Klebsiella aerogenes is a nosocomial pathogen associated with drug resistance and outbreaks in intensive care units. In a 5-month period in 2017, we experienced an increased incidence of cultures for carbapenem-resistant K. aerogenes (CR-KA) from an adult cardiothoracic intensive care unit (CICU) involving 15 patients.
KEYWORDS: AmpD, carbapenem-resistant Klebsiella aerogenes, MLST, ST4, cardiothoracic intensive care unit, genomic epidemiology, integrative conjugative element, outbreak, porins, yersiniabactin
ABSTRACT
Klebsiella aerogenes is a nosocomial pathogen associated with drug resistance and outbreaks in intensive care units. In a 5-month period in 2017, we experienced an increased incidence of cultures for carbapenem-resistant K. aerogenes (CR-KA) from an adult cardiothoracic intensive care unit (CICU) involving 15 patients. Phylogenomic analysis following whole-genome sequencing (WGS) identified the outbreak CR-KA isolates to group together as a tight monoclonal cluster (with no more than six single nucleotide polymorphisms [SNPs]), suggestive of a protracted intraward transmission event. No clonal relationships were identified between the CICU CR-KA strains and additional hospital CR-KA patient isolates from different wards and/or previous years. Carbapenemase-encoding genes and drug-resistant plasmids were absent in the outbreak strains, and carbapenem resistance was attributed to mutations impacting AmpD activity and membrane permeability. The CICU outbreak strains harbored an integrative conjugative element (ICE) which has been associated with pathogenic Klebsiella pneumoniae lineages (ICEKp10). Comparative genomics with global K. aerogenes genomes showed our outbreak strains to group closely with global sequence type 4 (ST4) strains, which, along with ST93, likely represent dominant K. aerogenes lineages associated with human infections. For poorly characterized pathogens, scaling analyses to include sequenced genomes from public databases offer the opportunity to identify emerging trends and dominant clones associated with specific attributes, syndromes, and geographical locations.
INTRODUCTION
Klebsiella aerogenes (formerly described as Enterobacter aerogenes) is a ubiquitous member of the Enterobacteriaceae family and a significant nosocomial pathogen associated with drug resistance and a wide variety of infections, including pneumonia, bacteremia, and urinary tract and surgical site infections (1–3). K. aerogenes infections can arise endogenously (gastrointestinal flora) or be acquired from surroundings in the facility where the patient is admitted (horizontal transmission through colonized health care workers, contaminated devices/shared equipment, other patients, etc.), with the most critical risk factor for acquiring infection being prolonged broad-spectrum antibiotic administration (4). Risk factors for K. aerogenes infections include prolonged stay at health care facilities, especially for patients who are immunosuppressed, on mechanical ventilation, or harbor foreign devices (4). Numerous hospital ward outbreaks in both pediatric and adult populations due to K. aerogenes have been described due to a common source (5, 6) or spread via patient-to-patient transmission (7–10). A particularly high frequency of hospital intensive care unit (ICU) outbreaks was continually reported from Western Europe in the period between the 1990s and early 2000s and was largely attributed to the spread and endemic establishment of a clonal K. aerogenes strain harboring the extended-spectrum β-lactamase (ESBL) TEM-24 (blaTEM-24) (2, 11, 12).
Within the United States and other regions across the globe, K. aerogenes has also been reported, along with Klebsiella pneumoniae, Enterobacter cloacae, and Escherichia coli, to be among the frequently isolated carbapenem-resistant Enterobacteriaceae (CRE) (13–15). Clinical carbapenem-resistant K. aerogenes (CR-KA) strains harboring plasmid-borne serine carbapenemases have been described in the United States and worldwide, while metallo-β-lactamases and OXA-48 have been reported in Europe, Asia, and Brazil (2, 16). However, the primary mechanisms underlying resistance to carbapenems in K. aerogenes are considered carbapenemase independent and attributed to chromosomal AmpC β-lactamase overexpression and mutations affecting membrane permeability (2). The latter has been well documented in K. aerogenes with reports describing mutations impacting porin function/expression that can arise in vivo during antibiotherapy (17), be reversible (2), and present complex diagnostic and therapeutic management challenges (18).
Despite the role of K. aerogenes as an important opportunistic pathogen and its epidemic potential, the clinical relevance of intraspecies genetic diversity and significance of specific sequence types (STs) remain unknown. In comparison, in the genomically more closely related K. pneumoniae and, to some extent, in E. cloacae, clonal complexes and STs associated with geographical distribution, multidrug resistance, hospital outbreaks, and disease syndromes have been defined (19, 20). Recently, a multilocus sequence typing (MLST) scheme has been developed for K. aerogenes that can help explore the above-mentioned issues; however, its performance in evaluating and discriminating clinical/environmental isolates has not yet been reported.
We pursued this study to investigate an outbreak of CR-KA in a cardiothoracic intensive care unit (CICU) at our hospital, which persisted for 5 months despite aggressive infection control measures. The primary goals of our study included whole-genome sequencing (WGS)-based investigation of the clonal relationships among the CR-KA strains isolated from patients in our hospital and defining putative loci associated with carbapenem resistance and virulence. In addition, the recently developed publicly available K. aerogenes MLST scheme afforded us the opportunity to delineate the population structure of CR-KA strains isolated from patients at our hospital. Our initial findings led us to broadly investigate the origin and significance of specific K. aerogenes sequence types identified in our hospital CR-KA strains by performing comparative genomics using publicly available global K. aerogenes genomes.
RESULTS
Epidemiological and genomic characterization of the CICU CR-KA cluster.
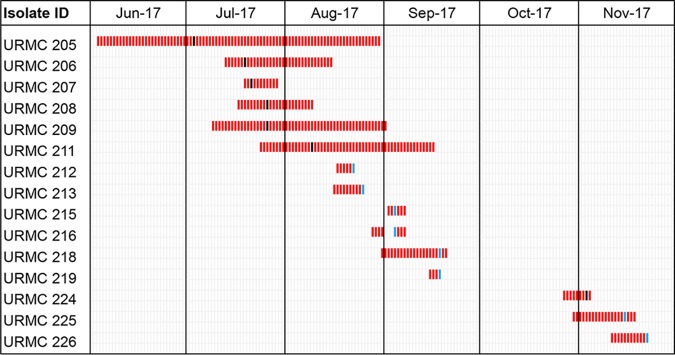
In July 2017, five CICU patients had CR-KA isolated from respiratory tract specimens (unit occupancy and patient demographics are detailed in Fig. 1 and in Table S1 in the supplemental material). The first identified case (patient A) had a past medical history significant for intravenous drug use and recurrent methicillin-resistant Staphylococcus aureus infections. Patient A’s CICU course is described in Fig. S1. The temporal association of subsequent positive cultures from patients in the CICU prompted an outbreak investigation.
FIG 1.
Patient occupancy and overlap in CICU ward during the K. aerogenes outbreak. Black bars, first positive CR-KA clinical cultures; blue bars, first positive CR-KA surveillance cultures.
In response to the event, the Infection Prevention team implemented universal contact precautions, weekly surveillance cultures, distribution of educational materials pertaining to hand hygiene, equipment disinfection, and the use of UV irradiation for all patient rooms as census allowed. Infected patients were separated into a cohort on one side of the unit with dedicated nursing staff. Environment-of-care rounds were performed in the CICU and all cardiac operating rooms. Limited environmental surveillance cultures (n = 4; hand-washing sinks near/inside the ward) were performed and were negative. Antibiotic stewardship efforts were also reinforced during the investigation.
Despite interventions, 10 additional CICU patients had positive CR-KA cultures between August and November 2017 (Fig. 1 and Table S1). Of the total 15 positive cultures from unique patients, 6 were clinical specimens (5 respiratory and 1 blood), and 9 were rectal surveillance cultures. Epidemiological investigation of possible risk factors among the CICU patients for developing CR-KA infection/colonization was performed. A retrospective case-control study (cases, n = 15; controls, n = 30) revealed no significant differences for gender, age, mortality, unit location, dialysis, ventilator support, transport from another hospital, or carbapenem exposure (Table S2). Infected patients were significantly more likely to have undergone a surgical procedure (P = 0.04) but were not found to be associated with a specific operating room (data not shown). After November 2017, no CR-KA strains were isolated in either clinical or surveillance specimens. Active surveillance in the ward was discontinued at the end of January 2018, and the outbreak was deemed to have subsided.
Antibiotic susceptibility testing (AST) of the CICU CR-KA strains revealed some differences in phenotypic susceptibilities to carbapenems and cephalosporins (Table 1 and Data Set S1). Sixty percent of the CICU cluster strains were resistant to three carbapenems (ertapenem, imipenem, and meropenem), while 93% showed phenotypic resistance to ertapenem only. Susceptibility to cefepime also varied, with a subset of outbreak strains displaying phenotypic resistance (Table 1; Data Set S1).
TABLE 1.
Phenotypic antibiotic susceptibility profiles of K. aerogenes strains in this studya
| URMC strain group and no.b | Amikacin | Gentamicin | Tobramycin | Ciprofloxacin | Moxifloxacin | Trimethoprim-sulfa | Piperacillin-tazobactam | Ceftriaxone | Cefepime | Ertapenem | Imipenem | Meropenem |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2017 CICU-associated CR-KA strains | ||||||||||||
| 205c | S | S | S | S | S | S | R | R | S | R | R | R |
| 206 | S | S | S | S | S | S | R | R | S | R | R | R |
| 207 | S | S | S | S | S | S | R | R | S | R | R | R |
| 208 | S | S | S | S | S | S | R | R | S | S | S | R |
| 209 | S | S | S | S | S | S | R | R | S | R | R | R |
| 211 | S | S | S | S | S | S | R | R | D | R | R | R |
| 212 | S | S | S | S | S | S | R | R | R | R | I | R |
| 213 | S | S | S | S | S | S | R | R | R | R | I | R |
| 215 | S | S | S | S | S | S | R | R | S | R | R | R |
| 216 | S | S | S | S | S | S | R | R | S | R | S | S |
| 218 | S | S | S | S | S | S | R | R | S | R | R | R |
| 219 | S | S | S | S | S | S | R | R | S | R | R | R |
| 224 | ND | S | S | S | S | S | R | R | R | R | ND | R |
| 225 | S | S | S | S | S | S | R | R | R | R | R | R |
| 226 | S | S | S | S | S | S | R | R | R | R | I | I |
| Other CR-KA strains | ||||||||||||
| 200 | S | S | S | S | ND | S | R | R | R | R | ND | R |
| 202 | S | S | S | S | S | S | R | R | S | R | R | R |
| 203 | S | S | S | S | S | S | R | R | S | R | R | R |
| 204 | S | S | S | S | ND | S | R | R | S | R | ND | R |
| 210 | S | S | S | S | ND | S | R | R | S | R | ND | S |
| 214 | S | S | S | S | ND | S | I | R | S | R | I | S |
| 217 | S | S | S | R | ND | S | R | R | S | R | I | S |
| 221 | S | S | S | S | ND | S | S | S | S | R | ND | S |
| 222 | S | S | S | S | S | S | R | R | R | R | I | S |
| Controlsd | ||||||||||||
| 201 | S | S | S | S | S | S | S | S | S | S | I | S |
| 223 | S | S | S | S | ND | S | S | S | S | S | ND | S |
R, resistant; S, sensitive; I, intermediate; D, dose dependent; ND, not determined. See Data Set S1 in the supplemental material for MIC values and Kirby-Bauer disk zone sizes and interpretations.
All strain numbers are prefixed by URMC.
Patient A (first case).
URMC 223, carbapenem susceptible; URMC 201, carbapenem intermediate.
To address the possibility of an outbreak, particularly in light of variable antibiotic susceptibilities, WGS was undertaken to identify phylogenetic relationships and potential transmission links. Clinical strains detected between July and August were sequenced retrospectively, while isolates from clinical specimens and surveillance cultures obtained between August and November were sequenced prospectively as part of the active-outbreak investigation. A total of 26 University of Rochester Medical Center (URMC) K. aerogenes isolates were sequenced using Illumina WGS. These included 15 CR-KA isolates from patients admitted in the CICU between June and November 2017 and an additional set of 9 CR-KA strains epidemiologically unlinked to the CICU cluster strains (patient isolates from between 2015 and 2017) (Table S3) for context and comparison. Two clinical isolates were included as controls, URMC 201 (intermediate susceptibility to imipenem) and URMC 223 (susceptible to all carbapenems tested). The AST profiles of the non-CICU CR-KA study strains are also described in Table 1.
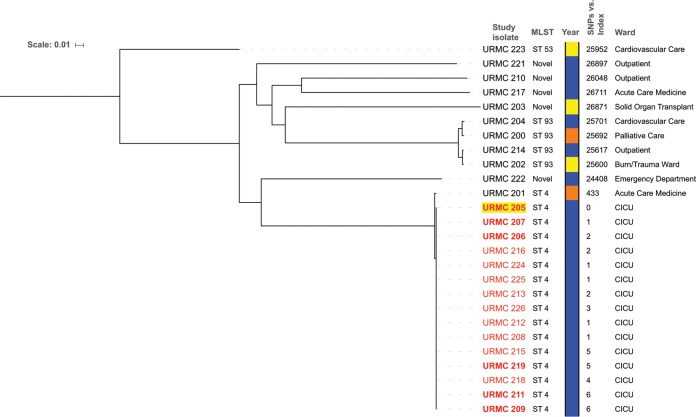
The 26 sequenced URMC K. aerogenes genomes showed high genomic coverage (>88%) relative to that of the K. aerogenes KCTC 2190 reference strain (ATCC 13048T; GenBank accession number NC_015663.1) (Data Set S2). Single nucleotide polymorphisms (SNPs) were identified across the study genomes relative to the reference sequence (pairwise SNP differences ranged from 1 to 28,170) (Data Set S3). MLST assignment indicated that all of the CICU clinical and surveillance isolates belonged to ST4, and SNP-based analyses grouped them in a tight cluster separately and distantly from nonoutbreak isolates (Fig. 2; Data Set S3). Within the CICU cluster, strains differed from URMC 205 (first case, patient A isolate) by no more than 6 SNPs. In addition, these isolates bore identical plasmid profiles (based on replicon and plasmid typing) (Table S5). In contrast, the 2017 non-CICU isolates were significantly distant from the CICU outbreak isolates (>20,000 SNPs). The most closely related non-CICU CR-KA strain was URMC 201 (isolated in 2015). This strain was also ST4, with 433 SNPs in a pairwise comparison to the sequence of URMC 205. The next most closely related non-CICU isolates, identified by pairwise SNP comparisons, belonged to ST93: the pair URMC 214 and URMC 202 (244 SNPs apart) and the pair URMC 200 and URMC 204 (517 SNPs apart) (Fig. 2; Data Set S3).
FIG 2.
Dendogram showing pairwise SNP differences based on the phylogenetic relatedness of URMC K. aerogenes strains. Whole-genome sequence of K. aerogenes KCTC 2190 (ATCC 13048) was used for reference mapping. Discriminatory high-quality SNPs in the core genomes obtained by the CFSAN SNP pipeline were used to plot the tree (excluding mobile elements and putative recombination sites). CICU outbreak strains are identified by number (in red); clinical isolates are in bold, and the patient A isolate (1st case) is highlighted in yellow. The year of strain isolation is color coded as follows: orange, 2015; yellow, 2016; blue 2017. SNP differences relative to patient A are shown. The scale bar indicates the number of nucleotide substitutions per site.
A potential limitation of mapping-based SNP identification approaches using a divergent reference and distantly related genomes is the possible underestimation of variation and decreased analyses resolution (21). To address this, intracluster comparative SNP analysis was performed for the CICU CR-KA isolates. The genome of CR-KA isolated from the earliest case (patient A, strain URMC 205) was de novo assembled and used as a mapping reference, and the rest of the closely related CICU CR-KA cluster strains were exclusively used as query genomes. The resulting phylogenetic analyses showed the strains to differ from URMC 205 by not more than 7 SNPs and from each other in a pairwise comparison by less than 11 SNPs (Table S4 and Data Set 4). These relationships, coupled with the epidemiological data, indicated the 2017 CICU CR-KA cluster to be a monoclonal outbreak.
Carbapenem resistance in the URMC CR-KA strains was driven by adaptive chromosomal gene alterations.
(i) WGS-based identification of acquired antibiotic resistance genes. Despite phenotypic carbapenem resistance, the CICU CR-KA strains did not harbor genes for carbapenemases, extended-spectrum β-lactamases, or plasmid-borne AmpC cephalosporinases. A single nonoutbreak CR-KA isolate (URMC 203) harbored a carbapenemase gene (blaNMC-A). No horizontally acquired genes conferring resistance to non-β-lactam antibiotics were identified among the study strains, consistent with their susceptibility profiles (Table 1).
(ii) Nonsynonymous sequence alterations in key chromosomal loci implicated in carbapenem resistance. In the absence of genes encoding carbapenemases and ESBLs in the CR-KA outbreak strains, variations in other genetic loci associated with carbapenemase-independent resistance mechanisms were investigated (22–24). By focusing on the AmpC cephalosporinase and outer membrane porins, sequence variations in ampD, ampG, ampR, omp35, omp36, and ompR genes in the study strains were assessed relative to the wild-type alleles in the carbapenem-susceptible reference type strain KCTC 2190 (25). The sequences were also compared to alleles in URMC 223 (carbapenem susceptible) and URMC 201 (intermediate susceptibility). For the omp genes, the upstream DNA sequences were also assessed. The ampG and ompR genes were wild type in all study strains. Variants were identified in all other loci and are described below.
(a) ampD. Mutations were identified in the ampD gene for each of the 24 CR-KA isolates in this study (Table 2), while the control strains URMC 223 and URMC 201 bore the wild-type ampD allele. The outbreak strains harbored single missense SNPs (either 284G→T or 482G→A) in ampD, resulting in a Trp95Leu or Arg161His substitution. Six nonoutbreak CR-KA strains harbored independent nonsynonymous single substitutions while two missense substitutions were identified in URMC 221. A single nonoutbreak strain, URMC 202, harbored a nonsense mutation resulting in a truncated AmpD protein (Table 2). DNA sequence corresponding to the ampD allele was absent in isolate URMC 203 (the only study strain to possess a carbapenemase-encoding gene blaNMC-A) due to a large deletion in the genomic region harboring ampD and the neighboring ampE gene.
TABLE 2.
Nonsynonymous SNPs in the ampD gene associated with carbapenem resistance in the URMC K. aerogenes isolates
| URMC strain group and no.a | SNP relative to ampD wild-type allelec | SNP effect on encoded AmpD sequence |
|---|---|---|
| CICU outbreak-associated CR-KA strains | ||
| 205,b 207, 209, 211, 212, 213, 215, 218, 219, 224, 225, 226 | 482G→A | Arg161His |
| 206, 208, 216 | 284G→T | Trp95Leu |
| Outbreak-unrelated CR-KA strains | ||
| 200 | 338T→G | Ile113Ser |
| 202 | 412C→Td | |
| 204 | 117C→T | Pro39Ser |
| 210 | 335C→T | Ser112Leu |
| 214 | 280G→A | Ala94Thr |
| 217 | 496G→C | Gly166Arg |
| 221 | 478A→G | Ile160Val |
| 501C→A | Ala168Asp | |
| 222 | 492T→G | Glu164Asp |
All strain numbers are prefixed by URMC. Control strains were URMC 201 and URMC 203; no sequencing reads mapped to the ampD gene in URMC 203. Isolates additionally resistant to cefepime are underlined.
Index patient isolate.
Unless noted otherwise, all SNPs produced missense mutations.
This nonsense mutation produced the substitution Gln138X, resulting in a premature stop codon.
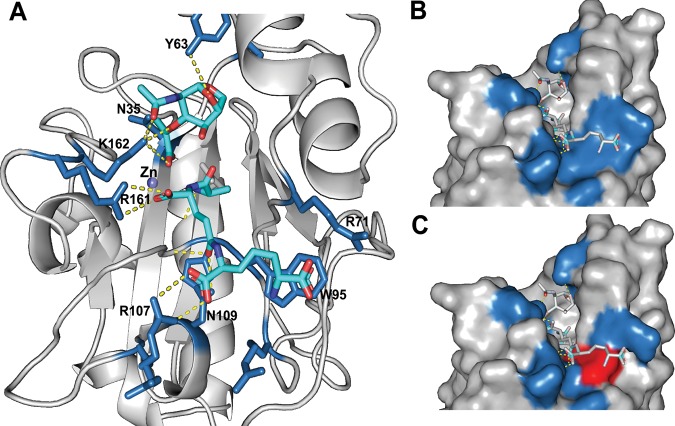
The potential impact of a Trp95Leu or Arg161His substitution on AmpD activity in the outbreak strains was investigated by homology-based structural modeling of K. aerogenes AmpD using the high-resolution crystal structure of Citrobacter freundii AmpD (26), a close homolog (83.33% amino acid sequence identity). AmpD contains a hydrophobic surface to accommodate its GlcNAc-anh-MurNAc (N-acetylglucosaminyl-β-1,4-anhydro-N-acetylmuramic acid) ligand, which is made up of tripeptide and glycan moieties. The tripeptide portion of GlcNAc-anh-MurNAc is coordinated through three salt bridges between carboxyl groups on the tripeptide and residues Arg71, Arg161, and Arg107, while the peptide backbone is oriented across the hydrophobic surface. Trp95 forms a planar surface at the end of the ligand-binding channel to position the diaminopimelate moiety at the distal end of the tripeptide.
Based on the in silico K. aerogenes AmpD model, the positively charged guanidinium group of Arg161 forms two strong electrostatic interactions with the carboxyl group of d-glutamine on the tripeptide portion of the ligand (Fig. 3). Mutation of this residue to histidine was predicted to weaken ligand binding, likely affecting the positioning of the ligand in the active site. In the Trp95Leu substitution, the hydrophobicity in the region is preserved, but the shorter length of the leucine side chain leaves a gap at the end of the binding channel, likely affecting the positioning of the entire ligand (Fig. 3). These observations lend support to the idea that the missense mutations observed in the CICU outbreak strains would alter ligand docking on AmpD, likely reducing/inhibiting its activity. Four out of the seven CR-KA nonoutbreak strains had substitutions within glycan- and peptide-interacting regions of AmpD (Table 2), suggesting that these alterations might also impact activity.
FIG 3.
Computational modeling of the impact of ampD mutations on AmpD in URMC outbreak CR-KA strains. (A) K. aerogenes AmpD modeled on the C. freundii AmpD structure. Key residues interacting with glycan and peptide portions of ligand are shown. (B and C) Surface models of K. aerogenes AmpD depicting the wild-type binding surface for the diaminopimelate moiety (B, W95) and the binding surface of AmpD containing the W95L mutation (C; blue surfaces indicate amino acid positions from panel A, and the altered surface is highlighted in red).
(b) ampR. All of the outbreak strains harbored the reference ampR allele. Several nonoutbreak CR-KA strains harbored two to five substitutions that likely represented variant alleles as they were also observed in the control carbapenem-susceptible strain URMC 223. A single nonoutbreak CR-KA strain, URMC 210, bore a nonsense mutation in the ampR gene, resulting in a premature stop codon (Trp117X) which likely resulted in a nonfunctional truncated AmpR protein.
(c) omp35 and omp36. Among the 15 outbreak CR-KA strains, three different omp36 variants were identified relative to the wild-type allele (Table 3). An identical profile of missense SNPs in these strains and the control strain, URMC 201 (intermediate carbapenem susceptibility), was observed relative to the reference genome allele. Seven outbreak strains had additional mutations that resulted in severely truncated proteins. Several nonoutbreak CR-KA strains also harbored missense SNPs of unclear significance. Two strains, URMC 202 and URMC 204, harbored distinct frameshift mutations yielding truncated Omp36 protein variants. A 42-bp region of high variation (nucleotides 680 to 724), corresponding to 15/16 amino acid substitutions in loop L5, was observed in all the clinical isolates relative to the reference sequence (Table 3; Fig. S2). The hypervariable region results in a different charge profile in the region and has been previously reported in a K. aerogenes study describing imipenem-resistant clinical isolates (harboring ESBL TEM-24) from patients in France (27). The predicted Omp36 proteins in clinical strain URMC 221 and the carbapenem-susceptible control strain (URMC 223) bore 87% identity relative to the sequence of the reference genome Omp36 and were considered significantly distant variants (not included in the comparative analyses).
TABLE 3.
Nonsynonymous genetic mutations in omp36 genes and resulting alterations in Omp36 sequence in the URMC K. aerogenes isolates
| URMC strain group and no.a | Type of mutation in omp36b
|
Effect of mutation on Omp36 sequencec
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| FS | HV region | NS SNP | MS SNP | FS | HV region | PS | Truncation | Single substitution(s) | |
| CICU outbreak associated CR-KA strains | |||||||||
| 205,d 207 | + | + | 175A→G, 564T→C, 566A→G | pAsp91ThrfsX12 | 102X | + | Ile59Val | ||
| 206, 208, 212, 213, 216, 224, 225, 226 | + | 175A→G, 564T→C, 566A→G | + | Ile59Val, Asp189Gly | |||||
| 209, 211, 215, 218, 219 | + | 184A→T | 175A→G, 564T→C, 566A→G | 62X | + | Ile59Val | |||
| Outbreak unrelated CR-KA strains | |||||||||
| 200, 203, 210, 214 | + | 564T→C, 566A→G, 615 T→G, 834T→C, 835A→G | + | Asp189Gly, Asp205Glu, Asn279Asp | |||||
| 202 | + | + | 564T→C, 566A→G, 615 T→G, 834T→C, 835A→G | pTrp77LysfsX2 | 78X | + | Trp77Lys | ||
| 204 | + | + | 564T→C, 566A→G, 615 T→G, 834T→C, 835A→G | pSer304ProfsX21 | + | 324X | + | Asp189Gly, Asp205Glu, Asn279Asp | |
| 217 | + | 564T→C, 566A→G, 569T→A | + | Asp189Gly, Phe190Tyr | |||||
| 222 | + | 175A→G,564T→C, 566A→G, 615 T→G | + | Ile59Val, Asp189Gly, Asp205Glu | |||||
| Control strain | |||||||||
| 201e | + | 175A→G, 564T→C, 566A→G | + | Ile59Val, Asp189Gly | |||||
All strain numbers are prefixed by URMC. Isolates additionally resistant to cefepime are underlined.
Changes shown are nucleotide positions relative to the wild-type sequence. FS, frameshift; HV, hypervariable region (nucleotides 680 to 724); NS, nonsense; MS, missense; SNP, single nucleotide polymorphism. Plus signs indicate the presence of the feature.
Changes shown are amino acid positions relative to the wild-type sequence. FS, frameshift; HV, hypervariable region (amino acids 226 to 241); PS, premature stop codon.
Index patient isolate.
Intermediate carbapenem resistance.
All but one of the study CR-KA strains had the wild-type allele of omp35. A single strain (URMC 202) bore a deletion in the 5' end of the gene. The DNA sequence upstream of the omp35 gene was investigated to identify mutations in the promoter sites of the strains, of which one (URMC 204) had a nucleotide difference of unclear significance at the −22 position.
Identification of a large pathogenicity-associated integrative and conjugative element, ICEKp10, in the CICU outbreak CR-KA strains.
The prolonged nature of the clonal CR-KA outbreak in our CICU led us to search for pathogenicity loci that could have promoted persistence and transmission. A cluster of chromosomally located genes encoding yersiniabactin (ybt) metallophore and colibactin (clb) genotoxin systems were identified in all of the outbreak strains (Fig. 4). These loci have been implicated in invasive infections due to pathogenic lineages of K. pneumoniae (28).
FIG 4.
Yersiniabactin- and colibactin-encoding gene loci on integrative conjugative element ICEKp10 in the CICU outbreak CR-KA strains. Blue arrows, integrase-encoding genes; brown arrows, Zn2+/Mn2+ modules; gray arrows, genes encoding mobilization proteins; pink arrows, vir-T4SS.
The ybt locus harbored putative genes involved in regulation as well as synthesis of the siderophore, corresponding transport-associated proteins, and a receptor protein for the uptake of metal-bound siderophore. The clb locus included putative homologs encoding enzymes, transferases, and transport proteins involved in production and secretion of the polyketide colibactin (Fig. 4; Data Set S5). Investigation of the genomic loci associated with the virulence factor gene cluster identified them to be present on a mobilizable integrative conjugative element (ICE) inserted in a tRNA-Asn site adjacent to a gene encoding the glycine cleavage system (Fig. 4). The ICE bore a modular arrangement of gene clusters encoding mobile elements, P4-like integrase, type IV secretion system (T4SS) conjugation machinery, and mobilization genes (Fig. 4; Data Set S5). The element was identified to be ICEKp10 using BLAST (99% identity to the ICEKp10 mobile element in K. pneumoniae strain 16703761; GenBank accession number KY454634) and a recently described virulence genomic typing scheme for Klebsiella spp. (28).
Among the URMC nonoutbreak K. aerogenes strains, these loci were identified in only 4/10 isolates, which were either ST4 or ST93. The control strain URMC 223 harbored the yersiniabactin locus exclusively. Detailed descriptions of these elements in the URMC K. aerogenes strains are given in Table S5.
Comparative analyses of URMC CR-KA and publicly available K. aerogenes genomes.
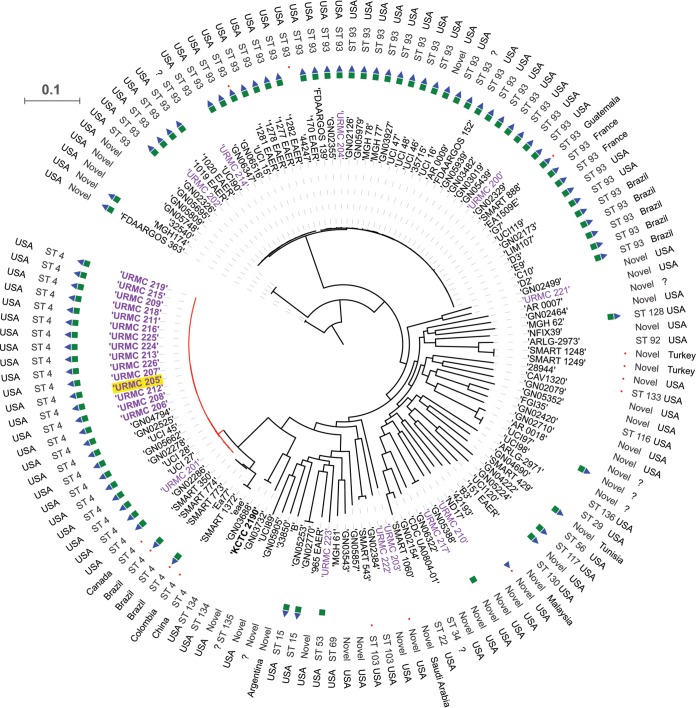
To gain insights into the emergence and epidemiology of the CICU outbreak clones and to place our hospital CR-KA strains in the broader context of global K. aerogenes strains, comparative phylogenomic analyses were performed using the Harvest genomics suite (29). Publicly available K. aerogenes genome assemblies (n = 110) were included in the analyses. These included 71 clinical and surveillance strains isolated from human specimens, 3 environmental strains, and 36 strains of unknown origin (Data Set S6). Based on the newly described MLST scheme, ST4 and ST93 strains were found to be markedly overrepresented in the available genomes (51.8%, or 57/110).
Excellent correlation was observed between Harvest-generated tree topologies (Fig. 5), as well as pairwise SNP differences (Data Set S7), and those generated by the CFSAN SNP pipeline for the URMC CR-KA study strains. Based on Harvest analyses, the CICU outbreak strains clustered closely with each other and with other global ST4 genomes compared to clustering of the other URMC CR-KA strains (outbreak unrelated), which were distantly dispersed throughout the phylogenomic distribution (Fig. 5). The MLST-based sequence types of URMC CR-KA strains and the global K. aerogenes genomes also correlated tightly with Harvest-generated core-genome-based topologies (Fig. 5).
FIG 5.
Harvest-based phylogenomic comparisons of URMC K. aerogenes genomes with global K. aerogenes genomes. Discriminatory SNPs based on core-genome comparisons were used to plot the tree. URMC study K. aerogenes strains are shown in purple (outbreak strains in bold; index patient strain highlighted in yellow). The presence of genes encoding a yersiniabactin siderophore system (green squares), colibactin synthesis cluster (blue triangles), and carbapenemases (red circles) in the assembled genomes is shown. Scale bar indicates the number of nucleotide substitutions per site.
Six publicly available assembled genomes grouped closely with the CICU outbreak strain genomes (<200 SNPs apart). These included ST4 strains UCI 27, UCI 28, and UCI 45, prospective isolates collected in the year 2013 from patients in Irvine, CA (described in a carbapenem resistance surveillance study by Cerqueria et al. [30]), and GN04794, GN05662, and GN02525, representing strains derived from various U.S. patient clinical specimens (blood, sputum, and wound drainage) in the years 2012, 2013, and 2007, respectively (Fig. 5; Data Set S7). The above-mentioned strains had fewer SNP differences in relation to the CICU outbreak strains than URMC 201, the closest and only nonoutbreak ST4 strain isolated in our hospital (Fig. 5; Data Set S7).
Carbapenemase-encoding genes were identified in a total of 13/110 global K. aerogenes genome assemblies (11.8%) (Fig. 5). These included genes encoding KPC-2 (n = 7), KPC-3 (n = 1), OXA-48 (n = 4), and NDM-6 (n = 1). The chromosomal serine carbapenemase, blaNMC-A, was solely present in URMC 203, a CR-KA strain isolated from a patient in our hospital in 2015. Among the 30 ST4 genomes, 4 strains (13%) harbored genes encoding carbapenemases (4/4, KPC-2), and these were clinical strains isolated from non-U.S. patients. The relative contribution of carbapenemase-mediated versus non-carbapenemase-mediated mechanisms of resistance to carbapenems in the global K. aerogenes strains could not be assessed due to the absence of antibiotic susceptibility metadata for most strains.
A characteristic genomic feature of the URMC CICU outbreak strains and a subset of non-outbreak-associated strains was the presence of the yersiniabactin siderophore and colibactin systems. Using the program Kleborate (28), we investigated the distribution and organization of these systems in the global K. aerogenes genomes to identify associations, if any, with specific STs and geographical regions (Fig. 5; Data Set S8). The prevalences of yersiniabactin- and colibactin-encoding systems in the global K. aerogenes genomes were found to be 53.64% (59/110) and 52.72% (58/110), respectively. A 100% association was found between the presence of colibactin in the genomes and the concurrent presence of yersiniabactin. Higher prevalence of the virulence cluster was observed in ST4 and ST93, with 85% (12/14) and 95% (42/44), respectively, although these two STs were also the most abundantly represented in the available set of genomes. The other STs were less well represented in the study set (n < 3), so the prevalence of these systems in them could not be accurately established. Interestingly, the strains exclusively designated environmental isolates (B3, FGI35, and B) did not harbor genes encoding the above-mentioned virulence systems (Fig. 5; Data Set S8). These trends correlated with analyses of our 26 hospital strains, where 75% of the ST93 strains (3/4) were positive for yersiniabactin and colibactin systems, while none of the strains with novel/unassigned STs harbored genes encoding the same (0%, 5/5).
DISCUSSION
This WGS study was initiated in order to establish the molecular epidemiology of CR-KA strains isolated from patients in our hospital following an outbreak event. Data from prospective WGS performed during the period between August and November 2017 informed the Infection Prevention team regarding the extent of the transmission event and effectiveness of infection control interventions. However, the source of the outbreak and how transmission occurred were not identified. The protracted nature of the outbreak and the significant number of cases at our institution highlight that non-carbapenemase-producing CRE also represent an important threat in health care systems. Additionally, K. aerogenes is a relatively poorly understood opportunistic pathogen. The population structure and clinically important clones are undefined while genetic attributes associated with multidrug resistance and virulence have not been detailed and led us to perform additional comparative analyses.
Infections due to carbapenem-resistant organisms present complex diagnostic and therapeutic management challenges, and a better understanding of how adaptive or acquired resistance emerges in the health care environment is needed (31). In clinical CR-KA strains, carbapenem resistance has been associated with either carbapenemase production or coupling of adaptive mutations affecting membrane permeability and AmpC hyperproduction (2). However, chromosomal mutations associated with AmpC overexpression in CR-KA strains have not been reported in the literature. In our study, 23/24 CR-KA isolates were found to harbor mutations in genes involved in the synthesis or regulation of the inducible AmpC cephalosporinase (ampD and ampR) and outer membrane porins (omp 35 and omp36). A majority of mutations were found to be within the open reading frames of ampD and omp36 (catalogued in Tables 2 and 3).
While the role of AmpD in AmpC expression in E. cloacae has been well established (32), there are limited and conflicting reports regarding the role of AmpD in adaptive carbapenem resistance in K. aerogenes strains (33, 34). In silico modeling of K. aerogenes AmpD using the C. freundii AmpD crystal structure (26) predicts that substitutions resulting from SNPs in the outbreak strains likely impact enzymatic activity (Fig. 3). Separate ampD mutations in several non-CICU CR-KA isolates were also found to result in substitutions that could affect substrate-enzyme interactions (Table 2). This signifies that missense mutations in ampD likely play an important role in AmpC-mediated carbapenem resistance in K. aerogenes. Studies have also reported a minority of E. cloacae clinical isolates to have chromosomal ampR mutations that can confer high-level constitutive AmpC expression (35, 36). To our knowledge, this is the first report of ampR mutation associated with carbapenem resistance in a clinical K. aerogenes isolate (URMC 210). A single non-CICU strain harbored a carbapenemase-encoding gene, blaNMC-A. NmcA has been reported in E. cloacae (37) but had not been described in K. aerogenes previously.
In clinical CR-KA strains, mutations in omp36 have been described, and functional studies investigating their impact have been reported previously (11, 17). A diverse array of mutations was identified in the omp36 gene among our study strains, including nonsynonymous mutations resulting in frameshifts or premature stop codons resulting in truncated and likely nonfunctional Omp36 variants (Table 3). Additional SNPs relative to the carbapenem-susceptible reference genome resulted in substitutions of unknown significance in predicted β-sheets and extracellular loop regions (Table 3; see Fig. S2 in the supplemental material). Interestingly, mutations resulting in substitutions in the “eyelet” region of internal loop 3 were not identified in our study CR-KA strains. A highly conserved region in the OmpC/Omp36 family of porins, the internal loop 3 forms a constriction pore that regulates β-lactam penetration, and mutations in this region have been reported to be associated with carbapenem resistance in K. aerogenes (27, 38, 39).
Within the CICU CR-KA monoclonal cluster, heterogeneity was observed in ampD and omp36 genes (Tables 2 and 3). These genetic loci likely represent mutational hot spots associated with adaptive and reversible carbapenem resistance in K. aerogenes. The testing and archiving of single isolated carbapenem-resistant colonies instead of multiple colonies during individual patient specimen workup likely represent a limitation that did not allow us to capture the full complement of CR-KA strain microdiversity associated with individual patients during the outbreak event.
The detailed significance of alleles and mutations described above in development of carbapenem resistance in K. aerogenes needs to be verified by additional genetic (allelic exchange and complementation) and biochemical approaches. Complex regulatory networks, including transcriptional activators, sensor kinases, and two-component systems, have been implicated in adaptive drug resistance in Enterobacteriaceae spp. (24), and their roles in contributing toward carbapenem resistance in our study strains also cannot be ruled out.
Despite the lack of horizontally acquired drug resistance elements, the outbreak strains in this study managed to persist for several months in the face of an active infection prevention effort, prompting us to assess determinants that could promote transmission/persistence. The outbreak CR-KA strains harbored an ICE encoding the metallophore yersiniabactin (Ybt) and genotoxin colibactin (Clb) systems (Fig. 4; Table S6), raising the intriguing possibility that the element was instrumental in the success of this clone. A Ybt-encoding pathogenicity island has been described in association with a prolonged nationwide outbreak in the Netherlands involving multiple hospitals and >100 patients due to a multidrug-resistant Enterobacter hormaechei clone (40). A recent study by Lam et al. described the prevalence of ybt to be higher among the global carbapenemase-associated K. pneumoniae clonal group CG258 (40%) and hypervirulent K. pneumoniae clonal group CG23 (∼87%) than in the wider K. pneumoniae population (∼32%) (28). The study also reported a significant association of Ybt with an increased risk of invasive infections (bacteremia, liver abscesses, etc.). Ybt was first described in pathogenic Yersinia spp., encoded by a chromosomal gene cluster, termed the high-pathogenicity island (HPI), with a critical role in iron scavenging during infection (41). Subsequent studies reported acquisition of the HPI in other clinical Enterobacteriaceae strains (28, 42), and additional functions ascribed to Ybt include evasion of host lipocalin-2 (43) and sequestration/import of heavy metals (44). The polyketide colibactin is frequently associated with yersiniabactin and has been shown to induce chromosomal instability and DNA damage in eukaryotic cells (45). CRE surveillance studies (carbapenem-resistant isolates, years 2013 to 2018) at our institution did not identify concurrent or recent outbreaks due to ICEKp10-harboring K. pneumoniae strains in our hospital CICU (data not shown). The outbreak clone could represent an entry of a K. aerogenes strain already harboring the ICE; however, recent acquisition from HPI-harboring K. pneumoniae strains silently colonizing patients cannot be ruled out. These findings present new avenues for research investigating the role of ICEs encoding Ybt and Clb in the pathogenicity and transmission of clinical K. aerogenes strains and other Enterobacteriaceae. While horizontal transmission of carbapenemases via mobile elements is increasingly recognized as a major public health issue (46), the transmission of mobilizable virulence factors presents an underappreciated threat in the health care environment that warrants more surveillance.
In order to set our hospital strains (outbreak and nonoutbreak) into a broader context, core-genome comparisons and MLST were used to examine the population structure of our strains relative to that of global K. aerogenes strains pulled from public databases (Fig. 5; Data Sets S6 and S8). This analysis is the first evaluation of the nascent K. aerogenes MLST scheme in discriminating clinical K. aerogenes isolates. The scheme was found to be robust, with distribution of STs correlating closely with topologies based on K. aerogenes strain core genomes. Our preliminary analyses suggest that ST4 and ST93 might be dominant global clones associated with K. aerogenes infections. Outbreak CR-KA strains clustered closest to other clinical U.S. ST4 strains, suggesting a clonal expansion of this ST (Fig. 5; Data Set S7). These strains were isolated from patients in the years 2007 to 2013 in Irvine, CA, and in undisclosed parts of the United States. It is noteworthy that the ST4 group also included carbapenemase-producing K. aerogenes isolates from international sites (Fig. 5; Data Set S8). These included CR-KA strains associated with drug-resistant intra-abdominal and urinary tract infections in patient samples from Brazil, Canada, Colombia, and China that had been sequenced as part of the SMART (Study for Monitoring Antimicrobial Resistance Trends) large surveillance study (47). ST93 was the most prevalent sequence type in the global K. aerogenes assembled genomes (43/110, or 39%), with wide global geographical distribution, including the United States (Fig. 5). Four of the 11 non-CICU CR-KA strains from our hospital belonged to this group. Two closely related ST93 isolates, K. aerogenes 1509E and G7, have been described as representatives of clonal strains associated with multiple multidrug-resistant K. aerogenes outbreaks in France (48, 49). Incidentally, global ST4 and ST93 isolates also had higher prevalences of HPI harboring ybt and clb (85% and 95%, respectively). Apart from the HPI, additional genetic traits or metabolic capabilities may also play a role in the success of these STs. These potentially high-risk clones need to be examined more closely by undertaking large-scale studies with strains from diverse global sites and patient populations. This will help in the examination of niche adaptation, emergence of antibiotic resistance, and evolution of pathogenicity, leading to a better understanding of K. aerogenes.
In summary, genomic approaches for surveillance and outbreak investigations have emerged as critical functions for infection prevention and diagnostic microbiology laboratories. Along with evaluating the effectiveness of infection measures and the dimension of transmission events, WGS applied across sets of local and global isolates is a powerful approach for identifying emerging clinically relevant trends.
MATERIALS AND METHODS
Setting, study design, K. aerogenes strains and metadata.
The University of Rochester Medical Center (URMC) is an 830-bed tertiary-care medical center, with a 14-bed cardiothoracic intensive care unit, serving the Greater Rochester Area, New York. Following approval by the University of Rochester Institutional Review Board (RSRB00068143), a total of 26 K. aerogenes strains isolated from patients at URMC in the course of regular clinical care and/or surveillance efforts were selected for the study. Each isolate corresponded to a single first CR-KA strain isolated during the course of hospitalization. Historical and contemporary patient K. aerogenes isolates epidemiologically unlinked to the CICU outbreak were also included in the study for context and comparison. Ward occupancy and pertinent clinical and epidemiological information were obtained through review of patient medical records and the laboratory information system and are described in Tables S1 and S3 in the supplemental material.
Clinical case-control study.
For investigating patient risk factors associated with CR-KA infection/colonization, a retrospective case-control analysis was performed for patients admitted in the CICU in the outbreak period (July to November 2017; CRE cases, n = 15; controls, n = 30). Patient demographics and mortality data, unit location, carbapenem exposure, transfer record, procedures, and surgical histories were collected by chart review (Table S2). Significance was determined by a two-tailed Student’s t test and z test for continuous and categorical variables, respectively. The significance level cutoff was set at a P value of ≤0.05.
AST of the study K. aerogenes strains.
Antibiotic susceptibility testing (AST) of the study strains was performed as part of routine diagnostics using a Vitek2 (bioMérieux, France) system and/or Kirby-Bauer disk diffusion methods. AST interpretations were based on interpretive criteria defined by the Clinical and Laboratory Standards Institute (50).
Sequencing library preparation and raw data acquisition.
The isolates were cultured on standard laboratory medium from archived frozen stocks, examined for purity, and reidentified by Vitek matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (bioMérieux, France). Dual-indexed sequencing libraries were prepared from genomic DNA extracted from single colonies and sequenced on an Illumina Miseq benchtop sequencer (Illumina, San Diego, CA) at the URMC Genomic Core Facilities (see the supplemental material for detailed methods).
Genomic analyses.
Analyses were performed using an in-house bioinformatics pipeline, URMC Bacterial Genomic Analysis Pipeline (version 2.0.6), run on a high-performance computer cluster at the Center for Integrated Research Computing at the University of Rochester (see the supplemental material for detailed methods).
In silico protein analyses.
For homology modeling, SWISS-MODEL (51) was used to thread K. aerogenes AmpD (GenBank accession number WP_015704411.1; amino acids 1 to 187) through the structure of C. freundii AmpD (26) (PDB accession number 2Y2C; amino acids 1 to 187). The overall quaternary structure of K. aerogenes AmpD was predicted with high precision (95% confidence, 99% coverage). Comparative analyses and imaging of protein structures were performed with PyMOL (52). JalView was used to create alignments (53).
Data availability.
WGS and metadata corresponding to the study URMC K. aerogenes isolates were deposited at NCBI under BioProject accession number PRJNA504784. Individual isolates were deposited under BioSample accession numbers SAMN10405413 to SAMN10405438 (Data Set S2).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the URMC Clinical Microbiology Laboratories and Infection Prevention staff in specimen processing, data collection, and epidemiological investigations. We acknowledge the URMC Genomics Research Center for support with WGS. We also thank Steve Gill (URMC, Genomics Research Center) for reviewing the manuscript draft.
Internal funding from the University of Rochester Department of Pathology and Laboratory Medicine supported this study.
We report no conflicts of interest relevant to this article.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02577-18.
REFERENCES
- 1.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 2.Davin-Regli A, Pagès J-M. 2015. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol 6:392. doi: 10.3389/fmicb.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang CI, Chung DR, Ko KS, Peck KR, Song JH, Korean N, For S, Of ID. 2012. Clinical predictors of Enterobacter bacteremia among patients admitted to the ED. Am J Emerg Med 30:165–169. doi: 10.1016/j.ajem.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Sanders WE Jr, Sanders CC. 1997. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin Microbiol Rev 10:220–241. doi: 10.1128/CMR.10.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards KE, Allen JR, Miller MJ, Yogev R, Hoffman PC, Klotz R, Marubio S, Burkholder E, Williams T, Davis AT. 1978. Enterobacter aerogenes primary bacteremia in pediatric patients. Pediatrics 62:304–306. [PubMed] [Google Scholar]
- 6.Loiwal V, Kumar A, Gupta P, Gomber S, Ramachandran VG. 1999. Enterobacter aerogenes outbreak in a neonatal intensive care unit. Pediatr Int 41:157–161. [DOI] [PubMed] [Google Scholar]
- 7.Salso S, Culebras E, Andrade R, Picazo JJ. 2003. Outbreak of TEM-24-producing Enterobacter aerogenes in a Spanish hospital. Microb Drug Resist 9:299–305. doi: 10.1089/107662903322286517. [DOI] [PubMed] [Google Scholar]
- 8.Piagnerelli M, Kennes B, Brogniez Y, Deplano A, Govaerts D. 2000. Outbreak of nosocomial multidrug-resistant Enterobacter aerogenes in a geriatric unit: failure of isolation contact, analysis of risk factors, and use of pulsed-field gel electrophoresis. Infect Control Hosp Epidemiol 21:651–653. doi: 10.1086/501704. [DOI] [PubMed] [Google Scholar]
- 9.Neuwirth C, Siebor E, Lopez J, Pechinot A, Kazmierczak A. 1996. Outbreak of TEM-24-producing Enterobacter aerogenes in an intensive care unit and dissemination of the extended-spectrum beta-lactamase to other members of the family Enterobacteriaceae. J Clin Microbiol 34:76–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davin-Regli A, Saux P, Bollet C, Gouin F, De Micco P. 1996. Investigation of outbreaks of Enterobacter aerogenes colonisation and infection in intensive care units by random amplification of polymorphic DNA. J Med Microbiol 44:89–98. doi: 10.1099/00222615-44-2-89. [DOI] [PubMed] [Google Scholar]
- 11.De Gheldre Y, Maes N, Rost F, De Ryck R, Clevenbergh P, Vincent JL, Struelens MJ. 1997. Molecular epidemiology of an outbreak of multidrug-resistant Enterobacter aerogenes infections and in vivo emergence of imipenem resistance. J Clin Microbiol 35:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertrand X, Hocquet D, Boisson K, Siebor E, Plesiat P, Talon D. 2003. Molecular epidemiology of Enterobacteriaceae producing extended-spectrum beta-lactamase in a French university-affiliated hospital. Int J Antimicrob Agents 22:128–133. doi: 10.1016/S0924-8579(03)00098-0. [DOI] [PubMed] [Google Scholar]
- 13.Guh AY, Bulens SN, Mu Y, Jacob JT, Reno J, Scott J, Wilson LE, Vaeth E, Lynfield R, Shaw KM, Vagnone PM, Bamberg WM, Janelle SJ, Dumyati G, Concannon C, Beldavs Z, Cunningham M, Cassidy PM, Phipps EC, Kenslow N, Travis T, Lonsway D, Rasheed JK, Limbago BM, Kallen AJ. 2015. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012–2013. JAMA 314:1479–1487. doi: 10.1001/jama.2015.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HJ, Choi JK, Cho SY, Kim SH, Park SH, Choi SM, Lee DG, Choi JH, Yoo JH. 2016. Carbapenem-resistant Enterobacteriaceae: prevalence and risk factors in a single community-based hospital in Korea. Infect Chemother 48:166–173. doi: 10.3947/ic.2016.48.3.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robert J, Pantel A, Merens A, Lavigne JP, Nicolas-Chanoine MH, Group O. 2014. Incidence rates of carbapenemase-producing Enterobacteriaceae clinical isolates in France: a prospective nationwide study in 2011-12. J Antimicrob Chemother 69:2706–2712. doi: 10.1093/jac/dku208. [DOI] [PubMed] [Google Scholar]
- 16.Franolić I, Bedenić B, Beader N, Lukić-Grlić A, Mihaljević S, Bielen L, Zarfel G, Meštrović T. 2019. NDM-1-producing Enterobacter aerogenes isolated from a patient with a JJ ureteric stent in situ. CEN Case Rep 8:38–41. doi: 10.1007/s13730-018-0360-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philippe N, Maigre L, Santini S, Pinet E, Claverie J-M, Davin-Régli A-V, Pagès J-M, Masi M. 2015. In vivo evolution of bacterial resistance in two cases of Enterobacter aerogenes infections during treatment with imipenem. PLoS One 10:e0138828. doi: 10.1371/journal.pone.0138828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 19.Wyres KL, Holt KE. 2016. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol 24:944–956. doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Gomez-Simmonds A, Annavajhala MK, Wang Z, Macesic N, Hu Y, Giddins MJ, O’Malley A, Toussaint NC, Whittier S, Torres VJ, Uhlemann A-C. 2018. Genomic and geographic context for the evolution of high-risk carbapenem-resistant Enterobacter cloacae complex clones ST171 and ST78. mBio 9:e00542-18. doi: 10.1128/mBio.00542-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuter S, Ellington MJ, Cartwright EJ, Koser CU, Torok ME, Gouliouris T, Harris SR, Brown NM, Holden MT, Quail M, Parkhill J, Smith GP, Bentley SD, Peacock SJ. 2013. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med 173:1397–1404. doi: 10.1001/jamainternmed.2013.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupont H, Choinier P, Roche D, Adiba S, Sookdeb M, Branger C, Denamur E, Mammeri H. 2017. Structural alteration of OmpR as a source of ertapenem resistance in a CTX-M-15-producing Escherichia coli O25B:H4 sequence type 131 clinical isolate. Antimicrob Agents Chemother 61:e00014-17. doi: 10.1128/AAC.00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pages JM, James CE, Winterhalter M. 2008. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 25.Lavigne J-P, Sotto A, Nicolas-Chanoine M-H, Bouziges N, Bourg G, Davin-Regli A, Pagès J-M. 2012. Membrane permeability, a pivotal function involved in antibiotic resistance and virulence in Enterobacter aerogenes clinical isolates. Clin Microbiol Infect 18:539–545. doi: 10.1111/j.1469-0691.2011.03607.x. [DOI] [PubMed] [Google Scholar]
- 26.Carrasco-López C, Rojas-Altuve A, Zhang W, Hesek D, Lee M, Barbe S, André I, Ferrer P, Silva-Martin N, Castro GR, Martínez-Ripoll M, Mobashery S, Hermoso JA. 2011. Crystal structures of bacterial peptidoglycan amidase AmpD and an unprecedented activation mechanism. J Biol Chem 286:31714–31722. doi: 10.1074/jbc.M111.264366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiolas A, Bornet C, Davin-Régli A, Pagès J-M, Bollet C. 2004. Resistance to imipenem, cefepime, and cefpirome associated with mutation in Omp36 osmoporin of Enterobacter aerogenes. Biochem Biophys Res Commun 317:851–856. doi: 10.1016/j.bbrc.2004.03.130. [DOI] [PubMed] [Google Scholar]
- 28.Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, Brisse S, Holt KE. 2018. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom 4:9. doi: 10.1099/mgen.0.000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerqueira GC, Earl AM, Ernst CM, Grad YH, Dekker JP, Feldgarden M, Chapman SB, Reis-Cunha JL, Shea TP, Young S, Zeng Q, Delaney ML, Kim D, Peterson EM, O’Brien TF, Ferraro MJ, Hooper DC, Huang SS, Kirby JE, Onderdonk AB, Birren BW, Hung DT, Cosimi LA, Wortman JR, Murphy CI, Hanage WP. 2017. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc Natl Acad Sci U S A 114:1135–1140. doi: 10.1073/pnas.1616248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodman KE, Simner PJ, Tamma PD, Milstone AM. 2016. Infection control implications of heterogeneous resistance mechanisms in carbapenem-resistant Enterobacteriaceae (CRE). Expert Rev Anti Infect Ther 14:95–108. doi: 10.1586/14787210.2016.1106940. [DOI] [PubMed] [Google Scholar]
- 32.Babouee Flury B, Ellington MJ, Hopkins KL, Turton JF, Doumith M, Loy R, Staves P, Hinic V, Frei R, Woodford N. 2016. Association of novel nonsynonymous single nucleotide polymorphisms in ampD with cephalosporin resistance and phylogenetic variations in ampC, ampR, ompF, and ompC in Enterobacter cloacae isolates that are highly resistant to carbapenems. Antimicrob Agents Chemother 60:2383–2390. doi: 10.1128/AAC.02835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzouvelekis LS, Tzelepi E, Kaufmann ME, Mentis AF. 1994. Consecutive mutations leading to the emergence in vivo of imipenem resistance in a clinical strain of Enterobacter aerogenes. J Med Microbiol 40:403–407. doi: 10.1099/00222615-40-6-403. [DOI] [PubMed] [Google Scholar]
- 34.Babouee Flury B, Ellington MJ, Hopkins KL, Turton JF, Doumith M, Woodford N. 2016. The differential importance of mutations within AmpD in cephalosporin resistance of Enterobacter aerogenes and Enterobacter cloacae. Int J Antimicrob Agents 48:555–558. doi: 10.1016/j.ijantimicag.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 35.Kuga A, Okamoto R, Inoue M. 2000. ampR gene mutations that greatly increase class C beta-lactamase activity in Enterobacter cloacae. Antimicrob Agents Chemother 44:561–567. doi: 10.1128/AAC.44.3.561-567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaneko K, Okamoto R, Nakano R, Kawakami S, Inoue M. 2005. Gene mutations responsible for overexpression of AmpC beta-lactamase in some clinical isolates of Enterobacter cloacae. J Clin Microbiol 43:2955–2958. doi: 10.1128/JCM.43.6.2955-2958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pottumarthy S, Moland ES, Juretschko S, Swanzy SR, Thomson KS, Fritsche TR. 2003. NmcA carbapenem-hydrolyzing enzyme in Enterobacter cloacae in North America. Emerg Infect Dis 9:999–1002. doi: 10.3201/eid0908.030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mallea M, Chevalier J, Bornet C, Eyraud A, Davin-Regli A, Bollet C, Pages JM. 1998. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology 144:3003–3009. doi: 10.1099/00221287-144-11-3003. [DOI] [PubMed] [Google Scholar]
- 39.De E, Basle A, Jaquinod M, Saint N, Mallea M, Molle G, Pages JM. 2001. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol Microbiol 41:189–198. doi: 10.1046/j.1365-2958.2001.02501.x. [DOI] [PubMed] [Google Scholar]
- 40.Paauw A, Caspers MP, Leverstein-van Hall MA, Schuren FH, Montijn RC, Verhoef J, Fluit AC. 2009. Identification of resistance and virulence factors in an epidemic Enterobacter hormaechei outbreak strain. Microbiology 155:1478–1488. doi: 10.1099/mic.0.024828-0. [DOI] [PubMed] [Google Scholar]
- 41.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. 1993. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol Microbiol 8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 42.Putze J, Hennequin C, Nougayrede JP, Zhang W, Homburg S, Karch H, Bringer MA, Fayolle C, Carniel E, Rabsch W, Oelschlaeger TA, Oswald E, Forestier C, Hacker J, Dobrindt U. 2009. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun 77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachman MA, Lenio S, Schmidt L, Oyler JE, Weiser JN. 2012. Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. mBio 3:e00224-11. doi: 10.1128/mBio.00224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson AE, Lowe JE, Koh EI, Henderson JP. 2018. Uropathogenic enterobacteria use the yersiniabactin metallophore system to acquire nickel. J Biol Chem 293:14953–14961. doi: 10.1074/jbc.RA118.004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bossuet-Greif N, Vignard J, Taieb F, Mirey G, Dubois D, Petit C, Oswald E, Nougayrède J-P. 2018. The colibactin genotoxin generates DNA interstrand cross-links in infected cells. mBio 9:e02393-17. doi: 10.1128/mBio.02393-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 47.Morrissey I, Hackel M, Badal R, Bouchillon S, Hawser S, Biedenbach D. 2013. A review of ten years of the Study for Monitoring Antimicrobial Resistance Trends (SMART) from 2002 to 2011. Pharmaceuticals (Basel) 6:1335–1346. doi: 10.3390/ph6111335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diene SM, Merhej V, Henry M, El Filali A, Roux V, Robert C, Azza S, Gavory F, Barbe V, La Scola B, Raoult D, Rolain JM. 2013. The rhizome of the multidrug-resistant Enterobacter aerogenes genome reveals how new “killer bugs” are created because of a sympatric lifestyle. Mol Biol Evol 30:369–383. doi: 10.1093/molbev/mss236. [DOI] [PubMed] [Google Scholar]
- 49.Thiolas A, Bollet C, La Scola B, Raoult D, Pages JM. 2005. Successive emergence of Enterobacter aerogenes strains resistant to imipenem and colistin in a patient. Antimicrob Agents Chemother 49:1354–1358. doi: 10.1128/AAC.49.4.1354-1358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing, 27th ed CLSI document M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 51.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seeliger D, de Groot BL. 2010. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WGS and metadata corresponding to the study URMC K. aerogenes isolates were deposited at NCBI under BioProject accession number PRJNA504784. Individual isolates were deposited under BioSample accession numbers SAMN10405413 to SAMN10405438 (Data Set S2).