Abstract
Long noncoding RNAs (lncRNAs) are involved in a variety of biological processes such as tumor proliferation and metastasis. A close relationship between hepatitis B virus X protein (HBx) and SMYD3 in promoting the proliferation and metastasis of hepatocellular carcinoma (HCC) was recently reported. However, the exact oncogenic mechanism of HBx-SMYD3 remains unknown. In this study, by performing lncRNA microarray analysis, we identified a novel lncRNA that was regulated by both HBx and SMYD3, and we named it lncIHS (lncRNA intersection between HBx microarray and SMYD3 microarray). lncIHS was overexpressed in HCC and decreased the survival rate of HCC patients. Knockdown of lncIHS inhibited HCC cell migration, invasion, and proliferation, and vice versa. Further study showed that lncIHS positively regulated the expression of epithelial mesenchymal transition (EMT)-related markers c-Myc and Cyclin D1, as well as the activation of the ERK- and AKT-signaling pathways. lncIHS exerted its oncogenic effect through ERK and AKT signaling. Moreover, results from transcriptome-sequencing analysis and mass spectrometry showed that lncIHS regulated multiple genes that were the upstream molecules of the ERK- and AKT-signaling pathways. Therefore, our findings suggest a regulatory network of ERK and AKT signaling through lncIHS, which is downstream of HBx-SMYD3, and they indicate that lncIHS may be a potential target for treating HCC.
Keywords: hepatocellular carcinoma, SMYD3, lncRNA-HIS, DUSP5, DUSP10, MAP3K8, ERK, AKT, metastasis, proliferation
Introduction
Hepatocellular carcinoma (HCC) is the third major cause of cancer mortality, and the global morbidity of the disease is increasing year by year.1 Among the therapeutic methods of HCC, surgical resection is the main treatment option, but about 80% of patients are preliminarily diagnosed with advanced stage and lose the operative chance.2, 3 Besides, the rate of postoperative recurrence and metastasis of HCC is high.4 Therefore, understanding the occurrence and development mechanism of HCC may provide new strategies for the diagnosis and treatment of this life-threatening disease.
Hepatitis B virus X protein (HBx) is a multifunctional regulator during HCC occurrence and development.5 Our previous studies have confirmed that HBx plays a key role in the molecular pathogenesis of hepatitis B virus (HBV)-HCC.6 On the other hand, our data and recent studies have indicated that SMYD3, an important methyltransferase in malignant tumor, is greatly associated with liver carcinogenesis and poor prognosis of HCC patients.7, 8, 9 Interestingly, HBx has been shown to not only upregulate SMYD3 expression in HCC but also interact with SMYD3 through a specific domain.10, 11 However, the exact mechanism of how HBx-SMYD3 promotes HCC occurrence and development remains unknown.
Long noncoding RNA (lncRNA) is a kind of RNA molecule that is over 200 nt, without a protein-encoding function.12 Studies have shown that lncRNAs can affect multiple biological processes of malignant tumor, such as cell proliferation and metastasis, and play an important role in the occurrence and development of HCC.13 Interestingly, HBx was recently reported to upregulate lncRNAs by recruiting EZH2, another important histone methyltransferase in malignant tumor.14 We hypothesize that HBx-SMYD3 may also promote HCC progression via lncRNAs.
In this study, by conducting microarray using HBx- and SMYD3-overexpressing HCC cells, we gained a novel lncRNA (LOC285740) and named it lncIHS (lncRNA intersection between HBx microarray and SMYD3 microarray). The role of lncIHS in HCC was then studied, which may provide a basis for clinical diagnosis and treatment of HCC.
Results
A Novel lncRNA Was Selected by Microarray
To explore lncRNAs regulated by both HBx and SMYD3, we first established HBx- and SMYD3-overexpressing HCC cells. We chose BEL-7402, Huh7, and SK-HEP-1 to establish SMYD3-overexpressing cell lines and MHCC-97H to establish the SMYD3 stable knockdown cell line (Figures S1A and S1B). SK-HEP-1 and HepG2 without HBV genome were chosen to establish HBx-overexpressing cell lines (Figure S1B). We confirmed the regulation of SMYD3 by HBx (Figure S1B).
Next, we used HBx- and SMYD3-overexpressing HCC cells to screen out lncRNAs that are regulated by both HBx and SMYD3 in HCC, and we found that five lncRNAs were upregulated by HBx-SMYD3, whereas three lncRNAs were downregulated by HBx-SMYD3 (Figure 1A). Considering the major function of SMYD3 is to trimethylate H3K4 and subsequently activate target gene expression,15 we chose lncRNAs upregulated by HBx-SMYD3 for the next investigation (Figure 1A). We first detected the expression of the five lncRNAs in HCC with tumor thrombus, HCC with non-tumor thrombus, and normal liver. We found that, compared with the other four lncRNAs, the expression of LOC285740 among the three groups was the most significant (Figure 1B), implying that LOC285740 was associated with HCC metastasis, which is the major oncogenic function of SMYD3 and HBx.8, 16 We therefore chose lncRNA LOC285740 for further investigation. Results from HBx-overexpressing, SMYD3-overexpressing, and SMYD3-knockdown HCC cells further confirmed that LOC285740 was positively regulated by HBx and SMYD3 (Figures 1C and S1C). Furthermore, knockdown of SMYD3 significantly reduced HBx-dependent expression of LOC285740 (Figure 1D). Because LOC285740 was initially screened out by both HBx microarray and SMYD3 microarray, we named LOC285740 as lncIHS (intersection between HBx microarray and SMYD3 microarray).
Figure 1.
A Novel lncRNA Is Selected by Microarray
(A) lncRNAs were screened out by microarray using HBx- and SMYD3-overexpresseing HCC cell lines. (B) The expression of lncIHS in HCC with tumor thrombus was significantly more than others. (C) lncIHS was positively regulated by HBx and SMYD3. (D) HBx promoted lncIHS expression through SMYD3. Data are presented as mean ± SD for three independent experiments. *p < 0.05.
lncIHS Is Overexpressed in HCC Tissues and Correlates with Patient Clinicopathological Features and Unfavorable Survival
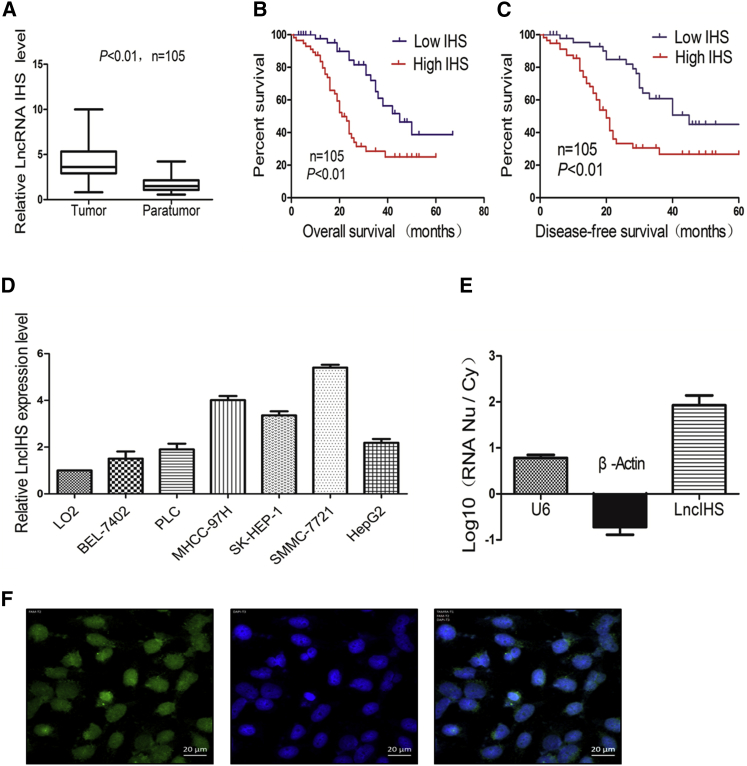
To explore the significance of lncIHS in HCC, we investigated its expression in HCC tissues. We observed that lncIHS expression was significantly increased in HCC tissues in comparison with para-tumor tissues (Figure 2A). High lncIHS expression in HCC was significantly associated with HBV infection, large tumor size, portal vein invasion, and high TNM stage (Table 1). Furthermore, HCC patients with high lncIHS expression had shorter overall and disease-free survival (Figures 2B and 2C). In addition, lncIHS expression was higher in HCC cell lines than normal liver cell line (Figure 2D). These observations indicated that lncIHS may act as an oncogene in HCC. The complete sequence of lncIHS was then identified by rapid amplification of cDNA ends (RACE) assay, and it is shown in Figure S2A. We used three different prediction tools, CPC2, coding potential assessment tool (CPAT), and PORTRAIT, to calculate the coding potential of lncIHS. lncIHS was classified as a noncoding sequence by the three prediction tools (coding probability, CPC2, 14%; CPAT, 17%; and PORTRAIT, 5.78%). In vitro translation analysis further confirmed the noncoding potential of lncIHS (Figure S2B).
Figure 2.
lncIHS Is Overexpressed in HCC and Correlates with Unfavorable Survival
(A) Compared with para-tumor tissues, the expression of lncRNA was significantly increased in HCC tissues. (B and C) Kaplan-Meier analysis of overall survival (B) and disease-free survival (C) based on lncIHS expression levels in 105 cases of patients with HCC. The median level of lncIHS is used as the cutoff. (D) The expression of lncRNA was detected in a normal liver cell line and different HCC cell lines. (E) The levels of lncIHS mRNA in purified nuclear or cytoplasmic RNAs were detected using real-time PCR. U6 and β-actin served as nuclear and cytoplasmic controls, respectively. (F) lncIHS was located in the cytoplasm and nucleus, but mainly in the nucleus, as determined by FISH assays. Data are presented as mean ± SD for three independent experiments. *p < 0.05, **p < 0.01.
Table 1.
Correlation between Expression of lncIHS and Clinicopathological Parameters in HCC Patients
| Characteristics |
lncIHS |
||||
|---|---|---|---|---|---|
| N = 105 | Low Expression | High Expression | χ2 | p Value | |
| Age (Years) | 0.06 | 0.801 | |||
| <59 | 50 | 17 | 33 | ||
| ≥59 | 55 | 20 | 35 | ||
| Gender | 1.53 | 0.216 | |||
| Male | 51 | 21 | 30 | ||
| Female | 54 | 16 | 38 | ||
| AFP | 2.6 | 0.105 | |||
| ≤25 μg/mL | 54 | 23 | 31 | ||
| >25 μg/mL | 51 | 14 | 37 | ||
| HBV Infection | 9.11 | 0.002* | |||
| (+) | 55 | 12 | 43 | ||
| (−) | 50 | 25 | 25 | ||
| Tumor Encapsulation | 3.46 | 0.063 | |||
| Present | 64 | 27 | 37 | ||
| Absent | 41 | 10 | 31 | ||
| Tumor Size (cm) | 5.51 | 0.018* | |||
| ≤5 | 49 | 23 | 26 | ||
| >5 | 56 | 14 | 42 | ||
| Tumor Stage | 9.34 | 0.002* | |||
| I, II stage | 47 | 24 | 23 | ||
| III, IV stage | 58 | 13 | 45 | ||
| Portal Vein Thrombus | 8.95 | 0.003* | |||
| Present | 52 | 11 | 41 | ||
| Absent | 53 | 26 | 27 | ||
*p < 0.05.
lncIHS Is Located in the Nucleus of HCC Cells
To further investigate the oncogenic function of lncIHS in HCC, we first detected the location of lncIHS in HCC cell lines. RNA was extracted respectively from the cytoplasm and nucleus of SMMC-7721 for detecting the expression level of lncIHS. By comparing with U6 and β-actin, lncIHS was determined to be located mainly in the nucleus (Figure 2E). Our fluorescence in situ hybridization (FISH) assays further showed that lncIHS was located in the cytoplasm and nucleus, but mainly in the nucleus (Figure 2F). These data suggested that lncIHS may exert its function mainly in the nucleus.
lncIHS Promotes Cell Migration, Invasion, and Proliferation In Vitro and In Vivo
The exact function of lncIHS in HCC cells is unknown. We therefore performed gain- and loss-of-function study. For loss-of-function study, we chose SMMC-7721 and MHCC-97H to establish lncIHS stable knockdown cell lines (Figure 3A; Figure S3A). Because the above results showed significantly high expression of lncIHS in HCC with tumor thrombus, we first focused on the association between lncIHS and the migration or invasive ability of HCC cells. Migration and invasion assays showed that knockdown of lncIHS significantly suppressed cell migration and invasion (Figure 3B; Figure S3B). We also measured the effect of lncIHS on the proliferation of HCC cells. Results from the MTT assay and colony formation assay showed that lncIHS knockdown significantly decreased the proliferation of SMMC-7721 and MHCC-97H (Figures 3C and 3D; Figures S3C and S3D). Furthermore, knockdown of lncIHS hindered cell cycle transition from G1 to S stage (Figure 3E; Figure S3E). We confirmed these results using transient transfection in both cell lines (Figures S4 and S5).
Figure 3.
Knockdown of lncIHS Inhibits SMMC-7721 Proliferation, Migration, and Invasion In Vitro
(A) lncIHS knockdown was confirmed by real-time PCR in SMMC-7721. (B) Knockdown of lncIHS suppressed the migration and invasion abilities of SMMC-7721, as determined by transwell assay. (C and D) In vitro proliferative ability was decreased in SMMC-7721 with lncIHS knockdown, as determined by MTT assay (C) and colony formation assay (D). (E) Knockdown of lncIHS inhibited cell cycle transition from G1 to S in HCC cells. Data are presented as mean ± SD for three independent experiments. *p < 0.05.
To further confirm the above in vitro results, in vivo experiments were performed. lncIHS knockdown decreased xenografted tumor growth, tumor weight, and tumor size in vivo (Figures 4A–4C). In addition, results from Ki67 staining of xenograft tumor tissues indicated that tumors derived from lncIHS-silenced cells showed significantly reduced proliferation (Figures 4D and 4E). In vivo effects of lncIHS on HCC metastasis were also detected. The incidence of lung metastasis in the lncIHS-knockdown group was significantly less than that in the control group (Figure 4F). These results indicated that lncIHS is required for HCC cell migration, invasion, and proliferation.
Figure 4.
Knockdown of lncIHS Inhibits Tumor Growth and Lung Metastasis of HCC Cells in Nude Mice
(A) SMMC-7721 cells (5 × 106) with stable knockdown of lncIHS were inoculated into nude mice, and the effect of lncIHS on HCC tumor growth was examined after 6 weeks (n = 5). A photograph of the tumors is presented. (B and C) Effect of lncIHS on HCC growth was described by tumor weight (B) and tumor size (C) in the two groups. (D) Representative images of immunohistochemistry (IHC) staining of Ki67 showed that lncIHS inhibition decreased tumor proliferation in xenografted tumors. (E) The expression of lncRNA in the tumor xenografts of the sh-lncIHS group was significantly lower than in the sh-NC group. (F) Representative H&E staining of lung metastases between SMMC-7721 sh-lncIHS cells and control cells. Data are presented as mean ± SD for three independent experiments. *p < 0.05, **p < 0.01.
Next, we questioned whether overexpression of lncIHS could strengthen the ability of migration, invasion, and proliferation of HCC cells. BEL-7402 and PLC/PRF/5 cells were transiently transfected with a vector containing the complete sequence of lncIHS (Figure 5A). Consistent with the results from lncIHS-knockdown cell lines, lncIHS overexpression significantly enhanced the migration, invasion, and proliferation capacities of BEL-7402 and PLC/PRF/5 cells (Figures 5B–5E). These results were also confirmed in stable lncIHS-overexpressing cells (Figures S6A–S6C). Taken together, these results suggested that lncIHS had an important role in promoting HCC cell migration, invasion, and proliferation.
Figure 5.
Overexpression of lncIHS Promotes HCC Cell Migration, Invasion, and Proliferation
(A) Overexpression of lncIHS was confirmed by PCR. (B and C) Overexpression of lncIHS promoted the migration and invasion abilities of HCC cells, as determined by wound-healing (B) and transwell assays (C). (D) In vitro proliferative ability of HCC cells was significantly promoted by lncIHS by MTT assay. (E) lncIHS overexpression stimulated cell cycle transition from G1 to S in HCC cells. Data are presented as mean ± SD for three independent experiments. *p < 0.05, **p < 0.01.
lncIHS Regulates the Expressions of the EMT and Cell Cycle-Associated Genes and the Activation of ERK- and AKT/GSK-3β-Signaling Pathways
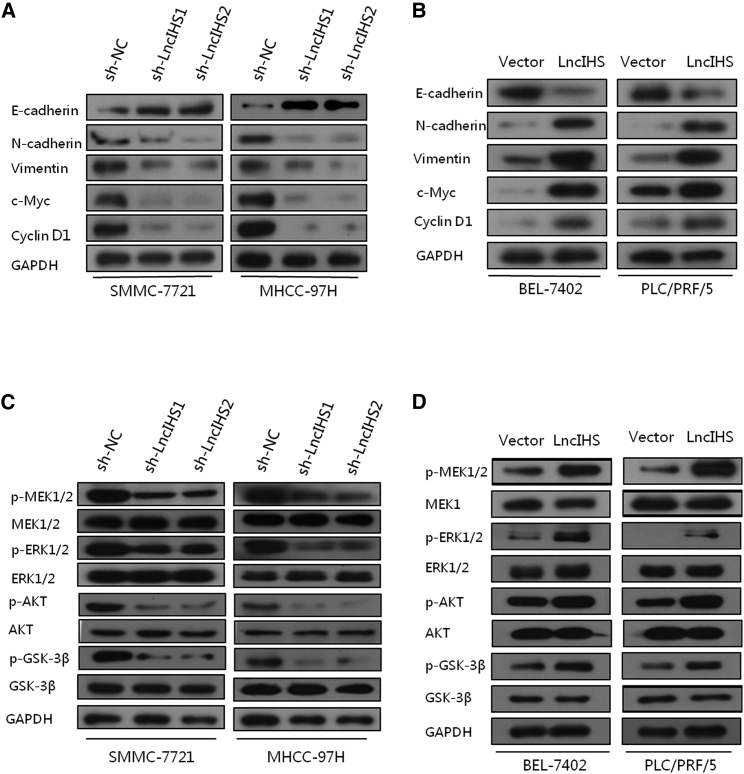
As our above results showed that lncIHS significantly promoted HCC cell growth, migration, and invasion, we sought to determine whether lncIHS regulated the expression of the epithelial mesenchymal transition (EMT) and cell cycle-associated genes in HCC cells. Knockdown of lncIHS decreased the expression of c-Myc, Cyclin D1, as well as mesenchymal markers N-cadherin and Vimentin, and it increased the expression of epithelial marker E-cadherin in SMMC-7721 and MHCC-97H (Figure 6A; Figure S7A). In contrast, overexpression of lncIHS increased the expression of c-Myc, Cyclin D1, N-cadherin, and Vimentin, and it deceased E-cadherin expression (Figure 6B; Figure S7B).
Figure 6.
lncIHS Regulates the Expression of the EMT and Cell Cycle-Associated Genes and the Activation of the ERK- and AKT/GSK-3β-Signaling Pathways
(A and B) Knockdown of lncIHS reduced the expression of c-Myc, Cyclin D1, N-cadherin, and Vimentin, and it increased the level of E-cadherin in SMMC-7721 and MHCC-97H (A), and vice versa in BEL-7402 and PLC/PRF/5 (B). (C and D) Knockdown of lncIHS decreased the expression of p-MEK1/2, p-ERK1/2, p-AKT, and p-GSK-3β in SMMC-7721 and MHCC-97H (C), and vice versa in BEL-7402 and PLC/PRF/5 (D).
AKT- and ERK-signaling pathways have been well known as the regulators of cell cycles and the EMT.17, 18, 19 We detected the effect of lncIHS on PI3K/AKT- and ERK-signaling pathways. Upon knockdown of lncIHS, the expressions of phosphorylated MEK1/2, ERK1/2, AKT, and GSK-3β were downregulated, whereas total MEK1/2, ERK1/2, AKT, and GSK-3β was kept unchanged (Figure 6C; Figure S7C). On the contrary, lncIHS overexpression enhanced the phosphorylation of MEK1/2, ERK1/2, AKT, and GSK-3β (Figure 6D; Figure S7D). These results indicated that lncIHS may promote HCC cell migration, invasion, and proliferation through the activation of ERK and AKT signaling and their downstream molecules, such as c-Myc, Cyclin D1, and EMT-associated genes.
lncIHS Promotes HCC Cell Migration, Invasion, and Proliferation through ERK- and AKT-Signaling Pathways
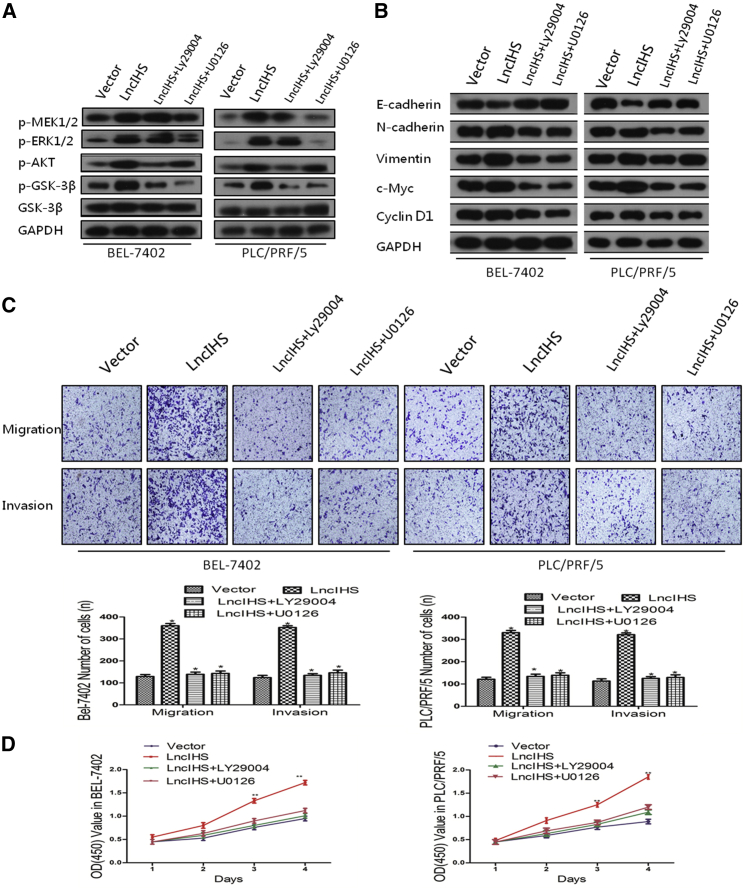
PI3K inhibitor Ly294002 and MEK1/2 inhibitor U0126 were applied to study the role of ERK and AKT signaling in lncIHS-mediated migration, invasion, and proliferation of HCC. The effects of Ly294002 and U0126 on the phosphorylation of AKT, GSK-3β, MEK1/2, and ERK1/2 were validated (Figure 7A; Figure S8A). Inactivation of ERK and AKT signaling in lncIHS-overexpressing cells not only reversed lncIHS-dependent decrease in E-cadherin expression and lncIHS-dependent increases in N-cadherin, Vimentin, c-Myc, and Cyclin D1 expression but also reduced lncIHS-enhanced cell migration, invasion, and proliferation (Figures 7B–7D; Figure S8B). To further verify the effects of ERK and AKT signaling on mediating the oncogenic function of lncIHS, we performed tumor xenograft experiments using stable lncIHS-overexpressing cell lines with or without LY294002 and U0126 treatment. ERK- and AKT-signaling inhibition significantly suppressed the tumor growth induced by lncIHS overexpression (Figures S6D–S6F). These data suggested that lncIHS promoted HCC cell migration, invasion, and proliferation through the MEK/ERK- and AKT/GSK-3β-signaling pathways.
Figure 7.
lncIHS Promotes HCC Cell Migration, Invasion, and Proliferation through the ERK- and AKT-Signaling Pathways
(A) The effects of Ly294002 and U0126 on the phosphorylation of AKT, GSK-3β, MEK1/2, and ERK1/2 were validated. (B) Treatments with Ly294002 and U0126 in HCC cells reversed the lncIHS-dependent decrease of E-cadherin expression and the lncIHS-dependent increase of N-cadherin, Vimentin, c-Myc, and Cyclin D1 expressions. (C and D) Inactivation of ERK and AKT signaling significantly reduced lncIHS-enhanced cell migration, invasion, and proliferation, as determined by transwell assay (C) and MTT assay (D). Data are presented as mean ± SD for three independent experiments. *p < 0.05, **p < 0.01.
Next-Generation Sequencing Reveals a lncIHS-Dependent Regulatory Network of ERK- and AKT-Signaling Pathways
Our above results showed that lncIHS positively regulated ERK- and AKT-signaling pathways. However, our above findings also indicated that lncIHS was mainly located in the nucleus of HCC cells, which suggested that lncIHS did not directly regulate ERK and AKT signaling. We hypothesized that lncIHS could regulate genes upstream of the ERK and AKT pathways on the mRNA level. We therefore performed next-generation sequencing (NGS) using SMMC-7721 sh-lncIHS and sh-NC cells to screen out genes that are regulated by lncIHS on the mRNA level. We constructed Gene Ontology (GO) trees and pathway-act-network based on the differentially expressed genes between the two groups.
Consistent with our above findings, results of the GO trees showed there were many genes regulated by lncIHS enriched in mitogen-activated protein kinase (MAPK) signaling, cell proliferation, and cell migration (Figure S9). Pathway-act-network revealed that the MAPK-signaling pathway and PI3K/AKT-signaling pathway were core nodes in the lncIHS-associated pathway network (Figure S10). Genes previously reported to be the upstream molecules of the ERK or AKT pathway were then selected and validated by real-time PCR in both lncIHS-knockdown and -overexpression HCC cells (Figures 8A and 8B).
Figure 8.
NGS Reveals a lncIHS-Dependent Regulatory Network of the ERK- and AKT-Signaling Pathways
(A) NGS screened out several genes that were upregulated or downregulated by lncIHS. (B) The lncIHS-related regulation of genes that may be the upstream molecules of the AKT- and ERK-signaling pathways was validated by real-time PCR. (C and D) Knockdown of lncIHS decreased MAP3K8 expression and increased DUSP5 and DUSP10 expressions in SMMC-7721 and MHCC-97H (C), and vice versa in BEL-7402 and PLC/PRF/5 (D). Data are presented as mean ± SD for three independent experiments. *p < 0.05.
We paid special attention to mitogen-activated protein kinase kinase kinase 8 (MAP3K8) and dual specificity phosphatase 5 and 10 (DUSP5 and DUSP10), which are kinase or phosphatase and have been well known as the upstream regulators of ERK- or AKT-signaling pathways.20, 21, 22, 23 The positive regulation of MAP3K8 and negative regulation of DUSP5/DUSP10 by lncIHS were further validated by western blot (Figures 8C and 8D; Figures S11A and S11B). We also observed a positive correlation between lncIHS and MAP3K8, as well as a negative correlation between lncIHS and DUSP5 or DUSP10 in HCC tissues (Figure S12). Furthermore, we employed lncRNA-WEE2-AS1, which we recently reported could promote HCC invasion through activating the PI3K/AKT/GSK-3β-signaling pathway,24 to see whether MAP3K8, DUSP5, and DUSP10 were regulated by lncRNA-WEE2-AS1. Overexpression of lncRNA-WEE2-AS1 did not affect the expressions of MAP3K8, DUSP5, and DUSP10 (Figure S13), suggesting the functional specificity of lncIHS. Taken together, our data suggested a complex regulation mechanism of the ERK- and AKT-signaling pathways by lncIHS in HCC.
lncIHS Regulated MAP3K8, DUSP5, and DUSP10 through Interacting with YBX1
To further identify the mechanisms underlying lncIHS-induced ERK and AKT pathway activation, we first examined whether lncIHS affected the mRNA expression levels of its cis genes. lncIHS is located on human chromosome 6q24.2, in antisense of the intron of a protein-coding gene PHACTR2 (Figure S14A). We found the expression of PHACTR2 was not regulated by lncIHS (Figures S14B and S14C). Besides, the expressions of other neighboring genes, such as FUCA2, LTV1, ADAT2, and PEX2, were also not regulated by lncIHS (Figures S14A–S14C). These results implied that lncIHS does not function in cis but in trans to activate the ERK and AKT pathways.
We then performed RNA pull-down assay using a biotin-labeled lncIHS RNA probe to identify potential binding proteins of lncIHS in HCC. The proteins pulled down by lncIHS were then analyzed by mass spectrometry. Among the proteins identified by mass spectrometry, we paid special attention to YBX1 (Figure 9A). YBX1 is a nuclear and cytoplasmic protein that can bind DNA and RNA by recognizing special nucleic acid sequences.25, 26 YBX1 has been demonstrated to serve as a transcriptional activator as well as mRNA stabilizer in cancer.27, 28 We further confirmed the interaction of lncIHS with YBX1 by RNA pull-down and RNA immunoprecipitation (RIP) assays (Figures 9B and 9C).
Figure 9.
lncIHS Interacts with YBX1 in HCC Cells
(A) YBX1 was a potential interactive candidate of lncIHS, as determined by RNA pull-down assay, and the enriched products were eluted and separated by SDS-PAGE and Coomassie blue staining. (B) Western blot of the proteins from antisense lncIHS and lncIHS pull-down assays. (C) The interactions of lncIHS with YBX1 were verified by an RIP assay. (D–F) The mRNA expressions of MAP3K8 (D), DUSP5 (E), and DUSP10 (F) were detected in BEL-7402-lncIHS and its control cells with or without YBX1 knockdown. (G–I) RNA stability assays were performed in BEL-7402-lncIHS and its control cells with or without YBX1 knockdown, and the degradation rates of the MAP3K8 (G), DUSP5 (H), and DUSP10 (I) mRNA were measured at the indicated time points. (J and K) YBX1 potential binding sites on the MAP3K8 gene promoter (J) and the DUSP5/DUSP10 3′ UTR (K) are shown. (L) YBX1 was mainly located in the nucleus in lncIHS-overexpressing cells. Data are presented as mean ± SD for three independent experiments. *p < 0.05, **p < 0.01.
Next, we questioned whether YBX1 mediates the lncIHS-dependent regulation of MAP3K8, DUSP5, and DUSP10. Results from real-time PCR showed that knockdown of YBX1 decreased the expressions of MAP3K8, DUSP5, and DUSP10 in HCC cells without lncIHS overexpression (Figures 9D–9F). On the other hand, in cells with lncIHS overexpression, YBX1 knockdown more significantly decreased MAP3K8 expression, but not DUSP5 and DUSP10 (Figures 9D–9F). We then employed actinomycin D to inhibit gene transcription. Consistent with the real-time PCR results, DUSP5 and DUSP10 mRNA half-lives were significantly decreased in cells either with lncIHS overexpression or with YBX1 konckdown (Figures 9H and 9I). Interestingly, the MAP3K8 mRNA half-life was influenced by neither lncIHS overexpression nor YBX1 knockdown (Figure 9G).
Considering the nuclear localization of lncIHS and the molecular function of YBX1, we hypothesized that lncIHS could bind and sequester YBX1 in the nucleus, facilitating the recruitment of YBX1 on the promoter of the MAP3K8 gene, leading to transcriptional activation of MAP3K8. Meanwhile, the cytoplasmic expression of YBX1 was downregulated, leading to the instability of DUSP5/DUSP10 mRNA. Consistently, we found many YBX1 potential binding sites on the MAP3K8 gene promoter region and DUSP5/DUSP10 3′ UTR (Figures 9J and 9K) and that overexpression of lncIHS resulted in YBX1 nuclear accumulation (Figure 9L). Taken together, our results suggested that lncIHS may upregulate MAP3K8 and downregulate DUSP5 and DUSP10 through its binding protein YBX1 in HCC.
Discussion
HCC is one of the most common malignant tumors worldwide. In China, nearly 80% of HCC cases are attributable to HBV.29 HBx, one of the central viral proteins produced by HBV, is now well established to be involved in hepatocarcinogenesis due to pleiotropic effects on several signaling pathways regulating cell death, proliferation, differentiation, invasion, and metastasis; oxidative stress; and DNA repair.5, 30 Recently, many lncRNAs (e.g., Dreh31 and HOTAIR32) have been reported to be associated with HBx in HCC, suggesting lncRNAs play an important role in the oncogenic process of HBx. SMYD3 is a methyltransferase, which mainly leads to trimethylation of the lysine 4 of histone H3 (H3K4), makes the spatial structure of chromosomes in an open state, and finally promotes target gene transcription.8, 33 SMYD3 is highly expressed in a variety of malignant tumors, such as HCC,34 leukemia,35 bladder cancer,36 and glioma.37 The overexpression of SMYD3 in malignant cells is associated with the proliferation, anti-apoptosis, autophagy, migration, and invasion abilities of tumor cells.38, 39
Given the potent roles of HBx and SMYD3 in HCC, it is important to enrich the oncogenic mechanism of both proteins. Interestingly, studies have shown that the expression of SMYD3 is regulated by HBx in HCC cells.10 Moreover, recent studies also indicate that HBx interacts with SMYD3 through a specific domain and the interaction between HBx and SMYD3 activates downstream signaling through AP-1.11 These studies suggest that HBx-SMYD3 is an important pathway in promoting tumorigenesis and progression of HCC. However, the downstream mechanism of HBx-SMYD3 in HCC needs to be further elucidated.
Many studies have shown that lncRNAs are closely associated with the occurrence, development, invasion, and metastasis of human malignant tumors.12, 13 However, there are few studies focusing on lncRNAs regulated by both HBx and SMYD3 in HCC. By conducting lncRNA microarray screening on HCC cell models overexpressing HBx or SMYD3 and subsequent validation in HCC cell lines, we found a novel lncRNA that was upregulated by HBx-induced overexpression of SMYD3, named it lncIHS, and studied its role in HCC.
It is well known that PI3K/AKT and MEK/ERK signaling are the major signaling pathways in the tumorigenesis and tumor progression of HCC.40 Numerous researchers have demonstrated that the activation of the PI3K/AKT- and ERK-signaling pathways is an important cause of EMT20 and cell cycle transition of G1/S in cancer.18 In this study, we showed that lncIHS could activate PI3K/AKT and ERK signaling. Moreover, by employing the inhibitors Ly294002 and U0126, we found that the activation of AKT and ERK was required for the lncIHS-dependent enhancement of HCC cell proliferation, migration, and invasion, as well as the EMT and cell cycle-related protein expressions, indicating that lncIHS is an important oncogenic noncoding RNA in HCC.
Regulation of the MEK/ERK and PI3K/AKT pathways is mediated by a series of kinases, phosphatases, and other factors.41 The synergistic alteration of these kinases or phosphatases will finally result in the pathway activation or inactivation. Based on this background knowledge, we applied NGS to screen out genes that are regulated by lncIHS on the mRNA level and, meanwhile, are the upstream regulators of the AKT- and ERK-signaling pathways. We found many genes that have been reported to be associated with the AKT- and ERK-signaling pathways in the NGS results. Among them, MAP3K8, DUSP5, and DUSP10 have been well studied as the upstream molecules of the ERK- and AKT-signaling pathways. MAP3K8 is a member of the serine or threonine protein kinase family. MAP3K8 controls several signaling pathways, including MEK/ERK and PI3K/AKT, and it functions as an oncogene in many human cancers.42 In contrast, DUSP5 and DUSP10, both of which are members of the DUSP family, mainly contribute to the dephosphorylation of ERK.23, 43 Both DUSP5 and DUSP10 serve as tumor suppressors in human cancers.23, 44 Hence, our results indicated that lncIHS positively regulates the AKT- and ERK-signaling pathways through multiple mechanisms.
lncRNAs function in a wide range of biological processes, and they regulate gene expression in cis or in trans via various mechanisms. Actually, many recent studies have reported a series of lncRNAs that function in trans, but not in cis.45, 46 Consistently, our data showed the funtions of lncIHS may be independent of its neighboring genes but dependent upon its binding protein, YBX1. YBX1 is not only a DNA-binding protein that recognizes the Y-box sequence in the nucleus but also a major component of cytoplasmic messenger ribonucleoprotein particles (mRNPs), with roles in pre-mRNA splicing and mRNA stability.47 Thus, different localization of YBX1 in cells exhibits different functions.
In this study, we found that YBX1 may be a functional partner of lncIHS according to the following findings: (1) lncIHS and YBX1 could bind to each other; (2) knockdown of YBX1 under the condition of low lncIHS expression only slightly influenced MAP3K8 mRNA expression, but it significantly downregulated DUSP5 and DUSP10 mRNA stability and expression; in contrast, under the condition of high lncIHS expression, knockdown of YBX1 significantly decreased MAP3K8 expression, but it had no effect on DUSP5 and DUSP10 mRNA stability and expression; (3) YBX1 did not regulate MAP3K8 mRNA stability; (4) many YBX1 potentail binding sites were found on the MAP3K8 gene promoter and DUSP5/DUSP10 3′ UTRs; and (5) overexpression of lncIHS accumulated YBX1 in the nucleus. All these clues lead to the hypothesis that lncIHS could bind and sequester YBX1 in the nucleus, thus leading to transcriptional activation of MAP3K8 and the instability of DUSP5/DUSP10 mRNA. These reasons could partly explain how lncIHS regulates ERK and AKT/GSK-3β signaling in HCC. However, further study is needed to illustrate a more detail mechanism.
In conclusion, our study reported a novel oncogenic lncRNA, lncIHS, which is positively regulated by HBx-induced overexpression of SMYD3 in HCC. The expression of lncIHS is upregulated in HCC tissues and is markedly associated with a poor prognosis in HCC patients. lncIHS promotes HCC cell proliferation and metastasis by activating the AKT/GSK-3β- and ERK-signaling pathways. More importantly, lncIHS, together with its binding protein YBX1, regulates ERK and AKT/GSK3-β signaling through multiple mechanisms, forming a regulatory network. Thus, lncIHS may be a candidate biomarker for HCC prognosis and a potential therapeutic target, especially in HBV-associated HCC with positive SMYD3 expression.
Materials and Methods
Cell Culture and Reagents
Human hepatocyte LO2 and HCC cell lines SMMC-7721, MHCC-97H, BEL-7402, PLC/PRF/5, SK-HEP-1, and HepG2 were purchased from the Chinese Academy of Sciences Cell Bank Type Culture Collection. The cells were cultured with DMEM and RPMI-1640 (Gibco, Carlsbad, CA) together with 10% fetal bovine serum (Gibco) at 37°C with a humidity of 5% CO2. PI3K inhibitor (LY294002) and ERK1/2 inhibitor (U0126) were purchased from Selleck Chemicals.
Patient Sample
105 fresh HCC tissues and para-tumor tissues were obtained from HCC patients at the time of 2010–2012 after surgery from Sun Yat-Sen Memorial Hospital. This study was conducted in accordance with the Declaration of Helsinki. The study protocol was approved by the research ethics committee of Sun Yat-Sen Memorial Hospital, and written informed consent was obtained from each patient. The diagnosis of HCC was based on pathology and dynamic contrast-enhanced imaging (computed tomography scan or MRI).
cDNA Preparation and Real-Time PCR
Total RNA was extracted from HCC cell lines, 105 fresh HCC tissues, and the para-tumor tissues with Trizol (Takara, China), according to the manufacturer’s instructions. Reverse transcription was performed using Prime Script RTase (Takara, China), according to the manufacturer’s protocol. The expression levels of mRNA were determined with real-time PCR using SYBR green (Takara, China), according to the manufacturer’s instructions. The primers used for real-time PCR are given in Table S1.
The Sequence of lncIHS
5′-RACE and 3′-RACE were performed using SMART RACE cDNA amplification kit (Clontech Laboratories, USA), according to the manufacturer’s instructions.
Establishment of Stable Knockdown and Overexpression Cell Lines
The HepG2 and SK-HEP-1 cell lines were used to construct HBx-overexpressing cells. The BEL-7402 cell line was used to construct SMYD3-overexpressing cells. The MHCC-97H cell line was used to construct SMYD3-knockdown cells. HEK293T packaging cells were transfected with the appropriate retroviral construct. Culture supernatants were collected at 48 h after transfection, and we infected the target cells. Cells were then selected with puromycin for 2 weeks. The target for SMYD3 small hairpin RNA (shRNA) was 5′-AGCCTGATTGAAGATTTGATTCT-3′.
lncIHS shRNA vectors were purchased from Obio Technology (Shanghai, China). The lentivirus of lncIHS shRNA was produced from HEK293T packaging cells and infected HCC cells (SMMC-7721 and MHCC-97H), as mentioned above. The targets for lncIHS shRNA were sh-IHS1, 5′-CCATCCTGTGGCTATGATT-3′; and sh-IHS2, 5′-GCACGAAACATCTCTCCAA-3′.
The full lengths of lncIHS sequences were sub-cloned into pEX-2 plasmid, and the recombined plasmid was referred to as pEX-IHS. pEX-IHS and pEX-2 were, respectively, transfected into BEL-7402 cells using the Lipofectamine 3000 reagent, according to the manufacturer’s instructions. Stably transfected cells were obtained after selection in culture medium containing 0.5 mg/mL G418 (Invitrogen, USA) for approximately 6 weeks.
Nuclear and Cytoplasmic RNA Isolation
HCC cells were prepared in a 150-mm plate, centrifuged, and the supernatant was discarded. RNA subcelluar isolation kit was purchased from Active Motif (Shanghai, 25501). Following the assay protocol, cytoplasmic RNA and nuclear RNA were extracted separately. The expression levels of RNA were determined by real-time PCR using SYBR green.
FISH Assays
4% paraformaldehyde was added to an SMCC-7721 cell sample; then the fixed cell was placed on the glass slide, immersed in ethanol solution, and dried. Further, 10 μL hybridization buffer and 1 μL probes were added to the sample on the sheet glass at the 46°C incubator. After 1.5 h of hybridization, the sheet glass was quickly put into the 48°C washing buffer for 30 min. Then the sheet glass was rinsed with ultrapure water and dried. The sample was covered with the proper amount of anti-fading reagent and the cover glass, and then it was placed under the confocal laser-scanning microscope for observation.
MTT Assays
2,000 HCC cells were added to each of the 5 wells of the 96-well plates. Then the cells were routinely cultured for 1–5 days according to the experimental time. At each time point, MTT solution was added to each well and incubated for 4 h at 37°C. DMSO was then used to dissolve the purple crystal substances. The absorbance at 570 nm was measured with a microplate reader, and the growth curve was drawn and analyzed.
Colony Formation Assay
1,000 HCC cells were placed in six-well plates. The cells were cultured with growth medium and 10% fetal bovine serum (FBS) for 2 weeks. The cells were fixed and stained with paraformaldehyde and crystal violet, respectively.
Cell Cycle Analysis
HCC cells were gathered, washed with PBS twice, fixed in 75% ice-cold alcohol, and then incubated with PBS containing propidium iodide (PI, 10 μg/mL) and RNase A (0.5 mg/mL). The cells were analyzed with a flow cytometer.
Wound-Healing Assay
HCC cells were cultured in six-well plates with 5% CO2 at 37°C. When the cells were full, three parallel scratch wounds were made across each well using a P-100 pipette tip. The cells were continuously incubated with fresh growth medium without FBS. Wound closure was observed at 0 and 48 h under a microscope.
Transwell Migration and Invasion Assays
For the migration assay, 1 × 105 cells in medium without FBS were placed on a polycarbonate membrane insert in a transwell apparatus (Corning, USA). After 24-h incubation, the bottom cells were removed with a cotton swab. The cells on the lower side of the chamber were analyzed under a microscope. The process for the invasion assay was the same as the migration assay, except that the transwell membranes were pre-treated with 24 mg/mL Matrigel (R&D Systems, USA).
In Vivo Experiments
The cells were placed in 10-cm-diameter plates, collected, and re-suspended with PBS. A total of 5 × 106 cells was subcutaneously injected into the right shoulder of BALB/c nude mice. Tumor growth was monitored with tumor weight and tumor volume, which was calculated using the formula V = W2 × L × 0.5. Meanwhile, 1 × 106 cells were injected into the caudal vein of nude mice. After 6 weeks, the mice were euthanized, tumors were excised and weighed, and the lung tissues were collected for H&E staining. To further explore the potential role of inhibitors in HCC progress, a dose of 10 mg/kg body weight U0126 or 75 mg/kg body weight LY294002 was injected intraperitoneally into mice 1 week after inoculation, three times per week for 4 weeks. The animal experiments of this study were approved by the animal ethics committee of Sun Yat-sen University.
Western Blot
All cells were lysed by cell lysis buffer (p0013 RIPA lysis, Beyotime). The BCA method was used to detect protein concentration. The proteins were separated by the SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, USA). Then, the PVDF membranes were incubated at 4°C overnight with the following primary antibodies: AKT, p-AKT, GSK-3β, p-GSK-3β, ERK, p-ERK, E-cadherin, Vimentin, N-cadherin (Cell Signaling Technology, 1:1,000), MAP3K8, DUSP5, DUSP10 (ABclone, 1:500), c-Myc, Cyclin D1 (Santa Cruz Biotechnology, 1:500), and GAPDH (Proteintech, 1:500). Then PVDF membranes were incubated with the appropriate secondary antibody (Santa Cruz Biotechnology, 1:10,000) for 1 h at room temperature. Exposure was detected by the chemiluminescence solution.
RNA Stability Assay
To analyze RNA stability, cells were treated with actinomycin D (1 μg/mL). Cells were collected at different time points, and the RNA was extracted using Trizol reagent and detected by real-time PCR.
RNA Pull-Down Assay
Cell nuclear lysates were incubated with biotinylated full-length lncIHS and streptavidin beads for RNA pull-down incubation. Then beads were collected by centrifugation. RNA-associated proteins were eluted and resolved by SDS-PAGE, followed by silver staining.
RIP
RIP assays were performed using an RNA-binding protein immunoprecipitation kit (Millipore), according to the manufacturer’s instructions. RIP products were analyzed by real-time PCR using total RNA as input controls.
Statistical Analysis
All data analysis was performed with SPSS software (version 18.0). The experimental data are represented by mean ± SD. Kaplan-Meier analysis and log rank test were used to determine cumulative survival. Student’s t test and Fisher’s exact test were used to analyze quantitative data and categorical data, respectively. p < 0.05 was considered statistically significant.
Author Contributions
C.H., Z.Z., and Z.X. conceived and designed the study. Z.C., W.Y., and Q.Z. carried out the experiments. H.J. and W.Y. collected and analyzed the clinical data. Z.C. and Z.Z. wrote the manuscript. D.H., J.W., and J.Z. revised the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors thank Xu Chen, Yongcong Yan, Kaishun Hu, and Yu Li for technical assistance. We apologize to the colleagues whose work was not cited due to space constraints. This work was supported by the National Natural Science Foundation of China (81672401, 81672405, 81572407, 81672403, and 81301768); the Key project of Natural Science Foundation of Guangdong Province, China (grant 4210016041); the project of Natural Science Foundation of Guangdong Province, China (grant 2018A030313809); the Science and Technology Program of Guangzhou, China (grant 4250016043); a grant from the Guangdong Science and Technology Department (2015B050501004); grant [2013] 163 from the Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology; and grant KLB09001 from the Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.04.021.
Contributor Information
Zhenyu Zhou, Email: zhouzhy26@mail2.sysu.edu.cn.
Chuanchao He, Email: hechch5@mail.sysu.edu.cn.
Zhiyu Xiao, Email: xzysurgeon@163.com.
Supplemental Information
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]; Torre, L.A., Bray, F., Siegel, R.L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87-108. [DOI] [PubMed]
- 2.Bellissimo F., Pinzone M.R., Cacopardo B., Nunnari G. Diagnostic and therapeutic management of hepatocellular carcinoma. World J. Gastroenterol. 2015;21:12003–12021. doi: 10.3748/wjg.v21.i42.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bellissimo, F., Pinzone, M.R., Cacopardo, B., and Nunnari, G. (2015). Diagnostic and therapeutic management of hepatocellular carcinoma. World J. Gastroenterol. 21, 12003-12021. [DOI] [PMC free article] [PubMed]
- 3.Yang L.Y., Fang F., Ou D.P., Wu W., Zeng Z.J., Wu F. Solitary large hepatocellular carcinoma: a specific subtype of hepatocellular carcinoma with good outcome after hepatic resection. Ann. Surg. 2009;249:118–123. doi: 10.1097/SLA.0b013e3181904988. [DOI] [PubMed] [Google Scholar]; Yang, L.Y., Fang, F., Ou, D.P., Wu, W., Zeng, Z.J., and Wu, F. (2009). Solitary large hepatocellular carcinoma: a specific subtype of hepatocellular carcinoma with good outcome after hepatic resection. Ann. Surg. 249, 118-123. [DOI] [PubMed]
- 4.Xia L., Huang W., Tian D., Zhu H., Qi X., Chen Z., Zhang Y., Hu H., Fan D., Nie Y., Wu K. Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology. 2013;57:610–624. doi: 10.1002/hep.26029. [DOI] [PubMed] [Google Scholar]; Xia, L., Huang, W., Tian, D., Zhu, H., Qi, X., Chen, Z., Zhang, Y., Hu, H., Fan, D., Nie, Y., and Wu, K. (2013). Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology 57, 610-624. [DOI] [PubMed]
- 5.Ringelhan M., O’Connor T., Protzer U., Heikenwalder M. The direct and indirect roles of HBV in liver cancer: prospective markers for HCC screening and potential therapeutic targets. J. Pathol. 2015;235:355–367. doi: 10.1002/path.4434. [DOI] [PubMed] [Google Scholar]; Ringelhan, M., O’Connor, T., Protzer, U., and Heikenwalder, M. (2015). The direct and indirect roles of HBV in liver cancer: prospective markers for HCC screening and potential therapeutic targets. J. Pathol. 235, 355-367. [DOI] [PubMed]
- 6.Huang P., Zhuang B., Zhang H., Yan H., Xiao Z., Li W., Zhang J., Tang Q., Hu K., Koeffler H.P. Hepatitis B Virus X Protein (HBx) Is Responsible for Resistance to Targeted Therapies in Hepatocellular Carcinoma: Ex Vivo Culture Evidence. Clin. Cancer Res. 2015;21:4420–4430. doi: 10.1158/1078-0432.CCR-14-2067. [DOI] [PubMed] [Google Scholar]; Huang, P., Zhuang, B., Zhang, H., Yan, H., Xiao, Z., Li, W., Zhang, J., Tang, Q., Hu, K., Koeffler, H.P., et al. (2015). Hepatitis B Virus X Protein (HBx) Is Responsible for Resistance to Targeted Therapies in Hepatocellular Carcinoma: Ex Vivo Culture Evidence. Clin. Cancer Res. 21, 4420-4430. [DOI] [PubMed]
- 7.He C., Xu J., Zhang J., Xie D., Ye H., Xiao Z., Cai M., Xu K., Zeng Y., Li H., Wang J. High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Hum. Pathol. 2012;43:1425–1435. doi: 10.1016/j.humpath.2011.11.003. [DOI] [PubMed] [Google Scholar]; He, C., Xu, J., Zhang, J., Xie, D., Ye, H., Xiao, Z., Cai, M., Xu, K., Zeng, Y., Li, H., and Wang, J. (2012). High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Hum. Pathol. 43, 1425-1435. [DOI] [PubMed]
- 8.Sarris M.E., Moulos P., Haroniti A., Giakountis A., Talianidis I. Smyd3 Is a Transcriptional Potentiator of Multiple Cancer-Promoting Genes and Required for Liver and Colon Cancer Development. Cancer Cell. 2016;29:354–366. doi: 10.1016/j.ccell.2016.01.013. [DOI] [PubMed] [Google Scholar]; Sarris, M.E., Moulos, P., Haroniti, A., Giakountis, A., and Talianidis, I. (2016). Smyd3 Is a Transcriptional Potentiator of Multiple Cancer-Promoting Genes and Required for Liver and Colon Cancer Development. Cancer Cell 29, 354-366. [DOI] [PubMed]
- 9.Zhou Z., Jiang H., Tu K., Yu W., Zhang J., Hu Z., Zhang H., Hao D., Huang P., Wang J. ANKHD1 is required for SMYD3 to promote tumor metastasis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019;38:18. doi: 10.1186/s13046-018-1011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhou, Z., Jiang, H., Tu, K., Yu, W., Zhang, J., Hu, Z., Zhang, H., Hao, D., Huang, P., Wang, J., et al. (2019). ANKHD1 is required for SMYD3 to promote tumor metastasis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 38, 18. [DOI] [PMC free article] [PubMed]
- 10.Yang L., He J., Chen L., Wang G. Hepatitis B virus X protein upregulates expression of SMYD3 and C-MYC in HepG2 cells. Med. Oncol. 2009;26:445–451. doi: 10.1007/s12032-008-9144-1. [DOI] [PubMed] [Google Scholar]; Yang, L., He, J., Chen, L., and Wang, G. (2009). Hepatitis B virus X protein upregulates expression of SMYD3 and C-MYC in HepG2 cells. Med. Oncol. 26, 445-451. [DOI] [PubMed]
- 11.Hayashi M., Deng L., Chen M., Gan X., Shinozaki K., Shoji I., Hotta H. Interaction of the hepatitis B virus X protein with the lysine methyltransferase SET and MYND domain-containing 3 induces activator protein 1 activation. Microbiol. Immunol. 2016;60:17–25. doi: 10.1111/1348-0421.12345. [DOI] [PubMed] [Google Scholar]; Hayashi, M., Deng, L., Chen, M., Gan, X., Shinozaki, K., Shoji, I., and Hotta, H. (2016). Interaction of the hepatitis B virus X protein with the lysine methyltransferase SET and MYND domain-containing 3 induces activator protein 1 activation. Microbiol. Immunol. 60, 17-25. [DOI] [PubMed]
- 12.Tsai M.C., Spitale R.C., Chang H.Y. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsai, M.C., Spitale, R.C., and Chang, H.Y. (2011). Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 71, 3-7. [DOI] [PMC free article] [PubMed]
- 13.Mattick J.S., Rinn J.L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]; Mattick, J.S., and Rinn, J.L. (2015). Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22, 5-7. [DOI] [PubMed]
- 14.Hu J.J., Song W., Zhang S.D., Shen X.H., Qiu X.M., Wu H.Z., Gong P.H., Lu S., Zhao Z.J., He M.L., Fan H. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci. Rep. 2016;6:23521. doi: 10.1038/srep23521. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hu, J.J., Song, W., Zhang, S.D., Shen, X.H., Qiu, X.M., Wu, H.Z., Gong, P.H., Lu, S., Zhao, Z.J., He, M.L., and Fan, H. (2016). HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci. Rep. 6, 23521. [DOI] [PMC free article] [PubMed]
- 15.Hamamoto R., Furukawa Y., Morita M., Iimura Y., Silva F.P., Li M., Yagyu R., Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]; Hamamoto, R., Furukawa, Y., Morita, M., Iimura, Y., Silva, F.P., Li, M., Yagyu, R., and Nakamura, Y. (2004). SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 6, 731-740. [DOI] [PubMed]
- 16.Hsieh A., Kim H.S., Lim S.O., Yu D.Y., Jung G. Hepatitis B viral X protein interacts with tumor suppressor adenomatous polyposis coli to activate Wnt/β-catenin signaling. Cancer Lett. 2011;300:162–172. doi: 10.1016/j.canlet.2010.09.018. [DOI] [PubMed] [Google Scholar]; Hsieh, A., Kim, H.S., Lim, S.O., Yu, D.Y., and Jung, G. (2011). Hepatitis B viral X protein interacts with tumor suppressor adenomatous polyposis coli to activate Wnt/β-catenin signaling. Cancer Lett. 300, 162-172. [DOI] [PubMed]
- 17.Sherr C.J., Beach D., Shapiro G.I. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov. 2016;6:353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sherr, C.J., Beach, D., and Shapiro, G.I. (2016). Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov. 6, 353-367. [DOI] [PMC free article] [PubMed]
- 18.Mahe M., Dufour F., Neyret-Kahn H., Moreno-Vega A., Beraud C., Shi M., Hamaidi I., Sanchez-Quiles V., Krucker C., Dorland-Galliot M. An FGFR3/MYC positive feedback loop provides new opportunities for targeted therapies in bladder cancers. EMBO Mol. Med. 2018;10:e8163. doi: 10.15252/emmm.201708163. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mahe, M., Dufour, F., Neyret-Kahn, H., Moreno-Vega, A., Beraud, C., Shi, M., Hamaidi, I., Sanchez-Quiles, V., Krucker, C., Dorland-Galliot, M., et al. (2018). An FGFR3/MYC positive feedback loop provides new opportunities for targeted therapies in bladder cancers. EMBO Mol. Med. 10, e8163. [DOI] [PMC free article] [PubMed]
- 19.Singh M., Yelle N., Venugopal C., Singh S.K. EMT: Mechanisms and therapeutic implications. Pharmacol. Ther. 2018;182:80–94. doi: 10.1016/j.pharmthera.2017.08.009. [DOI] [PubMed] [Google Scholar]; Singh, M., Yelle, N., Venugopal, C., and Singh, S.K. (2018). EMT: Mechanisms and therapeutic implications. Pharmacol. Ther. 182, 80-94. [DOI] [PubMed]
- 20.Gruosso T., Garnier C., Abelanet S., Kieffer Y., Lemesre V., Bellanger D., Bieche I., Marangoni E., Sastre-Garau X., Mieulet V., Mechta-Grigoriou F. MAP3K8/TPL-2/COT is a potential predictive marker for MEK inhibitor treatment in high-grade serous ovarian carcinomas. Nat. Commun. 2015;6:8583. doi: 10.1038/ncomms9583. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gruosso, T., Garnier, C., Abelanet, S., Kieffer, Y., Lemesre, V., Bellanger, D., Bieche, I., Marangoni, E., Sastre-Garau, X., Mieulet, V., and Mechta-Grigoriou, F. (2015). MAP3K8/TPL-2/COT is a potential predictive marker for MEK inhibitor treatment in high-grade serous ovarian carcinomas. Nat. Commun. 6, 8583. [DOI] [PMC free article] [PubMed]
- 21.Xu Z., Tong Q., Zhang Z., Wang S., Zheng Y., Liu Q., Qian L.B., Chen S.Y., Sun J., Cai L. Inhibition of HDAC3 prevents diabetic cardiomyopathy in OVE26 mice via epigenetic regulation of DUSP5-ERK1/2 pathway. Clin. Sci. (Lond.) 2017;131:1841–1857. doi: 10.1042/CS20170064. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu, Z., Tong, Q., Zhang, Z., Wang, S., Zheng, Y., Liu, Q., Qian, L.B., Chen, S.Y., Sun, J., and Cai, L. (2017). Inhibition of HDAC3 prevents diabetic cardiomyopathy in OVE26 mice via epigenetic regulation of DUSP5-ERK1/2 pathway. Clin. Sci. (Lond.) 131, 1841-1857. [DOI] [PMC free article] [PubMed]
- 22.Ng K.Y., Chan L.H., Chai S., Tong M., Guan X.Y., Lee N.P., Yuan Y., Xie D., Lee T.K., Dusetti N.J. TP53INP1 Downregulation Activates a p73-Dependent DUSP10/ERK Signaling Pathway to Promote Metastasis of Hepatocellular Carcinoma. Cancer Res. 2017;77:4602–4612. doi: 10.1158/0008-5472.CAN-16-3456. [DOI] [PubMed] [Google Scholar]; Ng, K.Y., Chan, L.H., Chai, S., Tong, M., Guan, X.Y., Lee, N.P., Yuan, Y., Xie, D., Lee, T.K., Dusetti, N.J., et al. (2017). TP53INP1 Downregulation Activates a p73-Dependent DUSP10/ERK Signaling Pathway to Promote Metastasis of Hepatocellular Carcinoma. Cancer Res. 77, 4602-4612. [DOI] [PubMed]
- 23.Png C.W., Weerasooriya M., Guo J., James S.J., Poh H.M., Osato M., Flavell R.A., Dong C., Yang H., Zhang Y. DUSP10 regulates intestinal epithelial cell growth and colorectal tumorigenesis. Oncogene. 2016;35:206–217. doi: 10.1038/onc.2015.74. [DOI] [PubMed] [Google Scholar]; Png, C.W., Weerasooriya, M., Guo, J., James, S.J., Poh, H.M., Osato, M., Flavell, R.A., Dong, C., Yang, H., and Zhang, Y. (2016). DUSP10 regulates intestinal epithelial cell growth and colorectal tumorigenesis. Oncogene 35, 206-217. [DOI] [PubMed]
- 24.Hu Z., Huang P., Yan Y., Zhou Z., Wang J., Wu G. Hepatitis B virus X protein related lncRNA WEE2-AS1 promotes hepatocellular carcinoma proliferation and invasion. Biochem. Biophys. Res. Commun. 2019;508:79–86. doi: 10.1016/j.bbrc.2018.11.091. [DOI] [PubMed] [Google Scholar]; Hu, Z., Huang, P., Yan, Y., Zhou, Z., Wang, J., and Wu, G. (2019). Hepatitis B virus X protein related lncRNA WEE2-AS1 promotes hepatocellular carcinoma proliferation and invasion. Biochem. Biophys. Res. Commun. 508, 79-86. [DOI] [PubMed]
- 25.Dolfini D., Mantovani R. YB-1 (YBX1) does not bind to Y/CCAAT boxes in vivo. Oncogene. 2013;32:4189–4190. doi: 10.1038/onc.2012.521. [DOI] [PubMed] [Google Scholar]; Dolfini, D., and Mantovani, R. (2013). YB-1 (YBX1) does not bind to Y/CCAAT boxes in vivo. Oncogene 32, 4189-4190. [DOI] [PubMed]
- 26.Dolfini D., Mantovani R. Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y? Cell Death Differ. 2013;20:676–685. doi: 10.1038/cdd.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dolfini, D., and Mantovani, R. (2013). Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y? Cell Death Differ. 20, 676-685. [DOI] [PMC free article] [PubMed]
- 27.Zhang E., He X., Zhang C., Su J., Lu X., Si X., Chen J., Yin D., Han L., De W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154–170. doi: 10.1186/s13059-018-1523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, E., He, X., Zhang, C., Su, J., Lu, X., Si, X., Chen, J., Yin, D., Han, L., and De, W. (2018). A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 19, 154-170. [DOI] [PMC free article] [PubMed]
- 28.Goodarzi H., Liu X., Nguyen H.C., Zhang S., Fish L., Tavazoie S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]; Goodarzi, H., Liu, X., Nguyen, H.C., Zhang, S., Fish, L., and Tavazoie, S.F. (2015). Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 161, 790-802. [DOI] [PMC free article] [PubMed]
- 29.Wang F.S., Fan J.G., Zhang Z., Gao B., Wang H.Y. The global burden of liver disease: the major impact of China. Hepatology. 2014;60:2099–2108. doi: 10.1002/hep.27406. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, F.S., Fan, J.G., Zhang, Z., Gao, B., and Wang, H.Y. (2014). The global burden of liver disease: the major impact of China. Hepatology 60, 2099-2108. [DOI] [PMC free article] [PubMed]
- 30.Xia Y., Cheng X., Li Y., Valdez K., Chen W., Liang T.J. Hepatitis B virus deregulates the cell cycle to promote viral replication and a premalignant phenotype. J. Virol. 2018;92 doi: 10.1128/JVI.00722-18. e00722-18. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xia, Y., Cheng, X., Li, Y., Valdez, K., Chen, W., and Liang, T.J. (2018). Hepatitis B virus deregulates the cell cycle to promote viral replication and a premalignant phenotype. J. Virol. 92, e00722-18. [DOI] [PMC free article] [PubMed]
- 31.Huang J.F., Guo Y.J., Zhao C.X., Yuan S.X., Wang Y., Tang G.N., Zhou W.P., Sun S.H. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57:1882–1892. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]; Huang, J.F., Guo, Y.J., Zhao, C.X., Yuan, S.X., Wang, Y., Tang, G.N., Zhou, W.P., and Sun, S.H. (2013). Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology 57, 1882-1892. [DOI] [PubMed]
- 32.Zhang H., Diab A., Fan H., Mani S.K., Hullinger R., Merle P., Andrisani O. PLK1 and HOTAIR Accelerate Proteasomal Degradation of SUZ12 and ZNF198 during Hepatitis B Virus-Induced Liver Carcinogenesis. Cancer Res. 2015;75:2363–2374. doi: 10.1158/0008-5472.CAN-14-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, H., Diab, A., Fan, H., Mani, S.K., Hullinger, R., Merle, P., and Andrisani, O. (2015). PLK1 and HOTAIR Accelerate Proteasomal Degradation of SUZ12 and ZNF198 during Hepatitis B Virus-Induced Liver Carcinogenesis. Cancer Res. 75, 2363-2374. [DOI] [PMC free article] [PubMed]
- 33.Liu C., Wang C., Wang K., Liu L., Shen Q., Yan K., Sun X., Chen J., Liu J., Ren H. SMYD3 as an oncogenic driver in prostate cancer by stimulation of androgen receptor transcription. J. Natl. Cancer Inst. 2013;105:1719–1728. doi: 10.1093/jnci/djt304. [DOI] [PubMed] [Google Scholar]; Liu, C., Wang, C., Wang, K., Liu, L., Shen, Q., Yan, K., Sun, X., Chen, J., Liu, J., Ren, H., et al. (2013). SMYD3 as an oncogenic driver in prostate cancer by stimulation of androgen receptor transcription. J. Natl. Cancer Inst. 105, 1719-1728. [DOI] [PubMed]
- 34.Li R.D., Tang Y.H., Wang H.L., Yang D., Sun L.J., Li W. The SMYD3 VNTR 3/3 polymorphism confers an increased risk and poor prognosis of hepatocellular carcinoma in a Chinese population. Pathol. Res. Pract. 2018;214:625–630. doi: 10.1016/j.prp.2018.04.005. [DOI] [PubMed] [Google Scholar]; Li, R.D., Tang, Y.H., Wang, H.L., Yang, D., Sun, L.J., and Li, W. (2018). The SMYD3 VNTR 3/3 polymorphism confers an increased risk and poor prognosis of hepatocellular carcinoma in a Chinese population. Pathol. Res. Pract. 214, 625-630. [DOI] [PubMed]
- 35.Oliveira-Santos W., Rabello D.A., Lucena-Araujo A.R., de Oliveira F.M., Rego E.M., Pittella Silva F., Saldanha-Araujo F. Residual expression of SMYD2 and SMYD3 is associated with the acquisition of complex karyotype in chronic lymphocytic leukemia. Tumour Biol. 2016;37:9473–9481. doi: 10.1007/s13277-016-4846-z. [DOI] [PubMed] [Google Scholar]; Oliveira-Santos, W., Rabello, D.A., Lucena-Araujo, A.R., de Oliveira, F.M., Rego, E.M., Pittella Silva, F., and Saldanha-Araujo, F. (2016). Residual expression of SMYD2 and SMYD3 is associated with the acquisition of complex karyotype in chronic lymphocytic leukemia. Tumour Biol. 37, 9473-9481. [DOI] [PubMed]
- 36.Shen B., Tan M., Mu X., Qin Y., Zhang F., Liu Y., Fan Y. Upregulated SMYD3 promotes bladder cancer progression by targeting BCLAF1 and activating autophagy. Tumour Biol. 2016;37:7371–7381. doi: 10.1007/s13277-015-4410-2. [DOI] [PubMed] [Google Scholar]; Shen, B., Tan, M., Mu, X., Qin, Y., Zhang, F., Liu, Y., and Fan, Y. (2016). Upregulated SMYD3 promotes bladder cancer progression by targeting BCLAF1 and activating autophagy. Tumour Biol. 37, 7371-7381. [DOI] [PubMed]
- 37.Dai B., Wan W., Zhang P., Zhang Y., Pan C., Meng G., Xiao X., Wu Z., Jia W., Zhang J., Zhang L. SET and MYND domain-containing protein 3 is overexpressed in human glioma and contributes to tumorigenicity. Oncol. Rep. 2015;34:2722–2730. doi: 10.3892/or.2015.4239. [DOI] [PubMed] [Google Scholar]; Dai, B., Wan, W., Zhang, P., Zhang, Y., Pan, C., Meng, G., Xiao, X., Wu, Z., Jia, W., Zhang, J., and Zhang, L. (2015). SET and MYND domain-containing protein 3 is overexpressed in human glioma and contributes to tumorigenicity. Oncol. Rep. 34, 2722-2730. [DOI] [PubMed]
- 38.Mazur P.K., Reynoird N., Khatri P., Jansen P.W., Wilkinson A.W., Liu S., Barbash O., Van Aller G.S., Huddleston M., Dhanak D. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510:283–287. doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mazur, P.K., Reynoird, N., Khatri, P., Jansen, P.W., Wilkinson, A.W., Liu, S., Barbash, O., Van Aller, G.S., Huddleston, M., Dhanak, D., et al. (2014). SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature 510, 283-287. [DOI] [PMC free article] [PubMed]
- 39.Tsai C.H., Chen Y.J., Yu C.J., Tzeng S.R., Wu I.C., Kuo W.H., Lin M.C., Chan N.L., Wu K.J., Teng S.C. SMYD3-Mediated H2A.Z.1 Methylation Promotes Cell Cycle and Cancer Proliferation. Cancer Res. 2016;76:6043–6053. doi: 10.1158/0008-5472.CAN-16-0500. [DOI] [PubMed] [Google Scholar]; Tsai, C.H., Chen, Y.J., Yu, C.J., Tzeng, S.R., Wu, I.C., Kuo, W.H., Lin, M.C., Chan, N.L., Wu, K.J., and Teng, S.C. (2016). SMYD3-Mediated H2A.Z.1 Methylation Promotes Cell Cycle and Cancer Proliferation. Cancer Res. 76, 6043-6053. [DOI] [PubMed]
- 40.Whittaker S., Marais R., Zhu A.X. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]; Whittaker, S., Marais, R., and Zhu, A.X. (2010). The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 29, 4989-5005. [DOI] [PubMed]
- 41.McCubrey J.A., Steelman L.S., Chappell W.H., Abrams S.L., Montalto G., Cervello M., Nicoletti F., Fagone P., Malaponte G., Mazzarino M.C. Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget. 2012;3:954–987. doi: 10.18632/oncotarget.652. [DOI] [PMC free article] [PubMed] [Google Scholar]; McCubrey, J.A., Steelman, L.S., Chappell, W.H., Abrams, S.L., Montalto, G., Cervello, M., Nicoletti, F., Fagone, P., Malaponte, G., Mazzarino, M.C., et al. (2012). Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget 3, 954-987. [DOI] [PMC free article] [PubMed]
- 42.López-Peláez M., Soria-Castro I., Boscá L., Fernández M., Alemany S. Cot/tpl2 activity is required for TLR-induced activation of the Akt p70 S6k pathway in macrophages: Implications for NO synthase 2 expression. Eur. J. Immunol. 2011;41:1733–1741. doi: 10.1002/eji.201041101. [DOI] [PubMed] [Google Scholar]; Lopez-Pelaez, M., Soria-Castro, I., Bosca, L., Fernandez, M., and Alemany, S. (2011). Cot/tpl2 activity is required for TLR-induced activation of the Akt p70 S6k pathway in macrophages: Implications for NO synthase 2 expression. Eur. J. Immunol. 41, 1733-1741. [DOI] [PubMed]
- 43.Kidger A.M., Rushworth L.K., Stellzig J., Davidson J., Bryant C.J., Bayley C., Caddye E., Rogers T., Keyse S.M., Caunt C.J. Dual-specificity phosphatase 5 controls the localized inhibition, propagation, and transforming potential of ERK signaling. Proc. Natl. Acad. Sci. USA. 2017;114:E317–E326. doi: 10.1073/pnas.1614684114. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kidger, A.M., Rushworth, L.K., Stellzig, J., Davidson, J., Bryant, C.J., Bayley, C., Caddye, E., Rogers, T., Keyse, S.M., and Caunt, C.J. (2017). Dual-specificity phosphatase 5 controls the localized inhibition, propagation, and transforming potential of ERK signaling. Proc. Natl. Acad. Sci. USA 114, E317-E326. [DOI] [PMC free article] [PubMed]
- 44.Ding J., Li J., Wang H., Tian Y., Xie M., He X., Ji H., Ma Z., Hui B., Wang K., Ji G. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8:e2997. doi: 10.1038/cddis.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ding, J., Li, J., Wang, H., Tian, Y., Xie, M., He, X., Ji, H., Ma, Z., Hui, B., Wang, K., and Ji, G. (2017). Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 8, e2997. [DOI] [PMC free article] [PubMed]
- 45.Chen X., Xie R., Gu P., Huang M., Han J., Dong W., Xie W., Wang B., He W., Zhong G. Long noncoding RNA LBCS inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin. Cancer Res. 2019;25:1389–1403. doi: 10.1158/1078-0432.CCR-18-1656. [DOI] [PubMed] [Google Scholar]; Chen, X., Xie, R., Gu, P., Huang, M., Han, J., Dong, W., Xie, W., Wang, B., He, W., Zhong, G., et al. (2019). Long noncoding RNA LBCS inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin. Cancer Res. 25, 1389-1403. [DOI] [PubMed]
- 46.Wang Y., Zhu P., Wang J., Zhu X., Luo J., Meng S., Wu J., Ye B., He L., Du Y. Long noncoding RNA lncHand2 promotes liver repopulation via c-Met signaling. J. Hepatol. 2018;69:861–872. doi: 10.1016/j.jhep.2018.03.029. [DOI] [PubMed] [Google Scholar]; Wang, Y., Zhu, P., Wang, J., Zhu, X., Luo, J., Meng, S., Wu, J., Ye, B., He, L., Du, Y., et al. (2018). Long noncoding RNA lncHand2 promotes liver repopulation via c-Met signaling. J. Hepatol. 69, 861-872. [DOI] [PubMed]
- 47.Mihailovich M., Militti C., Gabaldón T., Gebauer F. Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. BioEssays. 2010;32:109–118. doi: 10.1002/bies.200900122. [DOI] [PubMed] [Google Scholar]; Mihailovich, M., Militti, C., Gabaldon, T., and Gebauer, F. (2010). Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. BioEssays 32, 109-118. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.