Abstract
Background and Aims
To define pharmacodynamic and efficacy biomarkers in ulcerative colitis [UC] patients treated with PF-00547659, an anti-human mucosal addressin cell adhesion molecule-1 [MAdCAM-1] monoclonal antibody, in the TURANDOT study.
Methods
Transcriptome, proteome and immunohistochemistry data were generated in peripheral blood and intestinal biopsies from 357 subjects in the TURANDOT study.
Results
In peripheral blood, C-C motif chemokine receptor 9 [CCR9] gene expression demonstrated a dose-dependent increase relative to placebo, but in inflamed intestinal biopsies CCR9 gene expression decreased with increasing PF-00547659 dose. Statistical models incorporating the full RNA transcriptome in inflamed intestinal biopsies showed significant ability to assess response and remission status. Oncostatin M [OSM] gene expression in inflamed intestinal biopsies demonstrated significant associations with, and good accuracy for, efficacy, and this observation was confirmed in independent published studies in which UC patients were treated with infliximab or vedolizumab. Compared with the placebo group, intestinal T-regulatory cells demonstrated a significant increase in the intermediate 22.5-mg dose cohort, but not in the 225-mg cohort.
Conclusions
CCR9 and OSM are implicated as novel pharmacodynamic and efficacy biomarkers. These findings occur amid coordinated transcriptional changes that enable the definition of surrogate efficacy biomarkers based on inflamed biopsy or blood transcriptomics data.
ClinicalTrials.gov identifier
Keywords: Ulcerative colitis, inflammatory bowel disease, MAdCAM-1, PF-00547659, biomarkers
1. Introduction
Ulcerative colitis [UC] is a chronic, relapsing disease marked by inflammation and ulceration of the colonic mucosa that can significantly impact a patient’s quality of life.1 Whereas the physician’s therapeutic armamentarium continues to expand, including aminosalicylates, corticosteroids, immunosuppressants, and monoclonal antibodies against tumour necrosis factor [TNF]-α or α4β7 integrin, post-treatment remission remains a challenge, making the development of additional novel drugs a critical unmet medical need.2
PF-00547659 is a fully human IgG2k anti-human mucosal addressin cell adhesion molecule [MAdCAM] monoclonal antibody that limits intestinal inflammation through blockade of MAdCAM-dependent lymphocyte recruitment into the intestinal parenchyma.3 The efficacy of PF-00547659 was explored in the Phase 2b TURANDOT study in subjects with moderate to severe active UC.4 Four subcutaneous [s.c.] dose levels [7.5, 22.5, 75, and 225 mg] of PF-00547659 versus placebo were studied during a 12-week treatment period. The primary clinical efficacy end point—remission rates at Week 12 [a total Mayo score of ≤2 with no individual subscore >1 and a rectal bleed subscore of 0 or 1] were significantly greater in three of four active-treatment groups than in the placebo group (2.7% [2/73]): 7.5 mg (11.3% [8/71]), 22.5 mg (16.7% [12/72]), 75 mg (15.5% [11/71]) and 225 mg (5.7% [4/70]). However, the greatest remission rate was observed in the intermediate 22.5-mg dose arm, implicating a non-monotonic dose response.
Biomarkers that predict efficacy or act as robust surrogates for pharmacology or efficacy would be of immense value for drug development and eventual clinical management. To define such biomarkers, intestinal biopsy and peripheral blood samples were collected during the Phase 2b TURANDOT study for subsequent transcriptomic and proteomic interrogation. We also examined publicly available datasets to confirm our findings.
2. Materials and Methods
2.1. The clinical trial and clinical variables
The TURANDOT clinical trial was a Phase 2b randomized, double-blind, placebo-controlled, parallel-group, dose-ranging study to evaluate efficacy, safety, and pharmacokinetics of PF-00547659 in patients with moderate to severe UC. A total of 357 participants with moderate to severe UC, defined as total Mayo score of ≥6 points with an endoscopic subscore of ≥2 points, who had failed at least one standard therapy, were randomly assigned to receive placebo [n = 73] or PF-00547659 at doses of 7.5 mg [n = 71], 22.5 mg [n = 72], 75 mg [n = 71], or 225 mg [n = 70] for a 12-week treatment period.
Clinical efficacy of PF-00547659 was also evaluated using the secondary end points of clinical response, and mucosal healing. In terms of clinical response [a decrease from baseline of at least 3 points in Total Mayo score with at least a 30% change, accompanied by at least a 1 point decrease or absolute score of 0 or 1 in rectal bleeding subscore], three of four treatment groups [22.5 mg, 75 mg, and 225 mg] have shown statistical efficacy compared with placebo (28.8% [21/73]): 7.5 mg (38% [27/71]), 22.5 mg (54.2% [39/72]), 75 mg (45.1% [32/71]), and 225 mg (50% [35/70]). Mucosal healing [an absolute Mayo subscore for endoscopy of 0 or 1 at Week 12] rates for 22.5 mg and 75 mg over placebo are significantly different from placebo. The mucosal healing rates were observed as placebo (8.2% [6/73]), 7.5 mg (15.5% [11/71]), 22.5 mg (27.8% [20/72]), 75 mg (25.4% [18/71]), and 225 mg (14.3% [10/70]), respectively.
All endoscopic scores were determined by an adjudicated blinded central reading method.5 The central reading was used for the primary analysis.
2.2. Biospecimen collection and processing
Serum and whole blood samples were collected from enrolled subjects at baseline, Week 4, and Week 12. Serum protein concentration was measured. Whole blood samples were stored at –80°C in PAXgene Blood RNA tubes [PreAnalytiX GmbH, BD Biosciences, Mississauga, ON, Canada]. Inflamed and non-inflamed colon pinch biopsies were collected at baseline and Week 12 and placed into RNAlater or into formalin. The biopsies were taken from abnormal lesional mucosa, and from non-lesional mucosa and then completely submerged in the liquid. Labels specified whether the biopsy was from a lesional or non-lesional area. Obviously ulcerated areas were avoided. Endoscopists were instructed to take all the biopsies from the same area/segment taken at baseline. Biopsies were taken one at a time, and each one was placed immediately into a separate sample collection tube. RNA extraction and quality control from both blood and colon biopsies were performed by a contract laboratory and used to measure gene expression. Formalin-fixed colon biopsies were processed into paraffin blocks and used to assess cellular content by immunohistochemistry [IHC]. Consent was provided for the use of RNA, protein, and IHC data for all of the analyses. The numbers of samples for each molecular measurement are listed in Table 1.
Table 1.
Number of samples for each molecular measurement
| Screening | Week 4 | Week 12 | |
|---|---|---|---|
| Blood RNA | 320 | 256 | 320 |
| Biopsy RNA [inflamed] | 126 | 126 | |
| Biopsy RNA [non-inflamed] | 79 | 79 | |
| Serum protein | 331 | 275 | 331 |
| IHC samples | 30 | 30 |
2.3. Transcriptomic gene expression
Transcriptomic gene expression profiles from whole blood and tissue biopsies were examined using RNA sequencing [RNA-Seq] technology. RNA samples with an RNA Integrity Number [RIN] of >6 were sequenced at BGI [Hong Kong, China] using an Illumina Truseq stranded mRNA protocol. Globin RNA reduction was performed for whole blood samples prior to library construction. Each sample was sequenced at 100 bp and average 40 Mb-paired-end read depth. The clean raw sequence reads in FASTQ format were first mapped to the human reference genome version hg19 using STAR v2.3.0e.6 The uniquely mapped reads were counted towards individual genes by the program BEDTools.7 At first, a flattened bed file was generated based on GENCODE exon definition by merging overlapping exons for a gene into fused intervals, which can belong to more than one exon. Reads falling into these disjoined intervals were counted using BEDTools and then summarized into the gene level. The parameters for BEDTools v2.18.2 run were ‘-wo -split -bed -abam sample.bam -b hg19.flatten.bed’. Gene level counts per million were used in the analysis. To reduce false positives, genes with fewer than two reads in >80% samples and an average of fewer than three read counts across all samples were treated as not expressed, and thus omitted from downstream data analysis.
2.4. Serum protein
Profiling of protein concentrations in serum samples was measured at Olink Proteomics [Uppsala, Sweden] using the Proseek Multiplex Analysis platform and standard protocols.8 A total of 202 unique proteins were measured using the combined Proseek Multiplex ONC I v296×96, INF I96×96, and CVD I96×96 panels.
Protein concentration results are reported as normalized protein expression values [NPX, log2] for the serum samples. Briefly, the assay readout ‘quantification cycles’ [Cq values] from the real-time PCR step were pre-processed using a macro file provided by Olink and normalized, which produces NPX values. Assay run quality and sample data quality were assessed by monitoring a number of controls according to the manufacturer’s recommendations. Only results for samples that met these acceptance criteria were reported. Sample measurements below the limit of detection [LOD] for an analyte were reported as NaN [not a number] and substituted with one half of the LOD for that analyte during analysis.
2.5. Immunohistochemistry
Triple-label IHC was performed on colon pinch biopsies collected from inflamed and non-inflamed sites at baseline and Week 12 [i.e. four biopsies per subject] following the onset of dosing from subjects in the placebo, 22.5-mg, and 225-mg dose groups. Within each group, patients were ranked based on Week 12 MAYO scores and MAYO score change from baseline. Five subjects from both extremes of the ranking were picked [i.e. 10 subjects per dose group]; thus, a total of 120 biopsies were evaluated by IHC.
Immunoperoxidase and alkaline phosphatase reactions were combined in a triple-label IHC assay to identify regulatory T-lymphocytes, i.e. those that were simultaneously expressing CD3, CD25, and FoxP3 in 4-µm sections of formalin-fixed, paraffin-embedded [FFPE] colon biopsies. All IHC reactions were performed on a Discovery Ultra® automated staining platform [Roche/Ventana Medical Systems, Tucson, AZ], using Ventana OmniMap, UltraMap, and HQ detection reagents. Epitope retrieval was conducted using EDTA-based CC1 retrieval buffer [Ventana], and non-specific protein binding was blocked using Sblock [Ventana]. Rabbit monoclonal anti-CD25 [clone EP218, Epitomics, Burlingame, CA] was applied at a 1:100 dilution, followed by secondary anti-rabbit HQ, anti-HQ HRP, and ChromoMap diaminobenzidine [DAB] chromogen [Ventana]. Antibody denaturation and neutralization was performed to prevent cross-reactivity, followed sequentially by incubations with rabbit monoclonal anti-CD3 [clone SP7, Thermo Scientific, Waltham, MA] at 1:300 dilution, secondary OmniMap anti-rabbit HRP antibody, and Discovery Purple chromogen. Biopsy sections were then incubated with monoclonal mouse anti-FoxP3 [clone 236A/E7, Abcam, Cambridge, MA] at a 1:100 dilution, followed by UltraMap anti-mouse AP secondary antibody, and Discovery Yellow chromogen [Ventana]. Biopsy sections were counterstained with haematoxylin, treated with a bluing reagent [Ventana], dehydrated through graded alcohols, cleared in xylene, and cover-slipped with a synthetic mounting medium.
2.6. Image analysis
Computer image analysis was used to enumerate cells expressing CD3, CD25, and FoxP3, and the frequency of regulatory T-lymphocytes [Tregs] within the total T-lymphocyte population in the lamina propria of colon biopsy specimens was estimated by quantifying the number of triple-labelled cells [FoxP3+/CD3+/CD25+] and dividing by the total number of CD3+ cells.
Following multiparameter IHC, spectral images of singleplex and multiplex IHC sections were captured using a Vectra automated quantitative pathology imaging system [PerkinElmer, Waltham, MA], and image analysis, including phenotyping, was performed using inForm image analysis software [PerkinElmer]. Singleplex IHC for CD3 [Ventana Purple], CD25 [Ventana Brown], and FoxP3 [Ventana Yellow] and haematoxylin counterstain on biopsy sections were subjected to spectral analysis to generate a spectral library that facilitated multispectral unmixing of captured images. inForm software was trained to recognize and differentiate specific micro-anatomic regions of interest [e.g. lamina propria] from regions that were not to be evaluated [e.g. epithelium, crypt lumens, biopsy artefacts], and cell phenotypes were identified based upon the localization and association of membrane [e.g. CD3, CD25] and nuclear [e.g. FoxP3] spectral elements with haematoxylin-labelled cell nuclei. The numbers of cells expressing various combinations of single, double, and triple labelling for the target antigens [CD3, CD25, and FoxP3] was quantified, and the relative frequency of regulatory T-lymphocytes in the lamina propria (the region of interest [ROI]) was calculated as the percentage CD3+CD25+FoxP3+ cells / total CD3+ cells.
2.7. Expression data from colonic biopsy samples of infliximab-treated UC patients and vedolizumab-treated UC patients
We further examined identified clinical end point surrogate markers using several additional publicly available datasets.
Colonic biopsy data, GSE23597, was generated in a substudy of Active UC Trial 1 [ACT1], a placebo-controlled study of infliximab. Patients with moderate-to-severe, active UC were treated with infliximab or placebo at Weeks 0, 2, 6, and every 8 weeks thereafter. The primary end point was the proportion of patients in clinical response, defined as a decrease from baseline in the total Mayo score of ≥3 points and ≥30%, with an accompanying decrease in the subscore for rectal bleeding of ≥1 point or an absolute subscore for rectal bleeding of 0 or 1. Altogether, 33 patients had biopsy samples available at both baseline and Week 8 [primary time point]. Gene expression profiling was conducted using Affymetrix Human Genome U133 Plus 2.0 Array.9
Another set of colonic biopsy data, GSE16879, was generated in a study where mucosal biopsies were obtained at endoscopy in actively inflamed mucosa from 24 UC patients before, and 4–6 weeks after, their first infliximab infusion. The response to infliximab was assessed 4–6 weeks after the first infliximab treatment and was defined as a decrease to a Mayo endoscopic subscore of 0 or 1 with a decrease to Grade 0 or 1 in the histological score. Gene expression profiling was conducted using Affymetrix Human Genome U133 Plus 2.0 Array.10
A third endoscopic-derived biopsy dataset, GSE73661, was generated from UC patients during two Phase 3 trials of vedolizumab, GEMINI I, and GEMINI LTS.11 Gemini I was a Phase 3 study investigating clinical response and remission by vedolizumab in patients with moderate-to-severe UC, and Gemini LTS is a multicentre, open-label study on long-term safety and efficacy of vedolizumab. Forty-one patients who participated in these studies at the University Hospitals Leuven [Leuven, Belgium] and who were treated with vedolizumab were included in the study. Endoscopic mucosal healing was defined as a Mayo endoscopic subscore of 0 or 1. Biopsies were taken at Week [W] 0, W6, W12, and W52, or at study withdrawal. A total of 120 colonic mucosal biopsies were available for analysis. Whole-genome gene expression was generated using Affymetrix GeneChip Human Gene 1.0 ST arrays.11 As reported by Arijs et al., a maximal effect of vedolizumab on mucosal gene expression was seen at Week 52.11 In the current study, we only assessed gene expression change of 19 patients at W52.
2.8. Statistical analysis
Univariate statistical analysis was performed on current clinical and molecular data for the following goals: [1] identify pharmacodynamic [PD] biomarkers that respond to treatment; [2] identify markers whose change at Week 12 correlates with efficacy, including remission, response, or mucosal healing; [3] identify markers at baseline that predict subsequent efficacy, including remission, response, or mucosal healing at Week 12; and [4] determine whether Tregs in colon biopsies change differently across treatment arms. For the first three goals, linear and logistic regression models were employed, and a false discovery rate [FDR] of <0.1 was used to control for multiple comparisons.12 For the fourth goal, the Wilcoxon signed-rank test was employed, and a p-value of 0.05 was used as the significance cut-off. Enrichment analyses using Ingenuity Pathway Analysis [IPA; QIAGEN, Silicon Valley, CA] were performed on identified biomarker hits to further explore the pathways underlying the findings. The p-values were calculated using a right-tailed Fisher’s exact test to determine statistically significant over-representation of genes in identified networks. The z-scores as reported by IPA predict the activation [positive z-score] or repression [negative z-score] of networks based on the observed gene expression changes and the underlying knowledge in the database. A p-value of <0.05 and a z-score of |2| were established as significance cut-offs for identified networks. Receiver operating characteristic [ROC] analysis was performed to evaluate the ability of the identified markers to assess clinical end points.
We also built statistical machine learning models to identify combinations of transcripts that predict subsequent clinical remission or response at Week 12 based on baseline data, as well as to assess clinical remission or response based on Week 12 data. In each setting, we considered transcript data from blood, inflamed biopsies, and non-inflamed biopsies for a total of 12 contrasts [2 time points × 2 clinical end points × 3 transcript datasets].
For each contrast, we employed three state-of-the-art machine learning methods with different characteristics, namely logistic Elastic Net13 as a linear method, Random Forests14 as a non-linear method, and Response-CRE15 as a pathway-supported approach. All input data were normalized to a mean of 0 and a standard deviation of 1 before use. Each method was run under a 10-fold nested cross-validation scheme that separates hyperparameter optimization from performance estimation to give accurate estimates of predicted performance. For each of the 12 contrasts and each of the three employed methods, we report the area under the receiver operating curve [AUROC] or c-statistic with corresponding 95% confidence intervals. We considered a finding relevant if at least one of the methods had an estimated performance of >0.8 AUROC and the 95% confidence interval was bounded away from random performance. To gain insights into transcripts driving significant performance, we trained the elastic net linear classifier on the full dataset and assessed average rank of each transcript across cross-validation runs based on absolute magnitude of the regression coefficients. We investigated top transcripts for biological plausibility.
All analyses were performed using R [http://www.r-project.org/], with the exception of machine learning, which was carried out in Python using the scikit-learn library.14
3. Results
3.1. Unbiased molecular profiling reveals pharmacodynamic biomarkers
Biomarker changes from baseline were compared across different dose arms. From blood, we identified 1772 unique genes whose changes from baseline in at least one dose arm were significantly different from placebo [FDR < 0.1] [1701 upregulated and 71 downregulated]. The number of differentially expressed genes in blood suggests a dose-dependent effect [14, 32, 296, and 1599 genes from the 7.5, 22.5, 75, and 225 mg PF-00547659 arms, respectively]. Interestingly, no genes in biopsies or proteins in blood showed significant change compared with placebo [Table 2 and Supplementary Table S1].
Table 2.
Summary of identified pharmacodynamic biomarkers
| Number of patients | Number of hits [FDR <10%] | |||||
|---|---|---|---|---|---|---|
| Placebo | 7.5 mg | 22.5 mg | 75 mg | 225 mg | ||
| Blood RNA | 63 | 57 | 64 | 66 | 60 | 1772 [1701 upregulated and 71 downregulated] |
| Biopsy RNA [inflamed] | 29 | 22 | 18 | 25 | 23 | 0 |
| Biopsy RNA [non-inflamed] | 17 | 10 | 13 | 17 | 16 | 0 |
| Serum protein | 67 | 59 | 66 | 65 | 59 | 0 |
Pathway analysis on the differentially expressed genes in blood using IPA revealed a group of enriched signalling pathways [Supplementary Table S2]. At both 75-mg and 225-mg PF-00547659 doses, pathways that showed increased gene expression included CD28 signalling in T-helper cells, interleukin [IL]-8 signalling, IL-6 signalling, B-cell receptor signalling, integrin signalling, C-X-C-motif chemokine receptor 4 [CXCR4] signalling, and hepatocyte growth factor [HGF] signalling [Supplementary Table S2], implying increased immune cell signalling and accumulation in the blood as a result of the PF-00547659 treatment.
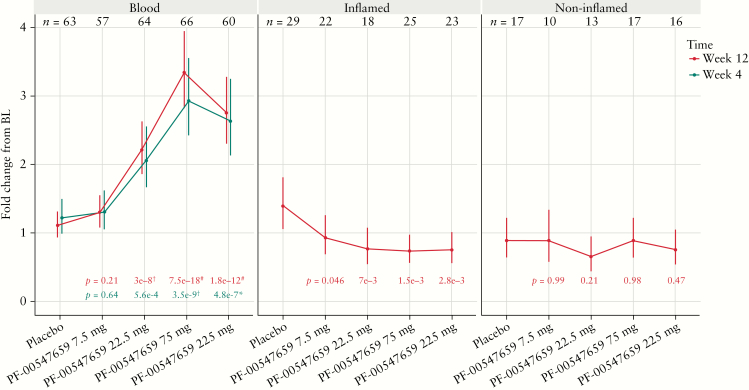
Among the 1772 differentially expressed genes in blood, C-C motif chemokine receptor 9 [CCR9, gut homing chemokine] showed the most significant changes across different doses. Compared with placebo, the change in CCR9 RNA expression at Week 12 from baseline demonstrated a 1.2-, 2.0-, 3.0-, and 2.5-fold increase in the 7.5, 22.5, 75, and 225 mg cohorts, respectively [p = 0.21, 3e-8, 7.5e-18, and 1.8e-12; FDR < 0.001 for the 22.5, 75, and 225 mg PF-00547659 groups; Figure 1]. The change in CCR9 expression in the blood was evident at Week 4 [Figure 1]. This suggests that more CCR9-expressing immune cells remain in the blood at higher doses. Among all gene expression changes in inflamed biopsies across different doses, CCR9 produced the smallest p-values, although it did not reach significance after the FDR correction. Conversely to the increase in CCR9 in blood, CCR9 mRNA expression change from baseline at Week 12 in inflamed biopsies demonstrated decreases of 1.5-, 1.8-, 1.9-, and 1.8-fold in the 7.5, 22.5, 75, and 225 mg PF-00547659 cohorts, compared with placebo, respectively [p = 0.046, 0.0070, 0.0015, 0.0028]. This suggests that fewer immune cells are migrating from the blood to the inflamed biopsy at higher doses. Also, CCR9 change from baseline remained constant across different doses in non-inflamed biopsies.
Figure 1.
Fold changes in CCR9 gene expression from baseline [BL] to Week 4 or Week 12 by treatment group. *0.01 < FDR ≤ 0.1; †0.00001 < FDR ≤ 0.01; #FDR ≤ 0.00001 vs placebo.
3.2. Unbiased molecular profiling reveals surrogate biomarkers for efficacy
Genome-wide gene expression data were analysed from blood and also inflamed and non-inflamed biopsies to identify novel efficacy biomarkers. A large number of potential efficacy biomarkers at Week 12 were identified [Table 3 and Supplementary Table S3]. Many of the genes that correlate with remission in inflamed biopsies and blood shared biological pathways. Most enriched pathways from both compartments were related to inflammation, tissue remodelling, and cell migration, such as CD28 signalling in T-helper cells, IL-8 signalling, B-cell receptor signalling, and leukocyte extravasation signalling [Supplementary Table S4]. Interestingly, these pathways show reversed patterns of being increased in blood and decreased in inflamed biopsies.
Table 3.
Summary of identified clinical efficacy biomarkers
| Remission | Response | Mucosal healing | ||||
|---|---|---|---|---|---|---|
| N [Yes/No] | Number of hits [FDR < 10%] | N [Yes/No] | Number of hits [FDR < 10%] | N [Yes/No] | Number of hits [FDR < 10%] | |
| Blood RNA | 37/273 | 2766 upregulated and 430 downregulated | 148/162 | 4975 upregulated and 1058 downregulated | 63/247 | 88 upregulated and 219 downregulated |
| Biopsy RNA [inflamed] | 14/96 | 1000 upregulated and 1126 downregulated | 55/55 | 3075 upregulated and 5018 downregulated | 26/84 | 3837 upregulated and 3146 downregulated |
| Biopsy RNA [non-inflamed] | 10/58 | 0 | 40/28 | 0 | 18/50 | 0 |
| Serum protein | 36/280 | 3 downregulated | 146/170 | 8 upregulated and 33 downregulated | 64/252 | 21 downregulated |
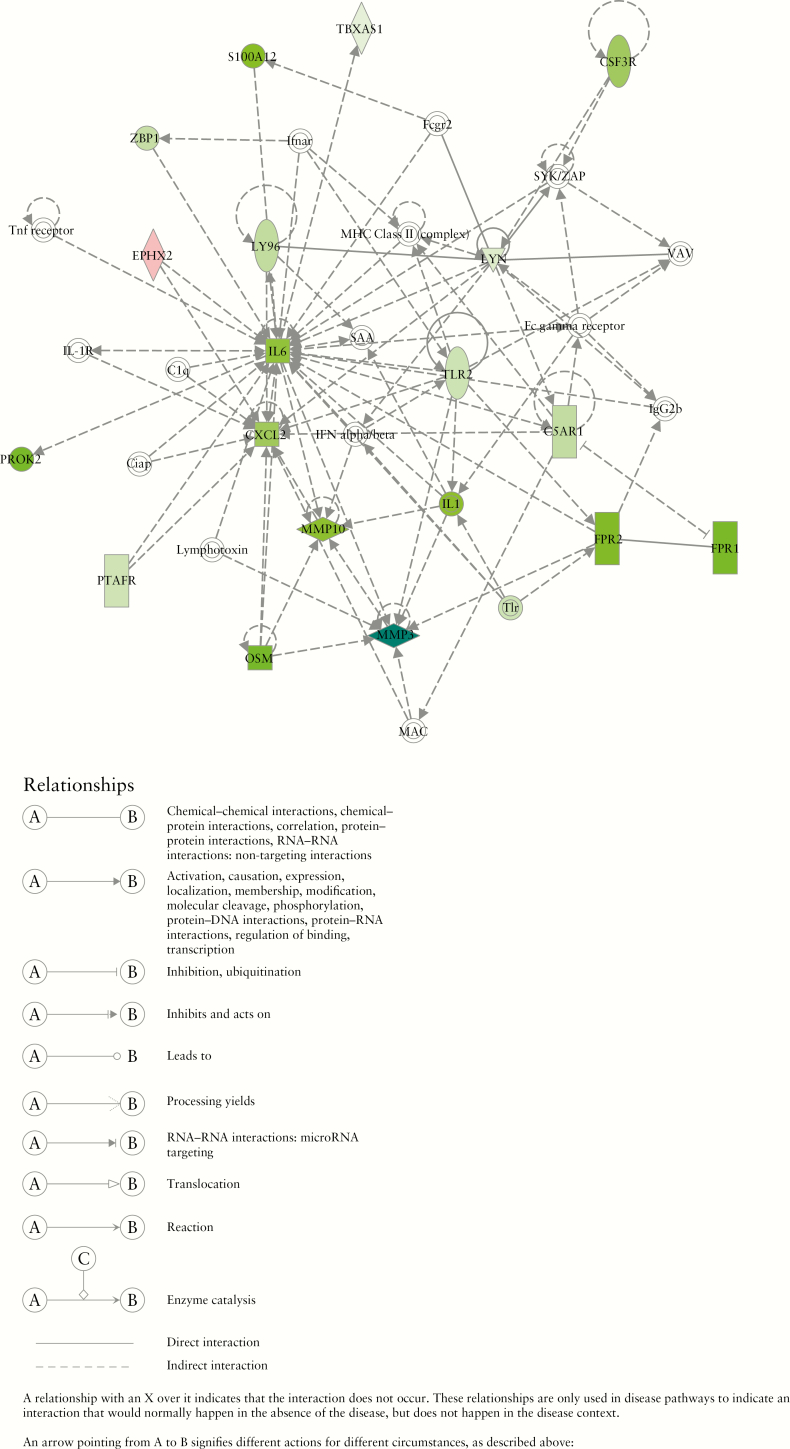
We further examined whether the associations between identified genes and clinical efficacy [remission, response, or mucosal healing] in the inflamed biopsies of the TURANDOT study patients could be replicated in three independent studies [GSE23597, GSE16879, and GSE73661],9,10,11 in which UC patients were treated with the TNF inhibitor infliximab [the first two studies] and vedolizumab [the third study]. As a result, 97 genes [20 upregulated and 77 downregulated] were confirmed to be associated with clinical efficacy in all four datasets after controlling for multiple comparisons using FDR < 0.1. To understand the underlying functions of these genes correlated with clinical efficacy and the roles they may have played in the physiological process and disease progression, we utilized IPA to construct a network by querying the IPA Knowledge Base [IPAKB] annotations for the identified 97 genes and their relationships and interactions [Figure 2]. The IL-6 cytokine family, including IL-6 and OSM, and metalloproteinases, including MMP3 and MMP10, showed reduced expression among patients who demonstrated efficacy, compared with those who did not. Among the 97 genes identified in the inflamed biopsies, MMP10 protein levels in serum were also associated with all three clinical efficacy end points with stronger reduction among the TURANDOT study patients who demonstrated efficacy compared with those who did not; OSM, IL6, and SELE protein levels were associated with two clinical efficacy end points, with stronger reduction among responders or patients who achieved mucosal healing [Supplementary Table S3].
Figure 2.
Network constructed by querying the Ingenuity Pathways Knowledge Base on 97 genes associated with clinical efficacy in the TURANDOT dataset and confirmed in all three public datasets [GSE23597, GSE16879, and GSE73661]. Genes coloured in green indicate greater decrease from baseline among patients who reached remission at 12 weeks compared with those who did not; genes coloured in red indicate opposite associations.
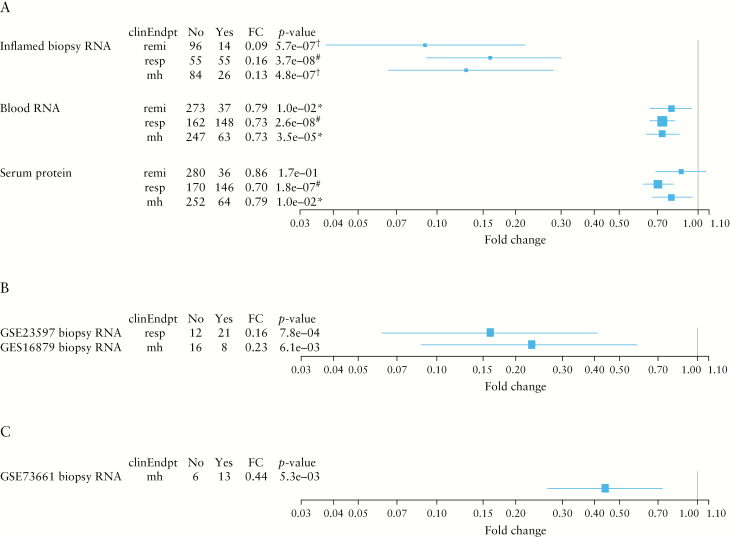
Looking closely at the univariate biomarker analysis, OSM produced the smallest p-value among coding genes in inflamed biopsies. Its strong correlation with patients who achieved remission was marked by an 11-fold stronger downregulation of expression at Week 12 from baseline compared with those who did not achieve remission [p = 5.7e-7, FDR = 0.0050; Figure 3A]. In blood, OSM expression at Week 12 demonstrated a 1.3-fold stronger downregulation from baseline among patients who achieved remission compared with those who did not [p = 0.010, FDR = 0.091]. Furthermore, the decrease in OSM expression also correlated with secondary end points of response and mucosal healing in both inflamed intestinal biopsies and peripheral blood [Figure 3A]: in inflamed biopsies, OSM expression at Week 12 demonstrated a 6.1- and 7.4-fold stronger downregulation from baseline among patients who achieved response [p = 3.7e-8, FDR = 5.0e-6] and mucosal healing [4.8e-7, FDR = 5.6e-5], respectively, compared with those who did not meet these clinical end points; in blood, OSM expression decrease was 1.4-fold greater among patients who achieved response [p = 2.6e-8, FDR = 5.0e-6] and mucosal healing [3.5e-5, FDR = 0.011] [Figure 3A].
Figure 3.
Associations between changes of OSM at Week 12 from baseline [RNA expression in the inflamed biopsies, RNA expression in blood and serum protein concentration], and clinical end points in the TURANDOT Trial [A], the GSE23597 and GSE16879 datasets [B], and the GSE73661 dataset [C]. remi, remission; resp, response; mh, mucosal healing. Blue boxes and lines indicate mean estimates and 95% confidence intervals, respectively. The p values were calculated by comparing those achieving remission vs those not achieving remission, responders vs non-responders, or patients who achieved mucosal healing vs those who did not, in terms of changes of OSM gene expression from baseline. Yes and No indicate in remission [Yes] and not in remission [No], responders [Yes] and non-responders [No], or patients who achieved mucosal healing [Yes] and who did not [No]. *0.01 < FDR ≤ 0.1; †0.00001 < FDR ≤ 0.01; #FDR ≤ 0.00001.
The correlation between OSM gene expression and clinical end points in inflamed intestinal biopsies was also confirmed in three independent studies [GSE23597, GSE16879, and GSE73661],9,10,11 [Figure 3B and C]. In these three studies, patients who achieved response [GSE23579],9 mucosal healing [GSE16879],10 and mucosal healing [GSE73661]11 had 6.3 [p = 0.00078], 4.4 [p = 0.0061], and 2.3 [p = 0.0053] -fold stronger OSM expression downregulation from baseline, which was consistent with the observation in our TURANDOT study data.
Further investigation of OSM protein measurement in serum revealed its correlation with secondary efficacy end points, with 1.4 [p = 1.8e-7, FDR = 7.3e-6] and 1.3 [p = 0.01, FDR = 0.099] -fold stronger downregulation from baseline in responders and patients who achieved mucosal healing as compared with those who did not meet these end points, respectively. OSM protein in serum was not significantly associated with remission, probably due to the small number of patients who achieved remission [36 over 316 patients] and reduced OSM effect in blood, compared with biopsies.
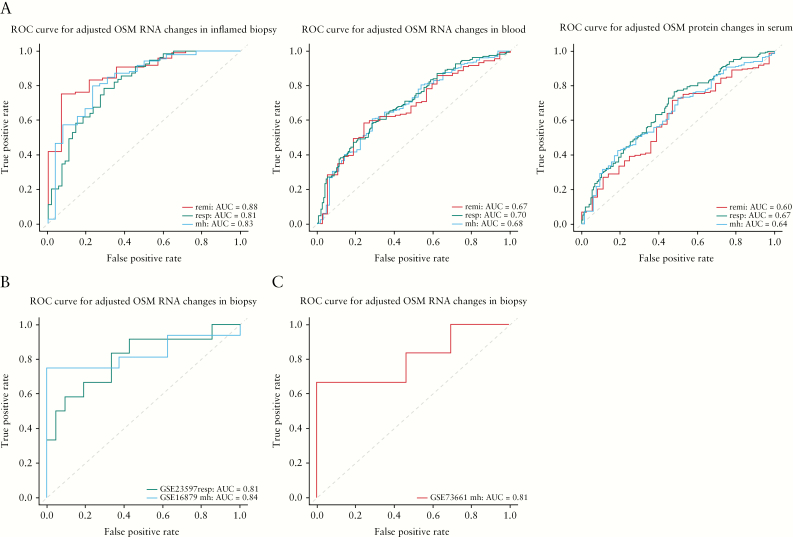
To assess the clinical utility of OSM as a surrogate efficacy biomarker, we further performed ROC analysis of OSM measurements. We found that baseline OSM contributed a large amount of variability to OSM change from baseline measurements. After adjusting for the baseline measurements, OSM changes in inflamed biopsies produced the best c-statistics [area under curve]: 0.88, 0.81, and 0.83 for remission, response, and mucosal healing, respectively, in the TURANDOT data [Figure 4]. Furthermore, the c-statistics were replicated in the GSE23579, GSE16879, and GSE73661 data: 0.81 for infliximab response, 0.84 for infliximab mucosal healing, and 0.81 for vedolizumab mucosal healing [Figure 4].9,10,11
Figure 4.
Receiver operator characteristic analysis of OSM [RNA expression in the inflamed biopsies, RNA expression in blood and serum protein level] changes at Week 12 after adjusting baseline levels, distinguishing patients who achieved clinical efficacy in the TURANDOT Trial [A], the GSE23597 and GSE16879 datasets [B], and the GSE73661 dataset [C]. remi, remission; resp, response; mh, mucosal healing.
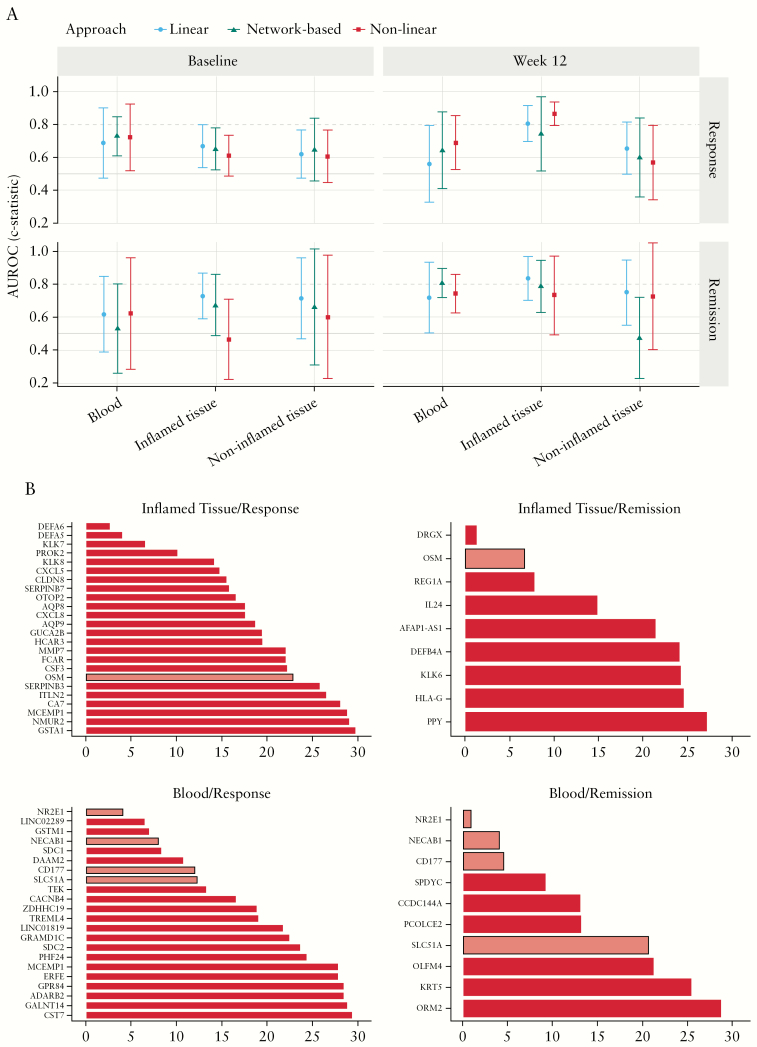
We also applied statistical machine learning models to identify combinations of transcripts to assess clinical response or remission based on Week 12 transcriptomics data. Models assessing clinical response based on inflamed tissue transcriptomics at Week 12 [AUROC of best method = 0.86], clinical remission based on inflamed tissue transcriptomics at Week 12 [AUROC of best method = 0.83], as well as clinical remission based on blood transcriptomics at Week 12 [AUROC of best method = 0.81], showed relevant estimated performance [Figure 5A]. Clinical remission end points generally had higher assessment variability because only a limited number of patients who achieved remission were present in the study. We observed generally consistent performance among our three representative machine learning methods. As outlined in the Methods section, we investigated the contribution of single transcripts to each model with relevant performance [Figure 5B]. Based on peripheral blood transcriptomic data at Week 12, we detected four transcripts that are concordant in assessing clinical response and clinical remission: NR2E1, NECAB1, CD177, and SLC51. Notably, OSM was the only transcript strongly contributing to the assessment of clinical remission as well as clinical response based on inflamed tissue biopsies.
Figure 5.
[A] Comparisons of model performance for three representative machine learning approaches [linear model in blue, network-based model in green, non-linear model in red] in all 12 contrasts of interest. Performance of each model is given as c-statistic or area under the receiver operating curve [AUROC] with 95% confidence intervals. Random performance of 0.5 is depicted by a grey line, and relevant performance is shown by a dashed line at an AUROC of 0.8. Three models for assessing clinical response or clinical remission reach relevant levels of performance [blood, remission; inflamed tissue, remission; inflamed tissue, response]. See text for more details. [B] Transcripts [out of a total of 14 000] contributing most strongly to performance in the linear models assessing clinical remission and clinical response based on whole blood or inflamed tissue biopsies at Week 12. Transcripts are ranked by relative importance as described in the Methods section. Transcripts that are shared between the clinical remission and clinical response contrasts are highlighted in lighter red to indicate their robust role in driving model performance [NR2E1, NECAB1, CD177, and SLC51 for blood and OSM for tissue biopsy data].
In addition to surrogate markers for efficacy, we further looked at baseline biomarkers that could predict efficacy. However, no individual biomarkers at baseline were predictive of subsequent clinical efficacy in the univariate analysis. Similarly, statistical machine learning models were not able to detect predictive combinations of transcripts [Figure 5A]. None of the predictive models based on baseline data met our criterion of relevant performance of >0.8 AUROC, though several models had estimated performance with 95% confidence intervals bounded away from 0.5, indicating non-random performance.
3.3. Frequency of Treg cells correlate with non-monotonic dose response
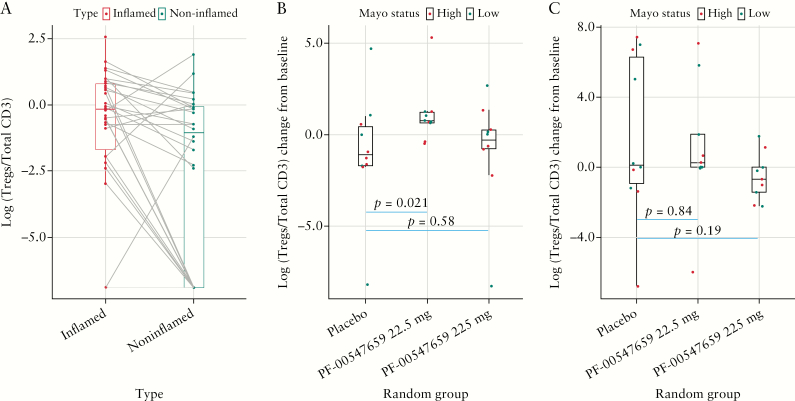
The Phase 2 TURNADOT study demonstrated the greatest efficacy in the intermediate 22.5-mg dose cohort.4 To assess whether the frequency of T regulatory cells correlates with this observed non-monotonic dose response, triple-label IHC was performed on inflamed and non-inflamed intestinal biopsies at baseline and Week 12 in the placebo, 22.5-mg, and 225-mg dose groups. Computer image analysis was used to enumerate Tregs [cells expressing CD3, CD25, and FoxP3], and their frequency within the total T-lymphocyte population in the lamina propria of intestinal biopsies. Based on this analysis, Tregs demonstrated a difference between inflamed and non-inflamed intestinal biopsies at baseline [Figure 6A]. The frequency of Treg cells was 4.6-fold higher in inflamed versus non-inflamed intestinal biopsies in the same subject at baseline [p = 0.0061]. In the inflamed intestinal biopsies, the change in Tregs from baseline was 5.4-fold higher in the 22.5-mg group compared with the placebo group at Week 12 [p = 0.021], whereas there was no significant difference between the change of Tregs in the 225-mg versus placebo groups [p = 0.58] [Figure 6B]. In the non-inflamed intestinal biopsies, there was no statistically significant difference in the change in CD3+/CD25+/FoxP3+ cells between the placebo and 22.5-mg [p = 0.84] or placebo and 225-mg groups [p = 0.19] at Week 12 [Figure 6C].
Figure 6.
Treg [CD3+/CD25+/FoxP3+] differences [A] at baseline between inflamed and non-inflamed biopsies; [B] between high and low MAYO score patients across treatment groups in the inflamed biopsies; [C] between high and low MAYO score patients across treatment groups in the non-inflamed biopsies.
4. Discussion
The recent Phase 2 TURANDOT study assessed the efficacy of MAdCAM blockade through the use of PF-00547659 in subjects with moderate to severe active UC.4 Interestingly, maximal clinical efficacy was observed in the intermediate 22.5-mg dose cohort. To further inform the clinical development of PF-00547659, we interrogated the intestinal biopsies and peripheral blood to define drug and disease biomarkers.
We have identified a large number of genes whose changes from baseline were significantly different from placebo in blood; however, we did not find any significant protein changes. The gene expression results could not be translated into protein. This might be because we examined a small portion of the proteins [202], compared with whole-genome RNA sequencing. Also, the levels of RNA expression can represent different cell types and be used to track their movements, but protein levels might not be sensitive enough. We show in the present study that gene expression of CCR9 in blood is a PD biomarker for lymphocytes entering the intestine that is dose dependent on inhibition of MAdCAM. Other studies have shown that CCR9 and the receptor for MAdCAM, α4β7, are expressed on lymphocytes. These cells target the intestine through CCR9–CCL25 and β7–MAdCAM-1 interactions, respectively. CCL25 and MAdCAM-1 are constitutively expressed on intestinal capillary venules and recruit CCR9+ and β7+ lymphocytes from the circulation.16 Our study has shown that blood CCR9 gene expression increases with increasing doses of PF-00547659 and strongly correlates with the number of circulatory α4β7 immune cells in subjects with moderate to severe active Crohn’s disease after PF-00547659 treatment.17 In the present study, CCR9 gene expression in blood also correlates with increasing doses of PF-00547659, and we further show an inverse correlation between CCR9 gene expression in peripheral blood and inflamed intestine biopsies, which is consistent with the proposed mechanism of PF-00547659 action through blockade of immune cell migration to inflamed intestinal parenchyma. An assessment of peripheral blood CCR9 gene expression is far more amenable than intestinal biopsies or even an assessment of peripheral blood α4β7 immune cell numbers. Therefore, further studies into the use of peripheral blood CCR9 gene expression as a potential PD biomarker for therapies that block immune trafficking is warranted.
A large number of genes are differentially expressed with respect to clinical remission or clinical response based on Week 12 data in both blood and inflamed intestinal biopsies. These genes fall into a number of pathways, of which the majority are inversely modulated in peripheral blood and inflamed intestinal biopsies. For instance, leukocyte extravasation signalling in inflamed biopsies is one of the top pathways enriched with genes that decreased more from baseline in patients who achieved remission compared with patients who did not achieve remission; however, in blood, the pattern is reversed and shows greater increases in those genes. The inflammation changes driven by inhibition of immune cell migration explain these inverse effects on the pathway. Machine learning–based modelling demonstrates that these differences in expression at Week 12 are robust enough to assess clinical response and clinical remission reasonably well based on transcriptomics alone [c-statistic of ~0.8]. These results can form the basis for future assessment tools that would have to incorporate appropriate penalties for the misclassification of responders and non-responders to arrive at concrete decision rules. Transcripts that strongly drive model performance differ between blood and inflamed tissue biopsies. This may indicate differences in cellular populations between these compartments and may reflect differential change following blockade of MAdCAM. The four genes that consistently contribute highly to model performance in blood represent different biological functions of signalling (transcription factor [NR2E1] and calcium-binding protein [NECAB1]), solute transporter for bile acids and dietary lipid adsorption [SLC 51], and a neutrophil activation marker involved in transmigration of neutrophils [CD177]. Some of the markers, namely NECAB1 and CD177, have been previously observed to be differentially expressed in the blood of children with severe UC resistant to corticosteroid therapy.18 The signatures from inflamed intestinal biopsies were more robust and reinforce the notion that samples from the active site of disease are crucial for biomarker studies in UC. The one transcript that is consistently driving model performance based on inflamed biopsy transcriptomics is the OSM gene.
In addition to a change in biopsy gene expression, OSM gene expression in blood and protein concentration in serum all correlate with multiple clinical end points of remission, response, or mucosal healing. The unbiased discovery of OSM in this study, and the confirmation of its implication in IBD disease pathology in other IBD gene expression profiling datasets independent of therapy make it a promising surrogate biomarker for clinical efficacy independent of the mechanism of action of the underlying therapeutic agent.
OSM is a member of the IL-6 cytokine family [IL-6, IL-11, LIF, CNTF, CT-1, CLC, IL-31], and signals through gp130/OSMRβ and gp130/LIFR. It is mainly produced by activated T cells, monocytes, neutrophils, and dendritic cells.19 Unique to OSM compared with other IL-6 family members is that it is expressed as a 26 kDa MW full-length protein that can be processed by proteolytic cleavage of 13 or 18 C-terminal amino acids by a trypsin-like protease.20 The longer full-length form binds to extracellular matrix [ECM] proteins and localizes this soluble protein to the extracellular domains of epithelial, endothelial, and stromal cells and may be enriched at the site of inflammatory infiltration in inflamed colon biopsies. OSM has pleiotropic effects in different cell types, which include regulation of cell growth and differentiation, acute-phase reaction, haematopoiesis, bone remodelling, and homeostasis of the ECM.21
Overproduction of OSM promotes disease pathologies, including skin and lung inflammation, atherosclerosis, and cancer.22 A single-nucleotide polymorphism [SNP] in the OSM locus has been reported to be strongly associated with risk of IBD development.23 However, the molecular mechanism of OSM in IBD has remained unclear. It was demonstrated that OSM promotes inflammatory activity in the intestinal stroma and that high OSM expression in intestinal biopsies is reproducibly associated with a high risk of resistance to anti-TNF therapy.24 However, in the current study, baseline OSM expression is not associated with any of the efficacy end points. This could be explained by the different mechanisms of TNF and MAdCAM inhibitors, where OSM synergizes with TNF to induce inflammatory signals in stromal cells in vitro, while MAdCAM may not function synergistically with OSM.23 This also suggests that PF-00547659 is equally effective regardless of patients’ baseline OSM expression and may provide a good treatment option for anti-TNF therapy failure patients.
In the current study, gene expression at baseline does not strongly correlate with efficacy of PF-00547659, either as single genes or a combination of baseline gene transcripts as a predictive signature. This negative result might be due to a lack of power to detect subtle signals, the heterogenous nature of the disease, lack of biologic-naïve patients in the study, or a confounding effect by the mixture of cell populations present in each sample. We expect future single-cell–based studies to provide further insights into the potential for predictive models based on transcriptomics data.
In the Phase 2 TURANDOT study, PF-00547659 treatment at the intermediate dose of 22.5 mg resulted in maximal efficacy.4 In the current study, we show that the relative ratio of T-regulatory to total T-cells is maximally increased in inflamed intestine after treatment with the intermediate 22.5-mg PF-00547659 dose. This finding could stem from a greater inhibition of T-effector relative to T-regulatory cell migration, resulting in the establishment of a local immunosuppressive milieu and eventual disease resolution. However, additional mechanistic studies are required to substantiate this hypothesis, which are beyond the scope of this current manuscript. If this does hold true, one could envision that at the highest 225-mg PF-00547659 dose, the antibody may be in greater excess, resulting in blockade of both T-effector and T-regulatory cells and failure to establish a local immunosuppressive milieu. These findings may suggest that to maximize the effectiveness of this therapeutic class, one must consider the effects on both immune-effector and -regulatory cells at a particular drug dose. At this time, it is unclear whether this optimal dosage would need to be individualized for a particular patient, stage of disease, concomitant medications, and so on. Furthermore, it is also unclear whether this ‘window’ of an optimal dosage will be applicable to other classes of drugs, because immune effectors and regulatory cells are often regulated by similar biochemical pathways.
In summary, our biomarker discovery effort has revealed biomarkers for UC and for the action of anti-integrin therapies. The results of these studies may have profound implications for further clinical development of PF-00547659.
Funding
This work was supported by Pfizer.
Conflict of Interest
HZ, LX, DZ, SO, JL, ZS, YZ [Zhan], SZ, YZ [Zhang], KP, AH, MM, BZ, KG, LF, VP, FC, MV, DvS, KH, and MHZ are employees of Pfizer or were employed by Pfizer at the time this study began.
Supplementary Material
Acknowledgments
The authors wish to thank the patients who participated in the trial and all the investigators and medical staff of all participating study centres. Editorial assistance in preparing the manuscript for submission was provided by Paul Hassan, PhD, of Engage Scientific Solutions, Horsham, UK, and was funded by Pfizer.
Glossary
Abbreviations:
- AUROC
area under the receiver operating curve
- CCR9
C-C motif chemokine receptor 9
- CXCR4
motif chemokine receptor 4
- DAB
diaminobenzidine
- ECM
extracellular matrix
- FDR
false discovery rate
- FFPE
formalin-fixed, paraffin-embedded
- IBD
inflammatory bowel disease
- IHC
immunohistochemistry
- IL
interleukin
- IPA
Ingenuity Pathway Analysis
- IPAKB
IPA Knowledge Base
- LOD
limit of detection
- MAdCAM
mucosal addressin cell adhesion molecule
- NaN
not a number
- NPX
normalized protein expression
- OSM
oncostatin M
- PD
pharmacodynamic
- PK
pharmacokinetic
- RIN
RNA Integrity Number
- ROC
receiver operating characteristic
- s.c.
subcutaneous
- SNP
single-nucleotide polymorphism
- TNF
tumour necrosis factor
- UC
ulcerative colitis
Author Contributions
HZ developed the analysis plan and performed a significant amount of the data analysis and overall drafting of the manuscript; LX and DZ contributed to pathway and machine learning analysis and drafting the related sections; SO, JL, ZS, and YZ performed some specific experiments; YZ and KP were involved in sample handling; AH and MM were involved in performing some specific machine learning analysis; SZ and BZ implemented the RNA-sequencing pipeline and quality control; KJG, LF, VP, FC, MV, DvS, and KH were involved in providing clinical and biological interpretation of the data and results; MHZ initiated, designed, and supervised the study, and was responsible for the overall content of the manuscript. All authors reviewed and revised the manuscript for intellectual content and approved the final version for submission.
References
- 1. Podolsky DK. Inflammatory bowel disease. N Engl J Med 2002;347:417–29. [DOI] [PubMed] [Google Scholar]
- 2. Duijvestein M, Battat R, Vande Casteele N, et al. . Novel therapies and treatment strategies for patients with inflammatory bowel disease. Curr Treat Options Gastroenterol 2018;16:129–46. [DOI] [PubMed] [Google Scholar]
- 3. Pullen N, Molloy E, Carter D, et al. . Pharmacological characterization of PF-00547659, an anti-human MAdCAM monoclonal antibody. Br J Pharmacol 2009;157:281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vermeire S, Sandborn WJ, Danese S, et al. . Anti-MAdCAM antibody [PF-00547659] for ulcerative colitis [TURANDOT]: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet 2017;390:135–44. [DOI] [PubMed] [Google Scholar]
- 5. Gottlieb K, Travis S, Feagan B, Hussain F, Sandborn WJ, Rutgeerts P. Central reading of endoscopy endpoints in inflammatory bowel disease trials. Inflamm Bowel Dis 2015;21:2475–82. [DOI] [PubMed] [Google Scholar]
- 6. Dobin A, Davis CA, Schlesinger F, et al. . STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Assarsson E, Lundberg M, Holmquist G, et al. . Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One 2014;9:e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Toedter G, Li K, Marano C, et al. . Gene expression profiling and response signatures associated with differential responses to infliximab treatment in ulcerative colitis. Am J Gastroenterol 2011;106:1272–80. [DOI] [PubMed] [Google Scholar]
- 10. Arijs I, De Hertogh G, Lemaire K, et al. . Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One 2009;4:e7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arijs I, De Hertogh G, Lemmens B, et al. . Effect of vedolizumab (anti-α4β7-integrin) therapy on histological healing and mucosal gene expression in patients with UC. Gut 2018;67:43–52. [DOI] [PubMed] [Google Scholar]
- 12. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 2003;100:9440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Series B Stat Methodol 2005;67:301–20. [Google Scholar]
- 14. Pedregosa F, Varoquaux G, Gramfort A, et al. . Scikit-learn: machine learning in python. J Mach Learn Res 2011;12:2825–30. [Google Scholar]
- 15. Zarringhalam K, Degras D, Brockel C, Ziemek D. Robust phenotype prediction from gene expression data using differential shrinkage of co-regulated genes. Sci Rep 2018;8:1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gorfu G, Rivera-Nieves J, Ley K. Role of beta7 integrins in intestinal lymphocyte homing and retention. Curr Mol Med 2009;9:836–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hassan-Zahraee M, Banerjee A, Cheng JB, et al. . Anti-MAdCAM antibody increases ß7+ T cells and CCR9 gene expression in the peripheral blood of patients with Crohn’s disease. J Crohns Colitis 2018;12:77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kabakchiev B, Turner D, Hyams J, et al. . Gene expression changes associated with resistance to intravenous corticosteroid therapy in children with severe ulcerative colitis. PLoS One 2010;5:e13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boniface K, Diveu C, Morel F, et al. . Oncostatin M secreted by skin infiltrating T lymphocytes is a potent keratinocyte activator involved in skin inflammation. J Immunol 2007;178:4615–22. [DOI] [PubMed] [Google Scholar]
- 20. Linsley PS, Bolton-Hanson M, Horn D, et al. . Identification and characterization of cellular receptors for the growth regulator, oncostatin M. J Biol Chem 1989;264:4282–9. [PubMed] [Google Scholar]
- 21. Radtke S, Hermanns HM, Haan C, et al. . Novel role of Janus kinase 1 in the regulation of oncostatin M receptor surface expression. J Biol Chem 2002;277:11297–305. [DOI] [PubMed] [Google Scholar]
- 22. Hermanns HM. Oncostatin M and interleukin-31: cytokines, receptors, signal transduction and physiology. Cytokine Growth Factor Rev 2015;26:545–58. [DOI] [PubMed] [Google Scholar]
- 23. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium [IIBDGC] Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. West NR, Hegazy AN, Owens BMJ, et al. ; Oxford IBD Cohort Investigators Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor–neutralizing therapy in patients with inflammatory bowel disease. Nat Med 2017;23:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.