P<0.001) and cell lines (P=0.008), and LINC00152 overexpression was related to lymphatic metastasis (P=0.03) and advanced pTNM classification (P=0.005). Furthermore, ESCC patients with LINC00152 overexpression had significantly shorter overall survival (P=0.007), and LINC00152 overexpression was an independent risk factor for overall survival of ESCC patients. LINC00152 knockdown inhibited the proliferation, migration and invasion of ESCC cells in vitro. In addition, mechanistic investigations through mRNA array and immunoblot analyses demonstrated that LINC00152 regulated the expression of several cell cycle-related proteins and SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) interactions in vesicular transport pathway proteins.
Conclusion: Our research indicated that LINC00152 exhibits oncogenic functions in ESCC and may represent a potential new target for ESCC therapy.
Keywords: LINC00152, esophageal squamous cell carcinoma, proliferation, metastasis
Introduction
Esophageal carcinoma is the seventh most common cancer worldwide, with approximately 509,000 deaths cases per year.1 Esophageal squamous cell carcinoma (ESCC) is the predominant type of esophageal cancer and comprises 90% of all esophageal cancer cases. In recent years, ESCC incidence has risen by 0.6% annually, particularly in men.2 Despite progress in medical and surgical treatments, most ESCC patients are diagnosed at advanced stage, leading to poor prognosis.3 According to the global surveillance of trends in cancer survival 2000–2014 (CONCORD-3), the 5-year survival rate of ESCC is 22–30% in China.4 Currently, the mechanism underlying esophageal cancer development remains unclear, resulting in limited clinical approaches for the early diagnosis and therapy of ESCC.
Protein-encoding genes account for only approximately 2% of genome, while noncoding genes account for the other 98% of genome, which was once thought to be transcriptional “noise”.5 In recent years, with the development of genomics, a large amount of evidence has shown that noncoding RNAs may play an important role in diseases. For example, several long noncoding RNAs, RNAs between 200 and 100,000 nucleotides,6 have shown significant roles in tumorigenesis and development, such as MALAT1, HOTAIR and TUG1.7
Long intergenic noncoding RNA 152 (LINC00152), an 828 bp lncRNA located at chromosome 2p11.2, was first discovered in hepatocellular carcinoma.8 LINC00152 has since been shown to function in the progression of many cancers, including gastric cancer, lung adenocarcinoma, tongue squamous cell carcinoma, gallbladder cancer and renal cell carcinoma.9–11 However, the expression and biological roles of LINC00152 in the development of ESCC remains unclear.
Herein, we examined the expression level of LINC00152 in ESCC tissues and analyzed its correlation with clinicopathological features and outcome. We also investigated the biological function and molecular mechanisms of LINC00152 in ESCC. Our results demonstrate that upregulation of LINC00152 promotes tumorigenesis of ESCC and suggest LINC00152 as an effective target for ESCC therapy.
Materials and methods
Patients and tissue specimens
This study was approved by the Ethics Committee of The First Affiliated Hospital of Wenzhou Medical University and was conducted in accordance with the Declaration of Helsinki. A total of 69 histologically diagnosed ESCC tissue samples and corresponding paracancerous normal tissues were obtained from patients who underwent surgery in the Department of Thoracic Surgery between 2014 and 2015. Paracancerous normal tissues were defined as normal mucosa tissue more than 5 cm away from the edge of cancer. None of the included patients had received any preoperative therapy. The tissues were collected immediately after surgical resection and stored at −80ºC until analysis. All patients provided written informed consent. The clinical and pathological characteristics for each patient were also collected.
Cell culture
The human esophageal carcinoma cell lines used in this study (TE1, TE13, ECA109 and KYSE-410) were obtained from the First Affiliated Hospital of Nanjing Medical University, and the human esophageal epithelial normal cell line (HEEC) was purchased from the laboratories of ScienCell Research (San Diego,Los Angeles, USA). All cell lines used in this study were approved by the Ethics Committee of The First Affiliated Hospital of Wenzhou Medical University. Three cell lines (TE1, TE13 and KYSE-410) were cultured in DMEM high glucose medium (Kaiji, China) containing 10% FBS. The ECA109 and HEEC cell lines were cultured in 1640 medium (Kaiji) containing 10% FBS. Cells were maintained in a 37ºC incubator containing 5% CO2.
RNA extraction, qRT-PCR and microarray hybridization
Total RNA was isolated from tissues and cells using TriZol (Life Technologies, Scotland, UK) following the manufacturer’s protocol. RNA (1.5 μg) was reverse transcribed using PrimeScript RT Master Mix (Takara, Osak , Japan). qRT-PCR was conducted using the SYBR Select Master Mix (Applied Biosystems, CA, USA) on the ABI7300 system (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was used as an internal standard for normalization. The PCR primer sequences are shown in Table S1. The 2−ΔΔCt method was used to calculate the relative mRNA expression levels.12 All samples were analyzed in triplicate.
Table S1.
The sequence of primers and synthetic siRNAs
| Primer | Forward | Reverse |
|---|---|---|
| LINC00152 | AAAATCACGACTCAGCCCCC | AATGGGAAACCGACCAGACC |
| GAPDH (glyceraldehyde-3-phosphate dehydrogenase) | AGGGCTGCTTTTAACTCTGGT | CCCCACTTGATTTTGGAGGGA |
| siRNAs | Sense(5’-3’) | Antisense(5’-3’) |
| siRNA1-LINC00152 | CCUCUUGACUCUUCUGUGATT | UCACAGAAGAGUCAAGAGGTT |
| siRNA2-LIINC00152 | GGAGAUGAAACAGGAAGCUTT | UUCUCCGAACGUGUCACGUTT |
| siRNA-NC | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
For the microarray, the processes of labeling, hybridization and scanning were carried out at CapitalBio Corporation (Beijing, China).
In situ hybridization (ISH)
The esophageal tissue microarray containing 105 cases of ESCC tissue was purchased from Outbo Biotech Co. Ltd (Shanghai, China). The sequence of the LINC00152 ISH probe (Outbo Biotech Co. Ltd.) was ATTAAGACACATAGAGACTGGCCA. The RNAscope 2.0 HD-Brown Manual Assay kit was used for ISH according to the manufacturer’s instructions (Advanced Cell Diagnostics, Newark, CA, USA). The staining results were visualized using the Dako EnVision FLEX detection system (Dako, Carpinteria, CA, USA). The LINC00152 expression level was scored according to staining intensity and positive staining as follows: the staining intensity score: 0 point (negative), 1 point (+), 2 points (++), 3 points (+++); and positive staining rate score: 0 point (negative), 1 point (1–25%), 2 points (26–50%), 3 points (51–75%), 4 points (76–100%). The total score was calculated as the staining intensity score multiplied by the positive staining rate score.
Cell proliferation assays
Cell Counting Kit-8 (CCK-8) assay and the real-time xCELLigence analysis system (RTCA) assay were used to assess cell proliferation. Each assay was performed three times independently. For CCK8 assays, cells transfected with siLINC00152 or negative control (NC) siRNA were plated in 96-well plates (2×103cells/well). Next, 10 μL CCK-8 reagent was added to each well, and plates were incubated for 2 hrs at 37ºC. Absorbance was measured at 450 nm spectrophotometrically every 24 hrs. For RTCA assay, the transfected cells were seeded in an E-Plate 16 plates (1×104cells/well) and cultured in 150 μL medium supplemented with 10% FBS. Cell proliferation was measured by the RTCA device.
Cell migration and invasion assays
Migration assay was performed using Transwell assay inserts (Millipore, Billerica, MA, USA). Approximately 2.5×105 transfected cells were seeded in the upper chamber with 200 μL serum-free DMEM medium. Conditioned medium was added to the lower chamber. For the invasion assay, the upper chamber was replaced by Matrigel invasion chambers (BD Biosciences, New Jersey, USA) and contained 4×105 cells suspended in 500 μL serum-free DMEM medium. The lower chamber was filled with 750 μL DMEM with 10% FBS. After 24 hrs or 48 hrs, cells that migrated across the membrane were fixed using methanol and stained by crystal violet. Each chamber was counted under the microscope in five random fields. All experiments were performed independently in triplicate.
Apoptosis and cell cycle analysis
For apoptosis evaluation, cells were harvested after transfection and then stained with the Annexin V-FITC/PI apoptosis detection kit (BD Biosciences) according to the manufacturer’s instructions. For cell cycle analysis, the cells were stained with propidium oxide (CycleTEST PLUS DNA Reagent Kit, BD Biosciences). Flow cytometry analysis of cells was performed by FACScan (BD Biosciences).
Western blot
Western blot assay was carried out as previously described.13 Total protein was extracted with RIPA lysis buffer (Solarbio, Bei Jing, China) supplemented with protease inhibitors (Roche Applied Science, Basel, Switzerland). The following primary antibodies were used: STX3 (Thermo Fisher, Waltham, Massachusetts, USA, PA5-31,208); STX12 (Thermo Fisher, PA5-65,939); GAPDH (Santa Cruz, sc-47,724); MAD2L1 (Thermo Fisher, PA5-18,090) and CDK6 (Thermo Fisher, MA5-13,330).
Statistical analysis
All statistical analyses involving Student’s test, Spearman test, and one-way ANOVA were performed using the SPSS software package (version 20.0, SPSS Inc.). Overall survival (OS) curves were analyzed using the Kaplan–Meier method and survival differences were evaluated by log-rank test. The significance of survival variables was evaluated by the Cox multivariate proportional hazards model. All P-values were two-sided, and a P-value less than 0.05 was considered statistically significant.
Results
LINC00152 is significantly overexpressed in ESCC
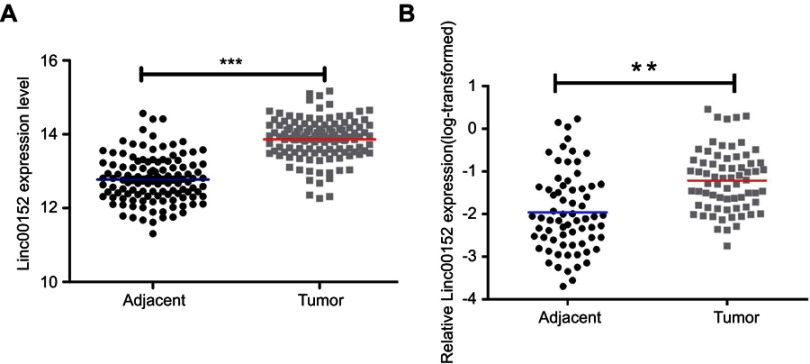
We first identified an ESCC tissue microarray from GEO DataSets (ID: GSE53624),14 in which the lncRNA expression profile was determined in tumor and corresponding paracancerous normal tissues from 119 ESCC patients. The results showed that LINC00152 was significantly overexpressed in ESCC tissues compared with paracancerous tissues (Figure 1A, P<0.001). We validated this result by performing qRT-PCR in 69 pairs of matched ESCC tissues and corresponding paracancerous normal tissues. LINC00152 was prominently overexpressed in 76.81% (53 of 69) of ESCC tissues, with an average 5.62-fold change in expression compared with normal tissues (Figure 1B, P=0.008).
Figure 1.
LINC00152 is overexpressed in ESCC tissues.
Notes: (A) LINC00152 is upregulated in ESCC tissue compared with paired normal tissues in the GEO database (GSE53624). (B) LINC00152 was detected in 69 pairs of ESCC tissues by qRT-PCR. The levels of LINC00152 in ESCC tissues were significantly higher than those in nontumorous tissues. **P<0.01, ***P<0.001.
Abbreviation: ESCC, esophageal squamous cell carcinoma.
LINC00152 overexpression is associated with clinicopathological characteristics and predicts poor prognosis of ESCC patients
We further evaluated LINC00152 expression in the microarray containing 105 cases of ESCC tissue using ISH and scored the samples as described in Methods. Based on the total score, the samples were divided into the low LINC00152 expression group (total score <4, n=19, Figure 2A and C) and high LINC00152 expression group (total score ≥4, N=86, Figure 2B and D). High expression of LINC00152 was closely associated with N stage (P=0.03) and advanced TNM stage (P=0.005) (Table 1). Furthermore, Kaplan–Meier analysis demonstrated that ESCC patients in the high LINC00152 expression group had a significantly shorter OS compared with patients in the low LINC00152 expression group (P=0.007, Figure 2E). Univariate Cox regression analyses indicated that LINC00152 expression, sex, p53, T, N and TNM stage were significantly correlated with OS of ESCC patients. Furthermore, multivariate Cox regression analysis showed that LINC00152 overexpression was an independent risk factor for OS of ESCC patients (Table 2).
Figure 2.
Relationship between LINC00152 expression and clinical features and prognosis in patients with esophageal cancer. LINC00152 expression was detected by ISH in 105 ESCC tissues.
Notes: (A and C) Low expression of LINC00152 in ESCC tissue, total score <4, n=19. (B and D) High expression of LINC00152 in ESCC tissue, total score ≥4, n=86. (E) ESCC patients with high LINC00152 expression showed shorter OS compared with patients with low LINC00152 expression by Kaplan–Meier survival curve according to the results of ISH (P=0.007).
Abbreviations: ESCC, esophageal squamous cell carcinoma; ISH, in situ hybridization.
Table 1.
Correlation between LINC00152 expression and clinicopathological characteristics
| Variables | LINC00152 expression | Total | rs | P-value | ||
|---|---|---|---|---|---|---|
| Low | High | |||||
| Age (years) | 0.013 | 0.908 | ||||
| ≤65 | 10 | 44 | 54 | |||
| >65 | 9 | 42 | 51 | |||
| Sex | 1.228 | 0.268 | ||||
| Female | 7 | 21 | 28 | |||
| Male | 12 | 65 | 77 | |||
| T stage | 1.434 | 0.231 | ||||
| T1/T2 | 5 | 11 | 16 | |||
| T3/T4 | 13 | 73 | 86 | |||
| Null | 3 | |||||
| N stage | 4.691 | 0.03* | ||||
| N0 | 13 | 34 | 47 | |||
| N1/N2/N3 | 6 | 49 | 55 | |||
| Null | 3 | |||||
| TNM stage | 7.798 | 0.005** | ||||
| I/II | 14 | 34 | 48 | |||
| III | 4 | 48 | 52 | |||
| Null | 5 | |||||
| Grade | 0.259 | 0.611 | ||||
| 1/2 | 16 | 65 | 81 | |||
| 3 | 3 | 21 | 24 | |||
| P53 | 0.521 | 0.47 | ||||
| Negative | 7 | 21 | 28 | |||
| Positive | 11 | 49 | 60 | |||
| Null | 17 | |||||
| Ki67 | 0.465 | 0.495 | ||||
| Negative | 6 | 32 | 38 | |||
| Positive | 12 | 44 | 56 | |||
| Null | 11 | |||||
Notes: *P<0.05, **P<0.01.
Table 2.
Univariate and multivariate analyses of factors correlated with overall survival of esophageal cancer patients
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| LINC00152 | 1.917 | 1.175–3.129 | 0.009** | 1.735 | 0.936–3.214 | 0.045* |
| Grade | 1.357 | 0.838–2.200 | 0.215 | |||
| Sex | 1.796 | 1.065–3.028 | 0.028* | 1.546 | 0.811–2.945 | 0.186 |
| Age (years) | 0.879 | 0.574–1.346 | 0.554 | |||
| T stage | 2.743 | 1.361–5.531 | 0.005** | 1.311 | 0.562–3.061 | 0.531 |
| N stage | 2.044 | 1.309–3.193 | 0.002** | 0.447 | 0.126–1.594 | 0.215 |
| TNM stage | 2.841 | 1.770–4.560 | 0.000*** | 4.978 | 1.247–19.871 | 0.023* |
| P53 | 1.826 | 1.072–3.110 | 0.027* | 1.269 | 0.699–2.302 | 0.433 |
| Ki67 | 1.085 | 0.683–1.722 | 0.731 | |||
Notes: *P<0.05, **P<0.01, ***P<0.001.
LINC00152 promotes proliferation and regulates the cell cycle of ESCC cells
To examine the function of LINC00152 in ESCC, we first evaluated LINC00152 expression in four human ESCC cell lines, TE13, TE1, ECA-109, KYSE-410 and a HEEC by qPCR. LINC00152 expression was upregulated in TE1, TE13 and ECA-109 cells but downregulated in KYSE-410 cells compared with HEEC cells (Figure 3A).
Figure 3.
LINC00152 promotes the proliferation of ESCC cells.
Notes: (A) LINC00152 expression in the indicated cell lines. (B) LINC00152 expression in ECSS cells after transfection of si-NC (negative control) or two siRNAs targeting LINC00152. siRNA-1 showed better inhibition efficiency. (C) Cell proliferation assay by CCK-8. Compared with NC, siRNA-mediated silencing of LINC00152 significantly inhibited cell proliferation. (D) Cell proliferation assay by RTCA. Depletion of LINC00152 reduced both TE1 and TE13 cell line proliferation. *P<0.05, **P<0.01.
Abbreviation: ESCC, esophageal squamous cell carcinoma.
To investigate the biological function of LINC00152, we used two siRNAs to knock down its expression in TE13 and TE1 ESCC cell lines. We confirmed downregulation of LINC00152 by qRT-PCR after transfection and found that siRNA1 showed a better silencing efficiency (Figure 3B). We next examined the role of LINC00152 in the proliferation ability of ESCC cells by performing CCK-8 assays in TE13 and TE1 cells transfected with siRNA1 for 24 hrs. The results showed that downregulation of LINC00152 inhibited cell proliferation of ESCC cells compared with controls (Figure 3C). Evaluation of cell proliferation using the xCELLigence assay showed similar results (Figure 3D).
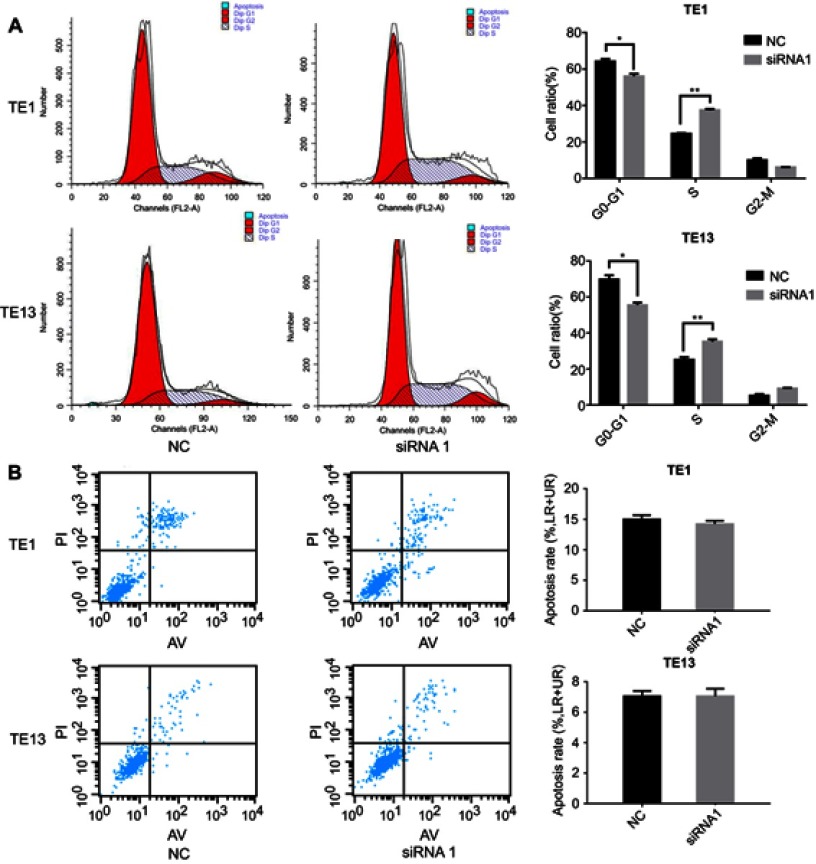
We also evaluated cell cycle distribution in ESCC cells downregulated for LINC00152 and found that both TE1 and TE13 cells with LINC00152 knockdown showed an S phase arrest (Figure 4A). However, silencing LINC00152 had no impact on the apoptosis of ESCC cells (Figure 4B).
Figure 4.
LINC00152 impacts the cell cycle but not apoptosis of ESCC cells.
Notes: (A) siRNA treatment significantly blocked TE1 and TE13 cells at S phase. (B) Silencing of LINC00152 had no impact on the apoptosis of ESCC cells. *P<0.05, **P<0.01.
Abbreaviation: ESCC, esophageal squamous cell carcinoma.
LINC00152 promotes the invasion and migration of ESCC cells
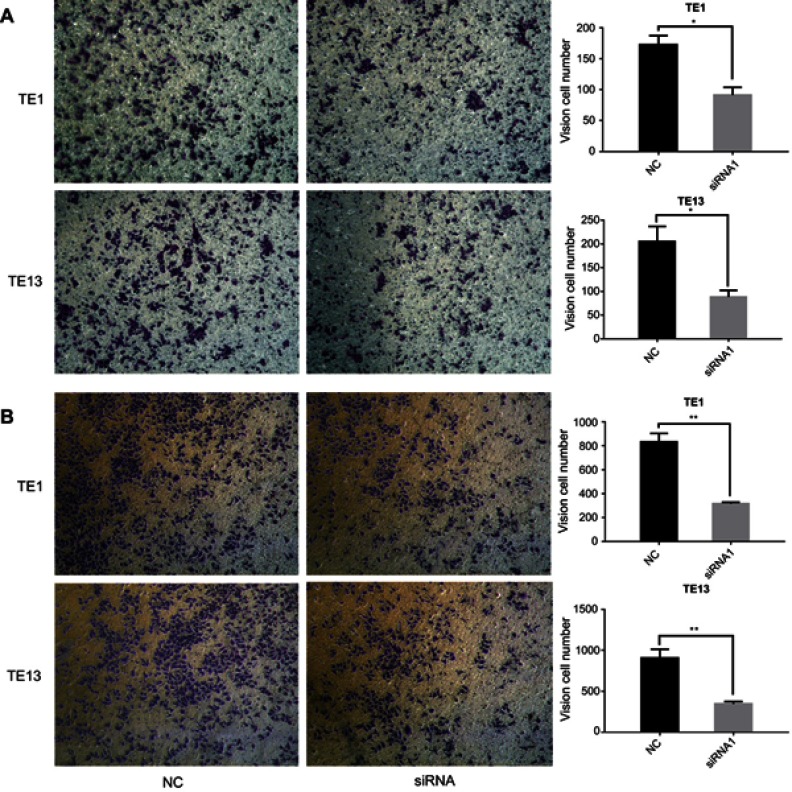
We next investigated the role of LINC00152 in ESCC cell migration and invasion. Transwell assay showed that transfection of ESCC cells with siRNA targeting LINC00152 significantly inhibited cell migration compared with cells transfected with NC siRNA (Figure 5A). In addition, Matrigel assay revealed that the invasion of ESCC cells was inhibited by LINC00152 knockdown compared with NC cells (Figure 5B). Taken together, these results demonstrated that LINC00152 may promote migration and invasion of ESCC cells.
Figure 5.
LINC00152 promotes the migration and invasion of ESCC cell lines.
Notes: (A) Compared with negative controls, ESCC cells transfected with siRNA against LINC00152 showed weaker migration ability in Transwell migration assays. **P<0.01. (B) In Matrigel invasion assay, ESCC cells transfected with siRNA against LINC00152 showed significantly impaired invasion capacity compared with controls. *P<0.05, **P<0.01.
Abbreviation: ESCC, esophageal squamous cell carcinoma.
Initial exploration of the oncogenic mechanism of LINC00152
Our results thus far indicate that LINC00152 may promote the proliferation, migration and invasion of ESCC cells, indicating it exhibits oncogenic functions. To uncover the potential oncogenic mechanism of LINC00152, we performed mRNA array analysis in LINC00152 knockdown ESCC cells compared with controls and analyzed the results by Gene Set Enrichment Analysis (GSEA). The differentially expressed genes in ESCC cells with LINC00152 knockdown were enriched in the cell cycle pathway and the SNARE interactions in vesicular transport pathway (Figure 6A).
Figure 6.
LINC00152 regulates ESCC progression by different ways.
Notes: (A) Gene set enrichment analysis (GSEA) of the microarray results of ESCC cells with LINC00152 knockdown. (B) Western blot assay showed that LINC00152 knockdown significantly increased the expression of MAD2L1, CDK6 and STX3, while the protein level of STX12 was decreased compared with controls. *P<0.05, **P<0.01.
Abbreviation: ESCC, esophageal squamous cell carcinoma.
Among genes with a differential expression of >2.5-fold, we selected two cell cycle-related genes (MAD2L1, CDK6) and two SNARE interactions in vesicular transport-related genes (STX3, STX12) for further analysis. Western blot analysis indicated that LINC00152 knockdown significantly increased protein levels of MAD2L1, CDK6 and STX3, while STX12 level was decreased compared with negative controls (Figure 6B).
Discussion
In recent years, the roles and mechanisms of lncRNAs in tumorigenesis and development have attracted more and more attention of researchers in the ESCC field. Multiple studies have shown that lncRNAs are involved in the progression of ESCC.15 Some researchers have reported significantly differentially expressed lncRNAs in ESCC cells or tissues. Several of these lncRNAs, such as UCA1, HOTAIR, SPRY4-IT1 and CBR3-AS1, have been demonstrated to exhibit oncogenic or tumor-suppressor functions in ESCC, suggesting these as potential diagnostic tools and molecular markers to determine the prognosis of patients.16–18 However, our understanding of the mechanisms underlying ESCC development and progression is still limited. Thus, in this study, we aimed to study the malignant biological behavior of lncRNA and ESCC metastasis. We report LINC00152 as an lncRNA related to the metastasis of ESCC, which may lead to a new biomarker for the diagnosis of esophageal cancer.
The dysregulation of LINC00152 has been found in various cancers including lung cancer, gastric cancer, hepatocellular cancer, pancreatic ductal adenocarcinoma, gallbladder cancer, renal cell cancer, glioma and tongue squamous cell cancer.19 LINC00152 has been shown to exhibit oncogenic functions and regulate other genes by epigenetic modifications and interactions with proteins and miRNAs.11,20 However, no study has examined the expression and potential function of LINC00152 in esophageal cancer. Herein, we examined the potential relationship between LINC00152 and esophageal cancer. Detection of LINC00152 expression level in 119 paired ESCC tissues by ISH showed that LINC00152 was upregulated in ESCC tissues compared with controls. Furthermore, patients with LINC00152 overexpression showed advanced N stage and TNM stage. We explored the function of LINC00152 in ESCC cell biological behavior and found that LINC00152 promoted the proliferation, invasion and migration of ESCC cells in vitro. These results suggested that LINC00152 plays an important role in ESCC metastasis and may serve as a diagnostic and prognostic factor for patients with ESCC.
A previous study reported that the circulating levels of LINC00152 were correlated with poor prognosis of ESCC patients and revealed that LINC00152 may be a potential target for early occurrence of ESCC.21 Gallbladder cancer with LINC00152 overexpression was inclined to have advanced TNM stage.22 A similar conclusion was demonstrated in gastric cancer23 and renal cell carcinoma.24 These studies are consistent with our results in ESCC.
The tumor pathogenesis process involves a complex gene regulation network. Although lncRNAs lack protein-coding capacity, they play critical roles in gene regulation via direct or indirect mechanisms.25,26 To elucidate the oncogenic mechanism of LINC00152, we conducted microarray and bioinformatics analyses. GSEA showed that differentially expressed genes in LINC00152 knockdown ESCC cells were associated with the cell cycle (such as MAD2L1 and CDK6) and the SNARE interactions in vesicular transport pathway (STX3 and STX12), which was validated by Western blotting. Notably, our flow cytometric analysis showed that ESCC cells knocked down for LINC00152 showed an S phase arrest, indicating LINC00152 might promote the proliferation of ECSS cells via regulating the cell cycle, which was consistent with the GSEA results.
The chief function of SNARE is the mediation of vesicle fusion. Multiple studies showed that the vesicular transport pathway functions in various aspects of cancer progression, such as angiogenesis, cell proliferation, cell metastasis and cell migration.27,28 Kean et al reported that STX12 promoted cell invasion by regulating the secretion of matrix metalloproteinases, thus degrading the extracellular matrix.29 STX3 also functions as a transcriptional regulator. Breast cancer patients with STX3 overexpression had advanced TNM stage and shorter overall and disease-free survival compared with patients with low STX3 expression.30 STX3 also promoted the proliferation of breast cancer cells by increasing activation of Akt-mTOR signaling.31 All these results indicated that LINC00152 may regulate the expression of genes involved in the cell cycle pathway and the SNARE interactions in vesicular transport pathway to promote migration, invasion and migration of ESCC.
LINC00152 may also be involved in the progression of ESCC through other mechanisms. For example, LINC00152 may act as a competing endogenous RNA to confer oxaliplatin resistance, leading to poor prognosis in colon cancer.11 In the progression of lung adenocarcinoma, LINC00152 inhibits the expression of IL24 by binding to EZH2.32 In addition, Ming et al found that LINC00152 could promote colon cancer epithelial–mesenchymal transition and metastasis by forming a positive feedforward loop between LINC00152 and Wnt/β-catenin signaling.33 In gastric cancer, LINC00152 also promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21.20 Further studies are required to determine the specific mechanisms that underlie the functions of LINC00152 in regulating the biological behavior of ESCC cells.
In conclusion, here we found that LINC00152 was closely associated with TNM stage and metastasis of ESCC. We also demonstrated functions for LINC00152 in promoting proliferation, invasion and migration of ESCC in vitro. The underlying mechanisms may involve MAD2L1 and CDK6 cell cycle proteins and STX3 and STX12 SNARE interactions in vesicular transport pathway proteins.
Acknowledgments
This research was supported by National Natural Science Foundation of China (81871966), Natural Science Foundation of Zhejiang Province, China (LY15H160062), and Science and Technology Project of Wenzhou City, China (Y20150125).
Disclosure
The authors report no conflicts of interest in this work.
Supplementary material
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64(3):381–387. doi: 10.1136/gutjnl-2014-308124 [DOI] [PubMed] [Google Scholar]
- 3.Smyth EC, Lagergren J, Fitzgerald RC, et al. Oesophageal cancer. Nature reviews. Dis Primers. 2017;3:17048. doi: 10.1038/nrdp.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svoboda M, Slyskova J, Schneiderova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35(7):1510–1515. doi: 10.1093/carcin/bgu055 [DOI] [PubMed] [Google Scholar]
- 6.Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khorshidi A, Dhaliwal P, Yang BB. Noncoding RNAs in tumor angiogenesis. Adv Exp Med Biol. 2016;927:217–241. doi: 10.1007/978-981-10-1498-7_8 [DOI] [PubMed] [Google Scholar]
- 8.Neumann O, Kesselmeier M, Geffers R, et al. Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology. 2012;56(5):1817–1827. doi: 10.1002/hep.25870 [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Wen J, Gu X, Wu D, Lu M, Zhao Q. Prognostic role of long non-coding RNA LINC00152 in Chinese cancer patients: a meta-analysis. Oncotarget. 2017;8(54):93227–93235. doi: 10.18632/oncotarget.21838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang T, Zeng H, Chen W, et al. Helicobacter pylori infection, H19 and LINC00152 expression in serum and risk of gastric cancer in a Chinese population. Cancer Epidemiol. 2016;44:147–153. doi: 10.1016/j.canep.2016.08.015 [DOI] [PubMed] [Google Scholar]
- 11. Yue B, Cai D, Liu C, Fang C, Yan D. Linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Mol Ther. 2016;24(12):2064–2077. doi: 10.1038/mt.2016.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. BioTechniques. 2008;44(5):619–626. doi: 10.2144/000112776 [DOI] [PubMed] [Google Scholar]
- 13. Qiu M, Xu Y, Wang J, et al. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis. 2015;6:e1858. doi: 10.1038/cddis.2015.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li J, Chen Z, Tian L, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63(11):1700–1710. doi: 10.1136/gutjnl-2013-305806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shen WJ, Zhang F, Zhao X, LncRNAs XJ. Esophageal squamous cell carcinoma - implications for pathogenesis and drug development. J Cancer. 2016;7(10):1258–1264. doi: 10.7150/jca.14869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7(11):7938–7944. [PMC free article] [PubMed] [Google Scholar]
- 17. Xie HW, Wu QQ, Zhu B, et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol. 2014;35(8):7743–7754. doi: 10.1007/s13277-014-2013-y [DOI] [PubMed] [Google Scholar]
- 18. Wang CM, Wu QQ, Li SQ, et al. Upregulation of the long non-coding RNA PlncRNA-1 promotes esophageal squamous carcinoma cell proliferation and correlates with advanced clinical stage. Dig Dis Sci. 2014;59(3):591–597. doi: 10.1007/s10620-013-2956-7 [DOI] [PubMed] [Google Scholar]
- 19. Zhang J, Yin M, Huang J, et al. Long noncoding RNA LINC00152 as a novel predictor of lymph node metastasis and survival in human cancer: a systematic review and meta-analysis. Clin Chim Acta. 2018;483:25–32. doi: 10.1016/j.cca.2018.03.034 [DOI] [PubMed] [Google Scholar]
- 20. Chen WM, Huang MD, Sun DP, et al. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget. 2016;7(9):9773–9787. doi: 10.18632/oncotarget.6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu HB, Jie HY, Zheng XX. Three circulating LncRNA predict early progress of esophageal squamous cell carcinoma. Cell Physiol Biochem. 2016;40(1–2):117–125. doi: 10.1159/000452529 [DOI] [PubMed] [Google Scholar]
- 22. Wu Y, Tan C, Weng WW, et al. Long non-coding RNA Linc00152 is a positive prognostic factor for and demonstrates malignant biological behavior in clear cell renal cell carcinoma. Am J Cancer Res. 2016;6(2):285–299. [PMC free article] [PubMed] [Google Scholar]
- 23. Pang Q, Ge J, Shao Y, et al. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35(6):5441–5447. doi: 10.1007/s13277-014-1709-3 [DOI] [PubMed] [Google Scholar]
- 24. Ji J, Tang J, Deng L, et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6(40):42813–42824. doi: 10.18632/oncotarget.5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang X, Zhou X, Hu Q, et al. Advances in esophageal cancer: A new perspective on pathogenesis associated with long non-coding RNAs. Cancer Lett. 2018;413:94–101. doi: 10.1016/j.canlet.2017.10.046 [DOI] [PubMed] [Google Scholar]
- 26. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews. Genetics. 2009;10(3):155–159. doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 27. Luga V, Zhang L, Viloria-Petit AM, et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151(7):1542–1556. doi: 10.1016/j.cell.2012.11.024 [DOI] [PubMed] [Google Scholar]
- 28. McCready J, Sims JD, Chan D, Jay DG. Secretion of extracellular hsp90alpha via exosomes increases cancer cell motility: a role for plasminogen activation. BMC Cancer. 2010;10:294. doi: 10.1186/1471-2407-10-663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kean MJ, Williams KC, Skalski M, et al. VAMP3, syntaxin-13 and SNAP23 are involved in secretion of matrix metalloproteinases, degradation of the extracellular matrix and cell invasion. J Cell Sci. 2009;122(Pt 22):4089–4098. doi: 10.1242/jcs.052761 [DOI] [PubMed] [Google Scholar]
- 30. Giovannone AJ, Winterstein C, Bhattaram P, et al. Soluble syntaxin 3 functions as a transcriptional regulator. J Biol Chem. 2018;293(15):5478–5491. doi: 10.1074/jbc.RA117.000874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nan H, Han L, Ma J, Yang C, Su R, He J. STX3 represses the stability of the tumor suppressor PTEN to activate the PI3K-Akt-mTOR signaling and promotes the growth of breast cancer cells. Biochimica et biophysica acta. Mol Basis Dis. 2018;1864(5 Pt A):1684–1692. doi: 10.1016/j.bbadis.2018.01.031 [DOI] [PubMed] [Google Scholar]
- 32. Chen QN, Chen X, Chen ZY, et al. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol Cancer. 2017;16(1):17. doi: 10.1186/s12943-017-0581-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yue B, Liu C, Sun H, et al. A positive feed-forward loop between LncRNA-CYTOR and Wnt/beta-catenin signaling promotes metastasis of colon cancer. Mol Ther. 2018;26(5):1287–1298. doi: 10.1016/j.ymthe.2018.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]