Polymyxins are nonribosomal peptide antibiotics used as the last-resort drug for treatment of multidrug-resistant Gram-negative bacteria. However, strains that are resistant to polymyxins have emerged in many countries.
KEYWORDS: Bacillus licheniformis, alkaline protease, polymyxin resistance, polymyxin-inactivating enzyme, polymyxins
ABSTRACT
Polymyxins are nonribosomal peptide antibiotics used as the last-resort drug for treatment of multidrug-resistant Gram-negative bacteria. However, strains that are resistant to polymyxins have emerged in many countries. Although several mechanisms for polymyxin resistance have been well described, there is little knowledge on the hydrolytic mechanism of polymyxin. Here, we identified a polymyxin-inactivating enzyme from Bacillus licheniformis strain DC-1 which was produced and secreted into the medium during entry into stationary phase. After purification, sequencing, and heterologous expression, we found that the alkaline protease Apr is responsible for inactivation of polymyxins. Analysis of inactivation products demonstrated that Apr cleaves polymyxin E at two peptide bonds: one is between the tripeptide side chain and the cyclic heptapeptide ring, the other between l-Thr and l-α-γ-diaminobutyric acid (l-Dab) within the cyclic heptapeptide ring. Apr is highly conserved among several genera of Gram-positive bacteria, including Bacillus and Paenibacillus. It is noteworthy that two peptidases S8 from Gram-negative bacteria shared high levels of sequence identity with Apr. Our results indicate that polymyxin resistance may result from inactivation of antibiotics by hydrolysis.
INTRODUCTION
Polymyxins are antibacterial peptides produced by the Gram-positive bacterium Paenibacillus polymyxa (formerly named Bacillus polymyxa). After their discovery, polymyxin B (PMB) and polymyxin E (colistin) were widely used clinically against Gram-negative bacteria. Because of their nephrotoxic and neurotoxic effects, polymyxins were replaced by new antibiotics (such as aminoglycosides) in the mid-1980s. However, polymyxins have been revived as the last-line therapy to treat multidrug-resistant Gram-negative bacteria (1–3).
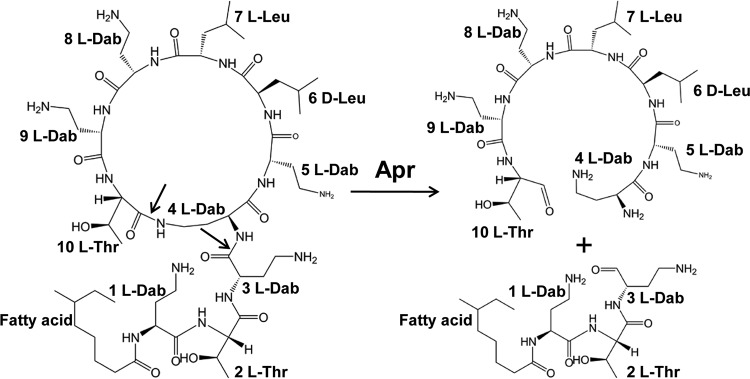
Both colistin and PMB are biosynthesized by nonribosomal peptide synthetases. They contain a cyclic heptapeptide ring and a tripeptide side chain which covalently binds to a fatty acid tail (Fig. 1). The only difference between colistin and PMB is the amino acid (aa) at position 6, d-Leu in colistin and d-Phe in polymyxin B. Both colistin and PMB are composed of two closely related components (i.e., colistin A and B for colistin and PMB1 and PMB2 for polymyxin B). The fatty acid in colistin A and PMB1 is 6-methyloctanoic acid, while the fatty acid in colistin B and PMB2 is 6-methylheptanoic acid (4, 5).
FIG 1.
Chemical structure and degradation pathway of colistin. The amino acid positions are numbered in accordance with the discussion in the text. The amino acid at position 6 is d-Phe in polymyxin B. Both colistin and PMB are mixtures of structurally related compounds. Fatty acid, 6-methyloctanoic acid for colistin A and PMB1 and 6-methylheptanoic acid for colistin B and PMB2; Dab, α-γ-diaminobutyric acid; Leu, leucine; Thr, threonine. The sites of cleavage by alkaline protease Apr are indicated by the arrows.
In Gram-negative bacteria, the electrostatic interaction between divalent cations (Mg2+ and Ca2+) and phosphate residues of lipid A is essential for the stability of lipopolysaccharide (LPS) and the outer membrane. Polymyxins contain six cationic l-α-γ-diaminobutyric acid (l-Dab) residues (Fig. 1). Due to the higher affinity between l-Dab and lipid A, polymyxins displace divalent cations and bind to lipid A molecules, resulting in destabilization of the outer membrane. The hydrophobic domains of polymyxins (including the fatty acyl chain and d-Phe/d-Leu-l-Leu) insert between the acyl chains of lipid A and then enter the periplasm via a self-promoted uptake mechanism. Subsequently, polymyxins penetrate into the phospholipid bilayer of the inner membrane (5–7). Although LPS is absent in Gram-positive bacteria, recent studies showed that polymyxins can also kill their producer, P. polymyxa, in a manner mediated by both cell membrane damage and oxidative stress (8–10).
As for other antibiotics, Gram-negative bacteria have developed several mechanisms of polymyxin resistance (11–14). The most important strategy is the modification of the outer membrane, especially the alteration of LPS. The addition of 4-amino-4-deoxy-l-arabinose (l-Ara4N), phosphoethanolamine (pEtN), or galactosamine to lipid A decreases the net negative charge of phosphate residues of lipid A, thus dramatically reducing the affinity of polymyxin to lipid A. A large number of studies have suggested that several two-component systems (PhoPQ, PmrAB, and CrrAB) are involved in the regulation of lipid A modification (13, 15, 16). Another mechanism of resistance to polymyxin is the expression of efflux pumps (17, 18).
Interestingly, studies from several decades ago showed that a colistin-inactivating enzyme (colistinase) was produced by P. polymyxa, resulting in the degradation of colistin (19–22). It is likely that colistinase is a kind of serine alkaline protease which cleaves the peptide bond between the tripeptide side chain and the cyclic heptapeptide ring (positions 3 and 4) (22). Colistin is also degraded by certain plant proteinases, such as papain, ficin, and bromelain (23). Curiously, there are no further studies in the literature about the hydrolytic mechanism of polymyxins. It is still unclear which genes are responsible for the degradation of polymyxins.
In this study, we identified the polymyxin-inactivating enzyme from Bacillus licheniformis strain DC-1. Heterogeneous expression suggested that this polymyxin-inactivating enzyme is the alkaline protease Apr, which has a catalytic domain similar to that of subtilisin Carlsberg from other Bacillus strains. Further studies demonstrated that Apr cleaves colistin at two peptide bonds: one is between the tripeptide side chain and the cyclic heptapeptide ring, and the other is between l-Thr and l-Dab within the cyclic heptapeptide ring (Fig. 1).
RESULTS
Polymyxins can be inactivated by Bacillus species.
Studies from several decades ago indicated that colistin-inactivating enzymes are probably produced and secreted by the producer of colistin, P. polymyxa (20–22). To identify the colistin-inactivating enzyme, the cylinder-plate diffusion method was performed to screen colistin-degrading bacteria from our laboratory stocks of Bacillus strains. Escherichiacoli strain DH5α was used as a colistin-sensitive indicator strain. The addition of colistin (10,000 U/ml) resulted in an obvious inhibition zone in a lysogeny broth (LB) agar plate. The inhibition zone diameter was positively correlated to the logarithm of the colistin concentration (Fig. S1 in the supplemental material). The residual activity of colistin was calculated according to a calibration curve. As shown by the results in Fig. 2, colistin was hardly degraded by the fermentation supernatant of Bacillussubtilis strain WB800, a protease-deficient strain (24) whose results were comparable to the results for the control. In contrast, colistin was efficiently degraded when supplemented with the fermentation supernatant of B. licheniformis DC-1. It has been reported that colistin activity is dependent on the culture medium and cation concentration (25). To exclude the effect of culture medium on colistin degradation, we repeated the experiments using cation-adjusted Mueller-Hinton medium (CAMHM) instead of LB agar. The results showed that colistin was also effectively inactivated by B. licheniformis DC-1 in CAMHM (Fig. 2; Fig. S1). Therefore, colistin-inactivating enzymes could exist in the fermentation broth of B. licheniformis DC-1.
FIG 2.
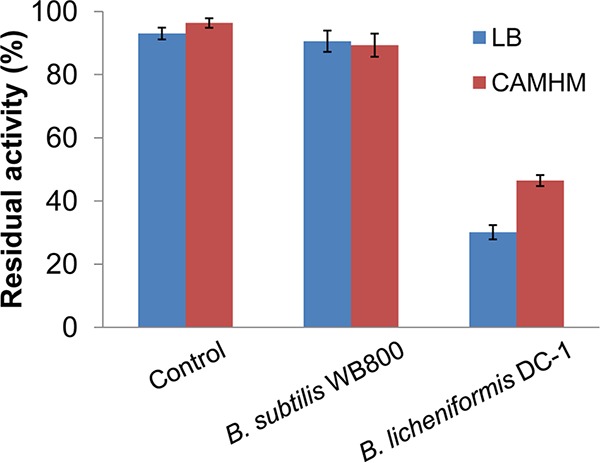
Colistin is degraded by the fermentation supernatant of Bacillus species bacteria. The residual antibacterial activity of colistin was determined by the cylinder-plate diffusion method in both LB agar and CAMHM. E. coli DH5α was used as the test strain. An amount of 200 μl of LB or CAMH broth (control) or fermentation supernatant of Bacillus bacteria was pipetted into stainless steel cylinder cups. The plates were left standing for 12 h at 4°C and then incubated at 37°C for 24 h. The diameters (in mm) of inhibition zones were accurately measured by vernier caliper. The residual antibacterial activity of colistin was calculated according to the following formula: residual activity (%) = (residual concentration/initial concentration) × 100. Each reaction was performed in triplicate. Error bars show standard errors of the means (SEM).
Next, we investigated the ability of B. licheniformis DC-1 to degrade other antimicrobial peptides (including PMB, bacitracin, and daptomycin). Among them, only PMB can be effectively degraded by B. licheniformis DC-1 (Fig. S2). These results suggest that the colistin-inactivating enzyme produced by B. licheniformis DC-1 is specific to polymyxins.
Polymyxin-inactivating enzyme is produced during entry into stationary phase.
To explore the relationship between cell growth and polymyxin-inactivating enzyme production in B. licheniformis DC-1, the residual antibacterial activity of colistin at various time points was determined. As shown by the results in Fig. 3, colistin was barely degraded by lag- and logarithmic-phase cultures. In contrast, colistin was rapidly degraded by stationary-phase cultures. Notably, more than 90% of colistin was degraded by bacterium-free culture supernatants after 26 h of fermentation. These results suggest that the polymyxin-inactivating enzyme may be produced and secreted by B. licheniformis DC-1 during entry into stationary phase.
FIG 3.
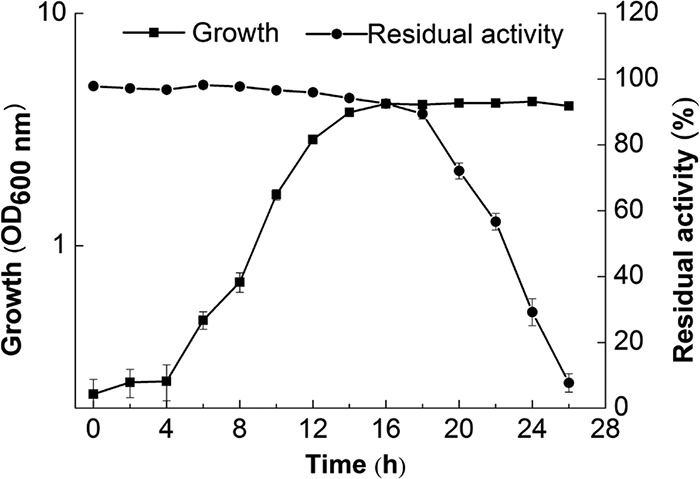
Colistin is degraded by B. licheniformis DC-1 during entry into stationary phase. Overnight culture was diluted 1:100 in 3 ml of fresh fermentation medium, and growth was recorded by measuring the OD600. Meanwhile, the residual antibacterial activity of colistin at various time points was determined by the cylinder-plate diffusion method. Data are shown as mean values ± standard errors of the means (SEM) from at least three experiments.
Isolation and identification of polymyxin-inactivating enzyme.
To isolate the polymyxin-inactivating enzyme from B. licheniformis DC-1, secreted proteins in fermentation supernatants were separated by SDS-PAGE. A large number of proteins were secreted by B. licheniformis DC-1 during logarithmic and stationary phases (Fig. 4A). It is notable that a band between ∼25 and 35 kDa (Fig. 4A, arrow) was observed in fermentation supernatant from stationary phase (Fig. 4A, lanes 1 and 4) but not in whole-cell lysate from stationary phase (Fig. 4A, lane 2) or fermentation supernatant from logarithmic phase (Fig. 4A, lane 3). Ammonium sulfate fractionation results showed that the fraction degrading colistin precipitated at 30% and 50% saturation (Fig. 4A, lanes 5 and 6; Fig. S3). The observation of this band is in accordance with the degradation of colistin as mentioned above (Fig. 3). Therefore, we speculated that this band might be involved in colistin degradation. The fraction containing the protein of interest was dialyzed, concentrated by centrifugal filters, and then subjected to a colistin degradation test. The addition of the concentrated fraction significantly decreased the inhibition zone of colistin, which is consistent with the results for the fermentation supernatant of B. licheniformis DC-1 (Fig. 4B). These data, collectively, indicate that the concentrated fraction possesses a component that inactivates colistin.
FIG 4.
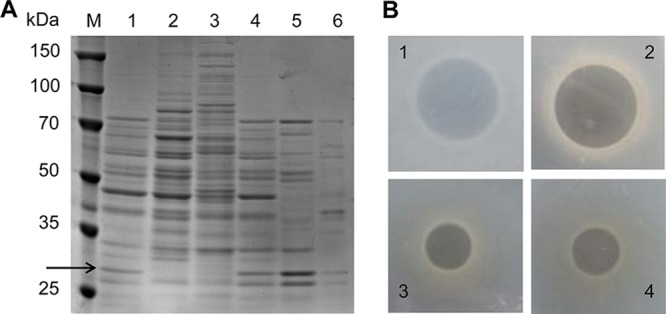
Isolation and purification of polymyxin-inactivating enzyme. (A) Analysis of secreted proteins by SDS-PAGE. Lane M, protein molecular weight marker; lanes 1 and 4, fermentation supernatant from stationary phase (incubation for 24 h); lane 2, whole-cell lysate from stationary phase (incubation for 24 h); lane 3, fermentation supernatant from logarithmic phase (incubation for 12 h); lane 5, 50% ammonium sulfate fraction; lane 6, 30% ammonium sulfate fraction. The arrow shows the predicted band. (B) Effects of different solutions on colistin inactivation. 1, phosphate-buffered saline (PBS); 2, LB broth; 3, fermentation supernatant of B. licheniformis DC-1 from stationary phase; 4, concentrated fraction. The residual antibacterial activity of colistin was determined by the cylinder-plate diffusion method.
To identify the protein of interest, the concentrated fraction was analyzed by nanoscale liquid chromatography-electrospray ionization-tandem mass spectrometry (NanoLC-ESI-MS/MS). In total, 17 proteins were predicted based on sequence alignment with the reference strain B. licheniformis ATCC 14580 (Table S1). However, nine of them were excluded because of high or low molecular weight (>35 kDa or <25 kDa). Among the remaining eight predicted proteins, Apr is a mature alkaline protease that contains a subtilisin Carlsberg-like catalytic domain. Given that polymyxins are polypeptides consisting of 10 amino acids, we speculated that the polymyxin-inactivating enzyme of B. Licheniformis DC-1 may be the alkaline protease Apr.
Amplification of the apr gene from B. licheniformis DC-1.
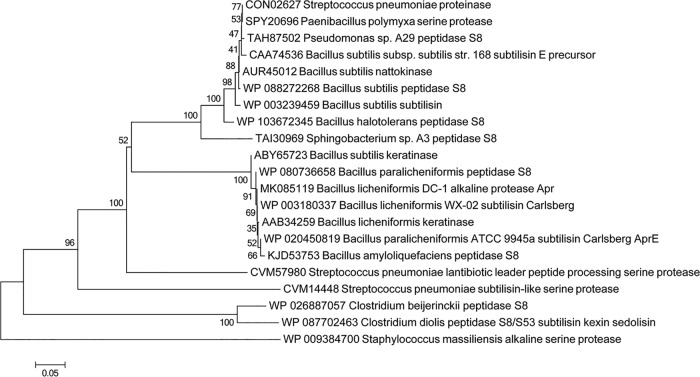
Although the genome of B. licheniformis DC-1 has not been sequenced, the 16S rRNA gene sequence is highly homologous to that of B. licheniformis strain WX-02, a halotolerant bacterium that produces poly-γ-glutamic acid (γ-PGA) (26). We designed primers according to the genome of B. licheniformis WX-02 to amplify the apr gene from B. licheniformis DC-1, together with its 5′ untranslated region (UTR). In total, a sequence of 1,521 bp was obtained, which contains an open reading frame (ORF) of 1,137 bp encoding a 379-amino-acid protein composed of a signal peptide (aa 1 to 29), a prosequence (aa 39 to 105), and a mature protein (aa 128 to 370). BLASTp alignment demonstrated that the sequence is identical to that of B. licheniformis WX-02 and shares extremely high homology with those of many strains of the genera Bacillus and Paenibacillus (Fig. 5). Apr homologs are also found in several other genera of Gram-positive bacteria, including the human pathogen Streptococcus pneumoniae. It is notable that two peptidases S8 from Gram-negative bacteria (Sphingobacterium sp. strain A3 and Pseudomonas sp. strain A29) share 64% sequence identity with the B. licheniformis Apr.
FIG 5.
Phylogenetic tree of B. Licheniformis DC-1 Apr. The tree was constructed using the neighbor-joining method. The data set was resampled 1,000 times by using the bootstrap option.
Heterogeneous expression of the apr gene.
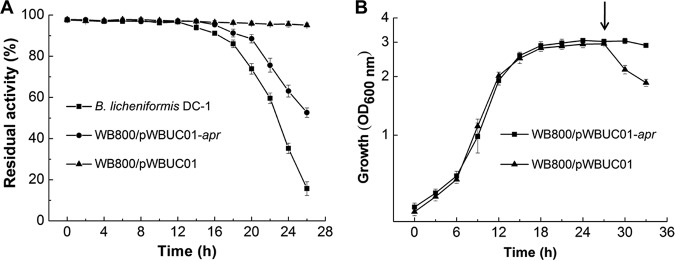
To confirm whether the Apr protein of B. licheniformis DC-1 is indeed a polymyxin-inactivating enzyme, the apr gene was expressed in the heterologous host B. subtilis WB800, which cannot degrade polymyxins, as mentioned above. pWBUC01, a shuttle plasmid for E. coli and B. subtilis, was employed to express exogenous genes under the control of P43, a strong constitutive promoter in B. subtilis (27). When we used this plasmid to clone B. licheniformis apr, we failed to obtain any B. subtilis transformants. It has been reported that overexpression of Apr is toxic and therefore lethal to the host cell (28). To avoid this, we removed the P43 promoter and sacB signal peptide from pWBUC01 and expressed B. licheniformis apr from its own promoter (Papr) and signal peptide (SP), resulting in recombinant strain B. subtilis WB800/pWBUC01-apr (Fig. S4). The ability to degrade colistin was assessed with bacterium-free culture supernatants of the recombinant strain (Fig. 6A). As expected, the inactivation of colistin did not occur in B. subtilis WB800 with empty plasmid under any test conditions. In contrast, colistin activity was significantly decreased by stationary-phase cultures of B. subtilis WB800/pWBUC01-apr, although the colistin degradation efficiency in the recombinant strain was slightly lower than that in B. licheniformis DC-1. Nearly 50% of colistin was inactivated by bacterium-free culture supernatant of B. subtilis WB800/pWBUC01-apr after 26 h of fermentation. These results suggest that the expression of B. licheniformis apr in B. subtilis WB800 produces a functional alkaline protease.
FIG 6.
The alkaline protease Apr is responsible for colistin inactivation. (A) Colistin is degraded by B. subtilis WB800/pWBUC01-apr. The residual antibacterial activity of colistin was determined by the cylinder-plate diffusion method. (B) Expression of the apr gene confers resistance to colistin during stationary phase. Growth was recorded by measuring the OD600. Colistin was added into the medium after incubation for 27 h, indicated by the arrow. Shown are either representative data or mean values ± SEM from at least three experiments.
To assess the role of Apr in colistin resistance, colistin was added to stationary-phase cultures (27 h) of these strains and the optical density at 600 nm (OD600) was measured (Fig. 6B). Upon exposure to colistin, the OD600 of B. subtilis WB800 harboring empty plasmid was substantially reduced, an indication of cell lysis. However, the addition of colistin did not affect the OD600 of B. subtilis WB800/pWBUC01-apr, indicating the inactivation of colistin by Apr. Therefore, the expression of the apr gene conferred the ability to grow in the presence of colistin. Based on these results, it can be concluded that alkaline protease Apr is responsible for the degradation of colistin in B. licheniformis DC-1.
Colistin is cleaved by Apr at two distinct sites.
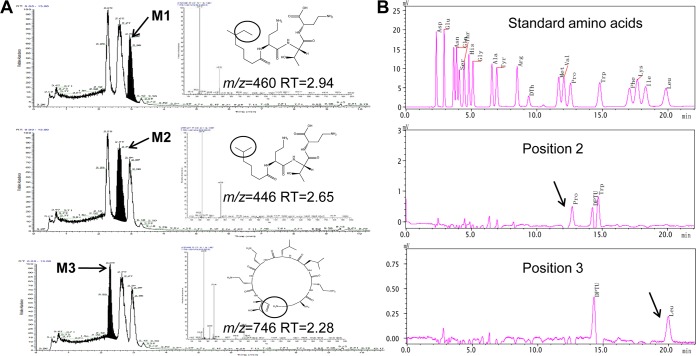
To determine the cleavage site(s) of Apr, the inactivation products of colistin were analyzed by LC-MS/MS. Colistin is composed of colistin A (m/z 1,169) (Fig. S5, peak A) and colistin B (m/z 1,155) (Fig. S5, peak B). The mass difference of 14 Da results from a methylene group at the fatty acid tail. After degradation by Apr for 4 h, three new inactivation products (peaks M1, M2, and M3) were detected (Fig. 7A). The masses were 460 Da for peak M1 and 446 Da for peak M2, corresponding to the tripeptide side chains of hydrolyzed colistin A and colistin B, respectively. Meanwhile, peak M3 (m/z 746) is consistent with the addition of a water molecule (18 Da) to the cyclic heptapeptide ring of colistin. These results suggest that Apr cleaves the peptide bond between the tripeptide side chain and the cyclic heptapeptide ring (positions 3 and 4). Besides this, Apr also cleaves another, unknown peptide bond within the cyclic heptapeptide ring moiety.
FIG 7.
Identification of the chemical structures of inactivation products of colistin. (A) Analysis of the three inactivation products by LC-MS/MS. (B) Analysis of the structure of the ring-opening heptapeptide by the Edman degradation method.
To explore the unknown cleavage site, the structure of the ring-opening heptapeptide was analyzed by the Edman degradation method (Fig. 7B). According to the 19-amino-acid standard (except for Cys), the third amino acid is easily determined to be Leu. Because of the structural similarity between Pro and l-Dab, the second amino acid is deduced to be l-Dab. Therefore, we can conclude that the ring-opening heptapeptide is l-Dab-l-Dab-d-Leu-l-Leu-l-Dab-l-Dab-l-Thr. In other words, Apr cleaves the peptide bond between l-Thr and l-Dab in the cyclic heptapeptide (positions 4 and 10) (Fig. 1).
DISCUSSION
Antibiotic resistance is mediated by several mechanisms, including prevention of access to targets, modification of targets, and inactivation of antibiotics by hydrolysis or modification (29). Polymyxins and other nonribosomal peptide antibiotics are highly effective against multidrug-resistant Gram-negative bacteria. Similar to the case for other types of antibiotics, modification of targets (outer membrane) and expression of efflux pumps are the two main strategies for polymyxin resistance. Here, we present evidence to demonstrate that polymyxins can be hydrolyzed by the alkaline protease Apr from B. licheniformis DC-1. Given that alkaline proteases are widely distributed in the genus Bacillus, polymyxin resistance could result from inactivation of antibiotics by hydrolysis.
A recent study based on global genome mining has demonstrated that many microorganisms harbor d-stereospecific peptidases which are involved in resistance to nonribosomal peptide antibiotics, such as polymyxin, vancomycin, and teixobactin (30). Consistently, daptomyxin and other bacterial cyclic peptides can be degraded by hydrolysis in Streptomyces spp. and Sphingosinicella spp. Although direct evidence is lacking, it is likely that serine proteases are responsible for inactivation of these antibiotics (31, 32). Furthermore, bacterial proteases are also active in degradation of defensins and antimicrobial peptides produced by the host. For example, extracellular protease OmpT is the enzyme that degrades the cationic antimicrobial peptide protamine in E. coli (33, 34). Collectively, these data indicate that hydrolytic cleavage by proteases may be a common mechanism underlying resistance to antimicrobial peptides.
It has been reported that colistin is specifically cleaved by colistinase II (purified from P. polymyxa) between l-Dab of the side chain and l-Dab in the adjacent cyclic heptapeptide (Dab-Dab bond) (19, 22). Our results confirm that the Dab-Dab bond in the side chain is the cleavage site of alkaline proteases. Moreover, we found that B. licheniformis Apr is able to cleave colistin at another site, the peptide bond between l-Thr and l-Dab in the cyclic heptapeptide (Thr-Dab bond) (Fig. 1). It is important to note that the former studies were carried out in vitro and that colistinase II is only one of the two components of the colistin-inactivating enzyme in P. polymyxa (19, 22). In this study, however, B. licheniformis Apr was successfully expressed in B. subtilis WB800 and the activity of the recombinant enzyme was determined in vivo. At present, we cannot distinguish whether the cleavage of two sites by Apr occurs sequentially or simultaneously. Except for the tripeptide side chain and the ring-opening heptapeptide, other intermediates are not observed in degradation products. Further work is required to determine the hydrolyzation process.
B. licheniformis Apr is produced only during the stationary growth phase (Fig. 2). This result is in agreement with several previous reports for other alkaline proteases (35, 36). In B. subtilis, the expression of extracellular proteases AprE and NprE is directly or indirectly regulated by multiple transcriptional regulators, including AbrB, DegU, Spo0A, ScoC, SinR, and CodY (36). Although the regulation of B. licheniformis apr remains unknown, it is possible that these regulators are also involved in the expression of Apr. The major function of AprE and NprE is to degrade extracellular proteins into amino acids for bacterial growth, but they are not essential. Additionally, the alkaline proteases play important roles in subtilin processing (37), poly-γ-glutamate synthesis (38), and quorum-sensing-signal production (39). Our results demonstrate that B. licheniformis Apr is required for bacterial survival upon colistin exposure during stationary phase. Given that a large number of antimicrobial peptides are produced by the genus Bacillus, especially at the beginning of stationary phase (40, 41), we suppose that alkaline proteases contribute to protection against antimicrobial peptides produced by themselves or their relatives.
Besides Bacillus, several other genera of Gram-positive bacteria possess proteins homologous to Apr, including the human pathogen S.pneumoniae (Fig. 5). Notably, two peptidases S8 from Gram-negative bacteria (Sphingobacterium sp. A3 and Pseudomonas sp. A29) share high sequence identity with Apr. The finding of inactivation of colistin by alkaline proteases is of great importance for clinical practice. It is possible that polymyxins will be destroyed by secreted proteases from other microorganisms, thus affecting the clinical efficacy of polymyxins. More importantly, it cannot be ruled out that the alkaline protease gene could be transferred to Gram-negative pathogens, resulting in inefficiency of this last-resort drug.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. For molecular cloning, bacterial cultures were grown in LB containing 5 g/liter yeast extract, 10 g/liter tryptone, and 10 g/liter NaCl at 37°C. For the polymyxin degradation test, Bacillus cells were grown in fermentation broth containing 60 g/liter d-glucose, 40 g/liter soya peptone, 3 g/liter Na2HPO4·12H2O, and 0.3 g/liter KH2PO4. When needed, the medium was supplemented with antibiotics at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| B. licheniformis DC-1 | Polymyxin-degrading bacterium | Laboratory stock |
| B. subtilis WB800 | Host for expression | 24 |
| B. subtilis WB800/pWBUC01 | B. subtilis WB800 with pWBUC01 | This study |
| B. subtilis WB800/pWBUC01-apr | B. subtilis WB800 with pWBUC01-apr | This study |
| E. coli DH5α | Colistin sensitive, host for cloning | Laboratory stock |
| Micrococcus luteus | Gramicidin and daptomyxin sensitive | Laboratory stock |
| Plasmids | ||
| pWBUC01 | Apr Kmr, E. coli-B. subtilis shuttle vectora | 27 |
| pWBUC01-apr | pWBUC01 carrying apr and its 5′ UTR | This study |
Apr, Ampicillin resistance; Kmr, kanamycin resistance.
Determination of the residual antibacterial activity of colistin.
The inactivation of colistin (0.05 μg/U; purchased from Zhejiang Qianjiang Biochemical Co., Ltd., Jiaxing, China) in bacterium-free culture supernatants was determined by the cylinder-plate diffusion method as described previously, with some modifications (42, 43). In brief, bacterial suspensions at various time points were centrifuged at 5,000 rpm for 5 min and then filtered through a 0.22-μm Millipore membrane. The E. coli DH5α test strain was grown in LB broth to an OD600 of 1.0. A total of 1.0 ml of bacterial suspension was inoculated into 20 ml of preheated LB agar medium. The medium was mixed evenly and poured into a petri dish. Colistin (final concentration of 10,000 U/ml) was added into the bacterium-free culture supernatants and the reaction solutions were allowed to stand at 37°C for 1 h. Next, 200 μl of each standard solution or reaction solution was pipetted into Oxford cups (stainless steel cylinders) placed on the agar surface. The plates were left standing for 12 h at 4°C and then incubated at 37°C for 24 h. The diameters (in millimeters) of inhibition zones were accurately measured by vernier caliper. All experiments were performed at least three times, and the results were corrected by the reference concentration (5,000 U/ml). A calibration curve for logarithms of concentrations (U/ml) of colistin versus zones of inhibition (mm) was plotted. The residual antibacterial activity (%) of colistin was calculated as follows: residual activity (%) = (residual concentration/initial concentration) × 100.
Characterization of B. licheniformis DC-1.
Because of its higher degradation rate, B. licheniformis DC-1 was chosen for further study. To determine bacterial growth, an overnight culture was diluted 1:100 in 50 ml of fresh fermentation medium, and the growth was recorded by measuring the OD600. To assess phylogeny, the genomic DNA was extracted using a bacterial genomic DNA extraction kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions, and then the 16S rRNA gene fragment was amplified by paired primers 27F (5′-AGAGTTTGATCATGGCTCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGACTT-3′). Afterwards, the PCR product was sequenced and analyzed using BLASTn.
Purification of polymyxin-inactivating hydrolase from B. licheniformis.
The overnight cultures were diluted into 50 ml of fresh fermentation broth for incubation at 37°C for 24 h. The bacterial suspension was centrifuged at 8,000 rpm for 10 min at 4°C. Anhydrous ethanol was added to the fermentation supernatant and the mixture centrifuged at 12,000 rpm for 20 min. The precipitate was dissolved into borate buffer (0.2 M, pH 9.0). Ammonium sulfate was added to the precipitating solution in different concentrations at 10, 20, 30, 40, 50, 60, and 70% saturation to obtain various fractions. Each precipitate was dissolved into borate buffer (0.2 M, pH 9.0) and subjected to the enzyme activity test. The protein fractions precipitated by (NH4)2SO4 at saturations of 30% and 50% were dissolved, and the solutions dialyzed for 24 h at 4°C in 0.2 M borate buffer. The fractions were concentrated with an Amicon ultra-15 centrifugal filter (10 kDa; Millipore). The concentrated fractions were analyzed by SDS-PAGE, digested with trypsin, and subjected to NanoLC-ESI-MS/MS analysis. Peptide mixtures were separated using the ThermoFisher Scientific Easy-nLC 1000 system and analyzed using a Thermo Scientific Q Exactive mass spectrometer.
Heterologous expression of the alkaline protease Apr.
To express the polymyxin-inactivating enzyme in B. subtilis WB800, the E. coli-B. subtilis shuttle vector pWBUC01 was employed (27). The recombinant plasmid was constructed using the ClonExpress II one-step cloning kit (Vazyme, Nanjing, China) (Fig. S4). Briefly, the backbone of plasmid pWBUC01 was amplified by paired primers WG01F (5′-CCGCTCGAGTTCAGTGCCGACCAAAACCA-3′) and WG01R (5′-CGCGGATCCTCCGCTCACAATTCCACACA-3′), and primers PaprF (5′-CCGCTCGAGATCTTTCACCCGTTTCTG-3′) and PaprR (5′-CGCGGATCCTTATTGAGCGGCAGCTTC-3′), designed according to the genome of B. licheniformis WX-02, were used to amplify the apr gene and its 5′ UTR from B. licheniformis DC-1 (XhoI/BamHI sites are underlined in both pairs of primers). This fragment was then cloned via XhoI/BamHI sites into the backbone of plasmid pWBUC01, resulting in the recombinant plasmid pWBUC01-apr. After being verified by sequencing, this plasmid was introduced by electroporation into B. subtilis WB800 as described previously (44).
Analysis of the inactivation products of colistin.
The purified polymyxin-inactivating hydrolase was added to the colistin solution at a final concentration of 10,000 U/ml, and the reaction solution was allowed to stand at 37°C for 1 h. Subsequently, the enzyme was separated by using the Amicon ultra-15 centrifugal filter unit (3 kDa; Millipore), and the filtrate containing inactivation products of colistin was collected. The products were analyzed by LC-MS/MS (Esquire 6000; Bruker, Bremen, Germany) (45). An aliquot of 20 μl of each sample was injected at a flow rate of 0.6 ml/min, using a YMC-Pack ODS-A reverse-phase C18 column (5-μm particle size, 150 mm by 4.6 mm; YMC Co. Ltd., Kyoto, Japan).
Accession number(s).
The amplified apr gene sequence from B. licheniformis DC-1 was deposited in GenBank under accession number MK085119.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by National Natural Science Foundation of China (grant number 31670114) and Provincial Natural Science Foundation of Zhejiang, China (grant number LY16C010002).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02378-18.
REFERENCES
- 1.Evans ME, Feola DJ, Rapp RP. 1999. Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant Gram-negative bacteria. Ann Pharmacother 33:960–967. doi: 10.1345/aph.18426. [DOI] [PubMed] [Google Scholar]
- 2.Falagas ME, Kasiakou SK, Saravolatz LD. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 4.Kwa A, Kasiakou SK, Tam VH, Falagas ME. 2007. Polymyxin B: similarities to and differences from colistin (polymyxin E). Expert Rev anti Infect Ther 5:811–821. doi: 10.1586/14787210.5.5.811. [DOI] [PubMed] [Google Scholar]
- 5.Velkov T, Thompson PE, Nation RL, Li J. 2010. Structure-activity relationships of polymyxin antibiotics. J Med Chem 53:1898–1916. doi: 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancock RE. 1997. Peptide antibiotics. Lancet 349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 7.Yu Z, Qin W, Lin J, Fang S, Qiu J. 2015. Antibacterial mechanisms of polymyxin and bacterial resistance. Biomed Res Int 2015:679109. doi: 10.1155/2015/679109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Z, Cai Y, Qin W, Lin J, Qiu J. 2015. Polymyxin E induces rapid Paenibacillus polymyxa death by damaging cell membrane while Ca2+ can protect cells from damage. PLoS One 10:e0135198. doi: 10.1371/journal.pone.0135198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Z, Zhu Y, Qin W, Yin J, Qiu J. 2017. Oxidative stress induced by polymyxin E is involved in rapid killing of Paenibacillus polymyxa. BioMed Res Int 2017:5437139. doi: 10.1155/2017/5437139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Z, Zhang L, Qin W, Yin J, Qiu J. 2019. Exogenous catalase stimulates the polymyxin E-induced rapid killing of Paenibacillus polymyxa. Int J Pept Res Ther 25:161–168. doi: 10.1007/s10989-017-9657-6. [DOI] [Google Scholar]
- 11.Falagas ME, Rafailidis PI, Matthaiou DK. 2010. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Update 13:132–138. doi: 10.1016/j.drup.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baron S, Hadjadj L, Rolain JM, Olaitan AO. 2016. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents 48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Jeannot K, Bolard A, Plésiat P. 2017. Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents 49:526–535. doi: 10.1016/j.ijantimicag.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 15.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 16.Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, Høiby N, Moskowitz SM. 2011. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 55:5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poole K. 2007. Efflux pumps as antimicrobial resistance mechanisms. Ann Med 39:162–176. doi: 10.1080/07853890701195262. [DOI] [PubMed] [Google Scholar]
- 18.Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki T, Hayashi K, Fujikawa K. 1963. Studies on the chemical structure of colistin. III. Enzymatic hydrolysis of colistin A. J Biochem 54:412–418. doi: 10.1093/oxfordjournals.jbchem.a127807. [DOI] [PubMed] [Google Scholar]
- 20.Ito M, Aida T, Koyama Y. 1966. Studies on the bacterial formation of a peptide antibiotic, colistin. Part I. On the enyzmatic inactivation of colistin by Bacillus colistinus. Agr Biol Chem 30:1112–1118. doi: 10.1080/00021369.1966.10858740. [DOI] [Google Scholar]
- 21.Woyczikowska B, Paśś L, Girdwoyń M, Raczyńska-Bojanowska K. 1973. Proteolytic degradation of polymyxins by the enzymes of Bacillus polymyxa. Acta Biochim Pol 20:285–296. [PubMed] [Google Scholar]
- 22.Ito-Kagawa M, Koyama Y. 1980. Selective cleavage of a peptide antibiotic, colistin by colistinase. J Antibiot (Tokyo) 33:1551–1555. doi: 10.7164/antibiotics.33.1551. [DOI] [PubMed] [Google Scholar]
- 23.Chihara S, Tobita T, Yahata M, Ito A, Koyama Y. 1973. Enzymatic degradation of colistin. Isolation and identification of α-N-acyl α,γ-diaminobutyric acid and colistin nonapeptide. Agr Biol Chem 37:2455–2463. doi: 10.1271/bbb1961.37.2455. [DOI] [Google Scholar]
- 24.Wu SC, Yeung JC, Duan Y, Ye R, Szarka SJ, Habibi HR, Wong SL. 2002. Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall-bound protease on antibody fragment production. Appl Environ Microbiol 68:3261–3269. doi: 10.1128/AEM.68.7.3261-3269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei X, Ji Z, Chen S. 2010. Isolation of halotolerant Bacillus licheniformis WX-02 and regulatory effects of sodium chloride on yield and molecular sizes of poly-gamma-glutamic acid. Appl Biochem Biotechnol 160:1332–1340. doi: 10.1007/s12010-009-8681-1. [DOI] [PubMed] [Google Scholar]
- 27.Shen M, Chen Z, Mao X, Wang L, Liang J, Huo Q, Yin X, Qiu J, Sun D. 2018. Two different restriction-modification systems for degrading exogenous DNA in Paenibacillus polymyxa. Biochem Biophys Res Commun 504:927–932. doi: 10.1016/j.bbrc.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 28.Tang XM, Shen W, Lakay FM, Shao WL, Wang ZX, Prior BA, Zhuge J. 2004. Cloning and over-expression of an alkaline protease from Bacillus licheniformis. Biotechnol Lett 26:975–979. doi: 10.1023/B:BILE.0000030042.91094.38. [DOI] [PubMed] [Google Scholar]
- 29.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 30.Li YX, Zhong Z, Hou P, Zhang WP, Qian PY. 2018. Resistance to nonribosomal peptide antibiotics mediated by d-stereospecific peptidases. Nat Chem Biol 14:381–387. doi: 10.1038/s41589-018-0009-4. [DOI] [PubMed] [Google Scholar]
- 31.Kato H, Tsuji K, Harada KI. 2009. Microbial degradation of cyclic peptides produced by bacteria. J Antibiot (Tokyo) 62:181–190. doi: 10.1038/ja.2009.8. [DOI] [PubMed] [Google Scholar]
- 32.D'Costa VM, Mukhtar TA, Patel T, Koteva K, Waglechner N, Hughes DW, Wright GD, De Pascale G. 2012. Inactivation of the lipopeptide antibiotic daptomycin by hydrolytic mechanisms. Antimicrob Agents Chemother 56:757–764. doi: 10.1128/AAC.05441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stumpe S, Schmid R, Stephens DL, Georgiou G, Bakker EP. 1998. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J Bacteriol 180:4002–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guina T, Yi EC, Wang H, Hackett M, Miller SI. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J Bacteriol 182:4077–4086. doi: 10.1128/JB.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kole MM, Draper I, Gerson DF. 2007. Production of protease by Bacillus subtilis using simultaneous control of glucose and ammonium concentrations. J Chem Technol Biotechnol 41:197–206. doi: 10.1002/jctb.280410305. [DOI] [Google Scholar]
- 36.Barbieri G, Albertini AM, Ferrari E, Sonenshein AL, Belitsky BR. 2016. Interplay of CodY and ScoC in the regulation of major extracellular protease genes of Bacillus subtilis. J Bacteriol 198:907–920. doi: 10.1128/JB.00894-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corvey C, Stein T, Düsterhus S, Karas M, Entian KD. 2003. Activation of subtilin precursors by Bacillus subtilis extracellular serine proteases subtilisin (AprE), WprA, and Vpr. Biochem Biophys Res Commun 304:48–54. doi: 10.1016/S0006-291X(03)00529-1. [DOI] [PubMed] [Google Scholar]
- 38.Kada S, Ishikawa A, Ohshima Y, Yoshida KI. 2013. Alkaline serine protease AprE plays an essential role in poly-γ-glutamate production during natto fermentation. Biosci Biotechnol Biochem 77:802–809. doi: 10.1271/bbb.120965. [DOI] [PubMed] [Google Scholar]
- 39.Lanigan-Gerdes S, Dooley AN, Faull KF, Lazazzera BA. 2007. Identification of subtilisin, Epr and Vpr as enzymes that produce CSF, an extracellular signalling peptide of Bacillus subtilis. Mol Microbiol 65:1321–1333. doi: 10.1111/j.1365-2958.2007.05869.x. [DOI] [PubMed] [Google Scholar]
- 40.Stein T. 2005. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol 56:845–857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- 41.Sumi CD, Yang BW, Yeo IC, Hahm YT. 2015. Antimicrobial peptides of the genus Bacillus: a new era for antibiotics. Can J Microbiol 61:93–103. doi: 10.1139/cjm-2014-0613. [DOI] [PubMed] [Google Scholar]
- 42.Costa MCN, Barden AT, Andrade JM, Oppe TP, Schapoval EE. 2014. Quantitative evaluation of besifloxacin ophthalmic suspension by HPLC, application to bioassay method and cytotoxicity studies. Talanta 11:367–374. doi: 10.1016/j.talanta.2013.10.051. [DOI] [PubMed] [Google Scholar]
- 43.Dafale NA, Semwal UP, Agarwal PK, Sharma P, Singh GN. 2015. Development and validation of microbial bioassay for quantification of levofloxacin in pharmaceutical preparations. J Pharm Anal 5:18–26. doi: 10.1016/j.jpha.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue GP, Johnson JS, Dalrymple BP. 1999. High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis. J Microbiol Methods 34:183–191. doi: 10.1016/S0167-7012(98)00087-6. [DOI] [Google Scholar]
- 45.Xu Y, Tian X, Ren C, Huang H, Zhang X, Gong X, Liu H, Yu Z, Zhang L. 2012. Analysis of colistin A and B in fishery products by ultra performance liquid chromatography with positive electrospray ionization tandem mass spectrometry. J Chromatogr B 899:14–20. doi: 10.1016/j.jchromb.2012.04.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.