Abstract
RNA polymerase (RNAP) from bacteriophage T7 is a representative single-subunit viral RNAP that can transcribe with high promoter activities without assistances from transcription factors. We accordingly studied this small transcription machine computationally as a model system to understand underlying mechanisms of mechano-chemical coupling and fidelity control in the RNAP transcription elongation. Here we summarize our computational work from several recent publications to demonstrate first how T7 RNAP translocates via Brownian alike motions along DNA right after the catalytic product release. Then we show how the backward translocation motions are prevented at post-translocation upon successful nucleotide incorporation, which is also subject to stepwise nucleotide selection and acts as a pawl for “selective ratcheting”. The structural dynamics and energetics features revealed from our atomistic molecular dynamics (MD) simulations and related analyses on the single-subunit T7 RNAP thus provided detailed and quantitative characterizations on the Brownian-ratchet working scenario of a prototypical transcription machine with sophisticated nucleotide selectivity for fidelity control. The presented mechanisms can be more or less general for structurally similar viral or mitochondrial RNAPs and some of DNA polymerases, or even for the RNAP engine of the more complicated transcription machinery in higher organisms.
Keywords: RNA polymerase, PPi release, Translocation, Nucleotide selection, Fidelity control
1. T7 RNA Polymerase as a Minimal Transcription Machine Model System
The RNA polymerase (RNAP) from bacteriophage T7 is regarded as one of the smallest transcription machines [[1], [2], [3]]. In bacteria and eukaryote species, RNAP II, the core engine of the transcription machinery, works with a variety of transcription factors to support gene expression [[4], [5], [6], [7], [8]]. RNAP II itself consists of multiple polypeptides, i.e., maintaining a complex molecular architecture. In comparison, T7 RNAP is a single-subunit enzyme with a simple hand-like structure [[9], [10], [11], [12]], and it is capable of transcribing with high promoter activity or processivity, self-sufficiently, without assistances from transcription factors, from initiation to elongation and to termination. Indeed, T7 RNAP structurally resembles a wide class of DNA polymerases (DNAPs), along with some other viral and mitochondrial RNAP species [10,13]. Hence, T7 RNAP makes a minimal model system to study transcription.
The high-resolution crystal structures of T7 RNAP had been initially determined by Sousa et al. [14] and then by Thomas A Steitz lab co-workers on its transcription initiation to elongation complexes since the late last century [[15], [16], [17], [18]]. In particular, several states of T7 RNAP elongation complex have been obtained, from nucleotide insertion or substrate state to catalytic product state, and to post-translocation state, together with an additional pre-insertion complex then resolved by Temiakov et al. [19]. Meanwhile, extensive bio-chemical studies [[20], [21], [22], [23]] along with single-molecule measurements on T7 RNAP elongation [[24], [25], [26], [27]] provide substantial quantitative features of the enzyme kinetics, from initiation to elongation. Accordingly, physical modeling and molecular dynamics (MD) simulation on this smallest transcription machine became feasible, so that to reveal underlying molecular mechanisms and essential structural dynamics details.
We have recently studied transcription elongation of T7 RNAP by combining physical modeling and all-atom MD simulations, addressing both mechano-chemical coupling and fidelity control mechanisms during elongation [[28], [29], [30], [31], [32], [33], [34], [35], [36]]. The mechanochemistry concerns about how the protein machine utilizes chemical free energy to generate mechanical or directional motions, referring to how the chemical synthesis of RNA couples with the polymerase translocation along DNA in the RNAP system. T7 RNAP had been suggested to work via a ‘power stroke’ (PS) mechanism [17,37,38], in which product release directly drives the RNAP translocation via simultaneous protein subdomain opening. Meanwhile, T7 RNAP along with RNAPs from bacteria and eukaryotic species had also been proposed to function in a ‘Brownian ratchet’ (BR) scenario [21,25,[39], [40], [41], [42]]. Below, we elaborate on how our studies actually support the BR working scenario of T7 RNAP, in which the translocation proceeds in Brownian motions after the product release, while the ratcheting part is fulfilled largely by cognate nucleotide incorporation to the growing end of the synthesizing RNA. Since non-cognate nucleotides unlikely support successful nucleotide incorporation or ratcheting, one regards that nucleotide selection plays a crucial role for the BR scenario such that an RNAP actually conducts ‘selective ratcheting’ along DNA. Accordingly, we illustrate then how T7 RANP achieves the nucleotide selection for the transcription fidelity control. Indeed, the mechanisms can be representative or apply in general to related enzymes on the catalyzed polymerization processes, in the presence of molecular template, though specific structural elements do vary for different polymerization machines.
2. PPi Product Release Unlikely Drives the Translocation of T7 RNAP
In previous structural studies of T7 RNAP, suggestions had been made on the PS mechanism such that the pyrophosphate (PPi) product release after catalysis directly drives the translocation via rotational opening of the fingers subdomain [17]. On the other hand, early [39] and immediately later single-molecule force measurements on T7 RNAP suggested alternatively the BR scenario [25]. Accordingly, we investigated the mechano-chemical coupling by studying the PPi release first, using atomistic MD simulation [31].
Indeed, the PPi release step along with the translocation of RNAP on DNA turns out to be too fast to be monitored directly by experiments. For example, the single molecule measurements had shown that forces implemented to hinder the RNAP movements on the DNA hardly slow down the overall elongation, suggesting that the translocation is not a rate-limiting step during an elongation cycle [25,26]. The elongation cycle of T7 RNAP, however, lasts tens of milliseconds or longer [22]. One can accordingly estimate that the fast steps of the product release and translocation happen from microseconds to sub-milliseconds [[43], [44], [45]], which are nevertheless too long for straightforward computational samplings by the atomistic MD simulations.
The all-atom simulation systems of T7 RNAP-DNA-RNA complex with explicit water solvent include over 100 K atoms. For systems of such a size, one can routinely simulate up to several microseconds under current high-performance computing technologies; yet it is still computationally prohibiting to further approach over tens of microseconds to millisecond time scale. Fortunately, by launching extensive sub-microseconds equilibrium simulations that spread around a wide range of conformation space for the relevant process, we were able to construct the Markov-state model (MSM) for the PPi release, and later for the translocation of T7 RNAP, which are estimated to happen at tens of microsecond time scale [31,34]. The strength and technical issues in building the MSM using MD can be found in abundant literature elsewhere [[46], [47], [48], [49], [50]].
The MSM we constructed (two hundreds micro-states according to structural root-mean-square deviations or RMSDs, and three macro-states for visualization) shows a jump-from-cavity PPi release mechanism (see Fig. 1A), in which the PPi-bound product state (S1a) and a pre-activation intermediate state (S1b) dominate the overall population (90%), while the PPi released state (S2) is achieved by thermally activating transitions S1a→S1b→S2 [31]. Inside the ‘cavity’ around the active site, PPi is hold by two aspartate residues (Asp527 and Asp812) that are crucial for the catalysis. Then PPi can shift away and associate more closely with positively charged residues aligning the product release channel (e.g. Lys631 and Arg627), particularly with Lys472 that is key to assist PPi to ‘jump’ out of the cavity via the lysine side-chain swing or fluctuations. Interestingly, there is always a lysine or a homologous arginine located at the exit of the product release channel in other polymerase species (including yeast RNAP II, bacterial RNAP, human mitochondrial RNAP, and several species of DNAPs), which appears to assist the PPi release in general [31]. Note that the jump-from-cavity happens comparatively slowly at the S1b→S2 transition, which is estimated above several microseconds at least.
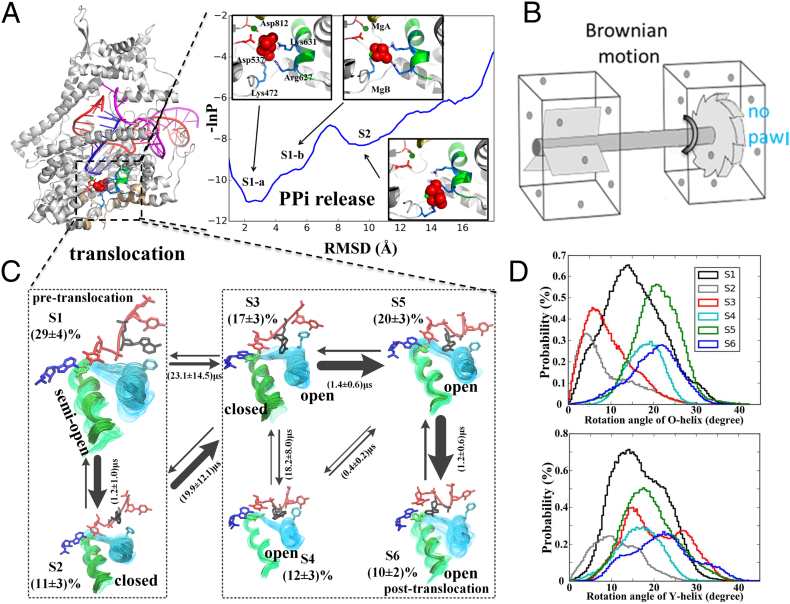
Fig. 1.
The PPi release and translocation mechanism of T7 RNAP revealed from extensive MD simulations and the MSM construction [31,34]. (A) Left panel: A molecular image of T7 RNAP elongation product complex with PPi bound (PDB:1S77) [17]; Right panel: The three-state MSM of the PPi release process derived from 100 × 20 ns MD simulations (by clustering ~106 conformations into 200 microstates etc.) [31]. Note that the PPi group is depicted in red spheres, while the O-helix is colored green. (B) The schematics of an incomplete Brownian-ratchet device, which is still lack of the ‘pawl’. (C) The six-state MSM of the RNAP translocation on the DNA (129 × 80 ns all-atom MD simulation, clustering ~ 9 × 105 conformations into 500 microstates etc.) [34]. The translocation starts after the PPi release from the product complex, or the pre-translocation state (S1), transiting all the way (via S2-S5 and mainly S3) to the post-translocation state (S6; PDB: 1MSW) [16] (populations and transition rates are labeled). Note that both the O-helix (green) and Y-helix (cyan) on the fingers subdomain are shown (with open/closed labeled), along with Y639 and F644 that are key residues in the translocation. The RNA and template DNA nucleotides are colored in blue and red, respectively. (D) The probability distributions of the rotational angles of the O-helix and the Y-helix during translocation process (from S1 to S6) are presented (as taken from [34]). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Essentially, the fingers subdomain does not show substantial conformational changes during the many short equilibrium simulation processes for the PPi release. Even in comparatively long simulations, the fingers subdomain or the O-helix on the subdomain shows no substantial opening, either in the PPi initially bound state, or upon PPi removal or its charge neutralization for control, up to a microsecond time scale. Anyhow, the rotational fluctuations of the O-helix increase in the control simulations, once having PPi or its charge removed. Hence, it seems that the PPi release does not necessarily couple with a progressive rotational opening of the O-helix or the fingers subdomain, which is essential to complete the RNAP translocation. Besides, the thermal activation of the PPi release also suggests that energetically it is unlikely for the release process to directly power or drive the translocation. Instead, the PPi release likely only enhances the rotational flexibilities of the fingers subdomain, which then facilitates the RNAP translocation thereafter.
3. Translocation Proceeds in Brownian Motions and is Facilitated bythe O-helix Fluctuation Opening at Pre-translocation that may also Prevent Backtracking
Then we employed even more extensive MD simulations in aggregation to ~ 10 μs to construct the MSM of the T7 RNAP translocation on DNA, by clustering a large amount of simulation snapshots into 500 micro-states, according to time-structure independent component analysis (tICA) [51,52]. The resulted model is further simplified for visualization into a six-state macro-state representation (see Fig. 1C) [34].
In the six-state translocation network model representation, both the O-helix and Y-helix on the fingers subdomain play significant roles, and they show rotational opening motions in non-synchronized manner. Importantly, in the initial pre-translocation state (S1), both the O-helix and Y-helix show a ‘semi-open’ conformation on average (i.e., the rotation angle peaked ~ 15°), with significant wide fluctuations spanning from the closed conformations to open ones ([34]. Microsecond transition into a less-populated pre-translocation configuration (S2) allows base un-stacking of the transition nucleotide (TN) from F644 (on the Y-helix) yet somehow quenches the O/Y-helix opening to the close status. The Y-helix opens first in the transition state (S3), after that the O-helix opens (S4 to S6). Essentially, Y639 (on the O-helix) pushes onto the 3′-end of the RNA to allow it to move ahead of the template DNA (S3). Hence, the O-helix opening seems to well couple with the DNA forward translocation. Overall, the free energy profile of translocation appears comparatively flat so that Brownian motions dominate. The slowest step of the translocation takes place in the transition to the key intermediate state (S3), which is estimated to last over tens of microseconds at least.
Interestingly, the O-helix along with the Y-helix (or say the fingers subdomain) seems to be able to open by enhanced rotational oscillations or fluctuations (after the PPi release) in the pre-translocation state. Note that it is NOT progressive opening yet until toward the post-translocation state. The product crystal structure with PPi bound was captured in the O-helix (or fingers subdomain) closed configuration [17], so that one expects that: (i) If the O-helix opens right after the PPi release, it is an indication that the translocation can be driven by the O-helix opening (as in the PS scenario), or else (ii) The O-helix remains closed at pre-translocation after the PPi release, and then opens only after the translocation (consistent with the BR scenario, but not necessarily the only situation). However, both the statements are more or less inconsistent with our observations. Our studies indicate that the rotational flexibility of the O-helix or the fingers subdomain becomes high immediately after the PPi release [30], so that one should treat the rotation angle as a highly fluctuating random variable (i.e., with a non-trivial probability distribution), rather than a fixed value. The occasional oscillations to open of the O-helix at pre-translocation appear to be crucial to facilitate the RNAP forward translocation, i.e., by lowering the activation barrier of the translocation. Meanwhile, we suspected that the occasional O-helix openings in the pre-translocation state might even prevent backtracking in T7 RNAP [34]. It is then the average rotational degree of the O-helix or the fingers subdomain that persistently shifts from the closed to open from the pre- to post-translocation state. Previous studies had also concerned about rotational flexibilities of the thumb subdomain [53,54]. We also noticed substantial rotational movements (~25°; non-published results) of the thumb subdomain from the pre- to the post-translocation.
Backtracking turns out to be an efficient way of coordination for RNAP to deal with errors of nucleotide incorporation, i.e., by proofreading or editing; or it supports necessary pauses during the transcription elongation, e.g., to coordinate with translation by ribosome [[55], [56], [57]]. Although backtracking had been identified in multi-subunit RNAPs or even the single subunit mitochondrial RNAP (mtRNAP) [58,59], it has not been detected for T7 RNAP. We then hypothesized that the very mechanism to facilitate the translocation, i.e., the O-helix fluctuation to opening in the pre-translocation state, may also prevent T7 RNAP backtracking. To test the hypothesis, we computationally designed a mutant T7 RNAP with several residues replaced on the O-helix to mimic the mtRNAP that is structurally similar to T7 RNAP [60]. Our simulation results showed that the mutant T7 RNAP would have the O-helix closed upon the 3′-end of the RNA being pulled to initiate the backtracking from the pre-translocation, while the O-helix opens for such a response in the wild-type T7 RNAP [34]. Hence, we considered that the mutant T7 RNAP we made might be able to backtrack to some extent. Preliminary experimental studies supported the hypothesis such that the mutant T7 RNAP maintained transcription activities, but lower than the wild-type system [34]. Further experimental investigation is still needed, e.g., at a single molecule level, to confirm on the mutant and wild-type T7RNAP capabilities of backtracking.
4. Selective Ratcheting Starts From the Nucleotide Pre-insertion Checkpoint
In an RNAP elongation cycle, the incoming nucleoside triphosphate (NTP) is recruited into the RNAP enzyme active site according to the Watson-Crick (WC) base pairing with the template DNA nucleotide. The RNAP translocation allows the incorporated nucleotide at the 3′-end of the synthesizing RNA to move upstream to vacant the active site. According to the BR scenario, prior to the NTP association, RNAP can keep moving forward and backward on DNA, due to the nearly flat free energy surface of the translocation. Once the incoming NTP binds and occupies the active site, the backward movement of RNAP can be prevented, so that forward translocation is finally biased upon the full nucleotide incorporation. That says, the NTP association and incorporation act as a ‘pawl’ in a ratchet device to achieve the BR process (see Figs. 2A and 1B) [61]. Nevertheless, non-cognate nucleotide species may also bind but likely dissociate prematurely before chemical synthesis, due to the nucleotide selectivity conducted by RNAP for the purpose of fidelity control. Consequently, only those successfully incorporated nucleotides, or in principle, the cognate nucleotide species, contribute to the ratcheting of RNAP along DNA.
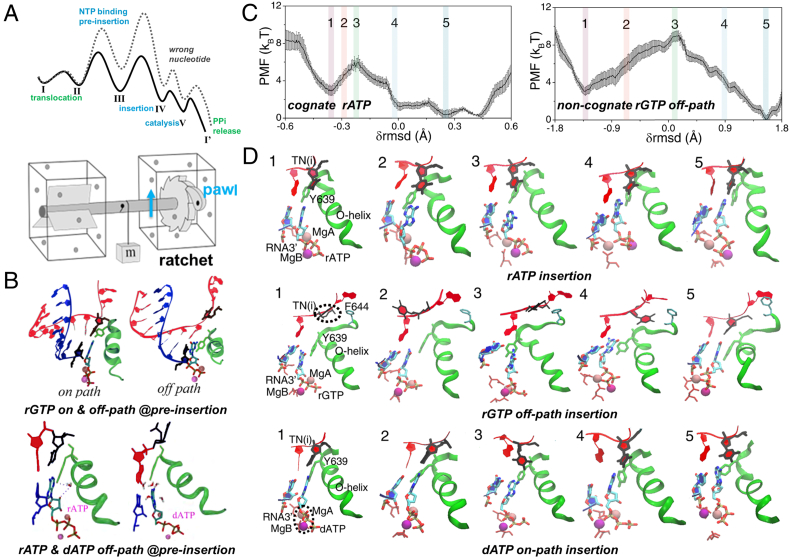
Fig. 2.
Selective ratcheting of T7 RNAP on DNA as nucleotides are differentiated and selected as being incorporated to the growing end of RNA in synthesis. (A) A schematics showing free energy profiles of the cognate/right and non-cognate/wrong nucleotide addition cycle (NAC), which includes translocation, nucleotide binding/pre-insertion, nucleotide insertion, catalysis, and PPi product release. The nucleotide pre-insertion, insertion, and catalysis together serve as a ‘pawl’ for the ratchet. (B) The molecular views around the active site of the pre-insertion complexes modeled for our simulation studies [30,64]. Upper row: the non-cognate rGTP pre-insertion complexes, made on-path and off-path [64], respectively; Lower row: the cognate rATP and the non-cognate dATP (off-path) pre-insertion complexes [30]. (C) The free energy profiles or PMFs calculated from umbrella sampling simulations of the cognate rATP and non-cognate rGTP off-path insertion [36]. (D) The molecular views around the active site of T7 RNAP from representative snapshots captured in the umbrella sampling MD simulations of cognate rATP, non-cognate rGTP (off-path) and dATP (on-path) insertion process [36]. See text for further illustration.
A prominent feature of the nucleotide addition cycle (NAC) of T7 RNAP and related single-subunit polymerase species is that a nucleotide pre-insertion state exists [12,19,62,63], and our studies confirm that the pre-insertion state serves well as an initial kinetic checkpoint to screen non-cognate nucleotide species out of the active site [30,64,36]. Indeed, the pre-insertion complex of T7 RNAP in a ‘semi-open’ conformation had been crystalized with a cognate nucleotide bound to a pre-insertion site, slightly away from the active site [19]. Although the WC base pairing had not been well captured between the pre-insertion nucleotide and the template counterpart in the crystal structure, our MD simulation on the pre-insertion complex revealed the WC base pairing formation after ~ 50 ns equilibrium simulation [30]. Besides, when we replaced the cognate nucleotide (rATP) by the non-cognate species (rGTP, dATP etc.) from the pre-insertion crystal structure complex and conducted equilibrium simulations accordingly, we found that the non-cognate nucleotide would be ‘grabbed’ by Y639, which actually blocks the insertion site of the DNA template nucleotide (i.e., the transition nucleotide or TN) in the pre-insertion complex. In such an equilibrated rGTP pre-insertion complex (see the rGTP off-path pre-insertion structure in Fig. 2B upper right, or Fig. 2D config 1 in the middle row), as the Y639 side chain grabs on rGTP, the template TN has its base rotated away from rGTP, as well as away from its post-translocation configuration [30,64,36]. In the dATP case (see the dATP off-path pre-insertion structure in Fig. 2B lower right), the template TN is even pushed ‘backward’ (to an intermediate configuration between the post- and pre-translocation state), while the Y639 side ring stacks with the upstream DNA template nucleotide and forms a ‘fake’ base pairing with dATP [30,64,36]. That is to say, by competing with the template TN in association with the incoming NTP, Y639 plays a critical role in deterring the non-cognate NTP at pre-insertion. Interestingly, comparing the rATP pre-insertion structure with that of dATP (off-path), one can see that a ‘bridge’ water molecule forms hydrogen bonds with the –OH group of Y639 side chain and that on the ribose of rATP, respectively; in contrast, no such a water bridge can be found in the dATP case due to lack of the ribose –OH for dATP [30] (see Fig. 2B lower panel).
Indeed, the non-cognate nucleotide pre-insertion complexes modeled above are denoted as the ‘off-path’ pre-insertion structures [64], which are at disadvantage for further nucleotide insertion but are easy for the non-cognate dissociation. In the off-path pre-insertion structure, Y639 directly associates with the non-cognate nucleotide to prevent it from closely interacting with the template TN. Energetically, it has been calculated from the MD simulations that the non-cognate nucleotide (rGTP e.g.) dissociation free energy almost vanishes at the off-path pre-insertion site, while the dissociation free energy for the cognate nucleotide from the pre-insertion site appears to be ~ 4 kBT [65]. Upon such a kind of cognate ‘trapping’, and non-cognate off-path Y639 ‘grabbing’ but energetically ‘non-trapping’ at pre-insertion, ~ 99% of the non-cognate nucleotide species can be already screened and removed from accessing further to the active site.
To compare, we also modeled the so-called on-path pre-insertion complexes for the non-cognate nucleotide species, by starting from the equilibrated pre-insertion complex of the cognate nucleotide, and then alchemically converting the cognate nucleotide into the non-cognate species (from rATP to rGTP and dATP e.g.) [64,36]. Accordingly, the modeled on-path pre-insertion structures appear similar to the cognate pre-insertion structure: In particular, the non-cognate nucleotide has its base closely associated with that of the template TN, even though in the absence of the WC base pairing (see Fig. 2B for rGTP on-path or Fig. 2D bottom config 1 for dATP on-path). From the alchemical free energy calculations, we got to see that the non-cognate rGTP on-path pre-insertion structure was energetically less stable (~3 kBT) than that of the cognate rATP [64], while the non-cognate dATP on-path pre-insertion structure was slightly more stable than that of the rATP structure (~ 1 kBT) [36]. Further calculations reveal that the rGTP on-path pre-insertion is indeed highly non-accessible due to a very high rGTP association barrier (with template TN a dTTP), while the dATP on-path pre-insertion does accommodate for a fairly easy dATP association, and allow for further nucleotide insertion and selection following the on-path (see Fig. 2D bottom).
5. Selective Ratcheting Proceeds Through Slow Nucleotide Insertion With Substantial Selection
The nucleotide insertion accompanied by a conformational transition from a ‘semi-open’ to the closed form has been suggested to be a slow step during the NAC in T7 RNAP from previous biochemical studies [22]. It is highly likely that the slow insertion step is employed by the enzyme to substantially scrutinize against non-cognate nucleotide species to achieve sufficiently high transcription fidelity [35]. In viral T7 RNAP elongation, an elongation error rate reaches to ~10−4 to 10−6 for the base mismatch incorporation (e.g. rGTP replacing rATP) [66,67], or ~10−2 for the deoxy-ribonucleotide mis-incorporation (e.g. dATP replacing rATP) [20]. Note that T7 RNAP has not been detected with proofreading or editing activities yet, hence, its fidelity control may fully rely on the nucleotide selectivity, from pre-insertion to insertion, and likely also during catalytic reaction.
To probe the insertion structural dynamics and energetics, we conducted systematical umbrella-sampling simulations in aggregation to microseconds for several nucleotides, along a highly collective coordinate for each nucleotide insertion [36]. The collective coordinate is defined according to the rmsd changes of a highly relevant set of atoms that are essentially involved from the nucleotide pre-insertion to insertion, which include five helices from the fingers subdomain, the insertion nucleotide, and the template DNA nucleotide. The free energy profiles along the δrmsd (in reference with the pre-insertion and insertion structures), or say, the potential of mean forces (PMFs) of the cognate rATP and non-cognate rGTP off-path insertion are shown representatively (see Fig. 2C). In the cognate rATP insertion, an activation barrier ~ 3 kBT occurs at the intermediate conformation (config 3 in Fig. 2D top), where the O-helix opens up to ~ 25° (~ 15° in config 1) to resist the nucleotide insertion. In the end, the insertion state is reached as ~ 3 kBT more stabilized than the pre-insertion state, while the O-helix closes to ~ 5° on average. It is worth pointing out that the thumb subdomain also rotates back (non-published results), along with the closing motions of the fingers subdomain, from the pre-insertion to the insertion state. The on-path insertion free energy profile of the non-cognate rGTP appears quite similar to that of the cognate rATP, although in reality, only a prohibitively low population (<1%) of non-cognate rGTP can be loaded at on-path pre-insertion for the further on-path insertion [36].
In comparison, the off-path insertion PMF of rGTP incurs a quite high barrier (~7 kBT; see Fig. 2C right), as the O-helix opens up ~ 30° in the intermediate state (config 3 in Fig. 2D middle). In addition, the template DNA nucleotide TN deviates its base from the incoming nucleotide all the way from the beginning (config 1–4), until finally the insertion state (config 5) is reached. Thus, the large deviation of the template base from the insertion nucleotide together with the O-helix full opening contributes to the significant high insertion barrier, which is ~ 4 kBT higher than that of the cognate rATP and serves strongly for the mismatch insertion inhibition. The non-cognate rGTP insertion state is also ~ 3 kBT more stabilized than the pre-insertion off-path, and ~ 3 kBT less stabilized than the cognate rATP insertion state. The O-helix closes ~ 5° into the rGTP insertion structure [36].
Upon the pre-insertion rejection and further insertion inhibition, the elongation error rate (i.e., for rGTP replacing rATP) would be reduced to ~ 10−3, according to our calculations via a chemical master equation (CME) approach [35,68]. Hence, to reduce the error rate further down to ~ 10−4, selection into the catalytic stage seems to be required, for which we estimate that a selection energy ~ 7 kBT is necessary. That means during the catalytic reaction, the non-cognate rGTP may face with an activation barrier ~ 7 kBT higher than that of the cognate rATP. Hence, even though our classic MD simulation approach cannot compute the catalytic energetics, we could still predict quantitatively the catalytic selection energetics, combining MD simulation results with experimentally measured error rate information [36].
On the other hand, the energetics for the insertion of deoxy-ribonucleotide dATP appears quite differently from the base mismatch rGTP. Firstly, we could infer that the pre-insertion on-path for dATP is quite accessible as the off-path pre-insertion configuration. Then the calculated PMF of the on-path dATP shows a significant high barrier (~ 6 kBT, or 3 kBT higher than that of rATP; see Fig. 2D bottom), which also corresponds to the O-helix opening above 30° at the transition intermediate (config 3). Interestingly, we could find that two magnesium ions switch positioning of leave and stay during the dATP on-path insertion, but not in other cases. According to a two magnesium catalysis mechanism suggested for the polymerase action [69,70], MgA remains close to the 3′-end of RNA before and after the catalysis, while MgB comes with the insertion NTP and leaves with the product PPi release. However, varying for several types of simulation force field settings, we could always find that MgA and MgB switch their positions during the on-path dATP insertion (Fig. 2D bottom), likely due to the lack of a negative charge on the dATP ribose, so that MgA soon drifts toward the beta-phosphate to ‘collide’ with MgB, which binds there early during the insertion [36]. Accordingly, the high energetics involved in the dATP on-path insertion appears to be related to the two‑magnesium ion switching, which may serve for the selectivity against the deoxy-ribonuleotide incorporation in T7 RNAP.
The off-path insertion of dATP, on the other hand, does not incur a very high barrier (~1 kBT higher than the cognate rATP); along this path, however, the O-helix cannot close below ~10° to reach a proper insertion state, until it possibly relaxes or merges with the on-path near the final insertion stage. Anyhow, due to the prominent selection coordinated by Y639 at the dATP pre-insertion off-path, together with the significant insertion inhibition on-path, the error rate (i.e. dATP replacing rATP) can be achieved at 10−2 as experimentally detected [20,71], according to our CME calculation. Hence, no further selection seems to be needed at the catalytic stage. That is to say, the non-cognate dATP, once being properly inserted, can be catalytically added to the growing end of the RNA as easy as the cognate rATP. Interestingly, previous experimental studies had found that by simply mutating Tyr to Phe at residue 639, the mutant Y639F RNAP can allow the dNTP incorporation as efficient as rNTP [20,71]. Our simulation study accordingly suggested the underlying mechanism by attributing the differentiation between dNTP and rNTP at pre-insertion to the –OH hydroxyl group of Tyr639 (see Fig. 2B bottom for the water bridge), which is lack of for Phe [30,64,36]. Early mutant studies indicated additional amino acids such as H784 in regulation of the nucleotide selectivity [20,72]. We also noticed that H784 could not approach sufficiently close to an incoming dNTP as to an rNTP into the insertion state in the MD simulations. How exactly H784 regulates the selectivity deserve further investigations by possibly studying the double mutant Y639F/H784A and/or including additional modifications to the non-cognate species.
6. Conclusion
Combining atomistic MD simulations with further quantitative analyses, we surveyed a complete NAC during T7 RNAP elongation and dissected detailed structural dynamics mechanisms of the Brownian movements of RNAP along DNA and the selective ratcheting processes for fidelity control. The Brownian-alike motions are thermally activated, with quasi-equal free energetics pre- and post-translocation at the equilibrium condition. The biased ratcheting then requires the system to work at non-equilibrium, i.e., driven by chemical free energy from the polymerization reaction. The template-based nucleotide selectivity is accordingly achieved at the non-equilibrium steady state during the RNAP elongation, which leads to substantially improved fidelity in comparison with the equilibrium condition or with the slow polymerization in the absence of the enzyme. The nucleotide selection is actually conducted stepwise from pre-insertion to insertion, and to catalytic reaction, prior to potential proofreading and further editing. For single-subunit polymerases similar to T7 RNAP, both the pre-insertion rejection and insertion inhibition of the non-cognate nucleotides play significant roles in the fidelity control, which are coordinated by the fingers subdomain opening and closing motions together with key residue interactions. Accordingly, only for those nucleotides that pass through the above selection checkpoints from initial association to final incorporation, successful ratcheting steps of the RNAP can be achieved. In summary, the nature's design of this small but highly capable viral transcription machine appears to be: The mechanical movements of RNAP along DNA mean to be thermally activated without much bias; the RNAP ratcheting forward along DNA is energetically supported by chemical reaction of polymerization, which is tightly coupled with genetic information interrogation for transcription fidelity control, and subjects to evolution pressure of biochemical synthesis. Furthermore, it is important to be aware that the translocation activation and ratcheting energetics involved in each NAC are closely tied to sequence stability variations upon displacing the transcription bubble along DNA [73,74]. Notably, genome-wide transcription dynamics and accuracy measurements could reveal sequence motifs for RNAP pausing, backtracking, or editing [[75], [76], [77]]. Comparative studies between non-backtracking and backtracking RNAP species, for example, may possibly contribute to elucidate consensus or enzyme-dependent motifs and mechanisms. As a minimum transcription machine, T7 RNAP had been laboratory evolved, engineered or redesigned for various purposes [[78], [79], [80]]. The redesigning of T7 RNAP variants with potential functions such as backtracking thus brings another trial of the synthetic approaches to the living systems.
Acknowledgements
Current work has been supported by National Natural Science Foundation of China (NSFC) grants nos. 11775016, 11635002 and 11275022. We acknowledge the computational support from the Beijing Computational Science Research Center (CSRC) and Special Program for Applied Research on Super Computation of the NSFC-Guangdong Joint Fund (the second phase) under grant no. U1501501.
References
- 1.Buc H., Strick T., editors. RNA polymerase as molecular motors. The Royal Society of Chemistry; Cambridge, UK: 2009. [Google Scholar]
- 2.Kochetkov S.N., Rusakova E.E., Tunitskaya V.L. Recent studies of T7 RNA polymerase mechanism. FEBS Lett. 1998;440:264–267. doi: 10.1016/s0014-5793(98)01484-7. [DOI] [PubMed] [Google Scholar]
- 3.Steitz T.A. The structural changes of T7 RNA polymerase from transcription initiation to elongation. Curr Opin Struct Biol. 2009;19:683–690. doi: 10.1016/j.sbi.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kornberg R.D. Eukaryotic transcriptional control. Trends Cell Biol. 1999;9:M46–M49. [PubMed] [Google Scholar]
- 5.Kettenberger H., Armacher K.-J., Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 6.Kornberg R.D. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svetlov V., Nudler E. Basic mechanism of transcription by RNA polymerase II. Biochim Biophys Acta (BBA) - Gene Regul Mech. 2013;1829:20–28. doi: 10.1016/j.bbagrm.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unarta I.C., Zhu L., Tse C.K.M., Cheung P.P.-H., Yu J., Huang X. Molecular mechanisms of RNA polymerase II transcription elongation elucidated by kinetic network models. Curr Opin Struct Biol. 2018;49:54–62. doi: 10.1016/j.sbi.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Sousa R. Structural and mechanistic relationships between nucleic acid polymerases. Trends Biochem Sci. 1996;21:186–190. [PubMed] [Google Scholar]
- 10.Steitz T.A. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 11.Cramer P. Common structural features of nucleic acid polymerases. BioEssays. 2002;24:724–729. doi: 10.1002/bies.10127. [DOI] [PubMed] [Google Scholar]
- 12.Johnson K.A. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim Biophys Acta (BBA)-Prot Proteom. 2010;1804:1041–1048. doi: 10.1016/j.bbapap.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAllister W.T., Raskin C.A. The phage RNA polymerases are related to DNA polymerases and reverse transcriptases. Mol Microbiol. 1993;10:1–6. doi: 10.1111/j.1365-2958.1993.tb00897.x. [DOI] [PubMed] [Google Scholar]
- 14.Sousa R., Chung Y.J., Rose J.P., Wang B.-C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 Å resolution. Nature. 1993;364:593. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- 15.Cheetham G.M., nbsp T., Steitz A.T. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 16.Yin Y.W., Steitz T.A. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 17.Yin Y.W., Steitz T.A. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 18.Durniak K.J., Bailey S., Steitz T.A. The structure of a transcribing T7 RNA polymerase in transition from initiation to elongation. Science. 2008;322:553–557. doi: 10.1126/science.1163433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Temiakov D., Patlan V., Anikin M., McAllister W.T., Yokoyama S., Vassylyev D.G. Structural basis for substrate selection by T7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 20.Brieba L.G., Sousa R. Roles of Histidine 784 and tyrosine 639 in ribose discrimination by T7 RNA polymerase. Biochemistry. 2000;39:919–923. doi: 10.1021/bi992324+. [DOI] [PubMed] [Google Scholar]
- 21.Guo Q., Sousa R. Translocation by T7 RNA polymerase: a sensitively poised Brownian ratchet. J Mol Biol. 2006;358:241–254. doi: 10.1016/j.jmb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Anand V.S., Patel S.S. Transient state kinetics of transcription elongation by T7 RNA polymerase. J Biol Chem. 2006;281:35677–35685. doi: 10.1074/jbc.M608180200. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y., Navaroli D.M., Enuameh M.S., Martin C.T. Dissociation of halted T7 RNA polymerase elongation complexes proceeds via a forward-translocation mechanism. Proc Natl Acad Sci. 2007;104:10352–10357. doi: 10.1073/pnas.0606306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skinner G.M., Baumann C.G., Quinn D.M., Molloy J.E., Hoggett J.G. Promoter binding, initiation, and elongation by bacteriophage T7 RNA polymerase. J Biol Chem. 2004;279:3239–3244. doi: 10.1074/jbc.M310471200. [DOI] [PubMed] [Google Scholar]
- 25.Thomen P., Lopez P.J., Heslot F. Unravelling the mechanism of RNA-polymerase forward motion by using mechanical force. Phys Rev Lett. 2005;94 doi: 10.1103/PhysRevLett.94.128102. [DOI] [PubMed] [Google Scholar]
- 26.Thomen P., Lopez P.J., Bockelmann U., Guillerez J., Dreyfus M., Heslot F. T7 RNA polymerase studied by force measurements varying cofactor concentration. Biophys J. 2008;95:2423–2433. doi: 10.1529/biophysj.107.125096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J.H., Larson R.G. Single-molecule analysis of 1D diffusion and transcription elongation of T7 RNA polymerase along individual stretched DNA molecules. Nucleic Acids Res. 2007;35:3848–3858. doi: 10.1093/nar/gkm332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J., Oster G. A small post-translocation energy Bias aids nucleotide selection in T7 RNA polymerase transcription. Biophys J. 2012;102:532–541. doi: 10.1016/j.bpj.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu J. Efficient fidelity control by stepwise nucleotide selection in polymerase elongation. Mol Based Math Biol. 2014;2 [Google Scholar]
- 30.Duan B., Wu S., Da L.-T., Yu J. A critical residue selectively recruits nucleotides for T7 RNA polymerase transcription Fidelity control. Biophys J. 2014;107:2130–2140. doi: 10.1016/j.bpj.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Da L.-T., Chao E., Duan B., Zhang C., Zhou X., Yu J. A jump-from-cavity pyrophosphate ion release assisted by a key lysine residue in T7 RNA polymerase transcription elongation. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J. Computational investigations on polymerase actions in gene transcription and replication: combining physical modeling and atomistic simulations. Chin Phys B. 2016;25 [Google Scholar]
- 33.E C., Yu J. Nucleotide selectivity at a Preinsertion checkpoint of T7 RNA polymerase transcription elongation. J Phys Chem B. 2017;121:3777–3786. doi: 10.1021/acs.jpcb.6b11668. [DOI] [PubMed] [Google Scholar]
- 34.Da L.-T., E C Shai Y., Wu S., Su X.-D., Yu J. T7 RNA polymerase translocation is facilitated by a helix opening on the fingers domain that may also prevent backtracking. Nucleic Acids Res. 2017;45:7909–7921. doi: 10.1093/nar/gkx495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long C., Yu J. Balancing non-equilibrium driving with nucleotide selectivity at kinetic checkpoints in polymerase Fidelity control. Entropy. 2018;20:306. doi: 10.3390/e20040306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long C., E C., Da L.-T., Yu J. Determining selection free energetics from nucleotide pre-insertion to insertion in viral T7 RNA polymerase transcription fidelity control. Nucleic Acids Res. 2019;47:4721–4735. doi: 10.1093/nar/gkz213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H., Oster G. Ratchets, power strokes, and molecular motors. Appl Phys A. 2002;75:315–323. [Google Scholar]
- 38.Bustamante C., Keller D., Oster G. The physics of molecular motors. Acc Chem Res. 2001;34:412–420. doi: 10.1021/ar0001719. [DOI] [PubMed] [Google Scholar]
- 39.Guajardo R., Sousa R. A model for the mechanism of polymerase translocation. J Mol Biol. 1997;265:8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- 40.Bar-Nahum G., Epshtein V., Ruckenstein A.E., Rafikov R., Mustaev A., Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 41.Abbondanzieri E.A., Greenleaf W.J., Shaevitz J.W., Landick R., Block S.M. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dangkulwanich M., Ishibashi T., Liu S., Kireeva M.L., Lubkowska L., Kashlev M. Complete dissection of transcription elongation reveals slow translocation of RNA polymerase II in a linear ratchet mechanism. eLIFE. 2013;2 doi: 10.7554/eLife.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Da L.-T., Pardo A.F., Wang D., Huang X. A two-state model for the dynamics of the pyrophosphate ion release in bacterial RNA polymerase. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Da L., Wang D., Huang X. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J Am Chem Soc. 2011;134:2399–2406. doi: 10.1021/ja210656k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silva D.A., Weiss D.R., Pardo Avila F., Da L.T., Levitt M., Wang D. Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc Natl Acad Sci U S A. 2014;111:7665–7670. doi: 10.1073/pnas.1315751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noé F., Fischer S. Transition networks for modeling the kinetics of conformational change in macromolecules. Curr Opin Struct Biol. 2008;18:154–162. doi: 10.1016/j.sbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 47.Lane T., Bowman G., Beauchamp K., Voelz V., Pande V. Markov state model reveals folding and functional dynamics in ultra-Long MD trajectories. J Am Chem Soc. 2011;133:18413–18419. doi: 10.1021/ja207470h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowman G.R., Pande V.S., Noé F. Springer Science & Business Media; 2013. An introduction to Markov state models and their application to long timescale molecular simulation. [Google Scholar]
- 49.Chodera J.D., Noé F. Markov state models of biomolecular conformational dynamics. Curr Opin Struct Biol. 2014;25:135–144. doi: 10.1016/j.sbi.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W., Cao S., Zhu L., Huang X. Constructing Markov state models to elucidate the functional conformational changes of complex biomolecules. Wiley Interdiscip Rev. 2018;8 [Google Scholar]
- 51.Naritomi Y., Fuchigami S. Slow dynamics of a protein backbone in molecular dynamics simulation revealed by time-structure based independent component analysis. J Chem Phys. 2013;139:12B605_601. doi: 10.1063/1.4834695. [DOI] [PubMed] [Google Scholar]
- 52.Pérez-Hernández G., Paul F., Giorgino T., De Fabritiis G., Noé F. Identification of slow molecular order parameters for Markov model construction. J Chem Phys. 2013;139:07B604_601. doi: 10.1063/1.4811489. [DOI] [PubMed] [Google Scholar]
- 53.Bonner G., Lafer E.M., Sousa R. The thumb subdomain of T7 RNA polymerase functions to stabilize the ternary complex during processive transcription. J Biol Chem. 1994;269:25129–25136. [PubMed] [Google Scholar]
- 54.Brieba L.G., Gopal V., Sousa R. Scanning mutagenesis reveals roles for helix n of the bacteriophage T7 RNA polymerase thumb subdomain in transcription complex stability, pausing, and termination. J Biol Chem. 2001;276:10306–10313. doi: 10.1074/jbc.M009866200. [DOI] [PubMed] [Google Scholar]
- 55.Nulder E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J., Palagnat M., Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dutta D., Shatalin K., Epshtein V., Gottesman M.E., Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Da L.-T., Pardo-Avila F., Xu L., Silva D.-A., Zhang L., Gao X. Bridge helix bending promotes RNA polymerase II backtracking through a critical and conserved threonine residue. Nat Commun. 2016;7 doi: 10.1038/ncomms11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zamft B., Bintu L., Ishibashi T., Bustamante C. Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proc Natl Acad Sci. 2012;109:8948–8953. doi: 10.1073/pnas.1205063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ringel R., Sologub M., Morozov Y.I., Litonin D., Cramer P., Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 61.Richard P., Feynman L., Sands M. Addinson-Wesley; 1963. Feynman lectures on physics. [Google Scholar]
- 62.Schlick T., Arora K., Beard W.A., Wilson S.H. Perspective: pre-chemistry conformational changes in DNA polymerase mechanisms. Theor Chem Acc. 2012;131:1287. doi: 10.1007/s00214-012-1287-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu E.Y., Beese L.S. The structure of a high Fidelity DNA polymerase bound to a mismatched nucleotide reveals an “ajar” intermediate conformation in the nucleotide selection mechanism. J Biol Chem. 2011;286:19758–19767. doi: 10.1074/jbc.M110.191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.E C., Duan B., Yu J. Nucleotide selectivity at a Preinsertion checkpoint of T7 RNA polymerase transcription elongation. J Phys Chem B. 2017;121:3777–3786. doi: 10.1021/acs.jpcb.6b11668. [DOI] [PubMed] [Google Scholar]
- 65.Wu S., Wang J., Pu X., Li L., Li Q. T7 RNA polymerase discriminates correct and incorrect nucleoside triphosphates by free energy. Biophys J. 2018;114:1755–1761. doi: 10.1016/j.bpj.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sultana S., Solotchi M., Ramachandran A., Patel S.S. Transcriptional fidelities of human mitochondrial POLRMT, yeast mitochondrial Rpo41, and phage T7 single-subunit RNA polymerases. J Biol Chem. 2017;292:18145–18160. doi: 10.1074/jbc.M117.797480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang J., Brieba L.G., Sousa R. Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce Misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry. 2000;39:11571–11580. doi: 10.1021/bi000579d. [DOI] [PubMed] [Google Scholar]
- 68.Gillespie D.T. A rigorous derivation of the chemical master equation. Phys A Stat Mech Appl. 1992;188:404–425. [Google Scholar]
- 69.Steitz T.A., Steitz J.A. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sosunov V., Sosunova E., Mustaev A., Bass I., Nikiforov V., Goldfarb A. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sousa R., Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Padilla R., Sousa R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res. 2002;30:e138. doi: 10.1093/nar/gnf138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bai L., Shundrovsky A., Wang M.D. Sequence-dependent kinetic model for transcription elongation by RNA polymerase. J Mol Biol. 2004;344:335–349. doi: 10.1016/j.jmb.2004.08.107. [DOI] [PubMed] [Google Scholar]
- 74.Mellenius H., Ehrenberg M. DNA template dependent accuracy variation of nucleotide selection in transcription. PLoS One. 2015;10 doi: 10.1371/journal.pone.0119588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Imashimizu M., Afek A., Takahashi H., Lubkowska L., Lukatsky D.B. Control of transcriptional pausing by biased thermal fluctuations on repetitive genomic sequences. Proc Natl Acad Sci. 2016;113:E7409–E7417. doi: 10.1073/pnas.1607760113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Traverse C.C., Ochman H. Conserved rates and patterns of transcription errors across bacterial growth states and lifestyles. Proc Natl Acad Sci. 2016;113:3311–3316. doi: 10.1073/pnas.1525329113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.James K., Gamba P., Cockell S.J., Zenkin N. Misincorporation by RNA polymerase is a major source of transcription pausing in vivo. Nucleic Acids Res. 2016;45:1105–1113. doi: 10.1093/nar/gkw969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chelliserrykattil J., Ellington A.D. Evolution of a T7 RNA polymerase variant that transcribes 2′-O-methyl RNA. Nat Biotechnol. 2004;22:1155. doi: 10.1038/nbt1001. [DOI] [PubMed] [Google Scholar]
- 79.Esvelt K.M., Carlson J.C., Liu D.R. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shis D.L., Bennett M.R. Synthetic biology: the many facets of T7 RNA polymerase. Mol Syst Biol. 2014;10:745. doi: 10.15252/msb.20145492. [DOI] [PMC free article] [PubMed] [Google Scholar]