Abstract
Background: MiR-1323 was identified in 2006. Until now, the roles and mechanisms of miR-1323 in the progression of cancers including hepatocellular carcinoma (HCC) remain unknown. The aim of this study was to investigate the expressions, roles and mechanisms of miR-1323 in HCC development.
Methods: QRT-PCR was used to evaluate the expressions of miR-1323, GAS5 and TP53INP1 in HCC tissues and cell lines. CCK-8 assay, transwell invasion assay and flow cytometry assay were conducted to evaluate the proliferation, invasion and apoptosis of HCC cells. Luciferase assay was used to identify microRNA-target interaction.
Results: Firstly, our results showed that miR-1323 promoted proliferation and invasion, and inhibited apoptosis of HCC cells. Secondly, we found that TP53INP1 was a direct target of miR-1323 and could reverse the effects of miR-1323 on proliferation, invasion and apoptosis of HCC cells. Thirdly, our results showed that long non-coding RNA (lncRNA) GAS5 and miR-1323 could interact with each other and affect biological processes of HCC cells. Furthermore, we identified the negative correlations between miR-1323 and TP53INP1, and between miR-1323 and GAS5 in tumor tissues of patients with HCC.
Conclusion: Taken together, our study revealed the important roles of GAS5/miR-1323/TP53INP1 axis in HCC progression. This study also provided promising strategies for targeted therapy of patients with HCC.
Keywords: miR-1323, TP53INP1, GAS5, HCC, cell proliferation and invasion
Introduction
Hepatocellular carcinoma (HCC) is one of the most common types of cancer and one of the leading causes of cancer‐related deaths all over the world. In the past few years, though many potential biomarkers and targets were revealed to improve the diagnosis and treatment for patients with HCC, the outcome of HCC remains unsatisfactory.1 Thus, it is important to explore some novel and practical diagnostic biomarkers and therapeutic targets for patients with HCC.
In recent years, non-coding RNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), have been reported to become potential diagnostic biomarkers and therapeutic targets for HCC.2–4 MiRNAs with the length of 18–25 nucleotides could bind to the 3′‐untranslated region (3′‐UTR) of target mRNA and regulate gene expression at the post-transcriptional level. Our previous studies showed that miR-23b and miR-766 were up-regulated in tumor tissues of HCC patients, and promoted proliferation and invasion of HCC cells.5,6 Though some miRNAs have been revealed to play important roles in the tumorigenesis of HCC by targeting various genes and signaling pathways, the roles and mechanisms of other miRNAs in HCC progression remain to be explored. MiR-1323 was reported to be up-regulated in radiation-resistant lung cancer cells and tumor tissues from esophageal squamous cell carcinoma patients which were resistant to neoadjuvant radiochemotherapy.7,8 In our previous study, small RNA sequencing identified miR-1323 as one of the up-regulated miRNAs in tumor tissues of patients with HCC.9 Law et al also found that miR-1323 level was increased in HCC tissues.10 However, the roles and mechanisms of miR-1323 remain unknown.
LncRNAs with more than 200 nucleotides in length could interact with RNA, DNA or protein, and play important roles in various biological processes including cell proliferation, invasion and apoptosis.11 Our previous study showed that lncRNA XIST could inhibit proliferation and invasion of HCC cells by interacting with miR-92b.9 Another lncRNA DSCR8 could also act as a molecular sponge for miR-485-5p to affect proliferation and apoptosis of HCC cells.12 However, more lncRNA–miRNA interactions should be revealed in the tumorigenesis of HCC.
In this study, we explored the functional roles and mechanisms of miR-1323 in HCC progression. Firstly, we examined the effects of miR-1323 on proliferation, invasion and apoptosis of HCC cells. Then the target gene and binding lncRNA of miR-1323 were also investigated. Finally, the expression of miR-1323 was detected in tumor tissues from patients with HCC. This study may provide a novel and promising strategy for diagnosis and treatment of patients with HCC.
Material and methods
Cell culture
Two HCC cell lines including SMMC-7721 and HepG2 were obtained from Cell Resource Center of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagles Medium (DMEM) containing 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and maintained in a humidified atmosphere with 5% CO2 at 37°C.
Patients and tissue samples
Tumor tissues and its adjacent non-tumorous liver tissues (ANLTs) were randomly collected from HCC patients who underwent surgical resection at the Department of Hepatobiliary and Pancreatic Surgery, the Affiliated Hospital of Qingdao University from January 2015 to March 2017. The clinical data of patients with HCC used in this study were listed in Table S1. The experimental protocols conformed to the Ethical Guidelines of the 1975 Declaration of Helsinki, revised in 2000. The study was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University and written informed consents were obtained from the participants.
Table S1.
Clinical data of patients with HCC in this study
| Factors | Number of patients |
|---|---|
| Age (<60/≥60) | 13/17 |
| Gender (male/female) | 20/10 |
| Hepatitis B virus (HBV) infection (Positive/Negative) | 29/1 |
| Serum α-fetoprotein (AFP) level (<20/≥20 μg/L) | 11/19 |
| Microvascular invasion (Absent/Present) | 20/10 |
| Number of tumors (1/≥2) | 29/1 |
| Tumor Size (<5/≥5 cm) | 21/9 |
RNA isolation and qRT-PCR
Total RNA was extracted using RNAiso (Takara, Otsu, Japan) according to the manufacturer’s instructions. SYBR-green premix (Takara) was used in the PCR reaction after reverse transcription. The expressions of miR-1323 and other genes were, respectively, normalized to U6 and β-actin expression. The data were analyzed by delta-delta Ct method. Primers for miR-1323 and U6 were obtained from Hairpin-itTM Quantitation PCR Kit (GenePharma, Shanghai, China). Primer sequences of TP53INP1, GAS5 and β-actin were listed as follows: TP53INP1: 5ʹ- TTCCTCCAACCAAGAACCAGA −3’ and 5ʹ- AAGCAGGAATCACTT GTATCAGC −3ʹ; GAS5: 5ʹ- ACTCCTGTGAGGTATGGTGCTGG −3’ and 5ʹ- GCTCTTTAGGACTTCAGTAGCTATTC −3ʹ; β-actin: 5′-CATCC TCACCCTGAAGTACCCC-3′ and 5′-AGCCTGGATGCAACGTACAT G-3′.
Cell counting kit-8 (CCK-8) assay
CCK-8 assay was used to evaluate the proliferation of HCC cells. Briefly, cells were seeded in a 96-well plate. 24 hrs later, cells were transfected with corresponding oligonucleotides. CCK-8 solution (Dojindo, Kumamoto, Japan) was added into the wells at indicated time points and incubated at 37°C for 1 hr. The absorption values at 450 nm were measured.
Transwell invasion assay
Cell invasion was evaluated using transwell chambers (24-well, 8.0 μm pore size, Corning Incorporated, Corning, NY, USA). Cells were cultured with DMEM medium in the upper chamber coated with matrigel (BD Biosciences, San Jose, CA, USA), and DMEM medium with 10% fetal bovine serum was added in the lower well. Then, non-invaded cells were removed from the upper membrane of chamber with cotton-tipped swabs at the appropriate time. The invaded cells on the lower surface of the membrane were fixed in methanol and stained with 0.1% crystal violet. Cells were counted under a light microscope.
Flow cytometry analysis for cell apoptosis
Cell apoptosis was evaluated using an Annexin V/propidium iodide (PI) apoptosis detection kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Cells were suspended in binding buffer and stained with annexin V/PI at room temperature. Then, annexin V-positive cells were detected using a flow cytometer (BD Biosciences). Data analysis was performed using FlowJo software (Tree Star, San Carlos, CA, USA).
Western blotting
Cells were lysed in RIPA buffer and a BCA Protein Assay Kit (Pierce Biotechnology, Rockford, IL, USA) was used to determine the protein concentration. Then the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred onto a PVDF membrane (Millipore, Billerica, MA, USA). The membrane was blocked with 5% non-fat milk and incubated with TP53INP1 antibody (Abcam, Cambridge, MA, USA) and β-actin antibody (Cell Signaling Technology, Danvers, MA, USA). Immunodetection was conducted using an Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA).
Transfection and luciferase reporter assay
Oligonucleotides, including miR-1323 mimics, miR-1323 inhibitor, TP53INP1 siRNA, GAS5 siRNA and NC Oligonucleotides, were obtained from GenePharma (Shanghai, China). TP53INP1 3′-UTR and GAS5 sequences including putative miR-1323 binding sites were fused to a modified pcDNA3.1 vector containing sequences encoding a luciferase reporter gene. Reporter plasmids with mutant sites were constructed using Mutagenesis Kit (Stratagene, La Jolla, CA, USA). Oligonucleotides and plasmids were transfected using Hiperfect transfection reagent (Qiagen, Valencia, CA, USA) and Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA), respectively. Luciferase reporter analysis was conducted using Dual Luciferase Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Statistical analysis
Statistical analysis was conducted using the GraphPad Prism 7 software (GraphPad Software, San Diego, CA, USA). Comparisons were performed using student’s t-test or one-way ANOVA. Pearson’s analysis was used in correlation analysis. P<0.05 was considered as statistically significant.
Results
MiR-1323 affected proliferation, invasion and apoptosis of HCC cells
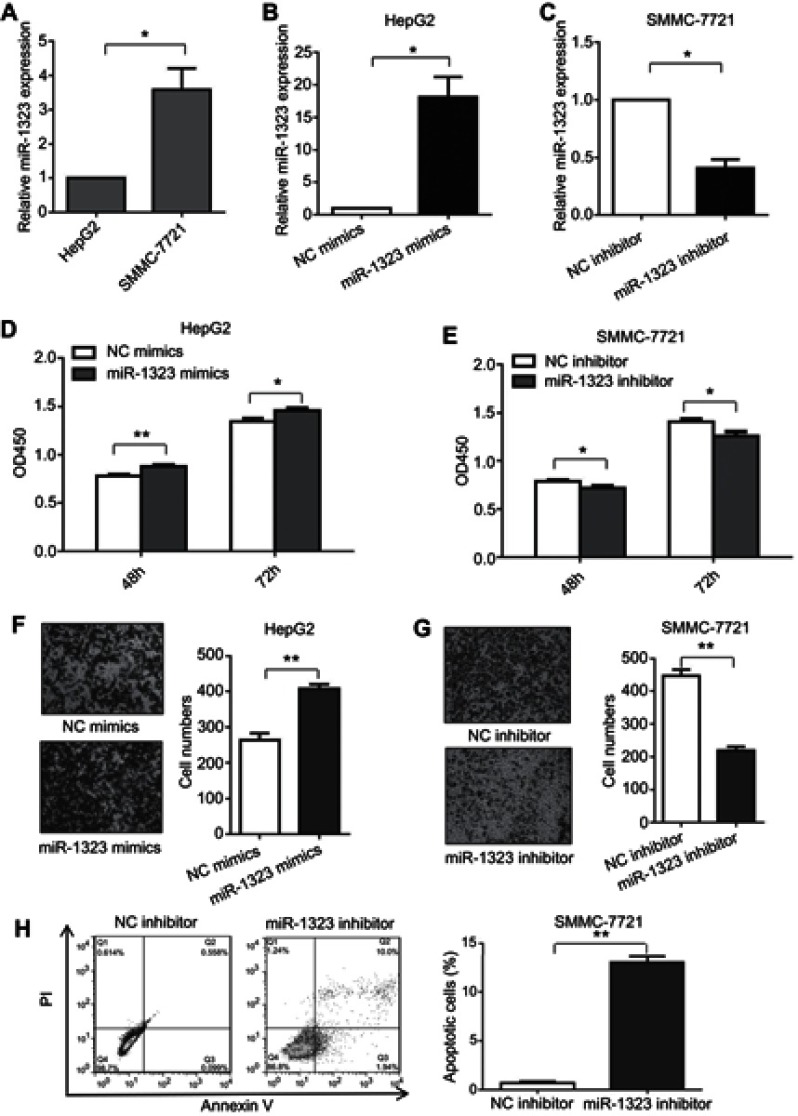
Firstly, we investigated whether miR-1323 could affect biological processes of HCC cells. The levels of miR-1323 were measured in two HCC cell lines including SMMC-7721 and HepG2 cells. Our results showed that the expression of miR-1323 was higher in SMMC-7721 cells than that in HepG2 cells (Figure 1A). Then we increased the miR-1323 expression in HepG2 cells using miR-1323 mimics and decreased its expression in SMMC-7721 cells using miR-1323 inhibitor (Figure 1B and C). CCK-8 assay, transwell invasion assay and flow cytometry with annexin V/PI staining were used to evaluate proliferation, invasion and apoptosis of HCC cells. Our results showed that miR-1323 over-expression could significantly promote proliferation and invasion of HepG2 cells, and knockdown of miR-1323 could inhibit proliferation and invasion of SMMC-7721 cells (Figure 1D–G). We also found that down-regulation of miR-1323 induced apoptosis of HCC cells (Figure 1H). These data suggested that miR-1323 could affect proliferation, invasion and apoptosis of HCC cells.
Figure 1.
MiR-1323 affects proliferation, invasion and apoptosis of HCC cells. (A) Relative miR-1323 expressions in SMMC-7721 and HepG2 cells were determined by qRT-PCR. The data were normalized to the expression level of miR-1323 in HepG2 cells. (B–C) miR-1323 was over-expressed in HepG2 cells and knocked down in SMMC-7721 cells. (D–E) CCK-8 assays were conducted. (F–G) Transwell invasion assays were conducted. The number of cells invaded was counted and compared in the right diagrams. (H) Flow cytometric assays of apoptosis were performed, and the percentage of apoptotic cells was calculated and compared in the right diagram. Results were represented as mean ± S.D. (n≥3). *P<0.05, **P<0.01.
TP53INP1 was a direct target of miR-1323
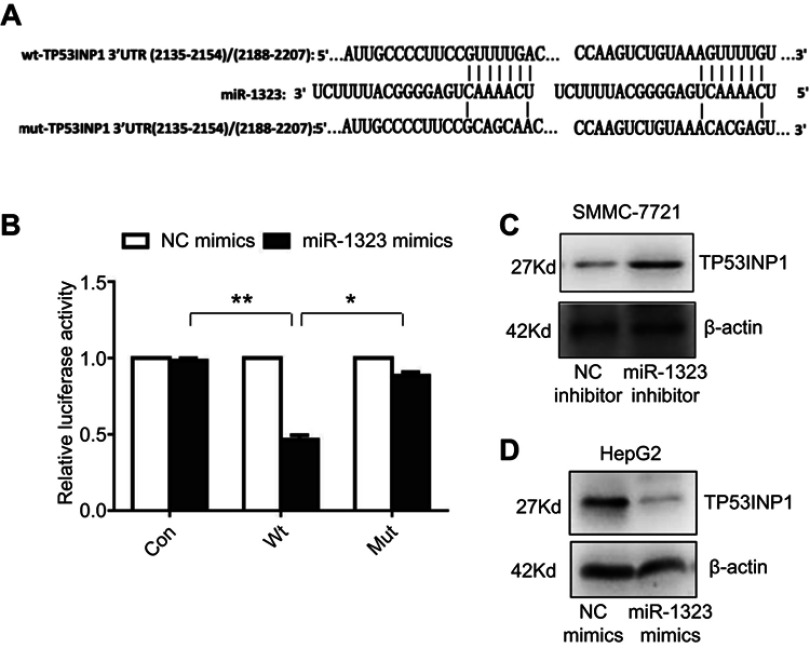
Then we explored the specific mechanism through which miR-1323 regulated biological processes of HCC cells. Three different microRNA-target prediction databases, including Targetscan, miRDB and miRMap, were used to screen the potential target genes for miR-1323. The results showed that TP53INP1 was the potential target gene of miR-1323 with high scores in all the three databases. To validate the binding ability between miR-1323 and 3ʹUTR of TP53INP1, we constructed the luciferase reporter plasmids with wide type (wt) or mutant (mut) 3ʹUTR of TP53INP1 (Figure 2A). Luciferase reporter gene assay revealed that miR-1323 could directly target 3′UTR of wt-TP53INP1 to negatively regulate the luciferase activity, but not target 3ʹUTR of mut-TP53INP1 (Figure 2B). We also found that miR-1323 inhibitor could increase the TP53INP1 level in SMMC-7721 cells and miR-1323 mimics could inhibit the expression of TP53INP1 in HepG2 cells (Figure 2C and D). Taken together, TP53INP1 was a direct target of miR-1323 in HCC cells.
Figure 2.
TP53INP1 is a direct target of miR-1323. (A) Diagram of two predicted binding sites and corresponding mutant sites for miR-1323 in the 3ʹ-UTR of TP53INP1 (wt, wild-type; mut, mutant type). (B) Effects of miR-1323 mimics on the activity of luciferase reporter genes containing wt and mut 3ʹ-UTR of TP53INP1. Each luciferase activity was normalized to the value of the cells transfected with NC mimics. (C–D) Western blot analysis of TP53INP1 expressions in SMMC-7721 cells transfected with miR-1323 inhibitor and HepG2 cells transfected with miR-1323 mimics. Results were represented as mean ± S.D. (n≥3). *P<0.05, **P<0.01.
Knockdown of TP53INP1 reversed the effects of miR-1323 inhibitor on biological processes of HCC cells
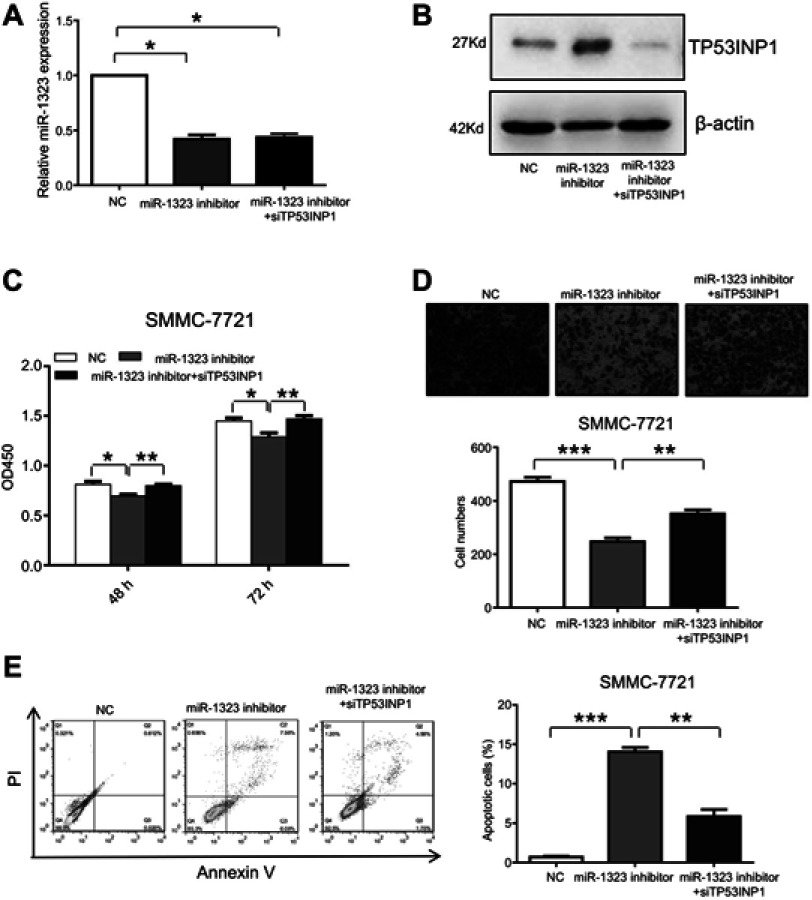
In this study, we also investigated whether miR-1323 regulated biological processes of HCC cells by targeting TP53INP1. SMMC-7721 cells were co-transfected with miR-1323 inhibitor and TP53INP1 siRNA, and the expressions of miR-1323 and TP53INP1 were shown in Figure 3A and B. The results of CCK-8 and transwell invasion assays showed that knockdown of TP53INP1 could significantly reverse the effects of miR-1323 inhibitor on the proliferation and invasion of HCC cells (Figure 3C and D). Flow cytometry with annexin V/PI staining also showed that knockdown of TP53INP1 could attenuate the apoptosis mediated by miR-1323 inhibitor (Figure 3E). These data suggested that TP53INP1 was essential for the effects of miR-1323 on proliferation, invasion and apoptosis of HCC cells.
Figure 3.
TP53INP1 attenuates the effects of miR-1323 on biological processes of HCC cells. (A–B) SMMC-7721 cells were co-transfected with miR-1323 inhinitor and TP53INP1 siRNA. MiR-1323 expression was measured by qRT-PCR and the level of TP53INP1 was measured by Western blot. (C) CCK-8 assays were conducted. (D) Transwell invasion assays were performed. The number of cells invaded was counted and compared in the diagram below. (E) Flow cytometric assays of apoptosis were performed, and the percentage of apoptotic cells was calculated and compared in the right diagram. Results were represented as mean ± S.D. (n≥3). *P<0.05, **P<0.01, ***P<0.001.
LncRNA GAS5 and miR-1323 interacted with and negatively regulated each other
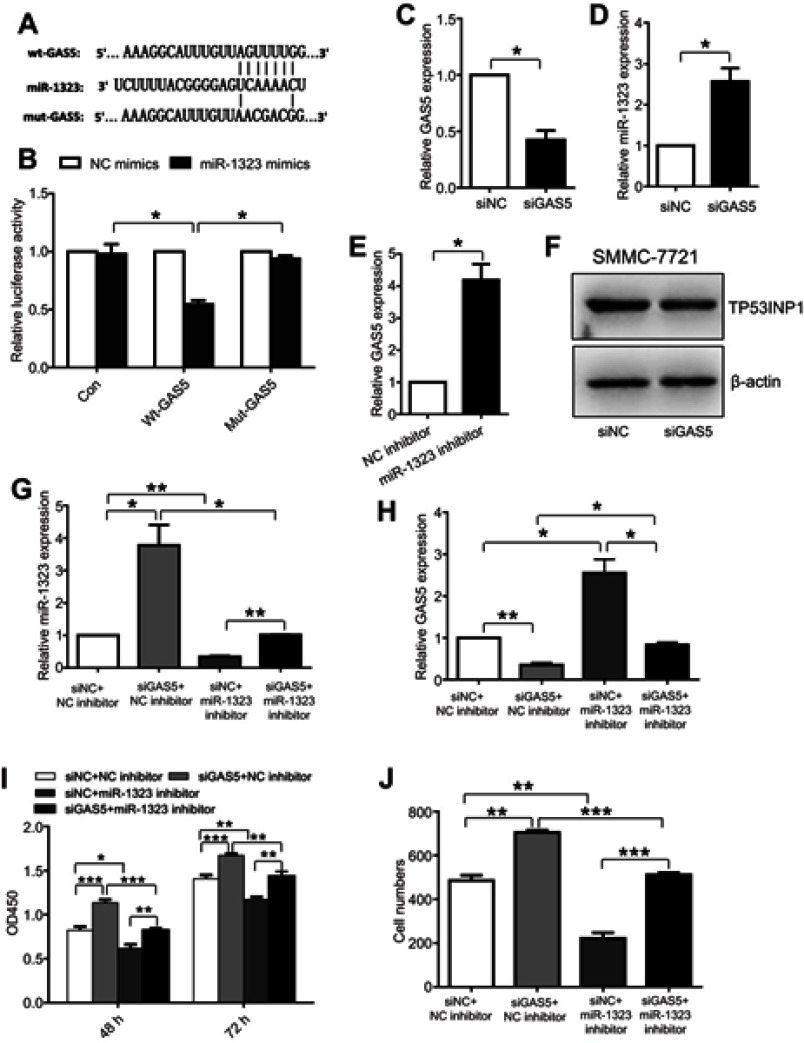
In recent years, more and more lncRNAs were reported to act as competing endogenous RNAs which could bind miRNAs to regulate biological processes of cancer cells.13 To explore whether the expression and function of miR-1323 were mediated by lncRNAs, we screened the predicted binding lncRNAs for miR-1323 using Starbase V3.0. The results showed that lncRNA GAS5 sequence contained the potential binding sites for miR-1323. To verify the binding ability between GAS5 and miR-1323, we constructed two luciferase reporter plasmids with wt- or mut-predicted miR-1323 binding sites of GAS5 sequence (Figure 4A). Luciferase reporter assays showed that miR-1323 over-expression could inhibit the luciferase activity of wt-GAS5 reporter vector, but not empty vector or mut-GAS5 reporter vector (Figure 4B). Moreover, the results of qRT-PCR assays indicated that miR-1323 was significantly up-regulated after GAS5 silencing in SMMC-7721 cells (Figure 4C and D). Knockdown of miR-1323 could also increase the expression of GAS5 in SMMC-7721 cells (Figure 4E). Western blot analysis showed that knockdown of GAS5 could inhibit TP53INP1 expression in SMMC-7721 cells (Figure 4F). Taken together, these results indicated that lncRNA GAS5 and miR-1323 could interact with and negatively regulate each other.
Figure 4.
The interaction between lncRNA GAS5 and miR-1323 affects proliferation and invasion of HCC cells. (A) Diagram of the predicted binding site and the corresponding mutant site for miR-1323 in the GAS5 sequence. (B) Effects of miR-1323 mimics on the activity of luciferase reporter genes containing wt or mut GAS5 sequence. Each luciferase activity was normalized to the value obtained from the cells transfected with NC mimics. (C–D) Relative GAS5 and miR-1323 expressions in SMMC7721 cells transfected with GAS5 siRNA were measured using qRT-PCR. (E) Relative GAS5 expression in SMMC7721 cells transfected with miR-1323 inhibitor was measured using qRT-PCR. (F) Western blot analysis of TP53INP1 expression in SMMC-7721 cells transfected with GAS5 siRNA. (G–H) Relative miR-1323 and GAS5 expressions in SMMC7721 cells co-transfected with GAS5 siRNA and miR-1323 inhibitor were measured using qRT-PCR. (I–J) CCK-8 and Transwell invasion assays were performed in SMMC-7721 cells co-transfected with GAS5 siRNA and miR-1323 inhibitor. Results were represented as mean ± S.D. (n≥3). *P<0.05, **P<0.01, ***P<0.001.
Reciprocal regulation between GAS5 and miR-1323 affected biological processes of HCC cells
Next, we investigated whether the interaction between GAS5 and miR-1323 could affect biological processes of HCC cells. SMMC-7721 cells were co-transfected with GAS5 siRNA and miR-1323 inhibitor, and the expressions of GAS5 and miR-1323 were shown in Figure 4G and H. The results from CCK-8 and transwell invasion assays showed that co-transfection with GAS5 siRNA and miR-1323 inhibitor could significantly reverse the effects of single knockdown of GAS5 or miR-1323 on proliferation and invasion of HCC cells (Figure 4I and J). These data suggested that the reciprocal regulation between GAS5 and miR-1323 could affect proliferation and invasion of HCC cells.
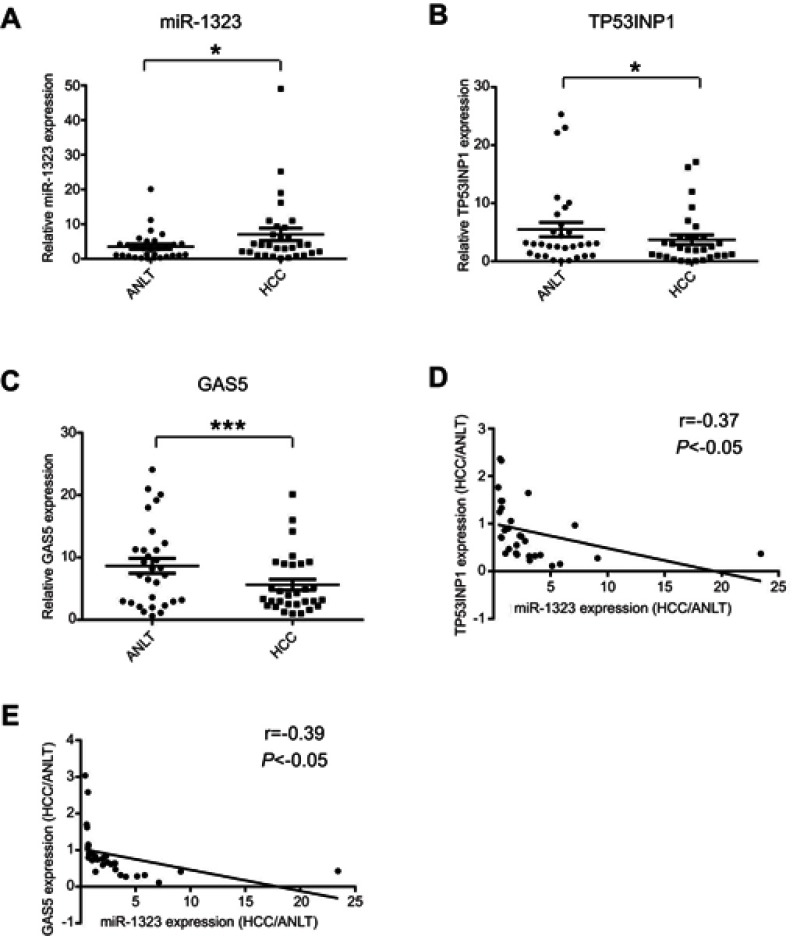
MiR-1323 level was inversely correlated with TP53INP1 or GAS5 expression in HCC tissues
In order to explore the association of miR-1323, GAS5 and TP53INP1 expression with HCC occurrence, we detected the expression levels of miR-1323, GAS5 and TP53INP1 in 30 pairs of tumor tissues and ANLTs from patients with HCC by qRT-PCR assays. As shown in Figure 5A, miR-1323 expression was significantly higher in HCC tissues than that in ANLTs. We also found that expressions of both TP53INP1 and GAS5 were lower in tumor tissues than that in ANLTs of patients with HCC (Figure 5B and C). Correlation analysis indicated that miR-1323 level was negatively associated with TP53INP1 or GAS5 expression in tumor tissues of patients with HCC (Figure 5D and E). These data suggested that GAS5/miR-1323/TP53INP1 axis might be related with the occurrence of HCC.
Figure 5.
Expression levels of miR-1323, TP53INP1 and GAS5 in HCC tissues. (A–C) Expression levels of miR-1323, TP53INP1 and GAS5 in 30 pairs of tumor tissues and ANLTs from patients with HCC were determined by qRT-PCR. (D) The correlation between miR-1323 and TP53INP1 expression in tumor tissues of patients with HCC was analyzed (n=30). (E) The correlation between miR-1323 and GAS5 expression in tumor tissues of patients with HCC was analyzed (n=30). *P<0.05, ***P<0.001.
Discussion
In recent years, miRNAs and lncRNAs have been considered as essential regulators in tumorigenesis of many cancers including HCC. Many miRNAs and lncRNAs have been revealed to be deregulated in HCC progression and affect biological processes of HCC cells.14,15 In this study, we found that miR-1323 was up-regulated and lncRNA GAS5 was down-regulated in tumor tissues of patients with HCC. Our results also showed that GAS5 and miR-1323 could interact with each other and affect proliferation and invasion of HCC cells. These data suggested that miR-1323 and GAS5 might be potential biomarkers and therapeutic targets for patients with HCC. We would also focus on the roles of miR-1323 and GAS5 in the prognosis of patients with HCC in our future work.
The gene encoding miR-1323 was located on chromosome 19. The sequence of miR-1323 was identified as one of the mammalian miRNA candidates in 2006 and was verified by miRNA cloning in 2008.16,17 Until now, the roles and mechanisms of miR-1323 in the progression of cancers including HCC remain to be explored. In this study, our results showed that miR-1323 could affect proliferation, invasion and apoptosis of HCC cells. Mechanical investigations showed that miR-1323 could interact with GAS5 and 3ʹ UTR of TP53INP1. These data for the first time revealed the oncogenetic roles and molecular mechanisms of miR-1323 in HCC progression.
The gene encoding lncRNA GAS5 was poorly conserved and was located on chromosome 1. GAS5 was widely expressed in human tissues and usually over-expressed in growth-arrest cells.18–20 GAS5 was reported to be down-regulated in many cancers and inhibit tumorigenesis. For example, up-regulation of GAS5 could inhibit cell proliferation, cell cycle progression and cell invasion in clear cell renal cell carcinoma.21 Another study revealed that GAS5 could repress the proliferation, migration and invasion of glioma cells.22 It was also reported that low GAS5 level was associated with poor prognosis of patients with HCC, and knockdown of GAS5 expression could promote proliferation and invasion of HCC cells.23–25 Moreover, GAS5 was investigated to bind and target many miRNAs, including miR-222, miR-196-5p and miR-365a-3p, to affect biological processes of many cell types.26–28 In this study, our results revealed a new interaction between GAS5 and miR-1323 in HCC cells. We also indicated that GAS5 could target miR-1323 to affect proliferation and invasion of HCC cells.
In this study, we also indicated that TP53INP1 was a direct target of miR-1323. TP53INP1 was a stress-induced gene and was located on chromosome 8.29 TP53INP1 was reported to be down-regulated in multiple cancers including gastric cancer, pancreatic cancer and advanced HCC.30–32 TP53INP1 was regulated by P53 and P73, and it could inhibit cell cycle and promote apoptosis by both P53-dependent and P73-dependent pathways.33,34 In recent years, many studies also showed that TP53INP1 could be regulated by miRNAs, including miR-221, miR-200a and miR-504, and act as a tumor suppressor gene in cancer progression.35–37 In this study, we revealed that miR-1323 could target 3ʹ UTR of TP53INP1 and decrease its expression in HCC cells, which indicated a new regulatory mechanism for TP53INP1 expression. We would also focus on the molecular mechanisms underlying how TP53INP1 affects biological processes of HCC cells in our future work.
Conclusion
In summary, our study investigated the expressions, roles and mechanisms of GAS5/miR-1323/TP53INP1 axis in HCC progression. Our findings also suggested that GAS5, miR-1323 and TP53INP1 might serve as potential diagnostic biomarkers and therapeutic targets for patients with HCC.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81501812, 81873976) (to LZ) and the Natural Science Foundation of Shandong Province (BS2015SW013) (to LZ).
Disclosure
The authors declare that they have no conflicts of interest in this work.
Supplementary material
References
- 1.Tang ZY, Ye SL, Liu YK, et al. A decade’s studies on metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2004;130(4):187–196. doi: 10.1007/s00432-003-0511-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plissonnier ML, Herzog K, Levrero M, Zeisel MB. Non-coding RNAs and hepatitis C virus-induced hepatocellular carcinoma. Viruses. 2018;10(11):591. doi: 10.3390/v10110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15(3):137–151. doi: 10.1038/nrgastro.2017.169 [DOI] [PubMed] [Google Scholar]
- 4.Xu X, Tao Y, Shan L, et al. The role of MicroRNAs in hepatocellular carcinoma. J Cancer. 2018;9(19):3557–3569. doi: 10.7150/jca.26350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang L, Wang X, Wang Z, et al. MicroRNA-23b functions as an oncogene and activates AKT/GSK3beta/beta-catenin signaling by targeting ST7L in hepatocellular carcinoma. Cell Death Dis. 2017;8(5):e2804. doi: 10.1038/cddis.2017.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang C, Ma X, Guan G, et al. MicroRNA-766 promotes cancer progression by targeting NR3C2 in hepatocellular carcinoma. Faseb J. 2019;33(1):1456–1467. doi: 10.1096/fj.201801151R [DOI] [PubMed] [Google Scholar]
- 7.Slotta-Huspenina J, Drecoll E, Feith M, et al. MicroRNA expression profiling for the prediction of resistance to neoadjuvant radiochemotherapy in squamous cell carcinoma of the esophagus. J Transl Med. 2018;16(1):109. doi: 10.1186/s12967-018-1492-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Han W, Ni TT, et al. Knockdown of microRNA-1323 restores sensitivity to radiation by suppression of PRKDC activity in radiation-resistant lung cancer cells. Oncol Rep. 2015;33(6):2821–2828. doi: 10.3892/or.2015.3884 [DOI] [PubMed] [Google Scholar]
- 9.Zhuang LK, Yang YT, Ma X, et al. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203. doi: 10.1038/cddis.2016.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Law PT, Qin H, Ching AK, et al. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58(6):1165–1173. doi: 10.1016/j.jhep.2013.01.032 [DOI] [PubMed] [Google Scholar]
- 11.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Sun L, Wang L, et al. Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485-5p to activate Wnt/beta-catenin signal pathway in hepatocellular carcinoma. Cell Death Dis. 2018;9(9):851. doi: 10.1038/s41419-018-0937-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song W, Miao DL, Chen L. Comprehensive analysis of long noncoding RNA-associated competing endogenous RNA network in cholangiocarcinoma. Biochem Biophys Res Commun. 2018;506(4):1004–1012. doi: 10.1016/j.bbrc.2018.10.186 [DOI] [PubMed] [Google Scholar]
- 14.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67(3):603–618. doi: 10.1016/j.jhep.2017.04.009 [DOI] [PubMed] [Google Scholar]
- 15.Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474(24):4219–4251. doi: 10.1042/BCJ20170079 [DOI] [PubMed] [Google Scholar]
- 16.Berezikov E, van Tetering G, Verheul M, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16(10):1289–1298. doi: 10.1101/gr.5159906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afanasyeva EA, Hotz-Wagenblatt A, Glatting KH, Westermann F. New miRNAs cloned from neuroblastoma. BMC Genomics. 2008;9:52. doi: 10.1186/1471-2164-9-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5ʹ-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18(12):6897–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54(6):787–793. [DOI] [PubMed] [Google Scholar]
- 20.Coccia EM, Cicala C, Charlesworth A, et al. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol Cell Biol. 1992;12(8):3514–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong X, Kong C, Liu X, et al. GAS5 functions as a ceRNA to regulate hZIP1 expression by sponging miR-223 in clear cell renal cell carcinoma. Am J Cancer Res. 2018;8(8):1414–1426. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, Liu Y, Zheng J, et al. GAS5 suppresses malignancy of human glioma stem cells via a miR-196a-5p/FOXO1 feedback loop. Biochimica Et Biophysica Acta. Mol Cell Res. 2017;1864(10):1605–1617. doi: 10.1016/j.bbamcr.2017.06.020 [DOI] [PubMed] [Google Scholar]
- 23.Chen CY, Chen CC, Shieh TM, et al. Corylin suppresses hepatocellular carcinoma progression via the inhibition of epithelial-mesenchymal transition, mediated by long noncoding RNA GAS5. Int J Mol Sci. 2018;19(2):380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang L, Li C, Lan T, et al. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep. 2016;13(2):1541–1550. doi: 10.3892/mmr.2015.4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu L, Ye H, Huang G, et al. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016;37(2):2691–2702. doi: 10.1007/s13277-015-4111-x [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Gu J, Lu H. The GAS5/miR-222 axis regulates proliferation of gastric cancer cells through the PTEN/Akt/mTOR pathway. Dig Dis Sci. 2017;62(12):3426–3437. doi: 10.1007/s10620-017-4831-4 [DOI] [PubMed] [Google Scholar]
- 27.Zhao H, Yu H, Zheng J, et al. Lowly-expressed lncRNA GAS5 facilitates progression of ovarian cancer through targeting miR-196-5p and thereby regulating HOXA5. Gynecol Oncol. 2018;151(2):345–355. doi: 10.1016/j.ygyno.2018.08.032 [DOI] [PubMed] [Google Scholar]
- 28.Liu YZ, Sun Y. High expression of GAS5 promotes neuronal death after cerebral infarction by regulating miR-365a-3p. Eur Rev Med Pharmacol Sci. 2018;22(16):5270–5277. doi: 10.26355/eurrev_201808_15726 [DOI] [PubMed] [Google Scholar]
- 29.Tomasini R, Samir AA, Pebusque MJ, et al. P53-dependent expression of the stress-induced protein (SIP). Eur J Cell Biol. 2002;81(5):294–301. [DOI] [PubMed] [Google Scholar]
- 30.Jiang PH, Motoo Y, Garcia S, Iovanna JL, Pebusque MJ, Sawabu N. Down-expression of tumor protein p53-induced nuclear protein 1 in human gastric cancer. World J Gastroenterol. 2006;12(5):691–696. doi: 10.3748/wjg.v12.i5.691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Wang L, Mo Q, Jia A, Dong Y, Wang G. A positive feedback loop of p53/miR-19/TP53INP1 modulates pancreatic cancer cell proliferation and apoptosis. Oncol Rep. 2016;35(1):518–523. doi: 10.3892/or.2015.4361 [DOI] [PubMed] [Google Scholar]
- 32.Ng KY, Chan LH, Chai S, et al. TP53INP1 downregulation activates a p73-dependent DUSP10/ERK signaling pathway to promote metastasis of hepatocellular carcinoma. Cancer Res. 2017;77(17):4602–4612. doi: 10.1158/0008-5472.CAN-16-3456 [DOI] [PubMed] [Google Scholar]
- 33.Okamura S, Arakawa H, Tanaka T, et al. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol Cell. 2001;8(1):85–94. [DOI] [PubMed] [Google Scholar]
- 34.Tomasini R, Seux M, Nowak J, et al. TP53INP1 is a novel p73 target gene that induces cell cycle arrest and cell death by modulating p73 transcriptional activity. Oncogene. 2005;24(55):8093–8104. doi: 10.1038/sj.onc.1208951 [DOI] [PubMed] [Google Scholar]
- 35.Liao D, Li T, Ye C, et al. miR-221 inhibits autophagy and targets TP53INP1 in colorectal cancer cells. Exp Ther Med. 2018;15(2):1712–1717. doi: 10.3892/etm.2017.5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu SJ, Yang L, Hong Q, Kuang XY, Di GH, Shao ZM. MicroRNA-200a confers chemoresistance by antagonizing TP53INP1 and YAP1 in human breast cancer. BMC Cancer. 2018;18(1):74. doi: 10.1186/s12885-018-4242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai Q, Zeng S, Dai X, Wu J, Ma W. miR-504 promotes tumour growth and metastasis in human osteosarcoma by targeting TP53INP1. Oncol Rep. 2017;38(5):2993–3000. doi: 10.3892/or.2017.5983 [DOI] [PubMed] [Google Scholar]