Salmonella enterica subsp. enterica serovar Dublin is a zoonotic pathogen that often leads to invasive bloodstream infections in humans that are multidrug resistant.
KEYWORDS: Salmonella, antimicrobial resistance, genomics
ABSTRACT
Salmonella enterica subsp. enterica serovar Dublin is a zoonotic pathogen that often leads to invasive bloodstream infections in humans that are multidrug resistant. Described here are the results of Canadian national surveillance of S. Dublin from 2003 to 2015 in humans and bovines, principally collected through the Canadian Integrated Program for Antibiotic Resistance Surveillance (CIPARS). An increase in human infections due to multidrug-resistant (MDR) S. Dublin was observed in 2010, many of which were bloodstream infections. Phylogenomic analysis of human and bovine isolates revealed a closely related network that differed by only 0 to 17 single nucleotide variants (SNVs), suggesting some potential transmission between humans and bovines. Phylogenomic comparison of global publicly available sequences of S. Dublin showed that Canadian isolates clustered closely with those from the United States. A high correlation between phenotypic and genotypic antimicrobial susceptibility was observed in Canadian isolates. IS26 replication was widespread among U.S. and Canadian isolates and caused the truncation and inactivation of the resistance genes strA and blaTEM-1B. A hybrid virulence and MDR plasmid (pN13-01125) isolated from a Canadian S. Dublin isolate was searched against NCBI SRA data of bacteria. The pN13-01125 coding sequences were found in 13 Salmonella serovars, but S. Dublin appears to be a specific reservoir. In summary, we have observed the rise of invasive MDR S. Dublin in humans in Canada and found that they are closely related to bovine isolates and to American isolates in their mobile and chromosomal contents.
INTRODUCTION
From its earliest descriptions, Salmonella enterica subsp. enterica serovar Dublin was noted for its propensity to cause septicemia (bloodstream infections) and sporadic outbreaks of salmonellosis in humans, often associated with the consumption of unpasteurized milk, and its association with cattle (1–3). S. Dublin has since been isolated from pigs, sheep, dogs, mice, and others, although decades of surveillance have shown a strong association with cattle and it is considered adapted to this host. S. Dublin infections in cattle are of concern because they are difficult to control and give rise to systemic infections that can spread through herds, precipitating outbreaks of spontaneous abortion (4). Control of S. Dublin is complicated because some infected cattle may asymptomatically shed bacteria, promoting long-term pathogen persistence within herds (5–7). Furthermore, inconsistent isolation of S. Dublin from bovine body sites complicates infection control practices to reduce pathogen spread (8, 9). Comprehensive surveillance of dairy herds in the United Kingdom has shown that S. Dublin is prevalent (10), and surveys of healthy animals at slaughter show that S. Dublin is also present in meat-producing animals (7, 11).
Infections of normally sterile body sites are termed invasive. These infections are difficult to treat and can be associated with high mortality rates. In nonindustrialized countries, the disease burden of invasive nontyphoidal Salmonella infections is higher than in industrialized countries (12, 13). Among nontyphoidal Salmonella infections, S. Dublin infections are highly invasive in humans, and a recent survey of U.S. infections showed that 75% of these infections required hospitalization, although the prevalence of S. Dublin compared to that of other serovars was low (14–18). Generally, S. Dublin is considered to be susceptible to antimicrobials in surveys of healthy animals (10, 19–22), while isolates recovered from sick humans and animals tend to exhibit decreased susceptibility (23–27). Recently, the U.S. Centers for Disease Control has observed an increase in multidrug-resistant (MDR) S. Dublin in a comprehensive survey of isolates from various surveillance streams (18). While S. Dublin human infections are rare, its propensity to cause invasive infections and the recent increase in incidence are of concern, especially where drug resistance may complicate treatment.
Public health surveillance and tracking of Salmonella has traditionally been done using pulsed-field gel electrophoresis, which has low discriminatory power for the highly related Salmonella serovars, like S. Dublin (28). Many national public health surveillance programs are switching to whole-genome sequencing (WGS)-based technologies for their higher discriminatory power, the wide range of phenotypes that can be inferred, and the ability to perform sophisticated retrospective analysis. In addition, genomic surveillance allows for the tracking of antimicrobial resistance (AMR) determinants and mobile genetic elements that may facilitate their spread. Three recent communications have investigated S. Dublin using WGS technologies, the work of Mohammed et al. (29), Ågren et al. (30), and Carroll et al. (31), who investigated 9, 28, and 21 isolates, respectively, from small outbreaks or state-level investigations of either strictly bovine or human-derived isolates.
We previously reported on a Canadian S. Dublin isolate bearing a novel hybrid resistance and virulence plasmid (pN13-01125), formed between an IncA/C2 resistance plasmid originally isolated from Salmonella enterica serovar Heidelberg (pSH696_135) and an S. Dublin-specific virulence plasmid (32). The hybridization was mediated by the action of an insertion element (IS26), which is a member of the IS6 family. Recently, the mechanism of IS26 movement was demonstrated to occur by both a replicative and a conservative mode of cointegrate formation, which can resolve to cause large-scale genomic rearrangements, deletions, and consolidation of genetic factors, such as antimicrobial resistance determinants (33–36). The replicative mode of action occurs through a novel translocatable unit, bearing a single copy of IS6 with passenger genes that can integrate in either a random or a targeted fashion. Recent work has demonstrated that IS26-driven genetic alterations can impact the host range of plasmids to facilitate replication in a new host (37) and drive rearrangement of clinically important AMR plasmids (38). Insertion sequence (IS) elements are implicated in the evolution of obligate and host-restricted organisms by a process of genome streamlining that follows a burst of IS activity and expansion (39).
In this work, we report the increased number of invasive S. Dublin isolates in Canada from 2003 to 2015 at a national scale in both humans and bovines. Concomitantly with the rise in the number of invasive infections, we report a rise in MDR among S. Dublin isolates. Through a WGS-based investigation of 120 Canadian S. Dublin isolates of human and bovine origins, we identified a close network of isolates, suggesting transmission between sources. We extended this analysis to demonstrate that Canadian and U.S. isolates are closely related, circulating similar plasmids and mobile elements. Further, we use a novel circulating plasmid to elaborate an S. Dublin-specific mobile genetic network that may aid in improved surveillance design. S. Dublin is an important public health concern because of its ability to cause invasive infections in humans that are increasingly difficult to treat due to rising rates of MDR.
RESULTS
Rise of Canadian S. Dublin MDR infections.
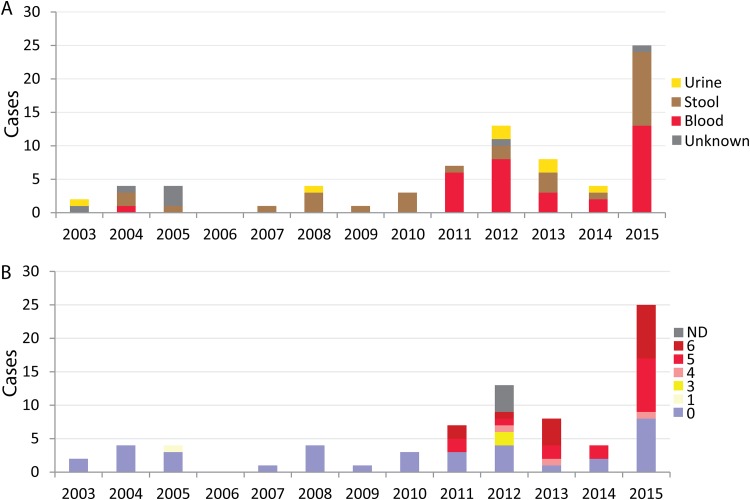
Seventy-six human S. Dublin isolates were identified in the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) database between 2003 and 2015. Of the 52 cases reporting gender, 29 (55.8%) were male, and where patient age was available (n = 28), the distribution was as follows: 0 to 4 years old (n = 3), 5 to 18 years old (n = 2), 18 to 29 years old (n = 4), 30 to 49 years old (n = 4), 50 to 69 years old (n = 11), and 70 years and over (n = 4). The total number of cases observed remained steady between 2003 and 2011, and no cases were observed in 2006. It was in 2012 when the first large increase was observed, with total cases increasing from 7 in 2011 to 13 in 2012. This was followed by an even larger increase in 2015, when a total of 25 cases were reported to CIPARS (Fig. 1A). During the earlier 8-year period (2003 and 2010), only one isolate was identified from blood (1/19; 5.3%), whereas between 2011 and 2015, the majority of isolates from known sources were from blood infections (32/57; 56.1%). The majority of blood isolates were from Québec (22/33; 65.6%), followed by Ontario (n = 7/33; 21.2%) and New Brunswick (n = 2/33; 6.1%), with a single case each from Alberta (3.0%) and British Columbia (3.0%). An increase in multidrug-resistant (≥3 classes of drugs) S. Dublin human isolates was observed over the study period. Between 2003 and 2010, all isolates were found to be pansusceptible, with the exception of one isolate, which was resistant to a single antimicrobial class. In contrast, between 2011 and 2015, 68.4% (n = 39/57) of isolates were MDR (Fig. 1B). The increase in MDR also coincided with an increase in blood infections (Fig. 1). Among the blood isolates, 63.6% (21 of 33) were resistant to four or more classes of antimicrobials.
FIG 1.
Infection type and antimicrobial resistance of S. Dublin human infections between 2003 and 2015. CIPARS results are presented by site of isolation (A) and number of antimicrobial drug classes to which isolates were resistant (B) for the indicated years (2003 to 2015). Invasive isolates are ones that were from blood. No S. Dublin isolates were detected in 2006. ND, not determined.
Genomic investigation of Canadian S. Dublin.
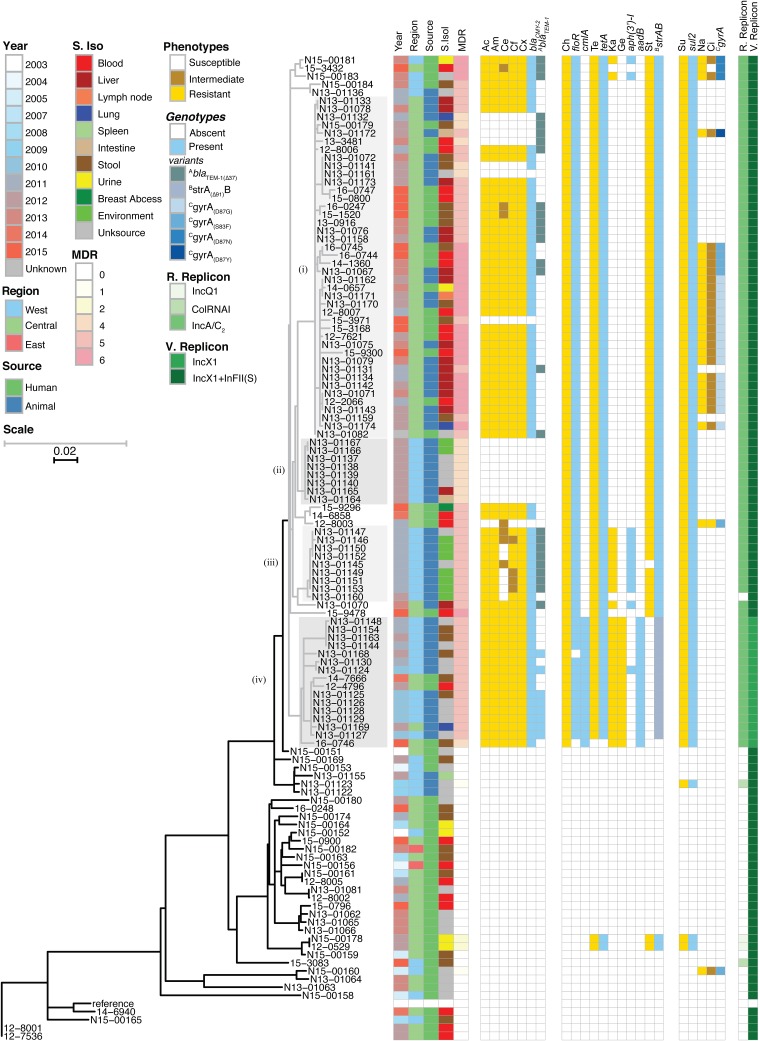
To investigate the molecular epidemiology of human cases and the possible zoonotic transmission of MDR S. Dublin to humans, we selected a convenience sample of isolates for WGS analysis from the CIPARS human and animal isolates collected between 2003 and 2015. Human isolates (n = 62) were obtained through the CIPARS program (n = 55) or submitted directly by the Laboratoire de Santé Publique du Québec (n = 7). Cattle isolates (n = 58) were collected through CIPARS (n = 50) and the Ministère de l'Agriculture, des Pêcheries et de l'Alimentation du Québec (n = 8). Multilocus sequence typing identified all isolates as of sequence type 10 (ST10). The plasmid replicons detected within isolates included IncFII(S), IncX1, IncA/C2, ColRNAI, and IncQ1. All MDR isolates had an IncA/C2 plasmid, while 4 isolates with resistance to 0 to 2 drugs (N13-01123,15-3083,12-0529, and N15-00178) were positive for replicons ColRNAI and IncQ1 (Fig. 2, right). The presence of both the IncFII(S) and IncX1 replicons was indicative of the S. Dublin virulence plasmid.
FIG 2.
Phenotypic and genomic comparison of Canadian S. Dublin isolates. (Left) HqSNV-based maximum-likelihood analysis-based dendrogram showing relatedness of human and animal S. Dublin isolates. The dendrogram is presented as a split scale, where the gray tree is 5 times the scale of the black tree, according to the scale bars in the image. (Right) Phenotypes were determined by broth microdilution, and genotypes were derived from resistance genes and mutations in whole-genome sequences. For each broad category of drugs, a phenotypic characterization (susceptible, intermediate, and resistant) is presented, followed by a genetic determination (absent and present). Results are organized into three blocks: cell wall synthesis inhibiting (left), protein translation inhibiting (middle), and DNA synthesis inhibiting (right). Abbreviations: S. Iso, site of isolation; MDR, number of antibiotic drug classes showing resistance; R. Replicon, replicon associated with mobile resistance; V. Replicon, replicon associated with mobile virulence; Unksource, unknown source; Ac, amoxicillin-clavulanic acid; Am, ampicillin; Ce, cefoxitin; Cf, ceftiofur; Cx, ceftriaxone; Ch, chloramphenicol; Ci, ciprofloxacin; Ge, gentamicin; Ka, kanamycin; Na, nalidixic acid; St, streptomycin; Su, sulfisoxazole; Te, tetracycline.
A comparative high-quality single-nucleotide variant (hqSNV) analysis of the core genome was performed using a reference genome of S. Dublin, CT_02021853. A phylogenetic analysis of the core genome using WGS of Canadian S. Dublin isolates is presented as a dendrogram (Fig. 2, left). Of the 120 S. Dublin isolates, 31 isolates were susceptible to all antimicrobials tested, 4 were resistant to 1 to 3 classes of antimicrobials, 14 were resistant to 4 classes, 46 were resistant to 5 classes, and 25 were resistant to 6 classes (Fig. 2, MDR group). The tree is bifurcated between closely related MDR strains (Fig. 2, gray subtree), with a median of 18 SNVs (range, 0 to 46) between isolates and distantly related susceptible isolates (Fig. 2, black subtree), with a median of 225 SNVs (range, 0 to 574) between isolates and a median of 284 SNVs (range, 0 to 567) between groups. The MDR isolates assorted into many closely related networks, differing by only 0 to 3 SNVs. Furthermore, the network of MDR isolates was comprised of both human and bovine isolates, whereas the network of susceptible isolates was primarily from human sources.
Four groups of MDR isolates were identified by inspection of the dendrogram based on geographic patterns and AMR genetic markers. A large group of closely related isolates with a median of 5 SNVs between them (range, 0 to 20) from central Canada was observed. This cluster included 42 isolates from both human (n = 19) and bovine (n = 23) sources (Fig. 2, group i). Furthermore, 62% of group i isolates were invasive in origin (lymph node [n = 1; 2%]), blood [n = 11; 26%], and liver [n = 14; 33%]. Isolates in groups ii and iii were isolated from western Canada and exclusively of animal origin; within each group, a median SNV difference of 1 was observed. All group ii isolates were from 2012, and the large majority of group iii isolates were from 2011. Group iv was principally made up of animal isolates, although 2 isolates were from humans, with a median of 10 SNVs (range, 0 to 20) separating isolates. Additionally, group iv is characterized by the loss of replicon IncF(II)s, which is indicative of plasmid hybridization between the S. Dublin-specific virulence plasmid pSVDR and an AMR resistance plasmid, as recently described by our group (32). Furthermore, group iv isolates were among the first MDR isolates to emerge, as 6 of these isolates were identified in 2010.
Among the generally susceptible isolates, there were two major groups. First, six isolates were identified as being intermediate in SNV differences between the MDR group and the bulk of the susceptible group (N13-01123, N13-01122, N15-00151, N15-00153, N15-00169, and N15-01155) (Fig. 2, bottom). This set of isolates was more related to the MDR group than to other susceptible isolates, with median SNV differences from the MDR group of 89 SNVs (range, 31 to 128) and from the rest of susceptible isolates of 289 SNVs (range, 229 to 567). This group was a mix of human and animal isolates, and two isolates were collected in 2003 to 2004; these were among the earliest observed isolates. The balance of primarily susceptible isolates (n = 29) is also found at the bottom of Fig. 2 All of these isolates are of human origin, and 28% were invasive blood isolates.
Patterns of AMR.
β-Lactam resistance was driven largely by the presence of blaCMY-2, which emerged in 2009. Isolates with this gene were generally resistant to all β-lactam antibiotics tested, including amoxicillin-clavulanic acid, ampicillin, cefoxitin, ceftiofur, and ceftriaxone (Fig. 2, center left). One exception was a group of cefoxitin-susceptible/intermediate isolates carrying an altered blaTEM-1B gene. Thirty-four isolates were positive for blaTEM-1B, which is known to contribute to ampicillin resistance; however, some of these isolates were susceptible to this antimicrobial (Fig. 2, blaTEM-1 column). Further investigation revealed that 26 TEM-1-positive isolates had a 37-nucleotide (nt) truncation at the 5′ end of the gene (blaTEM-1Δ37) caused by the replicative action of an IS26 transposon (Fig. 2, blaTEM-1 column, blue versus dark blue). These isolates were found among group i and group iii isolates, and in group iii, this genotype was often associated with cefoxitin susceptibility. One further exception was in a blaCMY-2-carrying isolate (N13-01141) that was susceptible to β-lactam antibiotics. This isolate was retested both phenotypically and by WGS (including PacBio long-read sequencing), and the initial results were confirmed. Furthermore, the integrities of blaCMY-2 loci, including bla’s promoter region and upstream gene, in the N13-01141 PacBio- and Illumina-based assemblies of its IncA/C2 AMR plasmid were compared by alignment to the other PacBio reference isolates in this study, and they were found to be identical. The reason for susceptibility to β-lactams could not be determined.
There was a nearly perfect correlation between phenotypic resistance to protein translation-targeting antibiotics and the presence of an antibiotic resistance enzyme (Fig. 2, center right). Chloramphenicol resistance was attributed to the presence of the floR or cmlA gene, with cmlA found exclusively in western Canadian isolates (Fig. 2, group iv). Similarly, gentamicin resistance was found exclusively in group iv and corresponded to the presence of aadB and the loss of the IncFII(S) sequence determinant. The presence of strAB was predictive of streptomycin resistance in all isolates except 15 isolates carrying a 91-nt truncation at the 5′ end of the gene (Fig. 2, strAΔ91 genotype). Sequence analysis revealed that the truncation was caused by insertion of IS26; these isolates were found exclusively in group iv. Tetracycline resistance and kanamycin resistance were attributed to the presence of tetA and aph(3′)-I, respectively.
A total of 88 isolates exhibited sulfisoxazole resistance and contained sul2, resulting in a perfect correspondence between phenotype and genotype (Fig. 2, center right). Only one isolate (15-9478) was resistant to the combination of trimethoprim-sulfamethoxazole; however, the target folA was wild type (data not shown). Downregulation of outer membrane porins (OmpK35 and OmpK36) has been shown to contribute to trimethoprim resistance in some Enterobacteriaceae (40). Orthologs of OmpK35 and OmpK36 (SED_RS05480, SED_RS12825) and the intergenic region immediately 5′ to each gene in the 15-9478 genome assembly were examined but were identical to the wild-type reference isolate.
Ciprofloxacin and nalidixic acid nonsusceptibility was found in 27 isolates, and in each case, an altered chromosomal gyrA gene was detected (Fig. 2, right). The mutations were found in the quinolone resistance-determining region; D87G, D87N, D87Y, and S83F were found in 17, 3, 1, and 6 isolates, respectively. The majority (n = 24) of these isolates were found in central Canada (Fig. 2, group i), and several sporadic instances were found in western Canadian isolates (n = 3).
Phenotypic and genotypic AMR determinations were highly correlated. A total of 1,653 of 1,681 phenotypic test results matched the genotype, for a concordance of 98.3%, and with the resistance phenotype as the reference result, the sensitivity for detection of resistance was 96.3%. When the strAΔ91 and blaTEM-1Δ37 genes, which result in potential inactive proteins, were incorporated into the analysis, then the observed concordance and sensitivity increased to 99.1% and 98.3%, respectively.
AMR plasmid analysis.
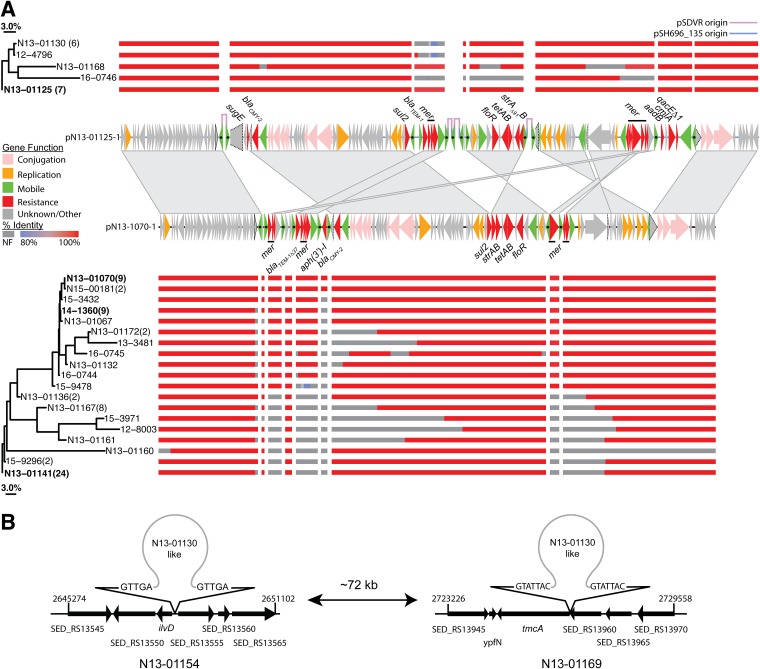
Multidrug resistance was driven by the presence of IncA/C2 plasmids, as all isolates that were resistant to more than three drug classes, with the exception of N13-01160 (see below), contained the sequence determinant for this resistance replicon (Fig. 2, R. Replicon column). Therefore, the structures of plasmids from these isolates were probed further by aligning contigs to reference plasmid sequences, with the IS26 sequence masked to simplify alignment (Fig. 3A). Three IncA/C2-positive isolates (N13-01141,14-1360-1, and N13-1070) sequenced on the PacBio platform for this work and one other sequence (pN13-01125) previously reported (32) were used to provide references for the plasmid analysis. A structural comparison of these plasmids is presented in Fig. S1 in the supplemental material, where the N13-01125-derived plasmid (pN13-01125) was previously reported as a novel hybrid of an AMR and virulence plasmid (32). Structural comparison of the AMR component of pN13-01125 and the remaining 3 reference plasmids show that they represent a related set of structurally similar but rearranged plasmids (Fig. S1). pN13-01070-1 (the plasmid derived from N13-01070) represented the largest plasmid that was not hybridized with the pSDVR virulence plasmid and was used as a reference sequence. The pN13-01125 plasmid was used as a reference for isolates that exhibited evidence of plasmid hybridization (see below).
FIG 3.
Plasmid structural analysis of Canadian S. Dublin isolates. (A) Alignment of contigs with high sequence identity to hybrid (top, pN13-01125; NCBI accession number KX815983.1) and nonhybrid (bottom, pN13-01070-1; NCBI accession number MK205416.1) plasmid reference sequences. Plasmid regions common to both pN13-01125 and pN13-01070 (>98% nucleotide identity) are linked by trapezoids, while regions with sequence inversions are linked by double triangles. The CDSs for each reference sequence are drawn as arrows and color coded by gene function, as indicated in the key on the left. The IS26 transposase is marked with a black dot. Truncated CDSs are outlined with a dotted line, and the full-length gene is outlined in a solid line (see Fig. S1 in the supplemental material for a structural comparison of plasmid reference sequences). The similarity of plasmid sequences was judged by the extent of coverage between isolates, expressed as a Jaccard distance, and displayed as a neighbor-joining tree; percent identity of aligned contigs is color coded according to the key on the left (NF, region not found). (B) The plasmids in two isolates (N13-01154 and N13-01169) were localized to the chromosome, and the plasmid insertion sites were mapped; repeat sequences at the boundaries of insertion sites are indicated.
Isolates were divided into two groups: hybrid and nonhybrid. The hybrid group contained evidence of IS26-driven plasmid hybridization determined by a reduction in S. Dublin virulence plasmid sequence. Previously, the formation of the pN13-01125 plasmid was concomitant with a loss of pSDVR genetic content by 23% (32). A further 15 isolates in this work were reduced by an average of 28.4% ± 5.5% and lost the virulence plasmid sequence, as judged by read alignment to pSDVR. All hybrid plasmid isolates were members of group iv, with a median SNV difference of 10 (Fig. 2, group iv). The nonhybrid group of isolates (n = 69) contained an IncA/C2 plasmid and had intact pSDVR sequences, with an average of reduction of 0.5% ± 0.6%; the whole-genome assemblies from this group were aligned to the pN13-01070-1 reference sequence. Similarity in plasmid structures is presented as a neighbor-joining tree produced from the pairwise Jaccard distances calculated from the extents of coverage for isolates within hybrid- and nonhybrid-plasmid-containing isolates, respectively (Fig. 3A).
The hybrid group assorted into 5 groups, where the largest group showed coverage identical to that of the pN13-01125 reference and represented 7 isolates (Fig. 3A, top). A further 6 isolates were found to have an arrangement similar to that of N13-01130; this group of isolates were missing a region that contains the blaTEM-1 β-lactamase and the heavy-metal-detoxifying operon mer. One isolate (N13-01124) was the sole hybrid-bearing isolate to contain the aph(3′)-I enzyme, but this insertion is not captured in the pN13-01125 reference. This gene was found in 11 nonhybrid isolates and appears to be the sole carrier gene of an adjacent IS26 element. The nonhybrid group was resolved into 19 profiles, 8 of which contained more than 2 members. The most numerous group consisted of 24 isolates that contained a profile similar to that of N13-01141 (Fig. 3A, bottom). This group has maintained blaCMY-2, sul2, strAB, tetAB, and floR and lost the genes associated with the mer operon. The signature of the nonhybrid groups, the varying maintenance of genes encoding functions in plasmid conjugation and mobility, was a key difference in the hybrid and nonhybrid groups (Fig. 3A). N13-01160 was found to not bear the IncA/C2 sequence determinant (Fig. 2, R. Replicon column), and alignment of the plasmid-bearing contig showed that a minimal plasmid sequence was found with a large continuous section missing at 3′ end of the reference sequence (Fig. 3A, bottom). Examination of the assembled contigs did not show evidence for plasmid hybridization. There was evidence that two isolates (N13-01134 and N13-01169) had a chromosomal insertion of their IncA/C2 plasmid (Fig. 3B). A pair of contigs that intersected at the same point in the chromosome was found for each hybrid; insertion of the plasmid resulted in the duplication of a 5- to 7-nt region.
AMR determinants and IS26 distribution in worldwide S. Dublin isolates.
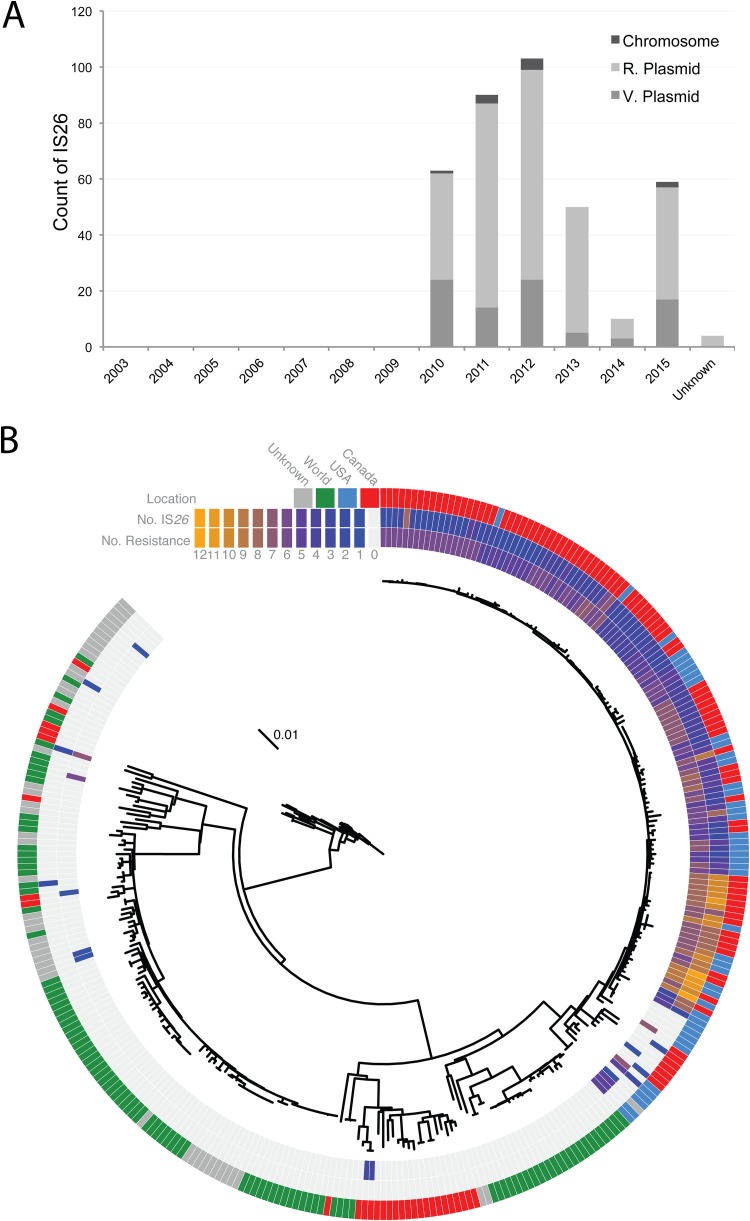
IS26 replication altered and inactivated the resistance genes strAΔ91 and blaTEM-1Δ37 and promoted plasmid hybridization in isolates expressing gentamicin resistance (e.g., pN13-01125; see reference 32). For the above-named reasons, the distribution of IS26 among isolates was investigated, and the data are presented in Fig. 4. IS26 elements were found in 86/120 isolates and mapped to assembled contigs classified as chromosome, resistance plasmid, or virulence plasmid, where 10, 282, and 87 IS26 elements were found, respectively (Fig. S2A). Nearly all IS26 transposons were found in isolates with resistance to >4 classes, the sole exception was a susceptible human isolate (N15-00169) where two IS26 elements were mapped to its chromosome (Fig. S2B). IS26 was first detected in a cluster of 6 isolates in 2010, and each of these isolates harbored a hybrid virulence and resistance plasmid. These were isolates of animal origin, isolated from western Canada, and all members belonged to group iv (Fig. 2) In subsequent years, there was a rise in IS26 occurrence, which peaked in 2012, followed by a second peak in 2015 (Fig. 4A).
FIG 4.
Distribution of IS26 elements and antimicrobial resistance in global S. Dublin isolates. (A) Distribution of IS26 elements among Canadian S. Dublin isolates by year. (B) Maximum-likelihood analysis-based dendrogram of Canadian and global isolates. MDR and IS26 status were derived from genome-based assemblies. The outer ring shows the geographic location, the middle ring shows the total number of IS26 elements identified, and the inner rings shows the total number of AMR determinants found in whole-genome assemblies.
Publicly available S. Dublin sequence data were queried to determine if the association of IS26 elements with the rise of MDR was localized solely to Canada or more globally distributed. Sequence data were acquired from the NCBI SRA database using the search term “Salmonella Dublin,” and 195 isolates were collected. A total of 101 were international isolates (99 from Europe, 1 from Africa, and 1 from South America), 46 were from North America (45 from the United States and 1 from Canada), and 48 were of unknown origin. A dendrogram of the phylogenetic relationships derived from a core hqSNV analysis of the above-named isolates and including Canadian S. Dublin, described here, is depicted in Fig. 4B. Additionally, each isolate was assembled and queried for IS26 elements and the number of AMR genes in the same manner as the Canadian S. Dublin isolate. Annotations reporting total IS26 elements and AMR elements accompany the dendrogram. Sequenced worldwide S. Dublin isolates are organized into two groups, one closely related MDR group and a loosely related and generally susceptible group (Fig. 4B). The MDR group is characterized by a median SNV difference of 19, while the susceptible group is separated by 387 SNVs. Furthermore, the group of MDR isolates is solely from Canada and the United States, and the IS26 elements are prevalent in this group of isolates, whereas currently sequenced global S. Dublin isolates are largely susceptible and do not contain IS26 elements.
S. Dublin plasmid gene network.
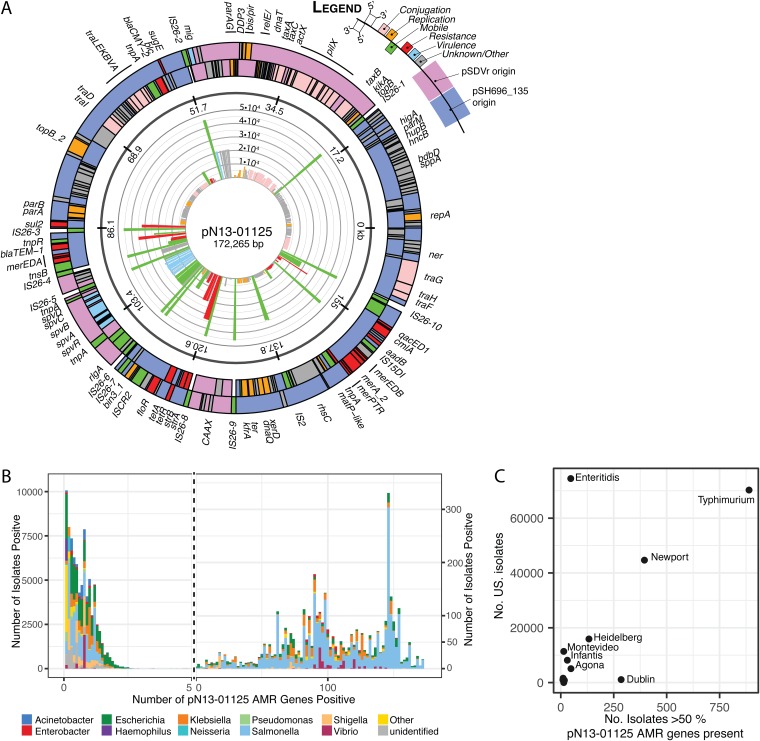
IS26 and MDR elements were prevalent and specific to North American S. Dublin isolates, and IS26 was mapped most frequently to MDR plasmids; thus, the origin of the circulating AMR S. Dublin plasmid was probed. We previously described a hybrid AMR and virulence plasmid, pN13-01125, isolated from a Canadian S. Dublin isolate in which IS26 replication was responsible for its formation. The pN13-01125 plasmid is a hybrid between an AMR plasmid (pSH696_135) and an S. Dublin-specific virulence plasmid (pSDVR, also known as pOU1115). Furthermore, the plasmid confers resistance to 5 antimicrobial classes in addition to bearing the mercury detoxification operon mer. To better understand the distribution of pN13-01125 and to gain insights into its origins, we mapped the distribution of coding DNA sequences (CDSs) from this plasmid by bacterial species and Salmonella serovar (Fig. 5). Complete CDSs of pN13-01125 were submitted individually to the BIGSI server, which indexes all viral and bacterial NCBI sequence read data and returns matches to a query sequence. The number of instances of each CDS is plotted in the inset of the plasmid map of pN13-01125 (Fig. 5A). The IS26 transposase was the most widely disturbed open reading frame (ORF) (4.5 × 104 instances), as were ORFs with functions in mobilizing DNA (Fig. 4A, green). AMR genes also were widely distributed, especially sul2 and blaTEM-1, tet, and qacED1 (>2 × 104 instances) (Fig. 5A, red). The spv virulence factor operon was the most widely distributed of genes derived from pSDVR, as were a group of poorly characterized genes (Fig. 5A, adjacent to mig, map position kbp 53), with >2 × 104 instances.
FIG 5.
Distribution of pN13-01125 hybrid AMR and virulence genes among NCBI SRA read data. (A) Plasmid map of pN13-01125 depicting the complete CDS color coded by function. (Inner ring) Radial bar graph depicting the number of instances of each gene in the NCBI SRA database. (B) Bar graph of the number of isolates with pN13-01125 genes derived from its pSH696_135 parental plasmid color coded by genus. The left axis and right axes apply to plot areas to the left and right of the vertical hashed line, respectively. (C) Numbers of isolates bearing >50% of pN13-01125 AMR-derived genes of U.S. origin (x axis) versus the number of isolates identified in the United States (y axis).
Since the signature of this cluster was the acquisition of AMR genes, we further investigated the origins of 136 genes derived from the pSH696_135 AMR component of the pN13-01125 hybrid plasmid. A total of 106,037 read files had at least 1 hit to this fraction of pN13-01125 CDSs, and the top 99.5% of species are presented in Fig. 5B. The distribution was highly tailed; 3,450 isolates harbored 50% of the query plasmid CDS (68/136), and of this group, Salmonella was the major genus reported (n = 2,494; 72%), followed by Klebsiella (n = 370; 11%), Escherichia (n = 325; 9%), and Vibrio (n = 119; 3%).
The Salmonella fraction of pN13-01125 AMR gene-positive isolates was further examined by serovar, and the results are presented in Fig. S3, where only the top 0.5% of serovars that contain at least 50% of the plasmid genes are depicted (n = 2,306), and 98% of these isolates were positive for the IncA/C2 sequence determinant. Salmonella enterica serovar Typhimurium, Salmonella enterica serovar Newport, and S. Dublin isolates were the top three serovars, with, respectively, 1,180, 414, and 295 isolates represented. S. Typhimurium exhibited a broad occupancy of genes, while S. Newport had 287 isolates concentrated at 123 pN13-01125 AMR genes. Most S. Dublin isolates had more than 100 out of 136 genes present (263/295 isolates), and this serovar also contained the most frequent instances of the complete gene complement of pN13-01125 AMR genes (n = 9).
To give context to the distribution of pN13-01125 AMR genes, the sum of each serovar with >50% of pSH696_135 genes of U.S. origin was plotted against the number of reported Salmonella isolates reported in the United States (see Materials and Methods), and these data are presented in Fig. 5C. Generally, the most common serovars had the greatest number of pSH696_135 genes, with the exception of S. Dublin, which was overrepresented in the number of isolates identified. Conversely, despite a high incidence of Salmonella enterica serovar Enteritidis in the United States, it was underrepresented in pN13-01125 AMR gene content.
DISCUSSION
The rise in the incidence of human Canadian S. Dublin is likely because of the acquisition of MDR or other epidemiological factors, rather than a change in intrinsic virulence. We queried the virulence factor database against our isolate’s WGS, in a manner previously described by our group (28), and no gain or loss of virulence determinants could be associated with invasive or MDR isolates (data not shown). Additionally, some two-thirds of human S. Dublin isolates described here were isolated from blood, which is in keeping with surveys of nontyphoidal Salmonella in humans from other national and local outbreaks (15, 41–44). What is distinctive is that the S. Dublin isolates presented here are MDR; indeed, recent regional increases in MDR S. Dublin strains have been described in American states that border Canada (23, 24). This finding is further supported by the observation here that S. Dublin isolates from the United States and Canada are highly similar. Alternatively, the relatedness of Canadian-U.S. isolates suggests that they may be spread by similar sources. For instance, the cross-border commercial distribution of animals might be responsible for the expansion of isolates. In sum, these data suggest that increases in MDR and/or epidemiological factors are the most parsimonious explanations for the rise in the incidence of Canadian S. Dublin isolates.
The work presented here is the first national WGS-based survey of S. Dublin, and we find that networks of MDR isolates show very little variation. The occurrence of MDR clusters containing both human and bovine isolates that differ by only 0 to 4 SNVs suggests possible zoonotic transmission. The interpretation criteria for SNV variation within the context of outbreak or epidemic events have not yet been clearly established for S. Dublin. PulseNet has suggested that 5 SNVs at a 90% core-genome threshold for verotoxigenic Escherichia coli O157:H7 is suggestive of outbreak-related strains in a study of Canadian isolates (45). Recently, Ågren et al. examined 28 bovine isolates of S. Dublin that were collected over a 4-year time span from a 20,000-km2 region of southern Sweden (30). From this data set, 537 variable positions survived quality filtering to support phylogenetic analysis where epidemiologically linked isolates ranged between 0 and 13 SNVs. Mohammed et al. compared S. Dublin isolates from a human outbreak collected over 2 months from an Irish hospital (29). The number of variable positions is not reported, but the authors found 1 and 9 SNVs between outbreak-related isolates and a 74- to 88-SNV difference between unrelated isolates collected from the same geographic area up to 3 years prior to the outbreak of interest. Finally, Carroll and coworkers examined 28 S. Dublin isolates from New York and Washington States from human and bovine sources (31). The number of variable positions within state clusters ranged from 4 to 48 SNVs. In the work presented here, our phylogeny of 120 isolates is supported by 2,517 high-quality-filtered SNVs drawn from a core genome that represents 4.5 × 106 nucleotides, or 92% of the reference genome length. For WGS-based phylogenomic reports, we suggest that the number of total variable sites and core-genome sizes are also reported to facilitate comparison of methods and data.
The close networks of mixed human and bovine isolates cannot suggest a specific source or inform on the transmission chain without detailed epidemiological investigations. Transmission may be associated with the handling of infected animals or environmental exposure on farms with infected herds, as has been previously described (46). The consumption of unpasteurized dairy products, including raw milk cheese, might be a potential source, although no information in Canada exists for this commodity as a risk for S. Dublin infections (47–49). A recent large multidecade epidemiological investigation of S. Dublin in the United States found that 99% of outbreak isolates were traced back to dairy sources (18). Currently sequenced Canadian and U.S. S. Dublin populations are genetically similar, and raw milk is legally available for sale in many northern U.S. states that border Canada. Future Canadian surveillance efforts should investigate the potential for a U.S.-Canada component to the increase in the incidence of S. Dublin infections described here.
WGS-based surveillance of pathogens is rapidly becoming standard practice for international public health surveillance programs. Public Health England has switched to genotypic tests for Salmonella, the U.S. CDC has recently published benchmarking data for genome-based AMR detection in Salmonella (50), and Canada implemented WGS for all Salmonella isolates in May 2017. Using a workflow similar to that of the U.S. CDC for AMR detection, our correspondence between phenotype and genotype are in agreement with the CDC study only when we correct for transposon-inactivated genes. Small changes at the beginning or end of gene boundaries and nucleotide changes can be frequent enough that the gains from the automation of WGS-based AMR detection must be balanced by the human intervention required to notice biologically significant variants. AMR detection relies on using thresholds of sequence identity and coverage because natural variation presents too many gene variants to be examined manually. The work presented here suggests that particularly in the validation stage, both the sequence and integrity of the gene should be considered when detecting AMR, as we found in S. Dublin that IS26 replication had compromised the integrity of blaTEM-1B and strA and altered the expression of their phenotypes.
The emergence of invasive S. Dublin is concomitant with the rise in MDR and the influence of IS26 in Canada. Our group has recently described how IS26 replication promoted the fusion of an S. Dublin virulence plasmid and an IncA/C2 MDR plasmid to form a hybrid plasmid (pN13-01125) that is 30 kbp smaller than the parental plasmids. Nearly half of resistant isolates (42/89) described here were influenced by IS26 replication through the truncation and inactivation. These alterations may serve to streamline the plasmids that likely promote adaptive benefits.
Recently, Porse and coworkers demonstrated that IS26 replication facilitated the long-term stability and alteration of the host range of a Klebsiella plasmid as it adapted to replication in Escherichia in an in vitro model of evolution (37). Interestingly, in that work, a large deletion of the plasmid of genes involved in conjugation was among a large-scale deletion that facilitated host range adaption. In the work presented here, we observe similar plasmid modifications, as some isolates bearing a nonhybrid plasmid have nonintuitively lost these regions as well. IS expansion is a documented driver of evolution and is observed in species that adopt host-restricted lifestyles. Here, vigorous replication of IS elements drive mutation and rearrangement of genetic material, which is then followed by IS-mediated curation of genetic material that promotes adaptive fitness (reviewed in reference 39). In the context of this work, we observed the initial expansion and consolidation of genetic material, and further surveillance is required to follow what genetic streamlining and adaptation may follow. Additionally, we see that the first plasmids to emerge in Canadian S. Dublin isolates are also those that carry the most numerous IS26 insertions, supporting the idea that a burst of IS26 activity facilitates plasmid host range expansion in S. Dublin. Finally, the pN13-01125 plasmid conjugates at a low frequency and is stably inherited (32), which suggests that AMR determinants may become fixed within the population of circulating MDR S. Dublin strains in North America, as these two populations appear to be homogenous.
The distribution of plasmid genes in this work suggest a plasmid gene network that is widely distributed among non-host-restricted serovars, such as S. Typhimurium and S. Newport. These prevalent serovars likely serve as conduits of horizontal gene transfer to host-adapted serovars, such as S. Dublin. Despite the fact that S. Enteritidis is the most frequently isolated human serovar in Canada and other countries, it appears to be excluded from this network of resistance- and plasmid-associated genes, and further work is warranted to understand the determinants of the actual conduits of transfer between Salmonella serovars and confirm that the plasmid gene network elaborated here reflects circulating plasmids. Finally, some caution is warranted, as the NCBI SRA is highly biased toward U.S. isolates and global sampling is similarly biased to isolates of medical importance. However, the plasmid network analysis is supported by a large number of isolates from a single continental area, and at least for the S. Dublin population, they are genetically similar.
Current public health surveillance of bacterial pathogens is focused on ascertaining genetic relatedness and AMR gene complementation to investigate outbreaks, monitor AMR trends, and inform antibiotic stewardship; however, tools to predict outbreaks and AMR expansions are lacking. This work suggests that examining the prevalence of mobile genetic elements, such as IS elements, might have predictive power in identifying pathogen subpopulations that are undergoing accelerated evolution and might warrant enhanced surveillance. Furthermore, future delineation of the plasmid networks of pathogens may better inform resource allocation and surveillance design to better capture the full complement of circulating plasmids.
Conclusions.
The number of Canadian human cases of S. Dublin infections is low but increasing, which is concerning because of the emergence of MDR isolates with resistance to 5 or 6 classes of antimicrobials and the increase in bloodstream infections. In addition, the small number of SNVs identified between human and cattle isolates suggests potential zoonotic transmission, although a clear source of these infections has not been identified. IS26 replication appears to have promoted alterations to the AMR patterns in S. Dublin, with unclear consequences. Canada continues to closely monitor this issue through CIPARS, and further targeted studies are planned to better understand the risk factors and source attributions of MDR S. Dublin infections in Canada.
MATERIALS AND METHODS
Antimicrobial susceptibilities were determined by broth microdilution using the Sensititre system (Sensititre automated microbiology system; Trek Diagnostic Systems Ltd., Westlake, OH, USA) and interpreted according to CLSI breakpoints (51).
Isolate collection.
The CIPARS is a national surveillance program for various bacterial genera from human, animal and animal-derived food sources coordinated by the Public Health Agency of Canada. The comprehensive sampling protocol for all surveillance streams is available at https://www.canada.ca/en/public-health/services/surveillance/canadian-integrated-program-antimicrobial-resistance-surveillance-cipars.html.
Briefly, human clinical isolates were submitted from hospital and private laboratories to provincial public health laboratories. Provinces with low populations (Yukon, Northwest Territories, Nunavut, Saskatchewan, Manitoba, New Brunswick, Nova Scotia, Prince Edward Island, and Newfoundland and Labrador) submitted all Salmonella isolates, whereas provinces with large populations (British Colombia, Alberta, Ontario, and Quebec) submitted isolates recovered only during the first 15 days of each month. Isolates were sent to the National Microbiology Laboratory in Winnipeg, Manitoba, Canada, for traditional serotyping and AMR testing.
CIPARS recovers Salmonella isolates from clinically sick bovine sources only. Animal clinical isolates were voluntarily submitted by veterinary diagnostic laboratories to the National Microbiology Laboratory in Guelph, Ontario, Canada, for traditional serotyping and AMR testing. These clinical isolates might have originated from dairy cattle, milk, grain-fed veal, beef cattle, animal feed, the animal’s environment, or nondiseased animals.
WGS was conducted on a convenience sample of 120 S. Dublin isolates from humans and animals collected through CIPARS between 2003 and 2015 to investigate the molecular characteristics of human cases and the possible zoonotic transmission of the MDR animal isolates. Sequence reads are available at https://www.ncbi.nlm.nih.gov/bioproject (BioProject accession number PRJNA402038). Worldwide S. Dublin sequences were downloaded from the NCBI SRA database using the search term “Salmonella Dublin”; sequences produced on the Illumina HiSeq or MiSeq instrument were retained for further analysis.
Multilocus sequence typing and AMR and IS26 detection.
Detection of resistance elements was performed by submission of de novo assemblies (median coverage, 89×) to the Centre for Genomic Epidemiology (CGE) website (https://cge.cbs.dtu.dk/services/) or a locally installed version of ResFinder (52) using a nucleotide threshold of 98% and length coverage of 60%. Chromosomal mutations were identified by a locally installed PointFinder script (53). Multilocus sequence typing was performed by submission of genome assemblies to the CGE website.
IS26 elements in assembled contigs were identified using the BLASTn protocol with default parameters and a threshold of 98% nucleotide identity to the reference sequence (inverted left repeat [IRL] to inverted right repeat [IRR]; NCBI accession number X00011). IS26 elements that were completely within the span of a contig were counted as a single instance, and partial elements at the boundaries of a contig were counted as a half. The identity of each contig was determined by querying a custom BLAST library comprised of the chromosomal sequence of S. Dublin (strain CT_02021853; NCBI accession number NC_011205.1), three virulence plasmids of S. Dublin (54) (pOU1113, pOU1114, and pSDVR/pOU1115; NCBI accession numbers AY517905.1, DQ115387.2, and DQ115388.2, respectively), and a series of related IncA/C2 plasmids, where one was found to have formed a hybrid with the virulence plasmid and found to be circulating within a Canadian S. Dublin isolate (32, 55) (pSH111_166, pSH163_120, and pSH686_135; NCBI accession numbers JN983043.1, JN983046.1, and JN983048.1, respectively). Each assembled contig was unambiguously matched to a reference chromosomal or plasmid sequence.
Phylogenomics.
Phylogenetic relationships were inferred by first identifying a common set of hqSNVs using a reference-based approach against S. Dublin CT_02021853, as previously described (56). Paired-end WGS was conducted on an Illumina MiSeq platform using the v3 MiSeq reagent kit (Illumina, Madison, WI, USA) with an insert size of ∼500 bp, generating approximately 1.5 × 106 reads and yielding 1.5 × 108 bp of DNA sequence. Briefly, repeat regions in the reference sequence were identified and masked from subsequent analysis using Mummer (v3.23). Reads were mapped to the masked reference using Smalt v0.7.5, and two sets of variants were generated using both Freebayes (v0.9.0.20) and SAMtools/Bcftools, independently. Variants were consolidated to a set of hqSNVs by evaluating concordance between calls, depth of coverage at position (>10× coverage), mapping quality, and filtering areas of high SNV density. An annotated pseudoalignment of all variant positions was produced and used for subsequent analysis. For analysis of Canadian isolates, 92% of positions in the reference genome met all quality and coverage criteria, and 2,517 variable sites were used for analysis. For worldwide isolates, a core genome covered 77% of the reference and had 4,671 variable positions. A pseudoalignment of variable positions was used to produce a maximum-likelihood phylogenetic tree using PhyML (3.1.1). A general time-reversible model was employed, with transition/transversion ratios and the gamma shape parameter determined empirically.
Plasmid analysis.
Four IncA/C2-positive isolates (N13-01125, N13-1070,14-1360, and N13-01141) were chosen for Pacific Biosciences sequencing using an RSII instrument based on their diverse AMR profiles, and the resultant sequence was assembled using the HGAP 3.0 protocol (32). Structural comparisons of reference plasmids were produced with Easyfig (57). Genome assemblies were aligned to two plasmid reference sequences representing a hybrid AMR virulence plasmid (pN13-01125) and the largest nonhybrid reference sequence (pN13-01070-1) using the BLASTn protocol. Plasmid reference sequences were divided into sections to exclude the repetitive IS26 sequences (accession number X00011), which simplified contig alignment. Contigs with sequence identity to chromosomal sequences were removed from the analysis (see “Multilocus sequence typing and AMR and IS26 detection” above). Overlapping and adjoining contigs were merged, and a weighted percent identity was calculated. The similarity of plasmid sequence was judged by the Jaccard distance between pairs of isolates calculated from the degree of overlap of aligned contig sequences. Two isolates (N13-01169 and N13-01154) were found to contain a pair of contigs with both plasmid and chromosomal sequences and were judged to contain chromosomal insertions of AMR plasmids. Insertion sites were mapped to the CT_02021853 chromosome.
pN13-01125 gene distribution analysis.
Individual CDSs from pN13-01125 plasmid (accession KX815983.1) were submitted to the BIGSI server with a cutoff set at 50% kmer coverage (58), which returned a set of SRA run numbers that were dereplicated by unique biosample id; biosample metadata, including species was retrieved using the NCBI E-utilities (https://www.ncbi.nlm.nih.gov/books/NBK25500). Salmonella serotyping data were downloaded through the Enterobase API (59); when serotyping was unavailable the predicted serotype is provided by the SISTR in silico typing tool (60). Total US Salmonella infections (2006 to 2016) were derived from appendix 3a of the National Enteric Disease Surveillance, Salmonella annual report 2016 (https://www.cdc.gov/nationalsurveillance/pdfs/2016-Salmonella-report-508.pdf).
Data availability.
IncA/C2-positive plasmid sequences were deposited in GenBank under the following accession numbers: for pN13-01125, KX815983.1; for pN13-1070-1, MK205416.1; for 14-1360, MK205418.1; and for pN13-01141-1, MK205417.1 (see Table 1 for a list of reference plasmids relevant to this work).
TABLE 1.
Major plasmids referred to in this work
| Name | Size (bp) | Origin | AMR gene(s) | Reference | Accession no. |
|---|---|---|---|---|---|
| pSH135_166a | 135,423 | IncA/C2 | blaTEM-1, blaCMY-2, floR, cmlA, aadB, strAB, sul2, tetA | 55 | JN983048.1 |
| pSDVR/pOU1115a ,b | 74,589 | IncFII(S), IncX1 | None | 54 | DQ115388.2 |
| pN13-01125c | 172,265 | IncX1, IncA/C2 | blaTEM-1, blaCMY-2, floR, cmlA, aadB, strAΔ91 B, sul2, tetA | 32 | KX815983.1 |
| pN13-01070-1c | 107,296 | IncA/C2 | blaTEM-1Δ37, blaCMY-2, floR, aph(3′)-I, strAB, sul2, tetA | This work | MK205416.1 |
| p14-1360-1 | 101,269 | IncA/C2 | blaTEM-1Δ37, blaCMY-2, floR, strAB, sul2, tetA | This work | MK205418.1 |
| pN13-01141-1 | 83,375 | IncA/C2 | blaCMY-2, floR, strAB, sul2, tetA | This work | MK205417.1 |
Parental plasmids of the pN13-01125 plasmid hybrid (see reference 32).
Virulence plasmid of S. Dublin.
Reference plasmids used for plasmid analysis.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Colleen Peterson, Melissa McCracken, Shaun Tyler, Morag Graham, and the staff at the NML Genomics Core Facility for whole-genome sequencing support. We acknowledge Gary Van Domselaar and the staff of the NML Bioinformatics group, particularly Aaron Petkau and Peter Kruczkiewicz, for helpful discussions. We acknowledge Andrea Desruisseau, Chad Gill, Laura Mataseje, Russell Mandes, and Stacie Langner for their contributions toward susceptibility testing of the isolates. Finally, we acknowledge Phelim Bradley and Zamin Iqbal of the Wellcome Trust Centre for Human Genetics for their assistance with BIGSI queries.
Members of the Canadian Integrated Program for Antimicrobial Resistance Surveillance Public Health Partnership were as follows: Linda Hoang, British Columbia Centres for Disease Control Public Health Microbiology & Reference Laboratory, Vancouver, British Columbia, Canada; Marie Louie, Provincial Laboratory for Public Health, Edmonton, Alberta, Canada; Roy Romanow, Saskatchewan Disease Control Centre Laboratory, Regina, Saskatchewan, Canada; David C. Alexander, Cadham Provincial Laboratory, Winnipeg, Manitoba, Canada; Vanessa Allen, Public Health Ontario, Toronto, Ontario, Canada; David Haldane, Queen Elizabeth II Health Sciences Centre, Halifax, Nova Scotia, Canada; Greg J. German, Queen Elizabeth Hospital, Charlottetown, Prince Edward Island, Canada; Sameh El Bailey, Saint John Regional Hospital, St. John, New Brunswick, Canada; and George Zahariadis, Newfoundland Public Health Laboratory, St. Johns, Newfoundland, Canada.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00108-19.
Contributor Information
Linda Hoang, British Columbia Centres for Disease Control Public Health Microbiology & Reference Laboratory, Vancouver, British Columbia, Canada;.
Marie Louie, Provincial Laboratory for Public Health, Edmonton, Alberta, Canada;.
Roy Romanow, Saskatchewan Disease Control Centre Laboratory, Regina, Saskatchewan, Canada;.
David C. Alexander, Cadham Provincial Laboratory, Winnipeg, Manitoba, Canada;
Vanessa Allen, Public Health Ontario, Toronto, Ontario, Canada;.
David Haldane, Queen Elizabeth II Health Sciences Centre, Halifax, Nova Scotia, Canada;.
Greg J. German, Queen Elizabeth Hospital, Charlottetown, Prince Edward Island, Canada;
Sameh El Bailey, Saint John Regional Hospital, St. John, New Brunswick, Canada;.
George Zahariadis, Newfoundland Public Health Laboratory, St. Johns, Newfoundland, Canada..
Collaborators: Linda Hoang, Marie Louie, Roy Romanow, David C. Alexander, Vanessa Allen, David Haldane, Greg J. German, Sameh El Bailey, and George Zahariadis
REFERENCES
- 1.White PB. 1930. Notes on organisms serologically related to S. enteritidis Gartner: I. The Dublin and Tokyo types of Salmonella. J Hyg (Lond) 29:443–445. doi: 10.1017/S0022172400010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith J, Scott WM. 1930. Continued fever due to a Gartner-like Salmonella of the type “Dublin.” J Hyg (Lond) 30:32–39. doi: 10.1017/S0022172400010263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Souper HR, Smith J, Stephen JA. 1930. Three sporadic cases of infection due to Salmonella type Dublin. Arch Dis Child 5:271–274. doi: 10.1136/adc.5.28.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen TD, Nielsen LR, Toft N, Houe H. 2010. Association between bulk-tank milk Salmonella antibody level and high calf mortality in Danish dairy herds. J Dairy Sci 93:304–310. doi: 10.3168/jds.2009-2528. [DOI] [PubMed] [Google Scholar]
- 5.Lawson GH, McPherson EA, Laing AH, Wooding P. 1974. The epidemiology of Salmonella Dublin infection in a dairy herd. I. Excretion and persistence of the organism. J Hyg (Lond) 72:311–328. doi: 10.1017/S0022172400023548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gronstol H, Osborne AD, Pethiyagoda S. 1974. Experimental Salmonella infection in calves. 1. The effect of stress factors on the carrier state. J Hyg (Lond) 72:155–162. doi: 10.1017/S0022172400023342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchner M, McLaren I, Clifton-Hadley FA, Liebana E, Wales AD, Davies RH. 2012. A comparison between longitudinal shedding patterns of Salmonella Typhimurium and Salmonella Dublin on dairy farms. Vet Rec 171:194. doi: 10.1136/vr.100865. [DOI] [PubMed] [Google Scholar]
- 8.Richardson A, Fawcett AR. 1973. Salmonella Dublin infection in calves: the value of rectal swabs in diagnosis and epidemiological studies. Br Vet J 129:151–156. doi: 10.1016/S0007-1935(17)36539-9. [DOI] [PubMed] [Google Scholar]
- 9.Veling J, Barkema HW, van der Schans J, van Zijderveld F, Verhoeff J. 2002. Herd-level diagnosis for Salmonella enterica subsp. enterica serovar Dublin infection in bovine dairy herds. Prev Vet Med 53:31–42. doi: 10.1016/S0167-5877(01)00276-8. [DOI] [PubMed] [Google Scholar]
- 10.Davison HC, Smith RP, Pascoe SJS, Sayers AR, Davies RH, Weaver JP, Kidd SA, Dalziel RW, Evans SJ. 2005. Prevalence, incidence and geographical distribution of serovars of Salmonella on dairy farms in England and Wales. Vet Rec 157:703–711. doi: 10.1136/vr.157.22.703. [DOI] [PubMed] [Google Scholar]
- 11.Webb HE, Brichta-Harhay DM, Brashears MM, Nightingale KK, Arthur TM, Bosilevac JM, Kalchayanand N, Schmidt JW, Wang R, Granier SA, Brown TR, Edrington TS, Shackelford SD, Wheeler TL, Loneragan GH. 2017. Salmonella in peripheral lymph nodes of healthy cattle at slaughter. Front Microbiol 8:2214. doi: 10.3389/fmicb.2017.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohmann EL. 2001. Nontyphoidal salmonellosis. Clin Infect Dis 32:263–269. doi: 10.1086/318457. [DOI] [PubMed] [Google Scholar]
- 13.Haselbeck AH, Panzner U, Im J, Baker S, Meyer CG, Marks F. 2017. Current perspectives on invasive nontyphoidal Salmonella disease. Curr Opin Infect Dis 30:498–503. doi: 10.1097/QCO.0000000000000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Threlfall EJ, Hall ML, Rowe B. 1992. Salmonella bacteraemia in England and Wales, 1981–1990. J Clin Pathol 45:34–36. doi: 10.1136/jcp.45.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vugia DJ, Samuel M, Farley MM, Marcus R, Shiferaw B, Shallow S, Smith K, Angulo FJ, Emerging Infections Program FoodNet Working Group. 2004. Invasive Salmonella infections in the United States, FoodNet, 1996–1999: incidence, serotype distribution, and outcome. Clin Infect Dis 38:S149–S156. doi: 10.1086/381581. [DOI] [PubMed] [Google Scholar]
- 16.Jones TF, Ingram LA, Cieslak PR, Vugia DJ, Tobin-D'Angelo M, Hurd S, Medus C, Cronquist A, Angulo FJ. 2008. Salmonellosis outcomes differ substantially by serotype. J Infect Dis 198:109–114. doi: 10.1086/588823. [DOI] [PubMed] [Google Scholar]
- 17.Wilkins EG, Roberts C. 1988. Extraintestinal salmonellosis. Epidemiol Infect 100:361–368. doi: 10.1017/S095026880006711X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harvey RR, Friedman CR, Crim SM, Judd M, Barrett KA, Tolar B, Folster JP, Griffin PM, Brown AC. 2017. Epidemiology of Salmonella enterica serotype Dublin infections among humans, United States, 1968–2013. Emerging Infect Dis 23:1493–1501. doi: 10.3201/eid2309.170136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wray C, McLaren IM, Beedell YE. 1993. Bacterial resistance monitoring of salmonellas isolated from animals, national experience of surveillance schemes in the United Kingdom. Vet Microbiol 35:313–319. doi: 10.1016/0378-1135(93)90156-2. [DOI] [PubMed] [Google Scholar]
- 20.Liebana E, Garcia-Migura L, Clouting C, Cassar CA, Clifton-Hadley FA, Lindsay EA, Threlfall EJ, Chappell SA, Davies RH. 2002. Investigation of the genetic diversity among isolates of Salmonella enterica serovar Dublin from animals and humans from England, Wales and Ireland. J Appl Microbiol 93:732–744. doi: 10.1046/j.1365-2672.2002.01737.x. [DOI] [PubMed] [Google Scholar]
- 21.Jones YE, Chappell S, McLaren IM, Davies RH, Wray C. 2002. Antimicrobial resistance in Salmonella isolated from animals and their environment in England and Wales from 1988 to 1999. Vet Rec 150:649–654. doi: 10.1136/vr.150.21.649. [DOI] [PubMed] [Google Scholar]
- 22.Khen BK, Lynch OA, Carroll J, McDowell DA, Duffy G. 2014. Prevalence and characteristics of Salmonella in the beef chain in the Republic of Ireland. Zoonoses Public Health 61:534–536. doi: 10.1111/zph.12099. [DOI] [PubMed] [Google Scholar]
- 23.Hong S, Rovira A, Davies P, Ahlstrom C, Muellner P, Rendahl A, Olsen K, Bender JB, Wells S, Perez A, Alvarez J. 2016. Serotypes and antimicrobial resistance in Salmonella enterica recovered from clinical samples from cattle and swine in Minnesota, 2006 to 2015. PLoS One 11:e0168016. doi: 10.1371/journal.pone.0168016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis MA, Hancock DD, Besser TE, Daniels JB, Baker KNK, Call DR. 2007. Antimicrobial resistance in Salmonella enterica serovar Dublin isolates from beef and dairy sources. Vet Microbiol 119:221–230. doi: 10.1016/j.vetmic.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 25.Zhao S, McDermott PF, White DG, Qaiyumi S, Friedman SL, Abbott JW, Glenn A, Ayers SL, Post KW, Fales WH, Wilson RB, Reggiardo C, Walker RD. 2007. Characterization of multidrug resistant Salmonella recovered from diseased animals. Vet Microbiol 123:122–132. doi: 10.1016/j.vetmic.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Ferris KE, Andrews RE, Thoen CO, Blackburn BO. 1992. Plasmid profile analysis, phage typing, and antibiotic sensitivity of Salmonella dublin from clinical isolates in the United States. Vet Microbiol 32:51–62. doi: 10.1016/0378-1135(92)90006-F. [DOI] [PubMed] [Google Scholar]
- 27.van Leeuwen WJ, Voogd CE, Guinee PA, Manten A. 1982. Incidence of resistance to ampicillin, chloramphenicol, kanamycin, tetracycline and trimethoprim of Salmonella strains isolated in The Netherlands during 1975-1980. Antonie Van Leeuwenhoek 48:85–96. doi: 10.1007/BF00399490. [DOI] [PubMed] [Google Scholar]
- 28.Edirmanasinghe R, Finley R, Parmley EJ, Avery BP, Carson C, Bekal S, Golding G, Mulvey MR. 2017. A whole-genome sequencing approach to study cefoxitin-resistant Salmonella enterica serovar Heidelberg isolates from various sources. Antimicrob Agents Chemother 61:e01919-16. doi: 10.1128/AAC.01919-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohammed M, Delappe N, O'Connor J, McKeown P, Garvey P, Cormican M. 2016. Whole genome sequencing provides an unambiguous link between Salmonella Dublin outbreak strain and a historical isolate. Epidemiol Infect 144:576–581. doi: 10.1017/S0950268815001636. [DOI] [PubMed] [Google Scholar]
- 30.Ågren ECC, Wahlström H, Vesterlund-Carlson C, Lahti E, Melin L, Söderlund R. 2016. Comparison of whole genome sequencing typing results and epidemiological contact information from outbreaks of Salmonella Dublin in Swedish cattle herds. Infect Ecol Epidemiol 6:31782. doi: 10.3402/iee.v6.31782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carroll LM, Wiedmann M, Bakker den H, Siler J, Warchocki S, Kent D, Lyalina S, Davis M, Sischo W, Besser T, Warnick LD, Pereira RV. 2017. Whole-genome sequencing of drug-resistant Salmonella enterica isolates from dairy cattle and humans in New York and Washington States reveals source and geographic associations. Appl Environ Microbiol 83:e00140-17. doi: 10.1128/AEM.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangat CS, Bekal S, Irwin RJ, Mulvey MR. 2017. A novel hybrid plasmid carrying multiple antimicrobial resistance and virulence genes in Salmonella enterica serovar Dublin. Antimicrob Agents Chemother 61:e02601-16. doi: 10.1128/AAC.02601-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harmer CJ, Moran RA, Hall RM. 2014. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5:e01801-14. doi: 10.1128/mBio.01801-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harmer CJ, Hall RM. 2015. IS26-mediated precise excision of the IS26-aphA1a translocatable unit. mBio 6:e01866-15. doi: 10.1128/mBio.01866-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harmer CJ, Hall RM. 2016. IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1:e00038-16. doi: 10.1128/mSphere.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harmer CJ, Hall RM. 2017. Targeted conservative formation of cointegrates between two DNA molecules containing IS26 occurs via strand exchange at either IS end. Mol Microbiol 106:409–418. doi: 10.1111/mmi.13774. [DOI] [PubMed] [Google Scholar]
- 37.Porse A, Schønning K, Munck C, Sommer M. 2016. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol Biol Evol 33:2860–2873. doi: 10.1093/molbev/msw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F. 2015. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6:e00762-15. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siguier P, Gourbeyre E, Chandler M. 2014. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gutmann L, Williamson R, Moreau N, Kitzis MD, Collatz E, Acar JF, Goldstein FW. 1985. Cross-resistance to nalidixic acid, trimethoprim, and chloramphenicol associated with alterations in outer membrane proteins of Klebsiella, Enterobacter, and Serratia. J Infect Dis 151:501–507. doi: 10.1093/infdis/151.3.501. [DOI] [PubMed] [Google Scholar]
- 41.Crump JA, Medalla FM, Joyce KW, Krueger AL, Hoekstra RM, Whichard JM, Barzilay EJ, Emerging Infections Program NARMS Working Group. 2011. Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob Agents Chemother 55:1148–1154. doi: 10.1128/AAC.01333-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang FC, Fierer J. 1991. Human infection with Salmonella dublin. Medicine (Baltimore, MD) 70:198–207. doi: 10.1097/00005792-199105000-00004. [DOI] [PubMed] [Google Scholar]
- 43.Taylor DN, Bied JM, Munro JS, Feldman RA. 1982. Salmonella dublin infections in the United States, 1979–1980. J Infect Dis 146:322–327. doi: 10.1093/infdis/146.3.322. [DOI] [PubMed] [Google Scholar]
- 44.Allerberger F, Liesegang A, Grif K, Khaschabi D, Prager R, Danzl J, Hock F, Ottl J, Dierich MP, Berghold C, Neckstaller I, Tschape H, Fisher I. 2003. Occurrence of Salmonella enterica serovar Dublin in Austria. Wien Med Wochenschr 153:148–152. doi: 10.1046/j.1563-258X.2003.03015.x. [DOI] [PubMed] [Google Scholar]
- 45.Rumore J, Tschetter L, Kearney A, Kandar R, McCormick R, Walker M, Peterson C-L, Reimer A, Nadon C. 2018. Evaluation of whole-genome sequencing for outbreak detection of verotoxigenic Escherichia coli O157:H7 from the Canadian perspective. BMC Genomics 19:870. doi: 10.1186/s12864-018-5243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mateus A, Taylor DJ, Brown D, Mellor DJ, Bexiga R, Ellis K. 2008. Looking for the unusual suspects: a Salmonella Dublin outbreak investigation. Public Health 122:1321–1323. doi: 10.1016/j.puhe.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 47.Vaillant V, Haeghebaert S, Desenclos J-C, Bouvet P, Grimont F, Grimont PAD, Burnens AP. 1996. Outbreak of Salmonella dublin infection in France, November–December 1995. Euro Surveill 1:9–10. doi: 10.2807/esm.01.02.00193-en. [DOI] [PubMed] [Google Scholar]
- 48.Centers for Disease Control. 1984. Salmonella dublin and raw milk consumption—California. MMWR Morb Mortal Wkly Rep 33:196–198. [PubMed] [Google Scholar]
- 49.Small RG, Sharp JC. 1979. A milk-borne outbreak due to Salmonella dublin. J Hyg (Lond) 82:95–100. doi: 10.1017/S0022172400025511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing, CLSI supplement M100S, 26th ed CLSI, Wayne, PA. [Google Scholar]
- 52.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. 2017. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu C, Feng Y, Chien A-C, Hu S, Chu C-H, Chiu C-H. 2008. Evolution of genes on the Salmonella virulence plasmid phylogeny revealed from sequencing of the virulence plasmids of S. enterica serotype Dublin and comparative analysis. Genomics 92:339–343. doi: 10.1016/j.ygeno.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 55.Han J, Lynne AM, David DE, Tang H, Xu J, Nayak R, Kaldhone P, Logue CM, Foley SL. 2012. DNA sequence analysis of plasmids from multidrug resistant Salmonella enterica serotype Heidelberg isolates. PLoS One 7:e51160. doi: 10.1371/journal.pone.0051160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petkau A, Mabon P, Sieffert C, Knox NC, Cabral J, Iskander M, Iskander M, Weedmark K, Zaheer R, Katz LS, Nadon C, Reimer A, Taboada E, Beiko RG, Hsiao W, Brinkman F, Graham M, Van Domselaar G. 2017. SNVPhyl: a single nucleotide variant phylogenomics pipeline for microbial genomic epidemiology. Microb Genom 3:e000116. doi: 10.1099/mgen.0.000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bradley P, Bakker den H, Rocha E, McVean G, Iqbal Z. 2017. Real-time search of all bacterial and viral genomic data. bioRxiv doi: 10.1101/234955. [DOI] [PMC free article] [PubMed]
- 59.Alikhan N-F, Zhou Z, Sergeant MJ, Achtman M. 2018. A genomic overview of the population structure of Salmonella. PLoS Genet 14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, Nash JHE, Taboada EN. 2016. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
IncA/C2-positive plasmid sequences were deposited in GenBank under the following accession numbers: for pN13-01125, KX815983.1; for pN13-1070-1, MK205416.1; for 14-1360, MK205418.1; and for pN13-01141-1, MK205417.1 (see Table 1 for a list of reference plasmids relevant to this work).
TABLE 1.
Major plasmids referred to in this work
| Name | Size (bp) | Origin | AMR gene(s) | Reference | Accession no. |
|---|---|---|---|---|---|
| pSH135_166a | 135,423 | IncA/C2 | blaTEM-1, blaCMY-2, floR, cmlA, aadB, strAB, sul2, tetA | 55 | JN983048.1 |
| pSDVR/pOU1115a ,b | 74,589 | IncFII(S), IncX1 | None | 54 | DQ115388.2 |
| pN13-01125c | 172,265 | IncX1, IncA/C2 | blaTEM-1, blaCMY-2, floR, cmlA, aadB, strAΔ91 B, sul2, tetA | 32 | KX815983.1 |
| pN13-01070-1c | 107,296 | IncA/C2 | blaTEM-1Δ37, blaCMY-2, floR, aph(3′)-I, strAB, sul2, tetA | This work | MK205416.1 |
| p14-1360-1 | 101,269 | IncA/C2 | blaTEM-1Δ37, blaCMY-2, floR, strAB, sul2, tetA | This work | MK205418.1 |
| pN13-01141-1 | 83,375 | IncA/C2 | blaCMY-2, floR, strAB, sul2, tetA | This work | MK205417.1 |
Parental plasmids of the pN13-01125 plasmid hybrid (see reference 32).
Virulence plasmid of S. Dublin.
Reference plasmids used for plasmid analysis.