The understanding of species distribution and inducible macrolide resistance in the Mycobacterium fortuitum complex (MFC) is limited. Of 90 mostly respiratory MFC clinical isolates, half were M. fortuitum, followed by M. peregrinum, M. porcinum, M. septicum, and M. conceptionense.
KEYWORDS: Mycobacterium fortuitum complex, clarithromycin, drug resistance, nontuberculous mycobacteria
ABSTRACT
The understanding of species distribution and inducible macrolide resistance in the Mycobacterium fortuitum complex (MFC) is limited. Of 90 mostly respiratory MFC clinical isolates, half were M. fortuitum, followed by M. peregrinum, M. porcinum, M. septicum, and M. conceptionense. Most M. fortuitum, M. porcinum, and M. septicum isolates were inducibly resistant to clarithromycin, whereas two-thirds of the M. peregrinum isolates were clarithromycin susceptible. Clarithromycin-resistant M. fortuitum isolates exhibited common mutations of erm(39), potentially involved in clarithromycin resistance.
INTRODUCTION
The Mycobacterium fortuitum complex (MFC), composed of several closely related species of nontuberculous mycobacteria (NTM), can cause human pulmonary and extrapulmonary infections. Based on identification of clinical isolates, the distribution of MFC species varies geographically. In Taiwan, Greece, and the United Kingdom, M. fortuitum was the second most frequently isolated NTM after members of the M. avium complex (23%, 21%, and 20%, respectively), whereas in South Korea and Japan, M. fortuitum accounted for only 8% and 2% of all clinical isolates, respectively (1). Currently, the MFC includes M. fortuitum, M. peregrinum, M. porcinum, M. septicum, M. conceptionense, M. boenickei, M. houstonense, M. neworleansense, M. brisbanense, M. farcinogenes, M. senegalense, and M. setense (2–4).
According to American Thoracic Society guidelines (5), 80% of M. fortuitum isolates are clarithromycin (CLR) susceptible. However, the guidelines recommend that macrolides be used with caution, due to the presence of the erythromycin-inducible methylase (erm) gene, which confers inducible resistance to macrolides in several NTM species (5). To date, only limited studies have reported that M. fortuitum clinical isolates harbor the erm(39) gene (6, 7). Moreover, there are no studies of the correlation between macrolide susceptibility and erm(39) sequevars of M. fortuitum. Additionally, it is not clear whether MFC species other than M. fortuitum possess inducible macrolide resistance.
The aims of this study were to elucidate the species distribution of MFC clinical isolates; to evaluate the isolates for the presence of macrolide resistance-related genes, such as erm and rrl; and to determine the association between macrolide susceptibility and erm(39) sequevars.
A total of 90 MFC clinical isolates collected from August 2011 to December 2013 at Samsung Medical Center were included in our study, which was approved by the Institutional Review Board (IRB) of Samsung Medical Center (IRB no. 2008-09-016). Multilocus sequencing analysis was carried out as described previously (8). Drug susceptibility testing (DST) for CLR was performed using the broth microdilution method (9). The MIC of CLR was determined on days 3 and 14 after incubation, and MFC isolates were considered susceptible (MIC, ≤2 μg/ml at days 3 and day 14), resistant (MIC, ≥8 μg/ml at day 3), or inducibly resistant (susceptible at day 3 but resistant at day 14) to CLR (9). We designed species-specific PCR primers for sequencing of the entire erm gene: erm(39)fo F (5′-GAAATTGAGTTGAGCGTCCG-3′) and erm(39)fo R (5′-TCTACATCGCCTGGACCATC-3′) for erm(39) in M. fortuitum and erm(39)po F (5′-CAGTGACCTACCTCCGCTTG-3′) and erm(39)po R (5′-CTACATCGCCTGGACCATCG-3′) for erm(39) in M. porcinum. The erm(39) sequences were trimmed using the CLUSTAL W program (10). Phylogenetic trees were obtained by the use of MEGA (version 6.0) software (11). To detect rrl mutations, PCR was performed as described previously (12).
Eighty-eight of the 90 MFC isolates were recovered from respiratory specimens, and 44 (49%) were reidentified as M. fortuitum, followed by 27 (30%) as M. peregrinum, 10 (11%) as M. porcinum, 7 (8%) as M. septicum, and 2 (2%) as M. conceptionense (Table 1). Among the 44 M. fortuitum isolates, 37 (84%) were inducibly resistant to CLR, 5 exhibited CLR resistance, and the remaining 2 were susceptible to CLR. Among the 27 M. peregrinum isolates, 18 (69%) were CLR susceptible and 8 (31%) were inducibly resistant to CLR, but DST results for CLR were not available for 1 isolate. Almost all M. porcinum and M. septicum isolates had inducible macrolide resistance, with only one of each species being resistant to CLR. Both M. conceptionense isolates were CLR susceptible. None of the 90 MFC isolates had rrl mutations, which can confer CLR resistance (12).
TABLE 1.
Reidentification and clarithromycin resistance of M. fortuitum complex clinical isolates
| Characteristic | Value for the following species: |
||||
|---|---|---|---|---|---|
| M. fortuitum | M. peregrinumb | M. porcinum | M. septicum | M. conceptionense | |
| No. (%) of isolates identified by MLSAa | 44 (49) | 27 (30) | 10 (11) | 7 (8) | 2 (2) |
| Patient data | |||||
| Median (interquartile range) age (yr) | 62 (54–72) | 58 (55–63) | 65 (56–76) | 49 (40–84) | 43, 59 |
| No. (%) of female patients | 21 (48) | 13 (48) | 3 (30) | 3 (43) | 2 (100) |
| No. (%) of isolates from the following specimens: | |||||
| Respiratory specimen | 42 (96) | 27 (100) | 10 (100) | 7 (100) | 2 (100) |
| Blood | 1 (2) | 0 | 0 | 0 | 0 |
| Joint fluid | 1 (2) | 0 | 0 | 0 | 0 |
| No. (%) of isolates with the following susceptibility to clarithromycin: | |||||
| Susceptible | 2 (5) | 18 (69) | 0 | 0 | 2 (100) |
| Inducibly resistant | 37 (84) | 8 (31) | 9 (90) | 6 (86) | 0 |
| Resistant | 5 (11) | 0 | 1 (10) | 1 (14) | 0 |
Abbreviation: MLSA, multilocus sequencing analysis.
Drug susceptibility testing results for clarithromycin were not available for one M. peregrinum isolate.
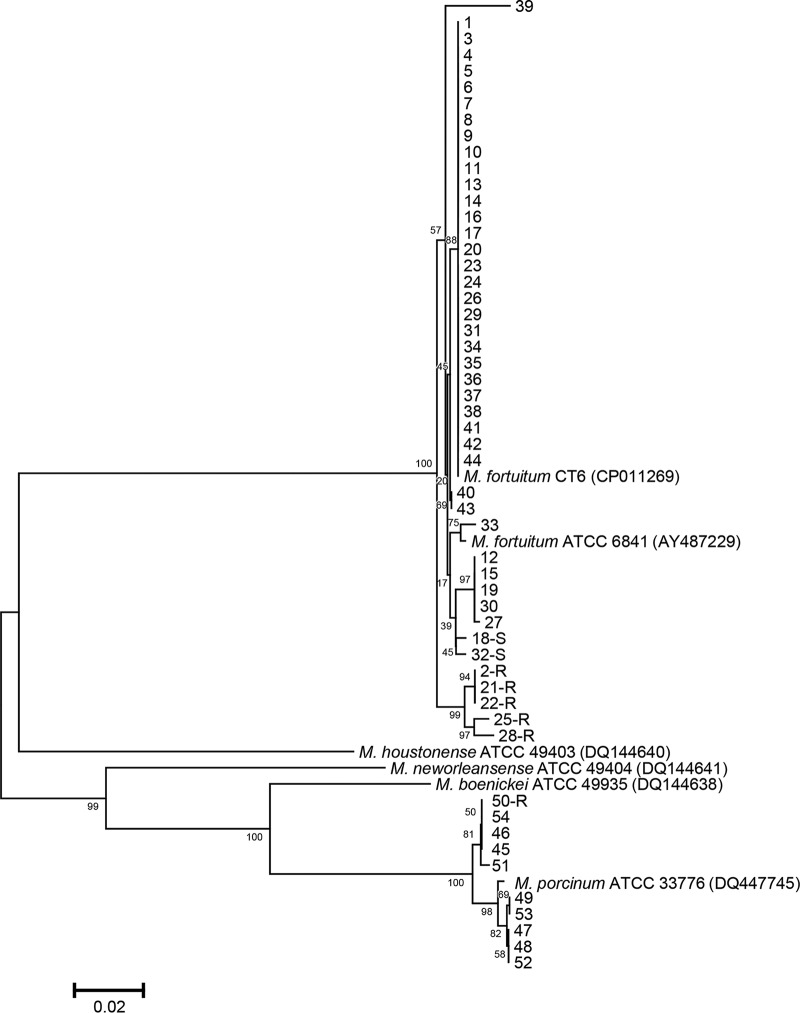
Whole erm gene sequencing was conducted for the 44 M. fortuitum and 10 M. porcinum clinical isolates, as these species are known for harboring erm(39) (13). Each of these sequences differed by at least 1 nucleotide from the erm(39) sequence of the M. fortuitum type strain DSM46621 (GenBank accession no. AY487229) (see Fig. S1 in the supplemental material). However, the sequences of 34 of the 44 isolates were identical to the erm(39) sequence of M. fortuitum strain CT6 (GenBank accession no. CP011269) (Fig. 1). Of the 45 single-nucleotide polymorphisms identified, 23 were synonymous (Fig. S1). The 16 amino acid substitutions are listed in Table 2. In all, nine erm(39) sequevars were identified and were numbered as sequevars 2 to 10. The sequevar of M. fortuitum type strain DSM46621 was designated sequevar 1. Thirty-seven of the 44 M. fortuitum isolates with inducible CLR resistance belonged to sequevars 2 to 5. Two CLR-susceptible isolates had sequevars 6 and 7. Five CLR-resistant isolates had common mutations at nucleotide positions 76, 78, 661, 707, and 729 which were not shared by isolates of the other sequevars; these five isolates were divided among sequevars 8, 9, and 10. Therefore, these mutations found only in CLR-resistant isolates are potentially involved in the CLR resistance of M. fortuitum. Sequence variation in erm has also been noted in M. abscessus, with some mutations resulting in the loss of inducible macrolide resistance (14).
FIG 1.
Phylogenetic tree for M. fortuitum complex (MFC) isolates derived using erm(39) sequences. Sequences were included for 44 M. fortuitum (isolate no. 1 to 44) and 10 M. porcinum (isolate no. 45 to 54) clinical isolates as well as other species belonging to the MFC. The clarithromycin-susceptible or -resistant isolates are indicated by an S or an R, respectively, after the isolate number. Sequences were compared with those of the type strains and other reference strains using the neighbor-joining method with Kimura’s two-parameter distance correction model. Bootstrap analyses determined from 1,000 replicates are indicated at the nodes. Bar, 2% difference in nucleotide sequence. GenBank accession numbers are given in parentheses.
TABLE 2.
The 10 erm(39) gene sequevar types identified in M. fortuitum
| Sequevar | No. of isolates | Susceptibility to CLRa | Nucleotide (amino acid)b
at the following base pair position (corresponding amino acid amino acid position): |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T11 (V4) | G19 (G7) | G64 (V22) | GTG 76-78 (V26) | G97 (A33) | G101 (G34) | A235 (I79) | G358 (A120) | T362 (I121) | C536 (A179) | G563 (R188) | G619 (A207) | G661 (A221) | C676 (P226) | G707 (R236) | T729 (D243) | |||
| 1c | 0 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | |
| 2 | 1 | IR | • | • | • | • | • | • | • | T (S) | • | • | • | • | • | • | • | • |
| 3d | 34 | IR | • | • | • | • | A (T) | • | G (V) | • | • | • | • | • | • | • | • | • |
| 4e | 1 | IR | C (A) | • | A (I) | • | A (T) | • | G (V) | • | • | • | • | • | • | • | • | • |
| 5 | 1 | IR | • | • | • | • | A (T) | • | G (V) | • | • | • | • | • | • | A (T) | • | • |
| 6 | 1 | Susceptible | • | A (S) | • | • | A (T) | • | G (V) | • | • | • | • | • | • | • | • | • |
| 7 | 1 | Susceptible | C (A) | • | • | • | A (T) | C (A) | G (V) | • | • | • | • | • | • | • | • | • |
| 8 | 3 | Resistant | C (A) | • | • | ATC (I) | A (T) | • | G (V) | • | • | • | • | • | A (T) | • | A (Q) | A (E) |
| 9 | 1 | Resistant | C (A) | • | • | ATC (I) | A (T) | • | G (V) | • | • | A (D) | • | A (T) | A (T) | • | A (Q) | A (E) |
| 10 | 1 | Resistant | C (A) | • | • | ATC (I) | A (T) | • | G (V) | • | C (T) | • | A (H) | • | A (T) | • | A (Q) | A (E) |
Abbreviations: CLR, clarithromycin; IR, inducibly resistant.
Nucleotides (amino acids) for sequevar type 1 are shown in the column subheads. •, identical nucleotides. For altered nucleotides, the corresponding amino acid change is also indicated.
Type strain DSM46621.
Identical to the sequence of strain CT6.
This isolate had the additional substitutions L99V, K156R, S158P, T174I, and P212S.
The erm(39) sequences of the 10 M. porcinum clinical isolates differed from the erm(39) sequence of the M. porcinum type strain ATCC 33776 (GenBank accession no. DQ447745) by 3 to 11 nucleotide mismatches (Fig. S2). Although we detected some shared mutations, there was no consistent association between a specific mutation and the macrolide resistance pattern.
The majority of M. peregrinum isolates were CLR susceptible in our study, but one-third had inducible macrolide resistance. M. peregrinum was originally reported to have no erm gene (13). However, a subsequent study revealed that 8 of 23 M. peregrinum clinical isolates had erm genes (7). Further studies are needed to resolve the role of the erm gene in this species.
The M. septicum type strain was reported to be susceptible to CLR using the Etest method with incubation for 3 days (15). However, we identified a putative erm gene in the M. septicum type strain (GenBank accession no. HG322951; the region from positions 2111635 to 2112375) with 86% identity to the sequences of erm(39) from M. boenickei and M. houstonense (GenBank accession no. DQ144638 and DQ144640, respectively). Additionally, six (86%) of our seven M. septicum clinical isolates had inducible resistance to CLR. These results provide further evidence that macrolide resistance in M. septicum can be inducible.
As in our results, M. conceptionense clinical isolates in two other studies were susceptible to CLR (16, 17). These findings suggest that M. conceptionense does not have a functional erm gene, but additional research is needed to determine whether this lack is a hallmark of the species.
In this study, we made several novel observations. First, 5% of the M. fortuitum clinical isolates were susceptible to CLR. Second, CLR-resistant M. fortuitum clinical isolates had several mutations in common in erm(39). Third, 30% of M. peregrinum isolates and most M. septicum isolates showed inducible macrolide resistance. To the best of our knowledge, this is the first report to investigate the association between erm(39) sequevars and resistance to CLR in M. fortuitum. This study highlights the importance of accurate species identification of MFC clinical isolates and of prolonged incubation during DST to screen for inducible macrolide resistance. Furthermore, as there can be variation in erm genes even within a species, such prolonged incubation may be of value even for NTM species not previously known to have inducible macrolide resistance.
Accession number(s).
The erm(39) sequences were deposited in GenBank under accession numbers MK468741 to MK468794.
Supplementary Material
ACKNOWLEDGMENTS
S.H.K. has been a consultant for Johnson and Johnson and received consultancy/speaker fees from Insmed, Inc., not associated with the submitted work. C.L.D. has received grants from Insmed, Inc., and served on advisory boards for Insmed Inc., Johnson and Johnson, Spero, and Horizon, not associated with the submitted work. W.-J. K. has received a consultation fee from Insmed, Inc., for the Insmed advisory board meeting, not associated with the submitted work. Otherwise, we have no conflicts of interest to declare.
This work was supported by the National Research Foundation of Korea (NRF) funded by the South Korea government (MSIT) (NRF-2018R1A2A1A05018309).
The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02331-18.
REFERENCES
- 1.Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, Beylis N, Boeree MJ, Cacho J, Chihota V, Chimara E, Churchyard G, Cias R, Daza R, Daley CL, Dekhuijzen PNR, Domingo D, Drobniewski F, Esteban J, Fauville-Dufaux M, Folkvardsen DB, Gibbons N, Gómez-Mampaso E, Gonzalez R, Hoffmann H, Hsueh P-R, Indra A, Jagielski T, Jamieson F, Jankovic M, Jong E, Keane J, Koh W-J, Lange B, Leao S, Macedo R, Mannsåker T, Marras TK, Maugein J, Milburn HJ, Mlinkó T, Morcillo N, Morimoto K, Papaventsis D, Palenque E, Paez-Peña M, Piersimoni C, Polanová M, Rastogi N, Richter E, et al. 2013. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J 42:1604–1613. doi: 10.1183/09031936.00149212. [DOI] [PubMed] [Google Scholar]
- 2.Brown-Elliott BA, Philley JV. 2017. Rapidly growing mycobacteria. Microbiol Spectr 5:TNMI7-0027-2016. doi: 10.1128/microbiolspec.TNMI7-0027-2016. [DOI] [PubMed] [Google Scholar]
- 3.Brown-Elliott BA, Wallace RJ Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev 15:716–746. doi: 10.1128/CMR.15.4.716-746.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tortoli E, Fedrizzi T, Meehan CJ, Trovato A, Grottola A, Giacobazzi E, Serpini GF, Tagliazucchi S, Fabio A, Bettua C, Bertorelli R, Frascaro F, De Sanctis V, Pecorari M, Jousson O, Segata N, Cirillo DM. 2017. The new phylogeny of the genus Mycobacterium: the old and the news. Infect Genet Evol 56:19–25. doi: 10.1016/j.meegid.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ Jr, Winthrop K. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 6.Li F, Li GL, Pang H, Liu HC, Xiao TY, Li SJ, Luo Q, Jiang Y, Wang RB, Wan KL. 2018. Preliminary study on drug susceptibility profile and resistance mechanisms to macrolides of clinical isolates of non-tuberculous mycobacteria from China. Biomed Environ Sci 31:290–299. doi: 10.3967/bes2018.037. [DOI] [PubMed] [Google Scholar]
- 7.Esteban J, Martín-de-Hijas NZ, García-Almeida D, Bodas-Sánchez A, Gadea I, Fernández-Roblas R. 2009. Prevalence of erm methylase genes in clinical isolates of non-pigmented, rapidly growing mycobacteria. Clin Microbiol Infect 15:919–923. doi: 10.1111/j.1469-0691.2009.02757.x. [DOI] [PubMed] [Google Scholar]
- 8.Kim SY, Shin SH, Moon SM, Yang B, Kim H, Kwon OJ, Huh HJ, Ki CS, Lee NY, Shin SJ, Koh WJ. 2017. Distribution and clinical significance of Mycobacterium avium complex species isolated from respiratory specimens. Diagn Microbiol Infect Dis 88:125–137. doi: 10.1016/j.diagmicrobio.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Clinical Laboratory Standards Institute. 2011. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard, 2nd ed CLSI document M24-A2 Clinical Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 10.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 11.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastian S, Veziris N, Roux AL, Brossier F, Gaillard JL, Jarlier V, Cambau E. 2011. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm(41) and rrl sequencing. Antimicrob Agents Chemother 55:775–781. doi: 10.1128/AAC.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nash KA, Andini N, Zhang Y, Brown-Elliott BA, Wallace RJ Jr. 2006. Intrinsic macrolide resistance in rapidly growing mycobacteria. Antimicrob Agents Chemother 50:3476–3478. doi: 10.1128/AAC.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nash KA, Brown-Elliott BA, Wallace RJ Jr. 2009. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother 53:1367–1376. doi: 10.1128/AAC.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adekambi T, Drancourt M. 2006. Isolation of Mycobacterium septicum from the sputum of a patient suffering from hemoptoic pneumonia. Res Microbiol 157:466–470. doi: 10.1016/j.resmic.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Shojaei H, Hashemi A, Heidarieh P, Ataei B, Naser AD. 2011. Pulmonary and extrapulmonary infection caused by Mycobacterium conceptionense: the first report from Iran. JRSM Short Rep 2:31. doi: 10.1258/shorts.2010.010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SY, Kim MS, Chang HE, Yim JJ, Lee JH, Song SH, Park KU, Song J, Kim EC. 2012. Pulmonary infection caused by Mycobacterium conceptionense. Emerg Infect Dis 18:174–176. doi: 10.3201/eid1801.110251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.