LETTER
The endemic spread of carbapenemase-producing Klebsiella pneumoniae in Taiwan has become problematic (1). Besides the predominance of Klebsiella pneumoniae carbapenemase (KPC)-2, oxacillinase (OXA)-48 emerged at the end of 2013 (2) and has become the second most prevalent (1). Previously, we demonstrated that KPC-2 and OXA-48 accounted for 36.2% and 12.6% of carbapenem-resistant K. pneumoniae isolates, respectively, and most of them belonged to sequence type (ST) 11 (3).
KPC160111, a KPC-2 ST11 isolate, was isolated from the urine culture of an 81-year-old female patient who suffered from urinary tract infection in November 2014. Besides KPC-2, this strain was found to coproduce OXA-48 and exhibited extensive resistance to almost all antimicrobials (Table 1). Considering the clinical significance of coproduction of KPC-2 and OXA-48 in a single K. pneumoniae ST11 strain, here we report the in-depth characterization of KPC160111.
TABLE 1.
Antimicrobial susceptibility test results for KPC160111
| Antimicrobial | MIC (μg/ml)a | Interpretationb |
|---|---|---|
| Aminoglycosides | ||
| Amikacin | >1,024 | R |
| Gentamicin | >1,024 | R |
| Beta-lactams | ||
| Ampicillin | >1,024 | R |
| Cefazolin | >1,024 | R |
| Cefepime | >1,024 | R |
| Ceftazidime | 64 | R |
| Ceftriaxone | >1,024 | R |
| Ertapenem | 256 | R |
| Imipenem | >1,024 | R |
| Meropenem | 256 | R |
| Piperacillin-tazobactam | >1,024/256 | R |
| Ampicillin-sulbactam | >1,024/512 | R |
| Ceftazidime-avibactam | 2/1 | S |
| Fluoroquinolones | ||
| Ciprofloxacin | 128 | R |
| Levofloxacin | 64 | R |
| Fosfomycin | ||
| Fosfomycin | >1,024 | R |
| Sulfonamide | ||
| Trimethoprim-sulfamethoxazole | >56/1,064 | R |
| Polymyxin | ||
| Colistin | 0.5 | S |
Antimicrobial susceptibility testing was performed with standard broth microdilution method and interpreted based on the criteria from the Clinical and Laboratory Standards Institute guidelines (see Table 2A in M100-ED29).
R, resistant; S, susceptible.
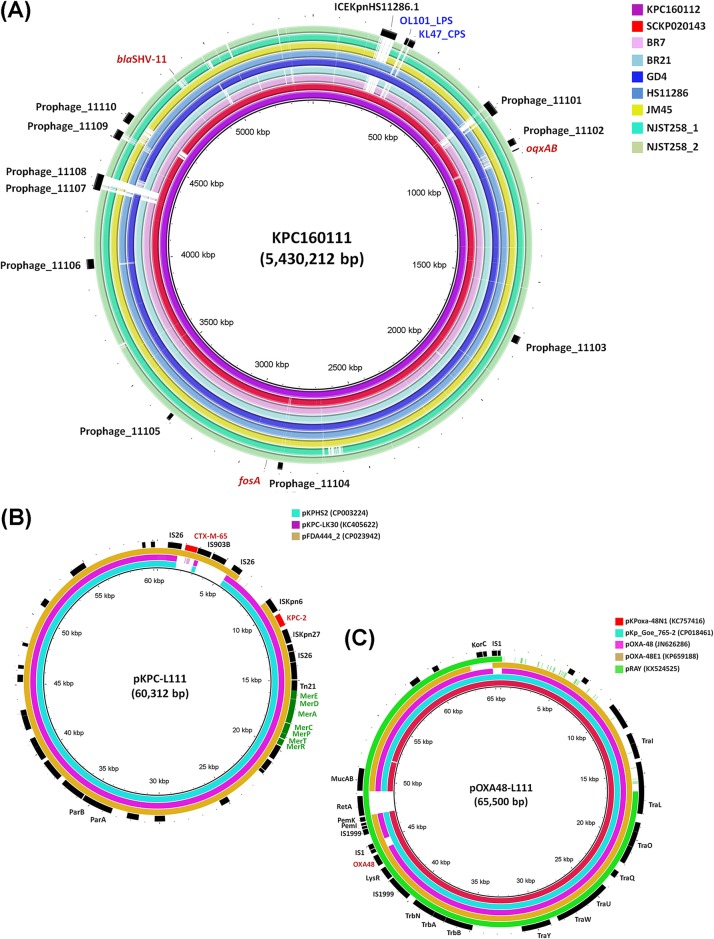
Whole-genome sequencing data (see the methods in the supplemental material) demonstrated that KPC160111 contained a 5.8-Mb genome, including a 5.43-Mb chromosome and six different plasmids. Ten β-lactamase-encoding genes (bla) and 21 antimicrobial resistance (AMR) genes were identified (Table 2). The KPC160111 chromosome carried three intrinsic AMR genes, blaSHV-11, oqxAB, and fosA (Fig. 1A). Based on analysis with Kaptive (4), the type of capsular polysaccharide (CPS) biosynthesis loci (KL-type) and the type of lipopolysaccharide (LPS) biosynthesis loci (OL-type) of KPC160111 were determined as KL47/OL101 (see the methods in the supplemental material), which were closely related to two ST11 KPC-2 strains, GD4 (NZ_CP025951.1) (5) and SCKP020143 (NZ_CP028548.1).
TABLE 2.
Genomic features of K. pneumoniae KPC160111
| Structure | Length (bp) | GC (%) | Antimicrobial resistance genes | Replicon type | Accession no. |
|---|---|---|---|---|---|
| Chromosome | 5,430,212 | 57.4 | blaSHV-11, fosA, oqxAB | CP029689 | |
| Plasmid | |||||
| pKPC-L111 | 60,312 | 55.4 | blaKPC-2, blaCTX-M-65 | IncR | CP030134 |
| pOXA48-L111 | 65,500 | 51.4 | blaOXA-48 | IncL | CP030135 |
| pIncAC2-L111 | 194,181 | 54.6 | aacA4, aph(3′)-Ia, aadA1, blaOXA-10, blaDHA-1, aac(6′)-lb-cr, catB3, cmlA1, arr2, sul1, dfrA14, aph(3′′)-lb, aph(6)-ld, aac(3)-lld, aadA2, rmtB, blaTEM-1B, blaTEM-2, erm, sul2, sul1, tet(G), dfrA12, blaCMY-2, blaCTX-M-14 | IncC | CP030132 |
| pIncFII-L111 | 39,248 | 54.4 | Not detected | IncFII | CP030133 |
| p10K-L111 | 10,060 | 55.1 | Not detected | ColRNAI | CP030130 |
| p5.6K-L111 | 5,596 | 55.1 | Not detected | ColRNAI | CP030131 |
FIG 1.
(A) Alignment of the KPC160111 chromosome and chromosomes of representative KPC-producing CG258 strains, including SCKP020143 (NZ_CP028548.1; ST11, KL47), GD4 (NZ_CP025951.1; ST11, KL47), JM45 (NC_022082.1; ST11, KL125), HS11286 (CP003200.1; ST11, KL103), BR7 (CP018883.1; ST437), BR21 (NZ_CP018885.1; ST437), NJST258.1 (NZ_CP006923.1; ST258), and NJST258.2 (NZ_CP006918.1; ST258). Notable features, including prophages (11101 to 11110), integrated conjugative element ICEKpnHS11286.1, the CPS biosynthesis region for KL-47, the LPS biosynthesis region for OL101, and AMR genes, oqxAB, fosA, and blaSHV11, are indicated in the outer ring. Circular maps were generated using the BLAST Ring Image Generator (BRIG). (B) The blaKPC-2- and blaCTX-M-65-cocarrying plasmid pKPC-L111. Alignment of pKPC-L111 with three closely related plasmids, pKPHS2 (CP003224), pKPC-LK30 (KC405622), and pFDA444_2 (CP023942), generated using BRIG. Genes associated with resistance to antimicrobials and heavy metals are highlighted in red and green, respectively. (C) The blaOXA-48 plasmid pOXA48-L111. Alignment of pOXA48-L111 with the related plasmids, pKPoxa-48N1 (KC757416), pKp_Goe_765-2 (CP018461), pOXA-48 (JN626286), pOXA-48E1 (KP659188), and pRAY (KX524525), generated using BRIG. The blaOXA-48 gene is indicated in red.
KPC160111 carried blaKPC-2 and blaOXA-48 by two distinct plasmids, named pKPC_L111 and pOXA48-L111, respectively. pKPC_L111 (Fig. 1B), a 60,307-bp IncR plasmid, captured blaKPC-2 by a Tn1722-based transposon unit with the core structure of ISKpn27-blaKPC-2-ΔISKpn6 as pKPC-LK30 (6) and acquired an additional IS26-based insertion of an ISEcp1-blaCTX-M-65-Δorf477 cassette downstream of blaKPC-2 (see Fig. S1). pOXA48-L111 (Fig. 1C) was a conjugative IncL plasmid (65,500 bp) and was transferred from KPC160111 to Escherichia coli J53-2 with an average frequency of 5.9 × 10−8 under standard laboratory conditions (see the methods in the supplemental material). pOXA48-L111 had blaOXA-48 as the only AMR gene enclosed on a Tn1999.2 composite transposon, similar to most of the blaOXA-48 carrying plasmids (7), and harbored an additional 1,911-bp fragment encoding RetA, a group II intron reverse transcriptase, upstream of the mucAB (see Fig. S2). Because KPC160111 was an ST11_KL47/OL101 strain related to other KPC-2 K. pneumoniae (3), the acquisition of pOXA48-L111, which brought the second carbapenemase OXA-48 into an already KPC-2 background, was suggested as a likely scenario for the emergence of this K. pneumoniae that carried both blaKPC-2 and blaOXA-48. Besides the carriage of the KPC-2 and OXA-48 plasmids, KPC160111 harbored a novel IncC plasmid (194,181 bp), which exhibited hybrid type 1 and type 2 IncC backbone features (8), and had 4 mosaic cassettes holding a total of 25 AMR genes (see Fig. S3).
To the best of our knowledge, this is the first report on the complete genome sequence of KPC-2-and OXA-48-coproducing K. pneumoniae. The uncommon cocarriage of genes encoding different classes of carbapenemases rendered KPC160111 extremely highly resistant to carbapenems (Table 1). Not restricted in our hospital, a total of 50 K. pneumoniae isolates coharboring blaKPC-2 and blaOXA-48 were also isolated from a regional hospital located in central Taiwan (9). For the better control of the endemic spread of carbapenemase-producing K. pneumoniae, horizontal spread of blaOXA-48 plasmids among KPC-2 strains needs to be actively monitored. The molecular mechanism underlying the superior capability of acquisition and maintenance of foreign DNA in ST11 K. pneumoniae deserves further studies.
This study was approved by the CSMUH Institute Review Board (IRB CS15022 and CS16104). Informed consent was obtained from all patients enrolled in this study. Patients were excluded if they were <18 years of age. All methods were carried out in accordance with relevant guidelines and regulations.
Data availability.
The genome sequence of K. pneumoniae KPC160111 has been deposited at NCBI under the BioSample number SAMN09279554 and the GenBank accession numbers CP029689 (KPC160111 chromosome), CP030134 (pKPC-L111), CP030135 (pOXA48-L111), CP030132 (pIncAC2-L111), CP030133 (pIncFII-L111), CP030130 (p10K-L111), and CP030131 (p5.6K-L111).
Supplementary Material
ACKNOWLEDGMENTS
We thank Yen-Yi Liu and You-Wun Wang for their assistance with genome assemblies.
This work was supported by Chung Shan Medical University Hospital, Taichung, Taiwan (grant number CSH-2016-C-017), China Medical University Hospital, Taichung, Taiwan (grant number DMR-106-194 and DMR-107-179), and the Ministry of Science and Technology of Taiwan (grant number MOST 106-2314-B-039-045).
The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. We declare no competing interests.
M.-C.L., C.-S.C., and Y.-C.L. conceived and designed the experiments; Y.-C.W., M.-C.L., and H.-L.T. collected clinical samples; M.-C.L., Y.-C.W., H.-L.T., Y.-T.C., and Y.-C.L. performed the experiments; H.-L.T., C.-S.C., Y.-T.C., and Y.-C.L. analyzed the data; H.-L.T. and Y.-C.L. prepared the figures and tables; M.-K.C. and Y-.C.L. wrote the manuscript; all authors read and approved the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02282-18.
REFERENCES
- 1.Chiu SK, Ma L, Chan MC, Lin YT, Fung CP, Wu TL, Chuang YC, Lu PL, Wang JT, Lin JC, Yeh KM. 2018. Carbapenem nonsusceptible Klebsiella pneumoniae in Taiwan: dissemination and increasing resistance of carbapenemase producers during 2012–2015. Sci Rep 8:8468. doi: 10.1038/s41598-018-26691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma L, Wang JT, Wu TL, Siu LK, Chuang YC, Lin JC, Lu MC, Lu PL. 2015. Emergence of OXA-48-producing Klebsiella pneumoniae in Taiwan. PLoS One 10:e0139152. doi: 10.1371/journal.pone.0139152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu MC, Tang HL, Chiou CS, Wang YC, Chiang MK, Lai YC. 2018. Clonal dissemination of carbapenemase-producing Klebsiella pneumoniae: two distinct sub-lineages of sequence type 11 carrying blaKPC-2 and blaOXA-48. Int J Antimicrob Agents 52:658–662. doi: 10.1016/j.ijantimicag.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 4.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE. 2016. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong N, Zhang R, Liu L, Li R, Lin D, Chan EW, Chen S. 8 February 2018. Genome analysis of clinical multilocus sequence Type 11 Klebsiella pneumoniae from China. Microb Genom doi: 10.1099/mgen.0.000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, Siu LK. 2014. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother 69:628–631. doi: 10.1093/jac/dkt409. [DOI] [PubMed] [Google Scholar]
- 7.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambrose SJ, Harmer CJ, Hall RM. 2018. Evolution and typing of IncC plasmids contributing to antibiotic resistance in Gram-negative bacteria. Plasmid 99:40–55. doi: 10.1016/j.plasmid.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Chen CM, Guo MK, Ke SC, Lin YP, Li CR, Vy Nguyen HT, Wu LT. 6 June 2018. Emergence and nosocomial spread of ST11 carbapenem-resistant Klebsiella pneumoniae co-producing OXA-48 and KPC-2 in a regional hospital in Taiwan. J Med Microbiol doi: 10.1099/jmm.0.000771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence of K. pneumoniae KPC160111 has been deposited at NCBI under the BioSample number SAMN09279554 and the GenBank accession numbers CP029689 (KPC160111 chromosome), CP030134 (pKPC-L111), CP030135 (pOXA48-L111), CP030132 (pIncAC2-L111), CP030133 (pIncFII-L111), CP030130 (p10K-L111), and CP030131 (p5.6K-L111).