Although macrolides are known as excellent antibacterials, their medical use has been significantly limited due to the spread of bacterial drug resistance. Therefore, it is necessary to develop new potent macrolides to combat the emergence of drug-resistant pathogens.
KEYWORDS: X-ray structure, antibiotic, dirithromycin, inhibitor, macrolides, nascent peptide exit tunnel, ribosomal protein uL4
ABSTRACT
Although macrolides are known as excellent antibacterials, their medical use has been significantly limited due to the spread of bacterial drug resistance. Therefore, it is necessary to develop new potent macrolides to combat the emergence of drug-resistant pathogens. One of the key steps in rational drug design is the identification of chemical groups that mediate binding of the drug to its target and their subsequent derivatization to strengthen drug-target interactions. In the case of macrolides, a few groups are known to be important for drug binding to the ribosome, such as desosamine. Search for new chemical moieties that improve the interactions of a macrolide with the 70S ribosome might be of crucial importance for the invention of new macrolides. For this purpose, here we studied a classic macrolide, dirithromycin, which has an extended (2-methoxyethoxy)-methyl side chain attached to the C-9/C-11 atoms of the macrolactone ring that can account for strong binding of dirithromycin to the 70S ribosome. By solving the crystal structure of the 70S ribosome in complex with dirithromycin, we found that its side chain interacts with the wall of the nascent peptide exit tunnel in an idiosyncratic fashion: its side chain forms a lone pair-π stacking interaction with the aromatic imidazole ring of the His69 residue in ribosomal protein uL4. To our knowledge, the ability of this side chain to form a contact in the macrolide binding pocket has not been reported previously and potentially can open new avenues for further exploration by medicinal chemists developing next-generation macrolide antibiotics active against resistant pathogens.
INTRODUCTION
Antibiotics have been successfully used for the last 70 years in the treatment of infectious diseases. More than half of all currently used antibiotics prevent the growth of pathogenic bacteria and, thereby, cure infections by selectively inhibiting their ribosomes, the central components of the protein synthesis apparatus (1). Prominent among this set of inhibitors are macrolide antibiotics, a large family of natural and semisynthetic compounds that consist of 14- to 16-membered macrolactone rings decorated with different side chains. All macrolide antibiotics bind in the nascent peptide exit tunnel (NPET) of the large ribosomal subunit and interfere with protein synthesis (2–5). By partially obstructing the NPET, macrolides create a physical obstacle for the progression of some nascent peptide chains. Macrolides also allosterically alter the properties of the ribosomal peptidyl transferase center (PTC) where amino acids are linked together to form a polypeptide chain (6).
The prototype macrolide antibiotic, erythromycin (ERY) (Fig. 1A), was discovered in 1952 (7). Due to its efficiency against a broad spectrum of bacterial pathogens and to its safety, it was introduced into the clinic in the same year (8). However, ERY is not stable and easily degrades under the acidic conditions in the stomach. As a result, a second generation of more stable and, thus, more efficient semisynthetic derivatives of ERY have been developed, such as clarithromycin(6-O-methyl erythromycin), azithromycin (a 15-membered azalide version of ERY), and dirithromycin [(9S)-9-deoxo-11-deoxy-9,11-(imino((1R)-2-(2-methoxyethoxy)ethylidene)oxy)erythromycin] (DIR) (Fig. 1A) (9), which possesses an extended hydrophobic (2-methoxyethoxy)-methyl side chain. We hypothesized that this chain potentially could encroach upon a new binding pocket in the upper part of the NPET and possibly provide a stronger anchoring of the drug on the ribosome.
FIG 1.
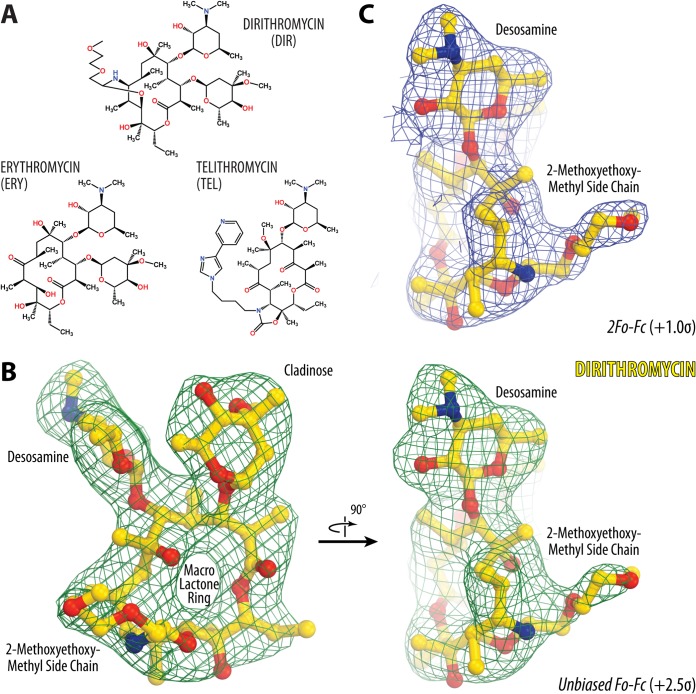
Electron density maps of ribosome-bound dirithromycin. (A) Chemical structures of the macrolides dirithromycin and erythromycin and the ketolide telithromycin. (B) Unbiased Fo − Fc (the observed and calculated structure factor amplitudes, respectively) electron density map of dirithromycin (DIR) in complex with the T. thermophilus 70S ribosome (green mesh) viewed from two different perspectives. (C) 2Fo − Fc electron density map of DIR viewed in the same perspective as the right side of panel B. The refined model of DIR is displayed in its respective electron density before (B) and after (C) the refinement contoured at 2.5σ and 1.0σ, respectively. Carbon atoms are shown in yellow, nitrogens are shown in blue, and oxygens are shown in red. Key chemical moieties of the drug are indicated. Note that the location of the (2-methoxyethoxy)-methyl side chain of DIR can be unambiguously determined from the electron density map.
Although macrolide drugs are known as excellent antibacterials, their medical use has been significantly limited due to the appearance of several resistance mechanisms among pathogens (10). The major mechanism of bacterial resistance to macrolides in the clinic is Erm-dependent methylation of nucleotide A2058 of the 23S rRNA (11, 12). This methylation results in displacement of the desosamine moiety, which is present in all currently used macrolides and ketolides, and in the inability of the drug to bind to such a ribosome. Due to the fast spread of Erm genes among pathogenic bacteria, there is a pressing demand for the development of new macrolides active against Erm-modified ribosomes of drug-resistant pathogens. One way to achieve this goal is to explore macrolides carrying side chains that interact with other parts of the NPET so that the anchoring of such a compound on the ribosome is less dependent on the presence of desosamine, a chemical group that is essential for binding and activity of all natural and semisynthetic macrolides studied to date. This idea was implemented previously while developing semisynthetic macrolides of a newer generation, called ketolides, such as telithromycin (TEL) (Fig. 1A) and solithromycin, which carry a keto-group instead of cladinose sugar and also have extended alkyl-aryl side chains that increase the affinity of these antibiotics for the ribosome by 10- to 100-fold (13) via stacking with the A752-A2609 base pair of the 23S rRNA (4, 5). Importantly, ketolides can bind to the A2058-methylated ribosomes, albeit with affinities which are too low to allow using these drugs for the treatment of infections caused by pathogens with constitutively expressed erm genes (14–16).
In this work, we have structurally explored DIR and obtained the high-resolution crystal structure of the 70S ribosome from Thermus thermophilus carrying mRNA and A-, P-, and E-site tRNAs in complex with DIR. We found that the distal oxygen of the (2-methoxyethoxy)-methyl side chain of DIR appears to form a lone pair-π stacking interaction with the histidine residue in ribosomal protein uL4. To our knowledge, this type of interaction was not known from the previously reported structures of macrolides in complex with bacterial ribosomes. Therefore, our study provides a structural basis for further exploration of this peculiar molecular contact by medicinal chemists in their pursuit to develop better macrolides. We believe that by combining DIR-like side chains with other known side chains in the same molecule, it will become possible to create a macrolide with high affinity to the Erm-modified ribosome due to desosamine-independent binding.
RESULTS AND DISCUSSION
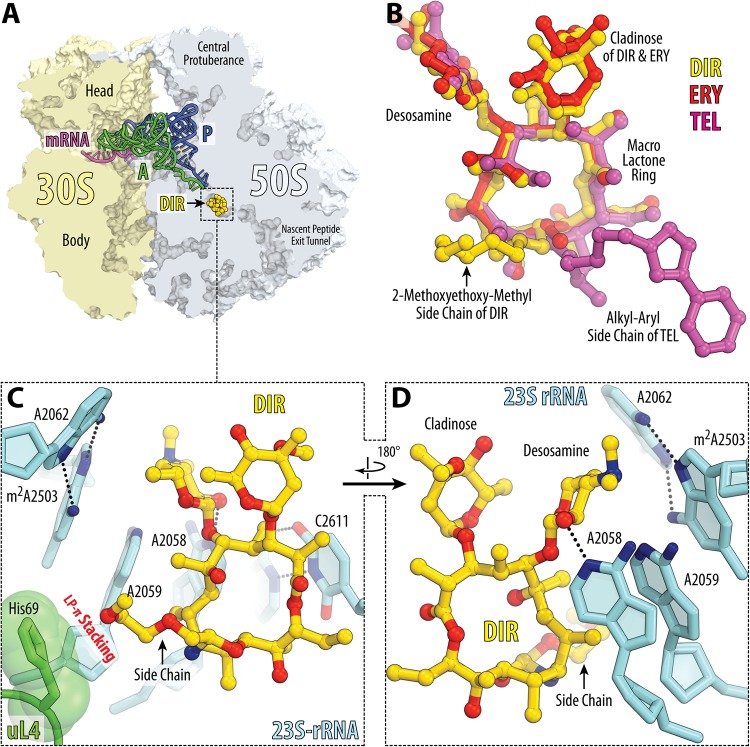
We have chosen DIR for the current study because of its extended hydrophobic (2-methoxyethoxy)-methyl tail (Fig. 1A), which other macrolides do not have and which could potentially endow the drug with unique binding properties. To determine the mode of DIR binding in the context of the ribosome functional complex, we crystallized Thermus thermophilus 70S ribosomes in the presence of DIR, mRNA, and A-, P-, and E-site tRNAs and determined the structure of the obtained complex by X-ray crystallography at 2.8-Å resolution (see Table S1 in the supplemental material). In this study, we used deacylated valine-specific tRNA as the A-site substrate and initiator methionine-specific tRNA as the P-site substrate. The E site of the ribosome contained tRNAVal. The unbiased difference Fourier map for DIR, which was calculated using the observed amplitudes from the crystal and the amplitudes and phases derived from the model of antibiotic-free ribosome (PDB accession number 4Y4P [17]), revealed positive electron density peaks (Fig. 1B) resembling characteristic features of the DIR chemical structure (Fig. 1A). A single binding site of DIR was observed in the peptide exit tunnel of the large ribosomal subunit (Fig. 2A). The observed DIR binding site overlaps sites of other known macrolides and ketolides, such as ERY and TEL (Fig. 2B). From the obtained structure it became evident that, similar to the binding of other macrolides, binding of DIR is mediated by the H-bonding of its desosamine sugar with the 23S rRNA residue A2058. Also, similar to other macrolides and most PTC-targeting antibiotics, DIR causes nucleotide A2062 of the 23S rRNA to rotate by ∼160° into a position where it forms a symmetric trans A-A Hoogsteen/Hoogsteen base pair with the residue m2A2503 (Fig. 2C and D; see also Fig. S1), which is favorable for the drug binding to the NPET.
FIG 2.
Structure of DIR in complex with the 70S ribosome and A- and P-site tRNAs. (A) Overview of the DIR binding site (yellow) in the T. thermophilus 70S ribosome viewed as a cross section through the nascent peptide exit tunnel. The 30S subunit is shown in light yellow, the 50S subunit is shown in light blue, the mRNA is shown in magenta, and the A- and P-site tRNAs are shown in green and dark blue, respectively. The E-site tRNA is omitted for clarity. (B) Superposition of ribosome-bound DIR (yellow) with ERY (red) (PDB accession number 6ND6 [35]) and TEL (magenta) (PDB entry 4V7Z [4]). All structures were aligned based on domain V of the 23S rRNA. (C and D) Close-up views of the DIR binding site shown in panel A. Potential H-bond interactions are indicated with dashed lines. Note that the side chain of DIR forms a lone pair-π (LP-π) stacking interaction with the imidazole ring of the uL4 His69 residue.
The most exciting finding of the current study is the location of the hydrophobic (2-methoxyethoxy)-methyl side chain of DIR, which encroaches upon the unexpected binding pocket within the NPET where it interacts with the imidazole ring of the His69 residue in the loop of the ribosomal protein uL4 (Fig. 2C and 3B). This loop together with the loop of another ribosomal protein, uL22, forms the uL4/uL22 constriction point in the NPET of the 70S ribosome, which plays a crucial functional role in the regulation of protein synthesis in response to the specific sequence motifs of the nascent polypeptide chains (18–21). Importantly, this additional contact with the wall of the NPET provided by the side chain of DIR is principally different from the contact established by the aromatic alkyl-aryl side chain of TEL, which forms standard π-π stacking interaction with the A752-U2609 base pair of the 23S rRNA (Fig. S2A). Based on the relative orientation of the distal oxygen in the (2-methoxyethoxy)-methyl side chain of DIR and the plane of the aromatic imidazole ring of the His69 residue in ribosomal protein uL4, it is likely that they form a lone pair-π stacking interaction (Fig. S2B). Similar types of contacts between lone pairs of electrons of the O4′ atom of ribose and aromatic nucleobases or aromatic amino acid side chains are abundant in the molecules of functional RNAs or RNA-protein complexes, respectively (22). The same region of the ribosomal protein uL4 was previously reported to interact with the forosamine moiety of the 16-membered macrolide spiramycin in the 50S subunit from Haloarcula marismortui (23). The exact nature of the interaction between the forosamine moiety of spiramycin and ribosomal protein uL4 is unclear due to poor electron density and the resulting uncertainty in forosamine orientation in the structure (23). However, the nature of the interaction between spiramycin and the ribosomal protein uL4 a priori cannot be the same as that observed here for DIR because the H. marismortui uL4 protein has a glycine residue in the position equivalent to His69 of the T. thermophilus uL4 protein and, therefore, cannot be involved in any type of stacking interactions.
FIG 3.
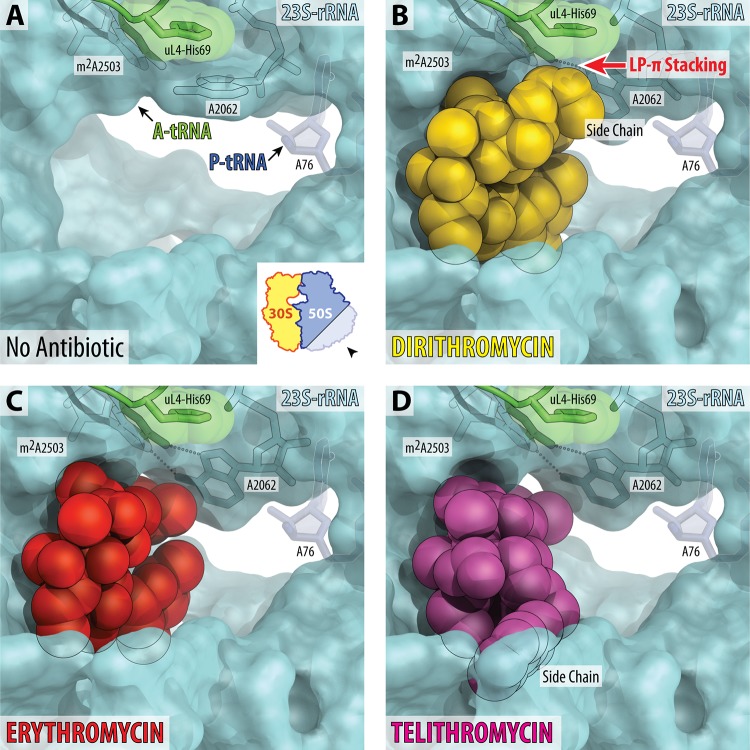
Binding sites of macrolides in the nascent peptide exit tunnel. (A) The lumen of the nascent peptide exit tunnel of the drug-free 70S ribosome (PDB accession number 4Y4P [17]). The view is from the wide-open part of the tunnel onto the PTC, as indicated by the inset. Nucleotide A76 of the P-site tRNA is shown. A-site tRNA is not visible in this view; however, its location is indicated by the arrow. Note that nucleotide A2062 of the 23S rRNA is pointed toward the viewer and is not involved in Hoogsteen-edge base pairing with the nucleotide m2A2503 in the wall of the NPET. (B to D) Occlusion of the nascent peptide exit tunnel by DIR, ERY, and TEL. Structures of ERY and TEL are from PDB entries 6ND6 (35) and 4X7Z (4), respectively. Note that the side chain of DIR forms LP-π stacking interaction with the His69 residue of the ribosomal protein uL4. Also note that binding of DIR, ERY, or TEL causes characteristic rotation of nucleotide A2062 by approximately 160° away from the viewer to form a symmetric trans A-A Hoogsteen/Hoogsteen base pair with the m2A2503 of the 23S rRNA.
Curiously, the residue equivalent to His69 of the T. thermophilus protein uL4 is nonconserved in other bacterial species (Fig. S3). Unlike T. thermophilus and mycoplasmas that contain histidine, the majority of bacteria (including Escherichia coli) possess glycine (Gly64 in E. coli) at this position (Fig. S3). However, even in the absence of this uL4 histidine residue, the side chain of DIR is likely to establish a stabilizing contact with the adjacent arginine residue in the loop of ribosomal protein uL4 in E. coli ribosome (Fig. S4). Also, it seems that the loop of protein uL4 plays an important role in modulation of the binding of macrolides to the ribosome via an allosteric mechanism because a spontaneous mutation of the conserved Lys63 residue to glutamic acid in protein uL4 renders E. coli cells resistant to ERY (Fig. S3 and S4) (24–26). This lysine residue precedes the position which is occupied by His69 in the loop of protein uL4 in T. thermophilus and based on the available structures is not expected to directly interact with the ribosome-bound macrolide molecule (Fig. S4). Thus, generation of macrolide derivatives with chemical groups replacing the (2-methoxyethoxy)-methyl side chain of DIR might be useful to create compensatory on-target interactions that would be sufficient to overcome the resistance. Moreover, the nonconserved nature of the uL4 protein residue equivalent to T. thermophilus His69 provides a possibility to create species-specific antibiotics. For example, to specifically and efficiently target mycoplasma ribosome, which possesses a histidine residue (similar to T. thermophilus), a suggestion might be to replace the (2-methoxyethoxy)-methyl side chain of DIR with an aromatic side chain (similar to alkyl-aryl group of TEL); this would potentially replace a relatively weak lone pair-π stacking interaction (22) with a stronger π-π stacking interaction between the uL4 His69 equivalent and a drug molecule and, thus, increase its affinity to the ribosome. Altogether, our structural data suggest that the (2-methoxyethoxy)-methyl side chain of DIR is a perfect candidate for further optimization by medicinal chemists in search of macrolide compounds active against species-specific or drug-resistant ribosomes. The observed additional contact provided by the side chain of DIR is unlikely to fully compensate for the loss of desosamine binding due to the A2058 methylation or A2058G substitution. Therefore, if combined with other already studied side chains (such as alkyl-aryl groups of synthetic ketolides), this previously unexplored contact of macrolides with the ribosome may pave the way toward the development of drugs whose binding does not depend on the presence of desosamine sugar. Such molecules are expected to become an effective cure against current macrolide-resistant pathogens.
MATERIALS AND METHODS
Materials for biochemical experiments.
Dirithromycin (DIR) was by Victor G. Kartsev from Interbioscreen, Ltd.
Crystallographic structure determination.
Ribosome complex containing mRNA and tRNAs was preformed by mixing 5 μM 70S T. thermophilus ribosomes with 10 μM mRNA and incubation at 55°C for 10 min, followed by addition of 20 μM P-site () and 20 μM A-site (tRNAVal) substrates (with minor changes from Polikanov et al. [17]). Each of these two steps was allowed to reach equilibrium for 10 min at 37°C in buffer containing 5 mM HEPES-KOH (pH 7.6), 50 mM KCl, 10 mM NH4Cl, and 10 mM Mg(CH3COO)2. Then, DIR, dissolved in the same buffer, was added to a final concentration of 250 μM to the preformed ribosome-mRNA-tRNA complex. Crystals were grown by vapor diffusion in sitting-drop crystallization trays at 19°C. Initial crystalline needles were obtained by screening around previously published ribosome crystallization conditions (27–29). The best-diffracting crystals were obtained by mixing 2 to 3 μl of the ribosome-mRNA-tRNA-DIR complex with 3 to 4 μl of a reservoir solution containing 100 mM Tris-HCl (pH 7.6), 2.9% (wt/vol) polyethylene glycol (PEG) 20000, 7 to 12% (vol/vol) 2-methyl-2,4-pentanediol (MPD), 100 to 200 mM arginine, and 0.5 mM β-mercaptoethanol (17). Crystals appeared within 3 to 4 days and grew to a size of 200 by 200 by 1,000 μm within 10 to 12 days. Crystals were cryo-protected stepwise using a series of buffers with increasing concentrations of MPD to a final concentration of 40% (vol/vol) MPD, in which they were incubated overnight at 19°C. In addition to MPD, all stabilization buffers contained 100 mM Tris-HCl (pH 7.6), 2.9% (wt/vol) PEG 20000, 50 mM KCl, 10 mM NH4Cl, 10 mM Mg(CH3COO)2, and 6 mM β-mercaptoethanol. DIR was added to the final cryo-protection solution to a concentration of 250 μM. After stabilization, crystals were harvested and flash frozen in a nitrogen cryo-stream at 80 K.
Diffraction data were collected on the beamlines 24ID-C and 24ID-E at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). A complete data set for each ribosome complex was collected using 0.979-Å wavelength at 100 K from multiple regions of the same crystal using 0.3° oscillations. The raw data were integrated and scaled using the XDS software package (30). All crystals belonged to the primitive orthorhombic space group P212121 with approximate unit cell dimensions of 210 Å by 450 Å by 620 Å and contained two copies of the 70S ribosome per asymmetric unit. Each structure was solved by molecular replacement using PHASER from the CCP4 program suite (31). The search model was generated from the previously published structure of the T. thermophilus 70S ribosome with bound mRNA and tRNAs (PDB accession number 4Y4P from Polikanov et al. [17]). The initial molecular replacement solutions were refined by rigid-body refinement with the ribosome split into multiple domains, followed by 10 cycles of positional and individual B-factor refinement using PHENIX (32). Noncrystallographic symmetry restraints were applied to four domains of the 30S ribosomal subunit (head, body, spur, and helix 44), and four domains of the 50S subunit (body, L1 stalk, L10 stalk, and C terminus of the L9 protein).
An atomic model of DIR was generated from its known chemical structure (Fig. 1A) using PRODRG online software (33), which was also used to generate restraints for energy minimization and refinement based on idealized three-dimensional (3D) geometry. Atomic model and restraints were used to fit/refine DIR into the obtained unbiased electron density map (Fig. 1B). The final model of the 70S ribosome in complex with DIR and mRNA/tRNAs was generated by multiple rounds of model building in COOT (34), followed by refinement in PHENIX (32). The statistics of data collection and refinement are compiled in Table S1 in the supplemental material. All figures showing atomic models were generated using PyMol software (www.pymol.org).
Accession number(s).
Coordinates and structure factors were deposited in the RCSB Protein Data Bank under accession number 6OF1 for the T. thermophilus 70S ribosome in complex with dirithromycin, mRNA, A-, P- and E-site tRNAs.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the A.L.K., I.A.O., and Y.S.P. laboratories for discussions and critical feedback. We thank Maxim Svetlov for critical reading of the manuscript and valuable suggestions. We thank the staff at Northeastern Collaborative Access Team (NE-CAT) beamlines 24ID-C and 24ID-E for help with data collection and freezing of the crystals, especially Kanagalakhatta Rajashankar, Malcolm Capel, Frank Murphy, Igor Kourinov, Anthony Lynch, Surajit Banerjee, David Neau, Jonathan Schuermann, Narayanasami Sukumar, James Withrow, Kay Perry, and Cyndi Salbego.
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165 to NE-CAT). The Pilatus 6M detector on the 24ID-C beamline is funded by an NIH-ORIP HEI (S10 RR029205 to NE-CAT). The Eiger 16M detector on the 24ID-E beamline is funded by an NIH-ORIP HEI grant (S10 OD021527 to NE-CAT). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract number DE-AC02-06CH11357.
This work was supported by Illinois State start-up funds (to Y.S.P.) and by the National Institutes of Health (grant R21-AI137584 to Y.S.P.), Russian Foundation for Basic Research (grant 17-00-00366 to P.V.S.), the Russian Science Foundation (grant 18-44-04005 to I.A.O.), and the Moscow University Development Program (PNR 5.13 to P.V.S.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02266-18.
REFERENCES
- 1.Lin J, Zhou D, Steitz TA, Polikanov YS, Gagnon MG. 2018. Ribosome-targeting antibiotics: Modes of action, mechanisms of resistance, and implications for drug design. Annu Rev Biochem 87:451–478. doi: 10.1146/annurev-biochem-062917-011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 3.Tu D, Blaha G, Moore PB, Steitz TA. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Bulkley D, Innis CA, Blaha G, Steitz TA. 2010. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci U S A 107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunkle JA, Xiong L, Mankin AS, Cate JH. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci U S A 107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sothiselvam S, Liu B, Han W, Ramu H, Klepacki D, Atkinson GC, Brauer A, Remm M, Tenson T, Schulten K, Vazquez-Laslop N, Mankin AS. 2014. Macrolide antibiotics allosterically predispose the ribosome for translation arrest. Proc Natl Acad Sci U S A 111:9804–9809. doi: 10.1073/pnas.1403586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGuire JM, Bunch RL, Anderson RC, Boaz HE, Flynn EH, Powell HM, Smith JW. 1952. Ilotycin, a new antibiotic. Antibiot Chemother (Northfield) 2:281–283. [PubMed] [Google Scholar]
- 8.Ying L, Tang D. 2010. Recent advances in the medicinal chemistry of novel erythromycin-derivatized antibiotics. Curr Top Med Chem 10:1441–1469. doi: 10.2174/156802610792232042. [DOI] [PubMed] [Google Scholar]
- 9.Counter FT, Ensminger PW, Preston DA, Wu CY, Greene JM, Felty-Duckworth AM, Paschal JW, Kirst HA. 1991. Synthesis and antimicrobial evaluation of dirithromycin (AS-E 136; LY237216), a new macrolide antibiotic derived from erythromycin. Antimicrob Agents Chemother 35:1116–1126. doi: 10.1128/AAC.35.6.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fyfe C, Grossman TH, Kerstein K, Sutcliffe J. 2016. Resistance to macrolide antibiotics in public health pathogens. Cold Spring Harb Perspect Med 6:a025395. doi: 10.1101/cshperspect.a025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutcliffe JA, Leclercq R. 2002. Mechanisms of resistance to macrolides, lincosamides, and ketolides, p 281–317. In Schönfeld W, Kirst HA (ed), Macrolide antibiotics. Birkhäuser, Basel, Switzerland. [Google Scholar]
- 12.Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother 43:2823–2830. doi: 10.1128/AAC.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen LH, Mauvais P, Douthwaite S. 1999. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol Microbiol 31:623–631. doi: 10.1046/j.1365-2958.1999.01202.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu M, Douthwaite S. 2002. Activity of the ketolide telithromycin is refractory to Erm monomethylation of bacterial rRNA. Antimicrob Agents Chemother 46:1629–1633. doi: 10.1128/AAC.46.6.1629-1633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llano-Sotelo B, Dunkle J, Klepacki D, Zhang W, Fernandes P, Cate JH, Mankin AS. 2010. Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis. Antimicrob Agents Chemother 54:4961–4970. doi: 10.1128/AAC.00860-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheinfeld N. 2004. Telithromycin: a brief review of a new ketolide antibiotic. J Drugs Dermatol 3:409–413. [PubMed] [Google Scholar]
- 17.Polikanov YS, Melnikov SV, Soll D, Steitz TA. 2015. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol 22:342–344. doi: 10.1038/nsmb.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenson T, Ehrenberg M. 2002. Regulatory nascent peptides in the ribosomal tunnel. Cell 108:591–594. doi: 10.1016/S0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- 19.Washington AZ, Benicewicz DB, Canzoneri JC, Fagan CE, Mwakwari SC, Maehigashi T, Dunham CM, Oyelere AK. 2014. Macrolide-peptide conjugates as probes of the path of travel of the nascent peptides through the ribosome. ACS Chem Biol 9:2621–2631. doi: 10.1021/cb5003224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson JH, Woolhead CA, Bernstein HD. 2010. The conformation of a nascent polypeptide inside the ribosome tunnel affects protein targeting and protein folding. Mol Microbiol 78:203–217. doi: 10.1111/j.1365-2958.2010.07325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence MG, Shamsuzzaman M, Kondopaka M, Pascual C, Zengel JM, Lindahl L. 2016. The extended loops of ribosomal proteins uL4 and uL22 of Escherichia coli contribute to ribosome assembly and protein translation. Nucleic Acids Res 44:5798–5810. doi: 10.1093/nar/gkw493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chawla M, Chermak E, Zhang Q, Bujnicki JM, Oliva R, Cavallo L. 2017. Occurrence and stability of lone pair-pi stacking interactions between ribose and nucleobases in functional RNAs. Nucleic Acids Res 45:11019–11032. doi: 10.1093/nar/gkx757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell 10:117–128. doi: 10.1016/S1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 24.Wittmann HG, Stoffler G, Apirion D, Rosen L, Tanaka K, Tamaki M, Takata R, Dekio S, Otaka E. 1973. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol Gen Genet 127:175–189. doi: 10.1007/BF00333665. [DOI] [PubMed] [Google Scholar]
- 25.Chittum HS, Champney WS. 1994. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol 176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovmar M, Nilsson K, Lukk E, Vimberg V, Tenson T, Ehrenberg M. 2009. Erythromycin resistance by L4/L22 mutations and resistance masking by drug efflux pump deficiency. EMBO J 28:736–744. doi: 10.1038/emboj.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selmer M, Dunham CM, Murphy FV IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 28.Korostelev A, Trakhanov S, Laurberg M, Noller HF. 2006. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 29.Polikanov YS, Blaha GM, Steitz TA. 2012. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuttelkopf AW, van Aalten DM. 2004. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Svetlov MS, Plessa E, Chen CW, Bougas A, Krokidis MG, Dinos GP, Polikanov YS 7 February 2019. High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition. RNA J doi: 10.1261/rna.069260.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.