Class 1 integrons accumulate antibiotic resistance genes by site-specific recombination at aatI-1 sites. Captured genes are transcribed from a promoter located within the integron; for class 1 integrons, the first gene to be transcribed and translated normally encodes an aminoglycoside antibiotic resistance protein (either an acetyltransferase [AAC] or adenyltransferase [AAD]).
KEYWORDS: DNA recombination, integrons, riboregulation, riboswitch, aminoglycosides, antibiotic resistance, translational control
ABSTRACT
Class 1 integrons accumulate antibiotic resistance genes by site-specific recombination at aatI-1 sites. Captured genes are transcribed from a promoter located within the integron; for class 1 integrons, the first gene to be transcribed and translated normally encodes an aminoglycoside antibiotic resistance protein (either an acetyltransferase [AAC] or adenyltransferase [AAD]). The leader RNA from the Pseudomonas fluorescens class 1 integron contains an aminoglycoside-sensing riboswitch RNA that controls the expression of the downstream aminoglycoside resistance gene. Here, we explore the relationship between integron-dependent DNA recombination and potential aminoglycoside-sensing riboswitch products of recombination derived from a series of aminoglycoside-resistant clinical strains. Sequence analysis of the clinical strains identified a series of sequence variants that were associated with class I integron-derived aminoglycoside-resistant (both aac and aad) recombinants. For the aac recombinants, representative sequences showed up to 6-fold aminoglycoside-dependent regulation of reporter gene expression. Microscale thermophoresis (MST) confirmed RNA binding. Covariance analysis generated a secondary-structure model for the RNA that is an independent verification of previous models that were derived from mutagenesis and chemical probing data and that was similar to that of the P. fluorescens riboswitch RNA. The aminoglycosides were among the first antibiotics to be used clinically, and the data suggest that in an aminoglycoside-rich environment, functional riboswitch recombinants were selected during integron-mediated recombination to regulate aminoglycoside resistance. The incorporation of a functional aminoglycoside-sensing riboswitch by integron recombination confers a selective advantage for the expression of resistance genes of diverse origins.
INTRODUCTION
The extensive use of antibiotics in the clinic, in commercial farming, and in veterinary applications has suffused them into the environment (1). This environmental contamination has led to significant selective pressures on commensal and pathogenic bacteria resulting in the evolution and proliferation of antibiotic resistance genes (2, 3). The emergence of antibiotic resistance is a prevalent and worsening threat to current medical practice (2). Environmentally, pathogen survival is driven by selective genetic adaptation; genetic variants are generated in response to host immune systems, environmental stresses, and antibiotic challenges, and these evolutionary changes to the pathogen are driven by mutation and genetic recombination (4). Genetic recombination is an important evolutionary driving force and plays a key role in the generation of genetic diversity.
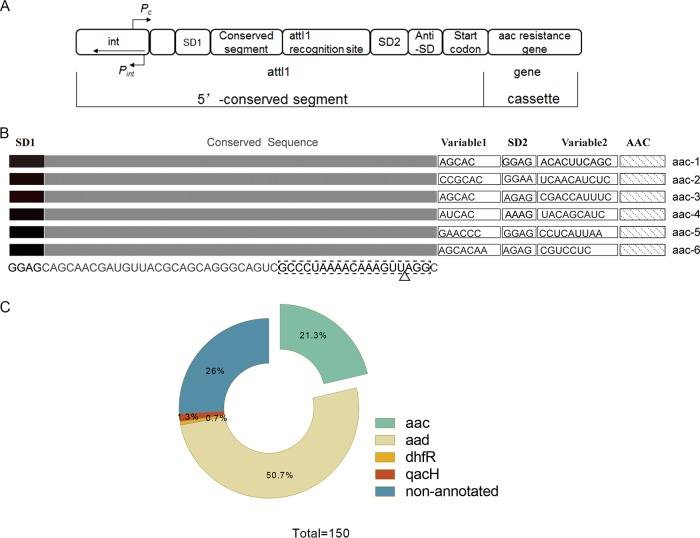
Integrons are a simple genetic apparatus that through site-specific recombination accumulate exogenous genes into a sequence context that ensures their accurate expression as functional proteins (reviewed in reference 5). In antibiotic-resistant bacteria, antibiotic resistance genes are accumulated on plasmids by a site-specific recombination mechanism of horizontal gene transfer which is mediated by integrons. The association of integrons with plasmids that carry multiple antibiotic resistance genes led to their original discovery. However, further investigation has identified that the integrons associated with antibiotic resistance had their origins as chromosomal elements that were recruited by transposition into plasmids and selected through exposure to antibiotics (5, 6). Three essential components are required for the sequestration of the exogenous genes, as follows: the first is an integrase (a site-specific recombinase, encoded by the int gene); the second is the target recognition sequence for the integrase gene (attI-1); and finally, the captured genes are efficiently transcribed from a promoter sequence (Pc) upstream of the attI-1 site and expressed (Fig. 1). The transcript mRNA has been shown to function as a regulatory RNA containing a small open reading frame consisting of a Shine-Dalgarno (SD) sequence and a putative leader peptide to coordinate efficient expression of the downstream (inserted exogenous) resistance genes (7–12).
FIG 1.
(A) The conserved 5′ segment of the class 1 integrons. The strong promoter Pc (8, 9, 35) transcribes the inserted gene cassette (i.e., the aac resistance gene), while the divergent promoter Pint transcribes the integrase (int) gene. Single-gene cassettes are inserted by int-mediated recombination between int recognition sites. The int expression is linked to the SOS response that can only be induced by nonaminoglycoside antibiotics in E. coli. (B) The leader aac riboswitch RNAs from the different pathogens, the positions of the ribosome binding sites SD1 and SD2, conserved sequences, and variable sequences are indicated; the anti-SD sequences are located within the variable 2 region. The RNA is located between the divergent int and aac genes at the conserved 5′ end of the integron. The dashed box shows the equivalent positions in the RNA of the core DNA attI-1 recognition site, and the triangle indicates the approximate position of the attC 3′ DNA insertion. (C) The BLAST search results for the aac and aad riboswitch sequence from P. fluorescens. A total of 111 of the top 150 BLAST sequences were annotated to identify the neighboring genes. The majority of neighboring resistance genes were aad (n = 76) and aac (n = 32), although one non-aminoglycoside resistance gene, that for dihydrofolate reductase (dhfR), as well as a nonfunctional multidrug exporter gene (qacH), were also noted.
The aminoglycoside antibiotics target the A site in the decoding region of the 30S ribosomal subunit and inhibit translocation during translation (13–15). Resistance to aminoglycosides occurs either through the enzymatic modification of the aminoglycosides, methylation of the target rRNA, or the overexpression of efflux pumps and is associated with mobile integron-containing plasmids (5, 16–18). Enzymatic modification of aminoglycosides by acetyltransferases (N-acetylation, AAC), nucleotidyltransferases or adenyltransferases (O-adenylylation [ANT or AAD] [10]), or phosphotransferases (O-phosphorylation [APH]) blocks aminoglycoside binding to their target site (the A site of the ribosome), leading to aminoglycoside resistance (19–21). Mobile plasmids carrying aac and aad genes, associated with the class I integrons, accumulate multiple antibiotic resistance genes and are a major contributor to the proliferation of antibiotic resistance.
The expression of resistance genes enables bacteria to survive in an antibiotic-rich environment; however, in the absence of antibiotics, their expression imposes a significant metabolic drain upon the cells (3, 22, 23). In order to reduce the fitness burden associated with antibiotic resistance, cellular mechanisms of antibiotic detection are deployed to regulate the expression of resistance genes and ensure an appropriate response upon exposure to the antibiotic (3, 22–24). For the antibiotic classes that target the bacterial ribosome, most of the sensing mechanisms are implemented at the RNA level and generally consist of strategies of translational and transcriptional attenuation that are exploited to detect antibiotic incursion (reviewed in references 25 and 26). Acting in cis, regulatory sequences in the 5′ untranslated regions (5′UTRs) of the antibiotic resistance genes are activated in the presence of antibiotics. Translational attenuation occurs when, in the absence of the antibiotic, the 5′UTR folds to mask the ribosome-binding site (RBS) of the resistance gene, leading to ribosome stalling at neighboring short open reading frame (μORF) sequences. Antibiotic ribosome interactions stimulate an alternative base-pairing scheme in the 5′UTR RNA, effectively derepressing gene expression by unmasking the RBS (27–30). For transcriptional attenuation, the 5′UTR RNA folds to adopt a transcriptional terminator structure that leads to the premature termination of transcription, and gene expression becomes derepressed when this structure becomes destabilized in the presence of the antibiotic (31).
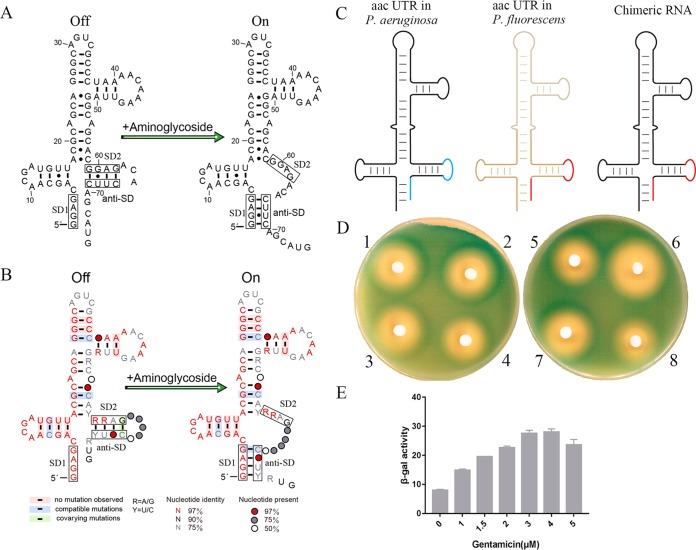
Riboswitches are noncoding regulatory RNAs that specifically bind to small metabolites to regulate the expression of downstream genes (reviewed in references 32 and 33). An aminoglycoside-sensing riboswitch in the 5′UTR RNA of the aac gene of a Pseudomonas fluorescens resistance plasmid has been identified that controls downstream aac gene expression (10). Reporter constructs expressing β-galactosidase (β-Gal) under the control of this 5′UTR demonstrated aminoglycoside-dependent induction of β-Gal gene expression in response to added 4,6-deoxystreptamine aminoglycosides compared to the control molecules ribostamycin, neamine, and 4,5-deoxystreptamine derivatives, which did not induce β-Gal expression. Essential functional features of the RNA secondary structure were confirmed by mutational analysis. There was also a correlation between the levels of induction measured and the aminoglycoside affinity for the leader RNA as measured by surface plasmon resonance (SPR). Higher-affinity molecules also induced changes in electrophoretic mobility and the chemical reactivity of the RNA consistent with a change in the RNA structure upon drug binding. Aminoglycoside-dependent β-Gal expression was also detected in strains that were resistant to aminoglycosides through ribosomal methylation or antibiotic acetylation, confirming that the induction observed was independent of aminoglycoside-ribosome interactions (10). This led to the proposal of a translation-based riboswitch model for the mechanism of gene regulation by the aminoglycoside-sensing RNA. The ribosome-binding site (SD2) (Fig. 1) of the resistance gene is sequestered by a complementary anti-SD sequence that blocks ribosome binding in the absence of the antibiotic (Fig. 4). Upon antibiotic binding, a structural transition unmasks SD2 for ribosome binding, leading to the translation of the resistance gene (10).
FIG 4.
(A) Secondary structure of the aac riboswitch in P. fluorescens (10). (B) The consensus sequence and secondary-structure model for the aac riboswitch. Conserved nucleotides are indicated by color, with red (at least 97%), black (90%), and gray (75%). Red, gray, and white circles denote positions where nucleotide identity is less conserved at 97%, 75%, and 50%, respectively. R and Y identify purines and pyrimidines, respectively. Base pairs shaded in red exhibit natural covariation, while those in blue and green exhibit compatible and covarying mutations. (C) Sketch of the domain swap between the leader RNA of aac-6 in P. aeruginosa (black and blue) and aac in P. fluorescens (brown and red). The chimeric RNA structures are colored black and red. (D and E) Agar diffusion assays (D) and β-Gal activity (in Miller units) (E) of cells transformed with the reporter plasmid containing the chimeric RNA. Each filter disc was spotted with the aminoglycoside antibiotics, as follows: 1, sisomicin; 2, gentamicin; 3, neamine; 4, ribostamycin; 5, kanamycin b; 6, amikacin; 7, paromomycin; and 8, tobramycin. The error bars are standard deviations of the results from at least three independent experiments.
At the RNA level, the aminoglycoside-sensing riboswitch functions to regulate aac gene expression and aminoglycoside resistance through acetylation of the drugs. At the DNA level, aminoglycoside resistance is acquired through class 1 integron-driven site-specific recombination of the aac gene. The DNA sequence of the aminoglycoside-sensing riboswitch RNA overlaps with and is an integral part of the attI-1 site, which is the recombination site of the class 1 integron-encoded integrase and extends into the neighboring gene (7, 11, 12, 34). In this study, we set out to investigate the relationship between integron-dependent DNA recombination and the potential aminoglycoside-sensing riboswitch products of recombination. Sequence analysis of clinical strains identified a series of sequence variants that were associated with class I integron-derived aminoglycoside-resistant (both aac and aad) recombinants. Representative sequences showed aminoglycoside-dependent regulation of reporter gene expression; RNA binding was confirmed by microscale thermophoresis (MST), and covariance analysis generated a secondary structure model for the RNA, suggesting a mechanism for the riboswitch that was consistent with previous models derived by chemical probing.
RESULTS
Analysis of the relationship between integron recombination and the riboswitch.
Of the 126 nucleotides (nt) of the P. fluorescens aminoglycoside-sensing riboswitch RNA, the minimal functional unit was demonstrated to be 75 nt (10). The DNA sequence that encodes the 75-nt aminoglycoside-sensing riboswitch RNA overlaps the site-specific attI-1 recombination insertion site of the class 1 integrons (Fig. 1B) (12, 35). In the class 1 integrons, the DNA sequence that encodes the leader riboswitch RNA is positioned between the (divergent) integrase and the AAC and ANT resistance genes, which encode the aminoglycoside acetyltransferases and adenyltransferases, respectively (36). A BLAST search of the 75-nt riboswitch leader RNA aac sequence in P. fluorescens revealed that in excess of 200 sequences have different levels of homology with the 75-nt aminoglycoside-sensing riboswitch. Moreover, 111 of the top 150 sequences in the BLAST search were next to annotated genes, which were predominantly aad (n = 76) and aac (n = 32) (Fig. 1C) (10, 37). In this study, we have focused on the analysis of the leader riboswitch RNA upstream of the aac genes. The study of the riboswitch RNAs upstream of the aad genes will be reported separately (our unpublished data). The riboswitch RNA of the aac gene is widely distributed over a range of antibiotic-resistant pathogens, such as Pseudomonas aeruginosa, Campylobacter jejuni, and Escherichia coli. Across all of the sequences, only two other non-aminoglycoside resistance genes, (i) dihydrofolate reductase (dhfR) and (ii) a nonfunctional multidrug exporter qacH, were identified.
Further analysis of these sequences revealed that the 5′ end of the RNAs consistently included SD1, a fully conserved region, two variable regions, and a second RBS (SD2). The fully conserved region corresponds to the DNA sequence overlapping the integrase binding site attI-1. Because each integron is the product of an independent genetic recombination event, the sequences between the attI-1 site and the aac genes show considerable sequence variation, as they contain remnants of the regulatory sequences of the inserted resistance genes. The strong upstream integron promoter (Pc) transcribes the captured aac resistance genes. Since they are the product of integron recombination at the attI-1 site, the promoters from the acquired resistance genes are absent (9, 34, 38, 39). The 3′ ends of the RNA sequences therefore vary considerably, extending to the aac gene coding sequence (Fig. 1B).
Different aac leader RNAs show aminoglycoside-dependent induction of reporter gene expression.
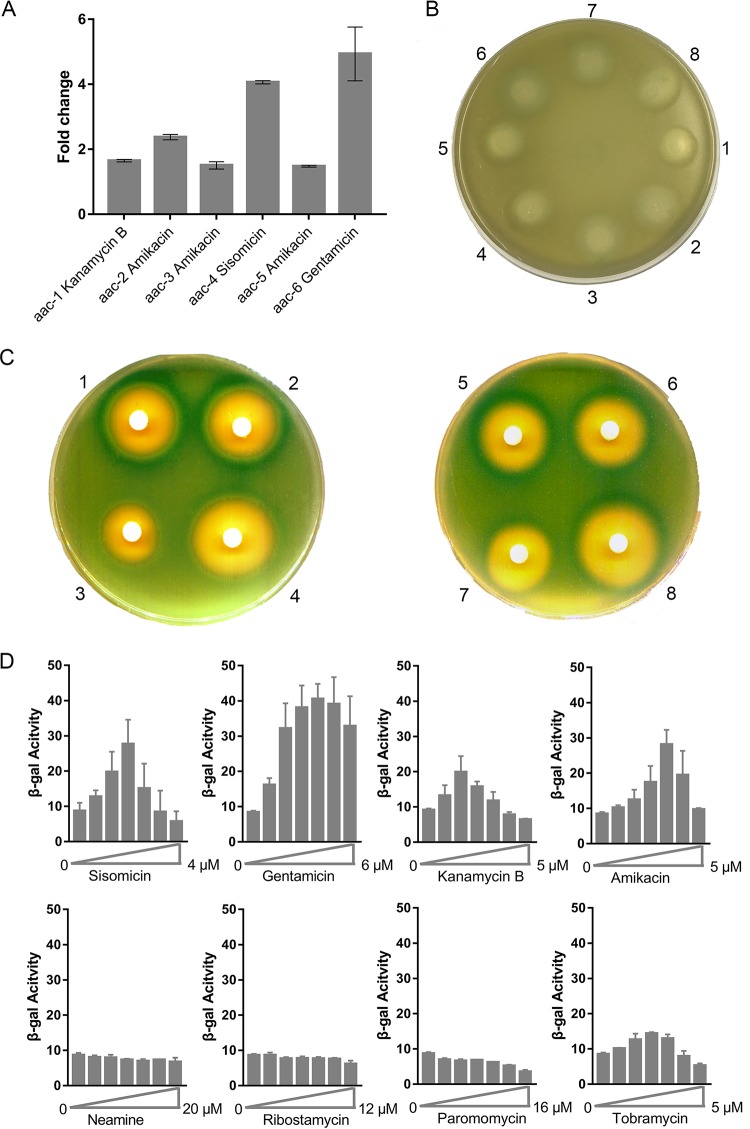
Certain aminoglycosides were shown to induce reporter gene expression by the interaction of the aminoglycosides with the 75-nt aac riboswitch RNA of P. fluorescens on the plasmid pGEX-leaderRNAaac/aad-lacZα by Miller assays in solution and by agar diffusion assays on solid medium. A number of leader RNA sequences were identified (Fig. 1 and Table S1) by BLAST sequence analysis, and these sequences contained regions that differed from the published riboswitch RNA in P. fluorescens. Mutations (even point mutations) in the P. fluorescens riboswitch RNA impaired the riboswitch (10). In contrast, the riboswitch RNAs in our study contained regions of significant local sequence variation. Thus, we examined the functions of these RNA sequences. We investigated the regulatory role of six riboswitch RNAs from the search and compared their function with that of the riboswitch RNA of P. fluorescens. The DNA coding sequence of the published riboswitch RNA was replaced in the reporter plasmid pGEX- leaderRNAaac/aad-lacZα by each of the six riboswitch RNAs, with the P. fluorescens sequence (aac-1) included for comparison (10, 37, 40). Then, each reporter construct was transformed into the E. coli strain JM109, and β-Gal activity was examined in the presence of aminoglycoside antibiotics by agar diffusion assays on plates or in solution by Miller assays, as previously described (10). Reporter gene expression was measured in the presence of the 4,6-deoxystreptamine aminoglycosides sisomicin, gentamicin, kanamycin B, amikacin, and tobramycin (20); the 4,5-deoxystreptamine derivatives ribostamycin, the fragment molecule neamine, and paromomycin were used as controls. Previous control experiments with analogous constructs containing the leader RNA of the cat-86 gene (encoding chloramphenicol acetyltransferase) (41) were shown to be unresponsive to added aminoglycosides (10) and verified that reporter gene expression was specific for the AAC leader RNA. As additional negative controls, the DNA sequence encoding the AAC leader RNA was replaced by two different leader RNAs, the UTR of the ermCL gene (erythromycin resistance methyltransferase gene) (29) and a randomly selected UTR (from a fragment of the E. coli ghoS gene) of similar nucleotide composition to the AAC leader RNAs. In these constructs (pGEX- leaderRNAermC-lacZα and pGEX- leaderRNAghoS-lacZα), reporter gene expression was also shown to be unresponsive to aminoglycoside antibiotics (Fig. 2B and S3). Because the control RNAs do not affect reporter gene expression in the presence of added aminoglycosides, this suggests that aminoglycoside-dependent effects on reporter gene expression are specific to the AAC leader RNAs. The results show that each of the six riboswitch RNAs could mediate the induction of reporter gene expression by 4,6-deoxystreptamine aminoglycosides but not by the 4,5-deoxystreptamine derivatives (Fig. 2B and C and S1), although very faint zones of induction were observed at the higher (100 μM) drug concentrations for ribostamycin and paromomycin in the diffusion assays. These findings are broadly similar to those observed for the published riboswitch in P. fluorescens. It was previously reported that mutations or even a single point mutation in the riboswitch in P. fluorescens were sufficient to impair its function. Although the six riboswitch RNAs examined in this study contained sequence variations, these RNAs could still function as riboswitches to activate downstream gene expression. Prolonged (30-day) incubation of the AAC-6 sequence with subinhibitory doses of gentamicin (3 μM) (42) showed the AAC-6 sequence to be stable toward mutagenesis (not shown). This suggests that not only were the specific RNA sequences required for the riboswitch to function but also that a common feature of the leader RNAs was preserved by the class I integron-mediated site-specific recombination event that generated each riboswitch. These variations in the leader RNA sequences of aac were originally introduced at the DNA level during the site-specific recombination by the class I integron when the bacteria were under survival pressure in an aminoglycoside-rich environment.
FIG 2.
(A) Miller assays of the six leader aac riboswitch RNAs. The fold change upon addition of the highest-inducing antibiotic is shown for aac-1 to aac-6 riboswitch RNAs. (B) Agar diffusion assays of cells transformed with the reporter plasmid containing the erm-C leader RNA grown on plates with isopropyl-β-d-thiogalactopyranoside (IPTG). Each filter disc was spotted with 1 ml of a 100 μM solution of the aminoglycosides, as follows: 1, sisomicin; 2, gentamicin; 3, neamine; 4, ribostamycin; 5, kanamycin b; 6, amikacin; 7, paromomycin; and 8, tobramycin. (C) Agar diffusion assays of cells transformed with the reporter plasmid containing the aac-6 leader RNA grown on plates with IPTG. Each filter disc was spotted with a solution of the aminoglycosides 1 to 8. (D) β-Gal activity (in Miller units) of the reporter gene upon titration of the aminoglycosides 1 to 8. Error bars are standard deviations of the results from at least three independent experiments.
The aminoglycosides induce reporter gene expression of diverse aac leader RNAs with distinct specificities.
It is also noteworthy that the leader RNAs of aac-3 and aac-5 induced the reporter gene expression by amikacin at levels similar or higher than those previously published for aac-1 by kanamycin B (Fig. 2A). Further, in the presence of gentamicin, the leader RNA of aac-6 mediated the induction of reporter gene expression 5-fold compared to that of aac-1 with kanamycin B (Fig. 2A). These results suggest that the sequence variants within the riboswitch RNAs may confer specificity for particular antibiotics to enhance the resistance response to certain aminoglycosides. Moreover, each of the riboswitch RNA sequences showed distinctive discrimination between different aminoglycosides. For example, for the leader RNA of aac-6, gentamicin induced the reporter gene expression by 5-fold, while for the leader RNA of aac-4, maximal (4-fold) induction of the reporter gene expression was observed, but with sisomicin instead. In these experiments, we used the published reporter construct for Miller and agar diffusion assays (10), but we replaced the P. fluorescens riboswitch with the six different riboswitch RNAs. From these results, we can conclude that the distinct preference for different aminoglycosides for each riboswitch was due to specific interactions between the riboswitch RNA sequences and particular aminoglycosides and not through interactions with other cellular components (like ribosomes, for example), which are invariant between the different constructs.
The aac leader RNA binds aminoglycosides.
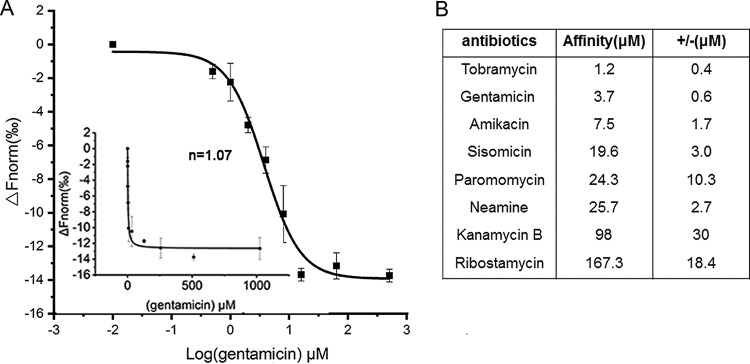
Surface plasmon resonance (SPR) (10, 43) was previously used to detect aminoglycoside binding to the P. fluorescens riboswitch RNA directly. The binding data were consistent with the results from the reporter assays. The aminoglycosides that induce reporter gene expression generally bind to the riboswitch RNA with higher affinity, and the control aminoglycosides that fail to induce reporter gene expression bind to the riboswitch RNA with low affinity. To obtain further biochemical evidence, based on the reporter assay results for the riboswitch RNAs, we employed an independent technique, that of microscale thermophoresis (MST) to measure the binding of aminoglycosides to RNA. We selected and prepared the aac-6 leader RNA by in vitro transcription using T7 RNA polymerase. Then, the RNA was labeled with fluorescein-5-thiosemicarbazide as previously described (44). Aminoglycoside-to-fluorescent-RNA-binding measurements were made on a Monolith NT.115 system by NanoTemper Technologies (45, 46); the results showed an increase in the measured response upon titration of the aminoglycosides, indicating the formation of an aminoglycoside-RNA complex. Figure 3B shows the dissociation constants (Kd) for the aminoglycoside-RNA complex formed, as measured by MST. Tobramycin, gentamicin, and amikacin showed the highest affinity for the RNA, at 1.2, 3.7, and 7.5 μM, respectively, and noncooperative binding behavior (with corresponding Hill constants [n] of 1.01, 1.07, and 0.80), which indicates the formation of a 1:1 complex (Fig. 3A and S2). In contrast, ribostamycin and neamine bound with lower affinities, at 98 and 167.3 μM, respectively (Fig. 3B and S2). Thus, the results showed that the aminoglycosides that induce reporter gene expression in the reporter assays in Fig. 2C and D also displayed the higher affinities for the riboswitch RNA in MST measurements. In contrast, the control aminoglycosides exhibited lower affinity for the riboswitch RNA. Thus, an alternative method, MST, confirmed aminoglycoside binding to the aac-6 riboswitch RNA and gave comparable values to previous SPR measurements for aminoglycoside binding to the P. fluorescens riboswitch RNA.
FIG 3.
(A) Microscale thermophoresis (MST) binding curve for the aac-6 leader riboswitch RNA with gentamicin. Inset shows the Hill plot and Hill coefficient (n) for gentamicin binding. Error bars are standard deviations of the results from at least three independent experiments. (B) Binding affinities (in micromolar) of the aminoglycoside antibiotics for the aac-6 riboswitch RNA. Error bars are standard deviations of the results from at least three independent experiments.
The integron preserves the aminoglycoside-sensing riboswitch function.
A model for the mechanism of the riboswitch of P. fluorescens was previously proposed on the basis of chemical probing data and mutational analysis. In the absence of aminoglycosides, the ribosome-binding site SD2 (GGAG) is sequestered by the anti-SD2 (CUUC) sequence, blocking the ribosomal access. In the presence of aminoglycosides, aminoglycoside binding induces a structural transition that allows the pairing of anti-SD2 with SD1 (GGAG), making SD2 available for the initiation of translation. This leads to translation of the downstream gene (Fig. 4A). In this study, analysis of the aminoglycoside riboswitch sequences showed that their SD2 and anti-SD2 sequences varied in some positions, and these sequence variations were generated by integron-dependent site-specific recombination (Fig. 1B). Despite the variations of the SD2 and anti-SD2 sequences in these riboswitches, they continued to function as aminoglycoside riboswitches in the reporter assay. To further investigate the mechanism of the riboswitch sequences and to uncover how the relationship between integron-mediated site-specific recombination preserves riboswitch function, the sequences from the BLAST search were further analyzed by covariance analysis.
Covariance analysis generated covariant RNA structures for 13 resistant bacterial strains that included Gram-positive and Gram-negative pathogens (Fig. 4 and Table S1). This is consistent with the model proposed for the P. fluorescens riboswitch RNA, which was based on chemical probing data (10). In the structures derived from covariance analysis, although the SD2 (RRAG) and anti-SD2 (YUNC) sequences were variable, in the absence of the aminoglycosides, SD2 remains sequestered by the anti-SD2 sequence and, upon aminoglycoside binding, undergoes a structural transition. This process allows the anti-SD2 sequence to pair with SD1 and consequently make SD2 available for ribosome binding. The variable sequences at SD2 and anti-SD2 of the riboswitch RNA are products of selective site-specific recombination into the integron DNA that in each case preserves riboswitch function so that aminoglycoside resistance can be controlled in an antibiotic-rich environment. In the situation where the recombination events take place in the absence of selection, the strain may become sensitive to the antibiotic because a functional riboswitch has not been preserved.
To test the covariant structure, we performed a domain swap experiment, illustrated in Fig. 4C. The domains from two variant sequences (AAC-6 [P. aeruginosa] and AAC [P. fluorescens]) were exchanged to form a chimeric RNA, which would be predicted to have a riboswitch function. The chimeric RNA construct (pGEX-leaderRNAaac/aad-lacZα) was tested on plates in the presence of aminoglycoside antibiotics or in solution using gentamicin as a representative aminoglycoside. The chimeric RNA was found to mediate reporter gene expression with aminoglycosides on plates, and titration of gentamicin induced a 3.6-fold increase in β-Gal expression (Fig. 4D and E) comparable to that observed for the original sequences (Fig. 2C). This result further substantiates the covariance analysis findings.
DISCUSSION
Here, we have studied the relationship between the class 1 integron, a site-specific DNA recombination system, at the DNA level and its attendant aminoglycoside-sensing riboswitch at the RNA level. The P. fluorescens aminoglycoside-sensing riboswitch RNA is associated with the site-specific DNA recombination system class 1 integron (8, 10). The DNA sequence corresponding to the 75-nt riboswitch RNA overlaps the recombinase binding site (attI-1). The 75-nt aminoglycoside-sensing riboswitch of P. fluorescens was used to identify homologous sequences in surviving aminoglycoside-resistant pathogen strains by BLAST searching. Analysis of six sequences that are the end products of site-specific recombination by class 1 integrons showed that there was considerable sequence variation compared to the P. fluorescens riboswitch. Despite the sequence variation for each of the six RNA sequences, 4,6-deoxystreptamine aminoglycosides induce reporter gene expression to retain the aminoglycoside-sensing riboswitch function. Direct binding between the aac-6 UTR and the aminoglycosides was also observed through MST measurements. Further sequence covariance analysis showed that the sequence from nucleotides 1 to 60 adopted the same secondary structure, and that binding of SD2 to anti-SD was maintained through complementarity between the variant SD2 and anti-SD sequences (Fig. 1B). Antibiotic binding results in a structural transition that unmasks SD2 for ribosome binding. The combination of reporter assays and covariance analysis suggests a structural transition model analogous to that of the P. fluorescens riboswitch in which antibiotic binding liberates SD2 to initiate translation of the reporter gene. In the presence of antibiotics, the selection of recombinants that maintain complementarity with the RBS of the resistance gene confers a survival advantage.
The integrase enzyme controls integration and resistance gene capture by integrons. The transcriptional repressor LexA controls expression of the integrase gene. Conditions that stress the cell and induce the bacterial SOS response lead to expression of the integrase gene through derepression of LexA. Subinhibitory doses of certain antibiotics induce the SOS response, which is highly mutagenic (47, 48), and the presence of the conserved regulatory RNA sequence also acts to stabilize and protect the attI-1 recombination site from accumulating deleterious mutations. The aac leader RNA sequences are maintained and function as regulatory RNAs alongside their established DNA function as a (attI-1) binding site for the integrase. This association of a concomitant inducible riboswitch that responds to a commonly used antibiotic class therefore provides an additional selective advantage to the integron cassette in an antibiotic-rich environment (34).
The leader RNA sequences (aac-1 to aac-6) may also be differentiated on the basis of their behavior toward individual aminoglycosides with, for example, gentamicin causing 5-fold induction of the aac-6 reporter gene (Fig. 2). These differences in antibiotic specificity are not consistent with possible regulatory models based upon aminoglycoside-ribosome interactions or with other global effects on translation, as has been suggested (49), because the individual strains expressing aac-1 to aac-6 have identical genetic backgrounds; the only difference between the strains occurs through the expression of the leader RNA. Therefore, the different specificities observed for the effect of the aminoglycosides on the induction of reporter gene expression are probably due to specific interactions between the individual riboswitch RNA molecules and the particular aminoglycosides.
The aac riboswitch sequences demonstrate progressive induction of reporter genes in response to increased doses of the antibiotic. After the initial induction, at higher antibiotic doses, a decline in reporter gene expression is observed. Normally, these conditions would induce expression of the aminoglycoside acetyltransferase resistance gene, leading to resistance to the drug. In the antibiotic-sensitive cells, however, inhibition of translation still takes place, leading to the observed reduction in reporter gene expression, which precedes cell death. All of the reporter constructs display significant background levels of reporter gene expression in the absence of aminoglycosides. The aminoglycosides are exemplary Michaelis-Menten substrates for the acetyltransferase enzymes (50, 51). Thus, although basal levels of gene expression are associated with the aac riboswitch sequences, because the catalytic rate of the enzyme is proportional to enzyme concentration, even a small increase in enzyme concentration will lead to a proportional increase in substrate turnover (we observed 2- to 5-fold induction, comparable to that observed in clinical strains [42]), such that at much higher enzyme concentrations, it is likely that the acetyl cofactor (acetyl coenzyme A) would become limiting; thus, no benefit from the production of an additional acetyltransferase enzyme would accrue. The riboswitch may therefore be regarded as a “dimmer switch” (52) that provides a response proportional to the dose of the drug. This mechanism allows the cells to respond rapidly to an antibiotic threat, and the commitment of cellular resources to the production of the resistance protein is only made when required. This mechanism reduces the metabolic load on the cells, reducing the fitness cost of maintaining the resistance plasmids and protecting their long-term stability.
In the class I integrons, the first gene of the integron cassette confers resistance to aminoglycoside antibiotics; each of the sequences tested came from a clinical strain and was associated with a downstream acetyltransferase gene, and in each case, the sequence was shown to function as an aminoglycoside-sensing riboswitch. Because each integron is the product of a unique recombination event, this suggests that the initial acquisition of the resistance gene by the class I integrons conferred a significant selective advantage. Genetic recombination rather than mutation accounts for the rapid development of resistance. Aminoglycoside resistance quickly emerged after their introduction in the clinic (16, 21, 53), and compared to the accumulation of mutations, the efficiency of the integron apparatus for the expression of the resistance genes confers a selective advantage through the aminoglycoside-sensing riboswitch.
The rapid progression of antibiotic resistance represents a continuing challenge in the clinic (2). Further detailed understanding of the regulation of resistance gene expression may identify novel antibiotic targets and supply new strategies for combination therapies. Significant progress has been made in elucidating the molecular details of the RNA-based regulatory processes that control ribosomal antibiotic resistance; however, the differences in the chemical structures of ribosomal antibiotics and consequent variety of antibiotic binding modes combined with the multiplicity of antibiotic binding sites and the range of ribosomal functions that are targeted by the antibiotics suggest that simple “one-size-fits-all” models for attenuation will necessarily need to be adjusted to accommodate different classes of antibiotics (54–56). These mechanisms were originally discovered on a gene-by-gene basis whereby the mechanism of resistance to the antibiotic and its mode of regulation were investigated in specific clinically resistant antibiotic strains. Recent advances in the application of genome-wide techniques for the analysis of transcription termination events have identified transcriptionally attenuated regulatory RNAs that control the expression of antibiotic resistance genes (54). Further genome-wide analysis will provide insights into translation- and transcription-dependent regulatory processes in the future.
It is becoming apparent that in addition to their established interactions with the ribosome and the resistance enzymes, the aminoglycosides have a wider role in their producer and other organisms. Like other natural product antibiotics, the aminoglycosides are secondary metabolites. Their biosynthesis involves multiple gene products and complex biosynthetic pathways (57). As such, they should not be regarded as simply inhibitors of bacterial growth; rather, they have been predicted to act as regulatory effector molecules for a range of biological activities (58). Novel functionalities for antibiotics continue to emerge such that they can be considered to be modulators as well as inhibitors (17, 56, 58, 65). The aminoglycosides have also recently been shown to induce T-box transcriptional riboswitch activity (59); they display organism-specific effects on the bacterial SOS response, inducing it in Vibrio cholerae but not in E. coli (47), and subinhibitory doses can induce biofilm formation (60). We speculate that aminoglycoside-sensing riboswitch mechanisms may have additional undiscovered cellular regulatory roles.
MATERIALS AND METHODS
Chemicals and reagents.
All aminoglycoside antibiotics and oligonucleotide primers were purchased from Sangon. SuperScript III was purchased from Invitrogen. T7 RNA polymerase was purified in-house.
Bioinformatics analysis.
Homologous riboswitch sequences of the 75-nt leader RNA were initially identified by a BLAST search (https://blast.ncbi.nlm.nih.gov) (10); subsequently, additional sequences were identified using Infernal (version 1.1) (61) to search the RefSeq database (version 85) (62). Covariance analysis was implemented using CMfinder (66, 67) and covariance models drawn using R2R (63).
Reporter construction and reporter assays.
Reporter plasmid pGEX-leaderRNAaac/aad-lacZα, as previously reported (10), was used as a vector to clone candidate riboswitch and control sequences (Table S1). Agar diffusion and Miller assays were performed as described previously (10, 64).
Microscale thermophoresis.
The riboswitch RNA was prepared by in vitro transcription using T7 RNA polymerase. Purified RNA was labeled with fluorescein-5-thiosemicarbazide, as previously described (44). The fluorescein-RNA and antibiotics were both prepared in 50 mM HEPES (pH 8.0), 100 mM NaCl, and 0.2 mM MgCl2. The RNA was annealed by heating to 95°C for 2 min and then cooled to room temperature. MST experiments were conducted in triplicate on a Monolith NT.115 system (NanoTemper Technologies) (45, 46) and the normalized fluorescence (△Fnorm) was determined from the MST traces for titration experiments.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Murchie laboratory for discussion.
This work was supported by grants from the Natural Science Foundation under the following grant numbers: for A.I.H.M., 2016YFA0500604, 31420103907, 31770873, and 31330022; for D.C., 31370107; and for S.W., 31470777.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00236-19.
REFERENCES
- 1.Kümmerer K. 2003. Significance of antibiotics in the environment. J Antimicrob Chemother 52:5–7. doi: 10.1093/jac/dkg293. [DOI] [PubMed] [Google Scholar]
- 2.WHO 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Wright GD. 2007. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol 5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 4.Arenas M, Araujo NM, Branco C, Castelhano N, Castro-Nallar E, Pérez-Losada M. 2018. Mutation and recombination in pathogen evolution: relevance, methods and controversies. Infect Genet Evol 63:295–306. doi: 10.1016/j.meegid.2017.09.029. [DOI] [PubMed] [Google Scholar]
- 5.Mazel D. 2006. Integrons: agents of bacterial evolution. Nat Rev Microbiol 4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 6.Rowe-Magnus DA, Guerout A-M, Mazel D. 2002. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol Microbiol 43:1657–1669. doi: 10.1046/j.1365-2958.2002.02861.x. [DOI] [PubMed] [Google Scholar]
- 7.Bouvier M, Demarre G, Mazel D. 2005. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J 24:4356–4367. doi: 10.1038/sj.emboj.7600898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanau-Berçot B, Podglajen I, Casin I, Collatz E. 2002. An intrinsic control element for translational initiation in class 1 integrons. Mol Microbiol 44:119–130. doi: 10.1046/j.1365-2958.2002.02843.x. [DOI] [PubMed] [Google Scholar]
- 9.Jacquier H, Zaoui C, Sanson-Le Pors M, Mazel D, Berçot B. 2009. Translation regulation of integrons gene cassette expression by the attC sites. Mol Microbiol 72:1475–1486. doi: 10.1111/j.1365-2958.2009.06736.x. [DOI] [PubMed] [Google Scholar]
- 10.Jia X, Zhang J, Sun W, He W, Jiang H, Chen D, Murchie A. 2013. Riboswitch control of aminoglycoside antibiotic resistance. Cell 152:68–81. doi: 10.1016/j.cell.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 11.MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN. 2006. Structural basis for broad DNA-specificity in integron recombination. Nature 440:1157–1162. doi: 10.1038/nature04643. [DOI] [PubMed] [Google Scholar]
- 12.Partridge SR, Recchia GD, Scaramuzzi C, Collis CM, Stokes HW, Hall RM. 2000. Definition of the attI1 site of class 1 integrons. Microbiology 146:2855–2864. doi: 10.1099/00221287-146-11-2855. [DOI] [PubMed] [Google Scholar]
- 13.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 14.Davies J, Davis BD. 1968. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. The effect of drug concentration. J Biol Chem 243:3312–3316. [PubMed] [Google Scholar]
- 15.Fourmy D, Recht MI, Blanchard SC, Puglisi JD. 1996. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science 274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 16.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liebert CA, Hall RM, Summers AO. 1999. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev 63:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikaido H. 2009. Multidrug resistance in bacteria. Annu Rev Biochem 78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swiatlo E, Kocka FE. 1987. Inducible expression of an aminoglycoside-acetylating enzyme in Providencia stuartii. J Antimicrob Chemother 19:27–30. doi: 10.1093/jac/19.1.27. [DOI] [PubMed] [Google Scholar]
- 20.Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. 1999. Aminoglycosides: activity and resistance. Antimicrob Agents Chemother 43:727–737. doi: 10.1128/AAC.43.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams JW, Northrop DB. 1976. Purification and properties of gentamicin acetyltransferase I. Biochemistry 15:125–131. doi: 10.1021/bi00646a019. [DOI] [PubMed] [Google Scholar]
- 22.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 23.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock L. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 24.Gupta P, Sothiselvam S, Vázquez-Laslop N, Mankin AS. 2013. Deregulation of translation due to post-transcriptional modification of rRNA explains why erm genes are inducible. Nat Commun 4:1984. doi: 10.1038/ncomms2984. [DOI] [PubMed] [Google Scholar]
- 25.Dar D, Sorek R. 2017. Regulation of antibiotic-resistance by non-coding RNAs in bacteria. Curr Opin Microbiol 36:111–117. doi: 10.1016/j.mib.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Dersch P, Khan MA, Mühlen S, Görke B. 2017. Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front Microbiol 8:803. doi: 10.3389/fmicb.2017.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubnau D. 1984. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem 16:103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- 28.Weisblum B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother 39:797–805. doi: 10.1128/AAC.39.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vazquez-Laslop N, Thum C, Mankin AS. 2008. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell 30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 30.Lovett PS, Rogers EJ. 1996. Ribosome regulation by the nascent peptide. Microbiol Rev 60:366–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwak JH, Choi EC, Weisblum B. 1991. Transcriptional attenuation control of ermK, a macrolide-lincosamide-streptogramin B resistance determinant from Bacillus licheniformis. J Bacteriol 173:4725–4735. doi: 10.1128/jb.173.15.4725-4735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. 2017. Riboswitch diversity and distribution. RNA 23:995–1011. doi: 10.1261/rna.061234.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D, Murchie A. 2014. An aminoglycoside sensing riboswitch controls the expression of aminoglycoside resistance acetyl and adenyl-transferases. Biochim Biophys Acta 1839:951–958. doi: 10.1016/j.bbagrm.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Collis CM, Grammaticopoulos G, Briton J, Stokes HW, Hall RM. 1993. Site-specific insertion of gene cassettes into integrons. Mol Microbiol 9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 36.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He W, Zhang X, Zhang J, Jia X, Zhang J, Sun W, Jiang H, Chen D, Murchie AI. 2013. Riboswitch control of induction of aminoglycoside resistance acetyl and adenyl-transferases. RNA Biol 10:1266–1273. doi: 10.4161/rna.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collis CM, Hall RM. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother 39:155–162. doi: 10.1128/AAC.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lévesque C, Brassard S, Lapointe J, Roy PH. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 40.Jia X, Zhang J, Sun W, He W, Jiang H, Chen D, Murchie A. 2013. Riboswitch regulation of aminoglycoside resistance acetyl and adenyl transferases. Cell 153:1419–1420. doi: 10.1016/j.cell.2013.05.050. [DOI] [PubMed] [Google Scholar]
- 41.Duvall EJ, Williams DM, Mongkolsuk S, Lovett PS. 1984. Regulatory regions that control expression of two chloramphenicol-inducible cat genes cloned in Bacillus subtilis. J Bacteriol 158:784–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaimee G, Halami PM. 2017. Conjugal transfer of aac(6′)Ie-aph(2″)Ia gene from native species and mechanism of regulation and cross resistance in Enterococcus faecalis MCC3063 by real time-PCR. Microb Pathog 110:546–553. doi: 10.1016/j.micpath.2017.07.049. [DOI] [PubMed] [Google Scholar]
- 43.Hendrix M, Priestley ES, Joyce GF, Wong CH. 1997. Direct observation of aminoglycoside-RNA interactions by surface plasmon resonance. J Am Chem Soc 119:3641–3648. doi: 10.1021/ja964290o. [DOI] [PubMed] [Google Scholar]
- 44.Wu TP, Ruan KC, Liu WY. 1996. A fluorescence-labeling method for sequencing small RNA on polyacrylamide gel. Nucleic Acids Res 24:3472–3473. doi: 10.1093/nar/24.17.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Entzian C, Schubert T. 2016. Studying small molecule-aptamer interactions using microscale thermophoresis thermophoresis (MST). Methods 97:27–34. doi: 10.1016/j.ymeth.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 46.Moon MH, Hilimire TA, Sanders AM, Schneekloth JS. 2018. Measuring RNA-ligand interactions with microscale thermophoresis. Biochemistry 57:4638–4643. doi: 10.1021/acs.biochem.7b01141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baharoglu Z, Mazel D. 2011. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob Agents Chemother 55:2438–2441. doi: 10.1128/AAC.01549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, Gonzalez-Zorn B, Barbé J, Ploy M-C, Mazel D. 2009. The SOS response controls integron recombination. Science 324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 49.Roth A, Breaker RR. 2013. Integron attI1 sites, not riboswitches, associate with antibiotic resistance genes. Cell 153:1417–1418. doi: 10.1016/j.cell.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Draker K, Northrop DB, Wright GD. 2003. Kinetic mechanism of the GCN5-related chromosomal aminoglycoside acetyltransferase AAC(6′)-II from Enterococcus faecium: evidence of dimer subunit cooperativity. Biochemistry 42:6565–6574. doi: 10.1021/bi034148h. [DOI] [PubMed] [Google Scholar]
- 51.Magalhaes MLB, Blanchard JS. 2005. The kinetic mechanism of AAC3-IV aminoglycoside acetyltransferase from Escherichia coli. Biochemistry 44:16275–16283. doi: 10.1021/bi051777d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baird NJ, Kulshina N, Ferré-D’Amaré AR. 2010. Riboswitch function: flipping the switch or tuning the dimmer? RNA Biol 7:328–332. doi: 10.4161/rna.7.3.11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamada T, Tipper D, Davies J. 1968. Enzymatic inactivation of streptomycin by R factor-resistant Escherichia coli. Nature 219:288–291. doi: 10.1038/219288a0. [DOI] [PubMed] [Google Scholar]
- 54.Dar D, Shamir M, Mellin JR, Koutero M, Stern-Ginossar N, Cossart P, Sorek R. 2016. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 352:aad9822. doi: 10.1126/science.aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knowles DJC, Foloppe N, Matassova NB, Murchie A. 2002. The bacterial ribosome, a promising focus for structure-based drug design. Curr Opin Pharmacol 2:501–506. doi: 10.1016/S1471-4892(02)00205-9. [DOI] [PubMed] [Google Scholar]
- 56.Vázquez-Laslop N, Mankin AS. 2018. Context-specific action of ribosomal antibiotics. Annu Rev Microbiol 72:185–207. doi: 10.1146/annurev-micro-090817-062329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sengupta S, Chattopadhyay MK, Grossart H-P. 2013. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol 4:47. doi: 10.3389/fmicb.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davies J. 1990. What are antibiotics? Archaic functions for modern activities. Mol Microbiol 4:1227–1232. doi: 10.1111/j.1365-2958.1990.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 59.Stamatopoulou V, Apostolidi M, Li S, Lamprinou K, Papakyriakou A, Zhang J, Stathopoulos C. 2017. Direct modulation of T-box riboswitch-controlled transcription by protein synthesis inhibitors. Nucleic Acids Res 45:10242–10258. doi: 10.1093/nar/gkx663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 61.Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pruitt KD, Tatusova T, Brown GR, Maglott DR. 2012. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinberg Z, Breaker RR. 2011. R2R–software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 12:3. doi: 10.1186/1471-2105-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Bremer H. 1995. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J Biol Chem 270:11181–11189. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
- 65.Schroeder R, Waldsich C, Wank H. 2000. Modulation of RNA function by aminoglycoside antibiotics. EMBO J 19:1–9. doi: 10.1093/emboj/19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. 2013. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yao Z, Weinberg Z, Ruzzo WL. 2006. CMfinder–a covariance model based RNA motif finding algorithm. Bioinformatics 22:445–452. doi: 10.1093/bioinformatics/btk008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.