Several emerging pathogens have arisen as a result of selection pressures exerted by modern health care. Klebsiella quasipneumoniae was recently defined as a new species, yet its prevalence, niche, and propensity to acquire antimicrobial resistance genes are not fully described.
KEYWORDS: environmental reservoir, infection control, KPC, Klebsiella, Klebsiella pneumoniae carbapenemase, Klebsiella quasipneumoniae, sink drains, carbapenemase, multidrug resistance, premise plumbing
ABSTRACT
Several emerging pathogens have arisen as a result of selection pressures exerted by modern health care. Klebsiella quasipneumoniae was recently defined as a new species, yet its prevalence, niche, and propensity to acquire antimicrobial resistance genes are not fully described. We have been tracking inter- and intraspecies transmission of the Klebsiella pneumoniae carbapenemase (KPC) gene, blaKPC, between bacteria isolated from a single institution. We applied a combination of Illumina and PacBio whole-genome sequencing to identify and compare K. quasipneumoniae from patients and the hospital environment over 10- and 5-year periods, respectively. There were 32 blaKPC-positive K. quasipneumoniae isolates, all of which were identified as K. pneumoniae in the clinical microbiology laboratory, from 8 patients and 11 sink drains, with evidence for seven separate blaKPC plasmid acquisitions. Analysis of a single subclade of K. quasipneumoniae subsp. quasipneumoniae (n = 23 isolates) from three patients and six rooms demonstrated seeding of a sink by a patient, subsequent persistence of the strain in the hospital environment, and then possible transmission to another patient. Longitudinal analysis of this strain demonstrated the acquisition of two unique blaKPC plasmids and then subsequent within-strain genetic rearrangement through transposition and homologous recombination. Our analysis highlights the apparent molecular propensity of K. quasipneumoniae to persist in the environment as well as acquire carbapenemase plasmids from other species and enabled an assessment of the genetic rearrangements which may facilitate horizontal transmission of carbapenemases.
INTRODUCTION
In the last 50 years, transformations in health care have created new niches for microorganisms such as Acinetobacter baumannii complex and Candida auris to arise from obscurity and emerge as important pathogens. Similarly, we have seen an increasing number of highly resistant Klebsiella pneumoniae strains which have been successfully transmitted worldwide (1). Klebsiella pneumoniae has proven to be an important contributor to the modern antibiotic resistance epidemic, with its ability to acquire and carry antimicrobial resistance plasmids, as well as being successful as a human pathogen. More recently, whole-genome sequencing has revealed that many isolates classified as K. pneumoniae actually encompass three related but distinct species: K. pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae (1, 2). K. quasipneumoniae was originally thought to be largely confined to agriculture and the environment; however, it appears that it may also be prominent in human disease (3), and several recent reports have demonstrated that it harbors virulence factors and acquires clinically relevant genes of antimicrobial resistance (4, 5). Although there have been relatively few reports of K. quasipneumoniae to date, the true prevalence of this organism is likely underestimated as it is not generally distinguished from K. pneumoniae in routine testing of clinical laboratories (2).
Bacterial evolution via horizontal gene transfer is central to the ongoing crisis of antimicrobial resistance among clinically relevant bacteria. Hospital wastewater is being increasingly recognized as an ideal reservoir for resistance gene exchange and amplification, with ongoing antimicrobial selection pressure exerted through antimicrobials excreted in patient waste (6). Premise plumbing can be seeded by antimicrobial resistance genes in diverse bacterial strains and species and represents a difficult-to-treat reservoir for ongoing gene exchange, creating successful drug-resistant bacteria that can thrive in both the environmental and human niches (7).
Whole-genome sequencing studies have demonstrated that our understanding of the interplay between antimicrobial resistance plasmids and their host strains/species is limited (8). The host range of a plasmid is critical for acquisition and persistence in specific species, but it appears that some bacterial strains are better equipped than others to prevent the acquisition of or destroy foreign plasmid DNA (9). The durability of plasmid acquisition events and the creation of new highly resistant strains reflect complex dynamics which depend on the characteristics of the plasmid in question as well as host strain tolerance (10, 11). Seldom do we have the opportunity to witness strains acquiring plasmids in vivo or in the environment, and inferences about genetic rearrangements are often highly speculative. However, understanding the mechanisms and frequency of resistance gene transfer events occurring in real world contexts can provide important insights into the wider evolutionary landscape creating modern multidrug-resistant bacteria which cannot be effectively modeled in lab experiments (12).
Within our institution, we have seen ongoing transmission of diverse carbapenemase-producing organisms for the last decade, driven by genetic exchange of the Klebsiella pneumoniae carbapenemase (KPC) gene (blaKPC) in patients and the environment (13, 14). This has enabled us to understand specific pathways of genetic mobility involving numerous different mobile genetic elements and host bacterial species (13, 15). Herein, we examine blaKPC acquisition and associated genetic rearrangements within K. quasipneumoniae as a real-life representation of an emerging pathogen associated with the hospital wastewater environment.
RESULTS
From our collection of blaKPC-positive isolates from patients (2007 to 2017) and the hospital environment (2013 to 2017), there were a total of 32 blaKPC-positive K. quasipneumoniae isolates, all of which were identified as K. pneumoniae in the clinical microbiology laboratory (Table 1). Twenty-three of these were K. quasipneumoniae subspecies quasipneumoniae (KpIIA) (10 patient isolates from four patients and 13 environmental isolates from seven rooms), and nine were K. quasipneumoniae subspecies similipneumoniae (KpIIB) (five patient isolates from four patients and four environmental isolates from four rooms). The KpIIA and KpIIB isolates were separated by >100,000 single nucleotide variants (SNVs). We identified a single strain of KpIIA and four strains of KpIIB differing from each other by >20,000 SNVs (Fig. 1). Many isolates have multiple virulence factors (see Data Set S1 in the supplemental material), including several genes involved in capsule production (16) and several fimbrial elements. A type VI secretion system was present in all KpIIA but not all KpIIB isolates. From a resistance gene standpoint, in addition to blaKPC, all isolates harbored fosA and blaOKP as well as a multidrug efflux transporter (oqxA-oqxB) (17).
TABLE 1.
All Sequenced blaKPC-Klebsiella quasipneumoniae isolates from patients and the hospital environment
| Label | Isolate | Subspecies of K. quasipneumoniae | Date (mo-yr) | Source | Infection/outcome |
|---|---|---|---|---|---|
| 1 | CAV1360 | KpIIA | Nov-09 | Sputum | Ventilator-associated pneumonia in complicated heart transplant recipient/expired |
| 2 | CAV2013 | KpIIA | Nov-13 | Perirectal surveillance | NAa |
| 2 | CAVp203 | KpIIA | Dec-13 | Bronchoscopy | Ventilator-associated pneumonia/successful treatment |
| 2 | CAVp26 | KpIIA | Apr-14 | Blood | Intraabdominal infection/expired |
| 2 | CAVp20 | KpIIA | Mar-14 | Perirectal surveillance | NA |
| 2 | CAVp64 | KpIIA | Aug-14 | Perirectal surveillance | NA |
| 2 | CAVp72 | KpIIA | Sep-14 | Perirectal surveillance | NA |
| 2 | CAVp103 | KpIIA | Nov-14 | Blood | Successful treatment |
| 3 | CAVp67 | KpIIA | Aug-14 | Perirectal surveillance | NA |
| 4 | CAVp275 | KpIIA | Jul-15 | Urine | Complicated urinary tract infection/successful treatment |
| 5 | CAV1142 | KpIIB | Aug-09 | Perirectal surveillance | NA |
| 6 | CAVp186 | KpIIB | Dec-13 | Perirectal surveillance | NA |
| 7 | CAV2009 | KpIIB | Feb-14 | Perirectal surveillance | NA |
| 8 | CAVp296 | KpIIB | Oct-15 | Perirectal surveillance | NA |
| 8 | CAVp360 | KpIIB | Dec-16 | Perirectal surveillance | NA |
| Room A (MICUb ) | CAV2244 | KpIIA | Jan-14 | Shower | |
| Room B (CTA) | CAV2279 | KpIIA | Jan-14 | Shower | |
| Room C (STBICUc ) | CAV1945 | KpIIA | Feb-14 | Drain swab (first sample after replacement) | |
| Room C (STBICU) | CAV1947 | KpIIA | Feb-14 | P-trap water (first sample after replacement) | |
| Room C (STBICU) | CAV1964 | KpIIA | Mar-14 | Drain swab | |
| Room C (STBICU) | CAV2018 | KpIIA | Apr-14 | P-trap water | |
| Room D (STBICU) | CAV2019 | KpIIA | Apr-14 | P-trap water | |
| Room C (STBICU) | CAV2397 | KpIIA | May-14 | Drain swab | |
| Room E (STBICU) | CAV2697 | KpIIA | Jul-14 | Drain swab | |
| Room F (MICU) | CAV2957 | KpIIA | Sep-15 | Drain swab | |
| Room G (STBICU) | CAV2983 | KpIIA | Oct-15 | P-trap water | |
| Room G (STBICU) | CAV2984 | KpIIA | Oct-15 | Drain swab | |
| Room G (STBICU) | CAV3444 | KpIIA | Feb-16 | P-trap water | |
| Room H (MICU) | CAV1880 | KpIIB | Dec-13 | Drain swab | |
| Room I (MICU) | CAV1895 | KpIIB | Dec-13 | Drain swab | |
| Room J (STBICU) | CAV1832 | KpIIB | Dec-13 | P-trap water | |
| Room K (STBICU) | CAV1887 | KpIIB | Dec-13 | P-trap water |
NA, not applicable.
MICU, medical intensive care unit.
STBICU, surgical, trauma, and burn intensive care unit.
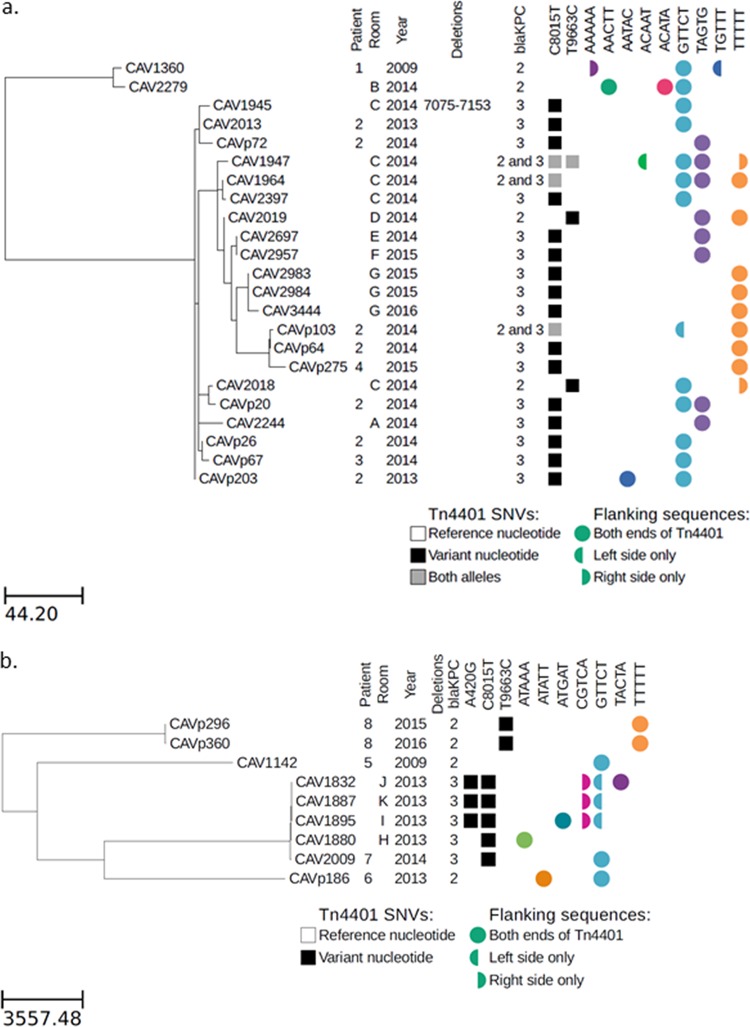
FIG 1.
Maximum likelihood phylogeny for KpIIA (a) and KpIIB (b) isolates, with Tn4401 variation and flanking genetic contexts. Branch lengths are shown as SNVs per genome.
Within the KpIIA strain, there were two subclades separated by ∼150 SNVs (Fig. 1a). The first subclade contained two isolates separated by 10 SNVs (Fig. 1a). CAV1360 was from patient 1 in November 2009, and CAV2279 was identified in early 2014 (shortly after environmental sampling began) from room B that patient 1 had occupied in May 2009.
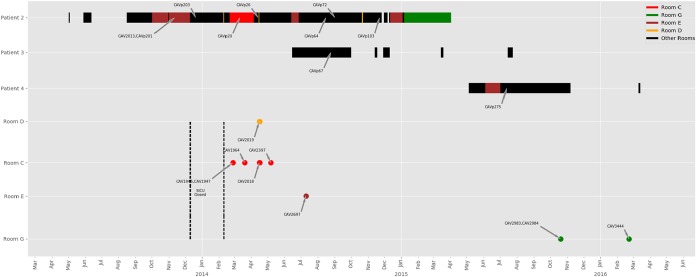
The second subclade of KpIIA contained isolates from three patients (patients 2 to 4) and six rooms (rooms A and C to G). The earliest of these was from patient 2 in November 2013. Patient 2 was in the hospital with a prolonged stay in the surgical, trauma, and burn intensive care unit (STBICU) following complications of a liver transplant (Fig. 2). Patient 2 was noted to be first colonized with blaKPC-positive KpIIA in November 2013. KpIIA was not found in the STBICU environment prior to closure for remediation of KPC contamination of the drains in December 2013. Following drain exchange and unit reopening, patient 2 was immediately moved back into the STBICU and subsequently occupied rooms C, D, E, and G in the STBICU, suggesting that the KpIIA isolates in these rooms originated from patient 2 (Fig. 2). Patient 3 was admitted to the STBICU at the same time as patient 2 and thus could have acquired KPC-KpIIA directly from patient 2 without environmental transmission. Patient 4 was later admitted to STBICU room E for 28 days and discharged before he was found to have KpIIA. He was never on a ward at the same time as any other patients known to carry KpIIA, suggesting acquisition from the hospital environment.
FIG 2.
Patient movements and positive environmental samples with a single strain of K. quasipneumoniae (KpIIA) in the STBICU. Colored bars for patients match rooms where environmental isolates were identified. Black bars represent rooms with no KpIIA identified. The dotted lines indicate STBICU closure with removal and new installation of sink drains and exposed sink plumbing. Patient 1 is not depicted, as there was no admission to the STBICU and no overlap in time or space with other patients carrying KpIIA.
There were four patients (patients 5 to 8) carrying four distinct strains of blaKPC-KpIIB seen over a 5-year period (Fig. 1b, Table 1). For patient 7, the same KpIIB strain (∼80 SNV differences) was also seen in sinks from two rooms in the medical intensive care unit (MICU) (rooms H and I) and two rooms in the STBICU (rooms J and K) in December 2013 when environmental sampling first began; this preceded the detection in the patient in February 2014. Patient 7 was admitted to the MICU (location of rooms H and I), but did not stay any of the rooms where the isolates within the same KpIIB were identified. The other three patients with KpIIB each had unique blaKPC strains, none of which were identified in another patient or the environment. Patient 6 also with a unique strain had a prolonged hospital stay and was also colonized/infected with another blaKPC-positive species (K. pneumoniae).
Three patients developed infections with KPC-KpIIA (Table 1). Patient 1 died of ventilator-associated pneumonia with KPC-KpIIA following a complicated heart transplant. Patient 2 had both ventilator-acquired pneumonia, which was successfully treated, and a subsequent untreatable intraabdominal infection with KPC-KpIIA bacteremia, which contributed to the patient’s death after a long hospital stay with a complicated liver transplant. Patient 4 had a successfully treated complicated KPC-KpIIA urinary tract infection. Patient 3 did not develop an infection with KpIIA. None of the patients with KpIIB developed K. quasipneumoniae infections; however, two of the patients did develop infections with other species carrying blaKPC (K. pneumoniae for patient 6 and Serratia marcescens for patient 8) (Table 2).
TABLE 2.
All additional blaKPC-positive isolates from patients with K. quasipneumoniae
| Patient | Isolate | Species | Date (mo-yr) | Source | Infection | Genetic information (Tn4401 isoform) | Flank sequence(s) (right/left) |
|---|---|---|---|---|---|---|---|
| 2 | CAVp202 | S. marcescens | Dec-13 | Urine | No | Tn4401b-8 | TTTTT/TTTTT |
| 2 | CAVp11 | S. marcescens | Feb-14 | Intraabdominal abscess | Yes | Tn4401b-8 | TTTTT/TTTTT |
| 2 | CAV1761 | S. marcescens | Mar-14 | Perirectal surveillance | Tn4401b-8 | TTTTT/TTTTT | |
| 3 | CAVp50 | Klebsiella pneumoniae | Jul-14 | Perirectal surveillance | NAa | Tn4401b-truncated (deletion 9299–10006) | —/TTGCA |
| 3 | CAVp57 | Klebsiella pneumoniae | Jul-14 | Perirectal surveillance | NA | Tn4401b-truncated | —/TTGCA |
| 3 | CAVp71 | Klebsiella pneumoniae | Aug-14 | Perirectal surveillance | NA | Tn4401b-truncated | —/TTGCA |
| 3 | CAVp104 | Klebsiella pneumoniae | Dec-14 | Perirectal surveillance | NA | Tn4401b-truncated | —/TTGCA |
| 6 | CAV1750 | Klebsiella pneumoniae | Dec-12 | Perirectal surveillance | NA | Tn4401b-1 | GTTCT/GTTCT |
| 6 | CAVp127 | Klebsiella pneumoniae | Feb-13 | Perirectal surveillance | NA | Tn4401b-1 | GTTCT/GTTCT |
| 6 | CAVp130 | Klebsiella pneumoniae | Mar-13 | Urine | Yes | Tn4401b-1 | GTTCT/GTTCT |
| 6 | CAVp139 | Klebsiella pneumoniae | Apr-13 | Perirectal surveillance | NA | Tn4401b-1 | GTTCT/GTTCT |
| 6 | CAVp151 | Klebsiella pneumoniae | Jul-13 | Perirectal surveillance | NA | Tn4401b-1 | GTTCT/GTTCT |
| 6 | CAVp152 | Klebsiella pneumoniae | Jul-13 | Perirectal surveillance | NA | Tn4401b-1 | GTTCT/GTTCT |
| 6 | CAVp177 | Klebsiella pneumoniae | Sep-13 | Perirectal surveillance | NA | Tn4401b-1 | GTTCT/GTTCT |
| 6 | CAVp180 | Klebsiella pneumoniae | Nov-13 | Perirectal surveillance | NA | Tn4401b-1 | GTTCT|TACCT/AGCAT|GTTCT |
| 6 | CAVp183 | Klebsiella pneumoniae | Nov-13 | Intraabdominal abscess | Yes | Tn4401b-1 | GTTCT/GTTCT |
| 6 | CAVp184 | Klebsiella pneumoniae | Nov-13 | Perirectal surveillance | NA | Tn4401b-1 | GTTCT/GTTCT |
| 6 | CAVp185 | Klebsiella pneumoniae | Nov-13 | Perirectal surveillance | NA | Tn4401b-1 | ATATT|GTTCT/ATATT|GTTCT |
| 6 | CAVp3 | Klebsiella pneumoniae | Jan-14 | Biliary drain | Yes | Tn4401b-1 | GTTCT/GTTCT |
| 8 | CAVp269 | Serratia marcescens | Jun-15 | Blood | Yes | Tn4401b-8 | TTTTT/TTTTT |
| 8 | CAVp270 | Serratia marcescens | Jun-15 | Perirectal surveillance | NA | Tn4401b-8 | TTTTT/TTTTT |
| 8 | CAVp361 | Escherichia coli | Dec-16 | Perirectal surveillance | NA | Tn4401b-8 | TTTTT/TTTTT |
| 8 | CAVp374 | Citrobacter freundii | Mar-17 | Perirectal surveillance | NA | Tn4401b-8 | TTTTT/TTTTT |
NA, not applicable.
Genetic variation and rearrangements within KpIIA.
All KpIIA isolates were closely related at the core chromosome level, with a maximum divergence of <180 SNVs. If blaKPC was acquired only once in this lineage, then any sequence variation within the 10-kb blaKPC transposon Tn4401 would be the result of mutational change, which is expected to be rare. Surprisingly, the Illumina sequence data revealed a great deal of sequence variation within Tn4401 (Fig. 1a). Two sites (positions 8015 and 9663 in the Tn4401b reference) showed variation at the single nucleotide level, and one isolate had a deletion at positions 7075 to 7153. Interestingly, several isolates showed mixtures at one or both of the variable sites, indicating two or more different versions of Tn4401 in the same isolate. This included mixtures at position 8015, which is located within the blaKPC gene and differentiates blaKPC-2 and blaKPC-3, indicating that there were isolates with both blaKPC alleles.
Similarly, if a single blaKPC plasmid was acquired and stably maintained within KpIIA, then we would expect to see a single flanking sequence context for Tn4401. On the contrary, there was significant diversity in Tn4401 flanking regions, with eight and seven different 5-bp sequences on the left and right sides of Tn4401, respectively, suggesting active transposition of Tn4401 within KpIIA and/or multiple plasmid acquisitions.
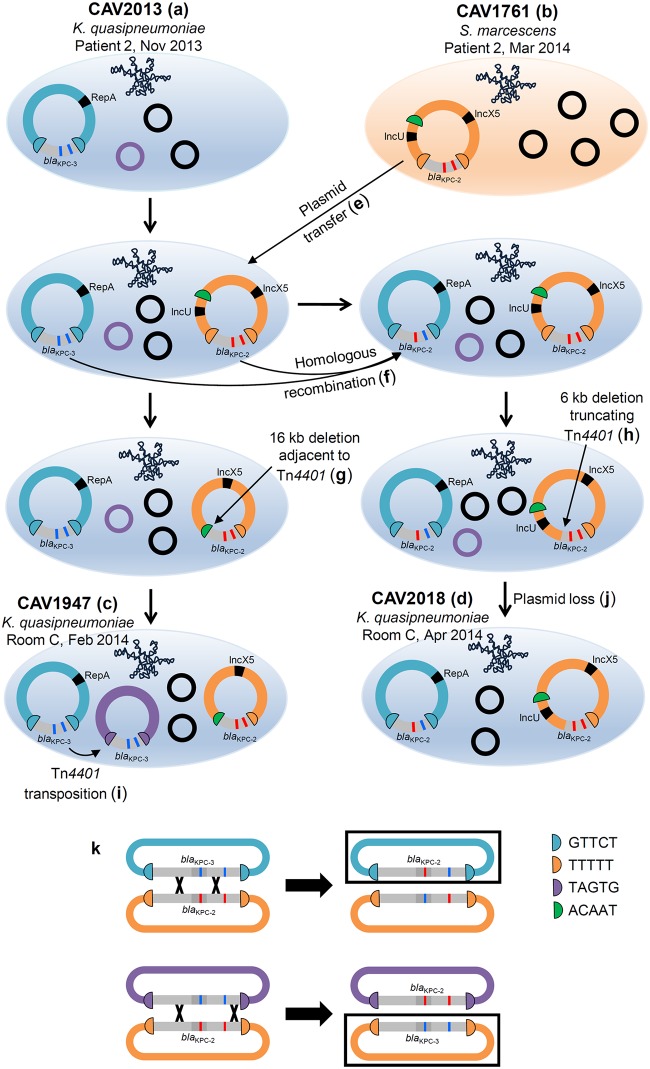
To better understand the origin of the genetic diversity within and surrounding Tn4401, we performed long-read PacBio sequencing on three of the KpIIA isolates (CAV2013 from patient 2, CAV1947 from room C, and CAV2018 from room C), as well as a S. marcescens isolate from patient 2 (CAV1761). The room C isolates were chosen because this room only became positive after the admission of patient 2 following sink trap exchange in the STBICU; hence, they are expected to be descended from the patient 2 KpIIA.
Both patient 2 isolates had a single blaKPC plasmid each (Fig. 3a and b). The KpIIA isolate had a 447,095-bp “RepA_CP011611” blaKPC-3 plasmid, and the S. marcescens isolate had a 69,158-bp IncU/IncX5 blaKPC-2 plasmid (18). Both plasmids contained Tn4401b; however, there were two SNV differences within the Tn4401b sequence, one at position 8015 (differentiating blaKPC-2 and blaKPC-3) and one at position 9663.
FIG 3.
Plasmid structures determined from long-read sequencing of four isolates and inferred intermediate blaKPC plasmid structures. (a to d) Sequenced isolates. (e to j) Inferred intermediate plasmid structures. Note that the ordering of deletion, homologous recombination, transposition, and plasmid loss events is arbitrarily represented, as the actual order of events is unknown. (k) Examples of crossover events leading to the generation of new combinations of SNVs within Tn4401 (top) or the complete swapping of Tn4401 variants between different plasmids (bottom). Black boxes indicate products of homologous recombination that were observed in long-read data (top) or Illumina data (bottom).
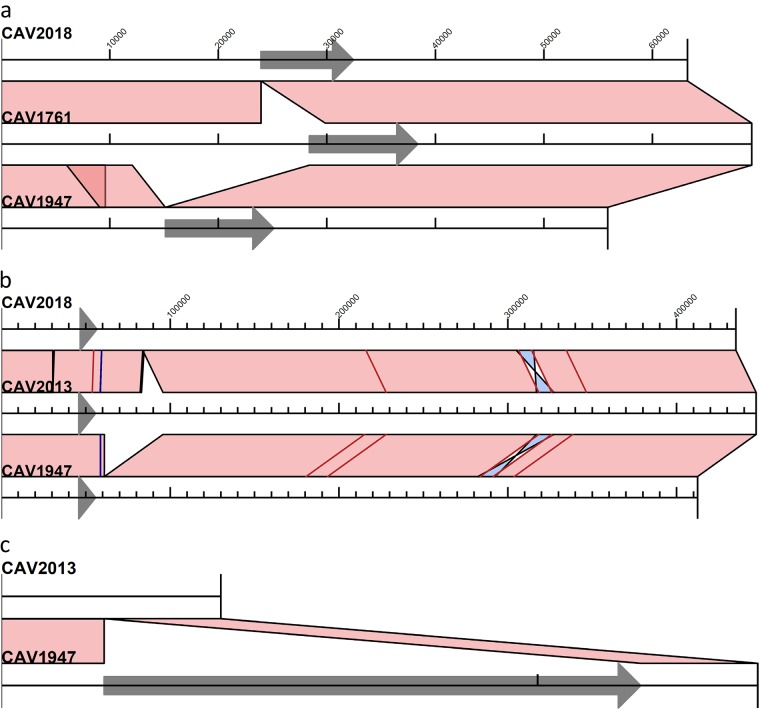
The KpIIA isolates from room C (CAV1947 and CAV2018) had three and two blaKPC plasmids, respectively (Fig. 3c and d). Both isolates harbored the IncU/IncX5 blaKPC plasmid from the patient 2 S. marcescens isolate, indicating likely blaKPC plasmid transfer from S. marcescens to K. quasipneumoniae (Fig. 3e). In CAV1947, the plasmid sequence was identical to that from the patient isolate, CAV1761, with the exception of two large indels (Fig. 4a). One of these was a 16,315-bp deletion immediately adjacent to Tn4401, presumably as a result of intramolecular transposition in cis, that converted the left flanking sequence from TTTTT to ACAAT and removed the IncU replicon sequence (Fig. 3g). In CAV2018, the plasmid sequence was identical to that in CAV1761, except for a single 5,923-bp deletion that truncated part of the Tn4401 sequence (Fig. 3h and 4a).
FIG 4.
Alignments of IncU/IncX5 (a), RepA_CP011611 (b), and nontypeable (c) blaKPC plasmid structures determined from long-read sequencing. Tn4401 is indicated by a gray arrow. Light pink shading indicates regions of identity, light blue shading shows inverted regions, and SNVs are indicated by red lines and short indels by blue lines.
Both isolates also harbored the ancestral RepA_CP011611 blaKPC plasmid from the patient 2 KpIIA isolate, with several SNVs and large indels (Fig. 4b). Interestingly, in CAV2018, one of the SNVs was located within Tn4401, such that the CAV2018 RepA_CP011611 plasmid contained blaKPC-2 rather than blaKPC-3. Given that there was plasmid transfer of the IncU/IncX5 blaKPC-2 plasmid from S. marcescens, we infer that the blaKPC-2-containing RepA_CP011611 plasmid most likely arose as a result of homologous recombination between these two different plasmids flanking the blaKPC region (Fig. 3f and k). The Illumina data also revealed similar patterns of homologous recombination in other isolates (notably CAV2983, CAV2984, CAV3444, CAVp64 and CAVp275, which all have the TTTTT IncU/IncX5 plasmid flanking sequences, but with the C8015T blaKPC-3 mutation and without the T9663C mutation), suggesting frequent exchange of Tn4401 variants between different blaKPC plasmids within the same host bacterium (Fig. 1 and 3k).
CAV1947 also harbored a third blaKPC plasmid, representing transposition of Tn4401 into a 4,095-bp nontypeable plasmid that was present in the CAV2013 ancestor from patient 2 (Fig. 3i and 4c).
K. quasipneumoniae has acquired blaKPC on multiple occasions.
The average unique plasmid Inc types per isolate was more than four according to PlasmidFinder (Data Set S1). Within KpIIB, there were four divergent strains separated by >20,000 SNVs, suggesting four separate acquisitions of blaKPC in this subspecies. Within KpIIA, there were two subclades separated by ∼180 SNVs. Given that Tn4401 variation and flanking sequences were different between the two subclades (apart from the GTTCT flanking sequence which is known to be present in many different blaKPC plasmids) (13) and that there was no epidemiological overlap, it is most likely that the subclades acquired blaKPC independently. Additionally, as described above, the second subclade likely acquired blaKPC on two occasions, with the second acquisition originating from S. marcescens. Therefore, overall, there were likely seven acquisitions of blaKPC by K. quasipneumoniae: three in KpIIA and four in KpIIB.
Interestingly, there was evidence that one of the acquisitions in KpIIB also originated from S. marcescens, indicating the compatibility of these two species in exchanging plasmids. This was in the patient 8 KpIIB lineage. Patient 8 was first colonized with blaKPC-S. marcescens carrying Tn4401b with a T9663C mutation and TTTTT/TTTTT flanking sequences. Four months later, blaKPC-KpIIB was identified with the same Tn4401 mutation and flanking sequences, suggesting plasmid transfer from S. marcescens to K. quasipneumoniae within this patient.
DISCUSSION
We describe the behavior of nosocomial blaKPC-positive K. quasipneumoniae strains within a single hospital setting, observing their propensity to take up multiple carbapenemase plasmids from other species and disseminate between patients and sink drains. Our study also suggests that rapid genetic rearrangement occurs in the mobile genetic elements carrying blaKPC in KpIIA.
There is increasing recognition that the hospital environment is an important reservoir in the transmission of carbapenemase-producing Enterobacteriaceae (CPE), but delineating transmission chains is often challenging (19, 20). Through our K. quasipneumoniae example, we provide compelling evidence for patient-to-drain and drain-to-patient transmission, as has been observed in other studies (7). We also provide evidence supporting the ability of K. quasipneumoniae to be maintained in the environment for a long period of time, with the first subclade of KpIIA detected in the environment on initial sampling, even though it had not been seen in a patient nor had that patient been in the room for more than 3 years. The costly closure of the STBICU and exchange of all the sink drain plumbing pipes had a limited effect on environmental contamination with CPE; instead, it appears to have provided an environment for immediate new seeding and establishment of previously unobserved carbapenem-resistant strains. There are potential other reservoirs to consider, but health care workers have not been identified as a source of CPE. We have a fairly robust screening program in place and have sequenced all patient isolates and included all K. quasipneumoniae in this series, making silent colonization less likely (21, 22). We were not sampling the toilets or hoppers during most of the study, and we have only sequenced a portion of environmental isolates which could provide an unidentified environmental source of K. quasipneumoniae (14). Understanding the dynamics and natural history of colonization of premise plumbing with CPE will be important in designing effective interventions to limit transmission (23).
Although there have only been a few reports of K. quasipneumoniae since its definition as a species in 2014, it appears that this organism is widespread (2, 5, 24, 25). As seen here, it is not readily distinguished from K. pneumoniae with current clinical microbiology techniques; thus, the true prevalence is unknown (2, 26). On the evolutionary time scale, modern medicine has provided a novel ecology with immunocompromised patients, widespread antimicrobial use, newly circulating antimicrobial resistance genes, and the design of the modern hospital providing new microbiologic niches for organisms to emerge (7, 27). We found several virulence factors in our collection, some of which have been identified in other K. pneumoniae or K. quasipneumoniae: capsule, fimbrial adhesion proteins, and a type VI secretion system (5, 16). As seen here, we provide evidence for K. quasipneumoniae to be sustained in both a human host and the environment, encountering several different species which may be relatively new in the evolutionary tree of Klebsiella sp. (1). As a consequence of these encounters, the transfer of mobile DNA occurs via traceable carbapenemase plasmids. We found evidence for seven independent acquisitions of blaKPC by K. quasipneumoniae, suggesting that this species is amenable to take up plasmids from other species of Enterobacteriaceae. Given the difficulties in accurately identifying K. quasipneumoniae, this species may therefore be more significant in the context of blaKPC dissemination than has previously been recognized.
Within K. quasipneumoniae, there was surprising variability in mobile elements carrying blaKPC, which was the result of several different processes observed among a limited number of highly related isolates (n = 23). We also found multiple acquired antimicrobial resistance genes, and every isolate had more than one plasmid incompatibility type (18). Specifically, there were multiple independent blaKPC plasmid acquisitions: homologous recombination between different blaKPC plasmids, transposition of Tn4401 into new plasmids, intramolecular transposition in cis of Tn4401, a deletion within Tn4401, and a deletion truncating Tn4401. This high degree of genetic mobility has been similarly observed in other small studies (28, 29) and highlights the difficulty in developing an accurate understanding of the transmission epidemiology of important drug resistance genes which can be rapidly mobilized by multiple independent genetic modalities.
Within KpIIA, there were multiple acquisitions of blaKPC within the same lineage, such that a blaKPC-positive KpIIA strain acquired a second unrelated blaKPC plasmid from S. marcescens. Consequently, there were then two different blaKPC plasmids, with different Tn4401 sequences and different blaKPC alleles, within the same host bacterium. This situation facilitated multiple rearrangements via homologous recombination between the different plasmids, resulting in the generation of new combinations of Tn4401 SNVs and host plasmids. Multiple acquisition of resistance plasmids followed by rearrangements between those plasmids is likely to be important in the generation of adaptive allelic combinations which contribute to the amplification of cross-class antimicrobial resistance within strains. High-risk clones with a propensity to take up antimicrobial resistance plasmids may represent important targets for intervention (30).
This study has several limitations. Most notably, it is a small retrospective series, preventing a full understanding of the role of the environment. Also, the order of genetic rearrangements is also not completely known, given the limited number of long-read sequenced isolates and inability to capture all isolates from the environment over time. We offer, however, that this is higher resolution than seen in many studies, and the analysis contributes to the greater understanding of rapid rearrangement and mechanisms at play regarding the mobility of genetic elements harboring genes of antibiotic resistance in Enterobacteriaceae.
In summary, we demonstrate the relevance of K. quasipneumoniae as a species fit for nosocomial transmission in the modern era that is capable of acquiring and maintaining relevant resistance elements.
MATERIALS AND METHODS
Setting.
Isolates were collected at the University of Virginia, a 619-bed tertiary care hospital, from August 2007 to May 2017. A robust K. pneumoniae carbapenemase-producing organism (KPCO) prevention program existed throughout the study period, as previously described (31), and included perirectal screening beginning in April 2009 in the medical intensive care unit (MICU) and surgical intensive care unit (STBICU) and weekly screening of all patients in the MICU and STBICU as well as units where any known KPCO-colonized patient was present (32). Screening was performed as previously described (32). Clinical Enterobacterales and Aeromonadaceae isolates, as identified by matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) or VITEK2 (bioMérieux, Durham, NC), with an elevated ertapenem or meropenem MIC by VITEK2 (bioMérieux, Durham, NC), immediately underwent CarbaR (Cepheid, Sunnyvale, CA) carbapenemase PCR testing. All species identification was performed using a combination of VITEK2 and Vitek-MS (bioMérieux, Durham, NC). Clinical data were gathered by retrospective electronic medical record review under University of Virginia Health Sciences institutional review board (IRB) number 13558 with waiver of consent.
In September 2013, sink trap sampling for KPCO began using previously described techniques (14) with a swab for drain collection and P-trap water. Following identification of KPCO in the hospital environment, the STBICU was closed to patient care in December 2013. Over the following 9 weeks, all sink drain pipes were removed and replaced with sink traps that eliminated overflows in the sink bowl. Patients were readmitted to the surgical intensive care unit in February 2014. Bleach, hydrogen peroxide, and ozone-impregnated water (2 ppm) were applied weekly from February to May 2014 in the STBICU (following drain exchange and sink bowl overflow closure and removal) and from March to May 2014 in the MICU (without drain exchange or sink bowl overflow removal).
Whole-genome sequencing and bioinformatics analysis.
Illumina sequencing was performed as described previously (33). PacBio long-read sequencing and assembly were performed as previously described (13).
Broad-level species classification was performed using Kraken (34). To identify K. quasipneumoniae isolates, we queried all isolates initially classified as K. pneumoniae against reference sequences representing each of the four clades described by Holt et al. (1). We arbitrarily selected a single reference sequence for each clade; these were ERR025521 (KpI), ERR025986 (KpIIA), ERR025528 (KpIIB), and ERR025573 (KpIII). We used mash v1.1.1 (35) with parameters “-r -m 5” to compare Illumina data for each of our isolates to these reference sequences. Each isolate was then assigned to one of the four K. pneumoniae clades according to the reference with the lowest distance value. All isolates assigned to KpIIA or KpIIB were included in the analysis. In addition, we also included any other KPCO isolates from patients carrying K. quasipneumoniae.
To identify chromosomal single nucleotide variants (SNVs), Illumina reads for each K. quasipneumoniae isolate were mapped to the CAV2013 chromosome sequence (derived from long-read sequencing), with mapping and variant calling performed as described previously (36). A phylogeny was generated using IQ-TREE v1.3.13 (37) from an alignment of variable sites where at least 70% of samples had a high-quality reference/variant call (i.e., we excluded sites where >30% of samples had an “N” call). This was run with parameters “-blmin 0.000000001 -nt 4 -m GTR,” with -fconst used to specify the number of invariant sites.
To identify Tn4401 variation and flanking sequences from Illumina data, we used TETyper with published parameters (38).
The Illumina paired-end short reads were de novo assembled using SPAdes assembler v 3.10.1 (35). Assembly statistics were evaluated using QUAST v4.0. (36) Plasmid Inc typing was performed using PlasmidFinder v2.0.1 against the Feb 2018 version of the Enterobacteriaceae database (16), with minimum identity of 80% and minimum coverage of 50%. Acquired antimicrobial resistance genes were screened from the assemblies using NCBI’s AMRFinder tool v1.0, which relies on a curated AMR protein database and a collection of hidden Markov models, with 90% minimum identity to translated amino acid residues and 50% minimum coverage of reference protein sequence. (37) Identification of bacterial virulence genes was performed using ABRicate v0.8.11 (https://github.com/tseemann/abricate), against the Virulence Factors Database (accessed on Feb 2019), with 80% minimum identity and 50% minimum coverage.
Data availability.
Illumina paired-end sequence data can be accessed from NCBI BioProject identifier (ID) PRJNA411762. The accession numbers for completed closed genomes from hybrid assembly of PacBio and Illumina are GCA_003146655.1 (CAV2013), GCA_003146685.1 (CAV1947), GCA_003146635.1 (CAV2018), and GCA_003146705.1 (CAV1761). All other relevant data for the manuscript are within Data Set S1 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by a contract from the Centers for Disease Control and Prevention (CDC) broad agency announcement (BAA 200-2017-96194). A.S.W. and D.C. are affiliated with the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Healthcare Associated Infections and Antimicrobial Resistance at University of Oxford in partnership with Public Health England (PHE) (grant HPRU-2012-10041) and are supported by the Oxford NIHR Biomedical Research Centre.
The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, the Department of Health, or Public Health England.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02513-18.
REFERENCES
- 1.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long SW, Linson SE, Ojeda Saavedra M, Cantu C, Davis JJ, Brettin T, Olsen RJ. 2017. Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2:e00290-17. doi: 10.1128/mSphereDirect.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brisse S, Passet V, Grimont PA. 2014. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol 64:3146–3152. doi: 10.1099/ijs.0.062737-0. [DOI] [PubMed] [Google Scholar]
- 4.Becker L, Fuchs S, Pfeifer Y, Semmler T, Eckmanns T, Korr G, Sissolak D, Friedrichs M, Zill E, Tung ML, Dohle C, Kaase M, Gatermann S, Russmann H, Steglich M, Haller S, Werner G. 2018. Whole genome sequence analysis of CTX-M-15 producing Klebsiella isolates allowed dissecting a polyclonal outbreak scenario. Front Microbiol 9:322. doi: 10.3389/fmicb.2018.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolas MF, Ramos PIP, Marques de Carvalho F, Camargo DRA, de Fatima Morais Alves C, Loss de Morais G, Almeida LGP, Souza RC, Ciapina LP, Vicente ACP, Coimbra RS, Ribeiro de Vasconcelos AT. 2018. Comparative genomic analysis of a clinical isolate of Klebsiella quasipneumoniae subsp. similipneumoniae, a KPC-2 and OKP-B-6 beta-lactamases producer harboring two drug-resistance plasmids from southeast Brazil. Front Microbiol 9:220. doi: 10.3389/fmicb.2018.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hocquet D, Muller A, Bertrand X. 2016. What happens in hospitals does not stay in hospitals: antibiotic-resistant bacteria in hospital wastewater systems. J Hosp Infect 93:395–402. doi: 10.1016/j.jhin.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Weingarten RA, Johnson RC, Conlan S, Ramsburg AM, Dekker JP, Lau AF, Khil P, Odom RT, Deming C, Park M, Thomas PJ, NISC Comparative Sequencing Program, Henderson DK, Palmore TN, Segre JA, Frank KM. 2018. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9:e02011-17. doi: 10.1128/mBio.02011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki H, Yano H, Brown CJ, Top EM. 2010. Predicting plasmid promiscuity based on genomic signature. J Bacteriol 192:6045–6055. doi: 10.1128/JB.00277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price VJ, Huo W, Sharifi A, Palmer KL. 2016. CRISPR-Cas and restriction-modification act additively against conjugative antibiotic resistance plasmid transfer in Enterococcus faecalis. mSphere 1:e00064-16. doi: 10.1128/mSphere.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loftie-Eaton W, Yano H, Burleigh S, Simmons RS, Hughes JM, Rogers LM, Hunter SS, Settles ML, Forney LJ, Ponciano JM, Top EM. 2016. Evolutionary paths that expand plasmid host-range: implications for spread of antibiotic resistance. Mol Biol Evol 33:885–897. doi: 10.1093/molbev/msv339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loftie-Eaton W, Bashford K, Quinn H, Dong K, Millstein J, Hunter S, Thomason MK, Merrikh H, Ponciano JM, Top EM. 2017. Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat Ecol Evol 1:1354–1363. doi: 10.1038/s41559-017-0243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardiman CA, Weingarten RA, Conlan S, Khil P, Dekker JP, Mathers AJ, Sheppard AE, Segre JA, Frank KM. 2016. Horizontal transfer of carbapenemase-encoding plasmids and comparison with hospital epidemiology data. Antimicrob Agents Chemother 60:4910–4919. doi: 10.1128/AAC.00014-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheppard AE, Stoesser N, Wilson DJ, Sebra R, Kasarskis A, Anson LW, Giess A, Pankhurst LJ, Vaughan A, Grim CJ, Cox HL, Yeh AJ, Modernising Medical Microbiology (MMM) Informatics Group, Sifri CD, Walker AS, Peto TE, Crook DW, Mathers AJ. 2016. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 60:3767–3778. doi: 10.1128/AAC.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathers AJ, Vegesana K, German Mesner I, Barry KE, Pannone A, Baumann J, Crook DW, Stoesser N, Kotay S, Carroll J, Sifri CD. 2018. Intensive care unit wastewater interventions to prevent transmission of multispecies Klebsiella pneumoniae carbapenemase-producing organisms. Clin Infect Dis 67:171–178. doi: 10.1093/cid/ciy052. [DOI] [PubMed] [Google Scholar]
- 15.Mathers AJ, Stoesser N, Chai W, Carroll J, Barry K, Cherunvanky A, Sebra R, Kasarskis A, Peto TE, Walker AS, Sifri CD, Crook DW, Sheppard AE. 2017. Chromosomal integration of the Klebsiella pneumoniae carbapenemase gene, blaKPC, in Klebsiella species is elusive but not rare. Antimicrob Agents Chemother 61:e01823-16. doi: 10.1128/AAC.01823-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin MH, Hsu TL, Lin SY, Pan YJ, Jan JT, Wang JT, Khoo KH, Wu SH. 2009. Phosphoproteomics of Klebsiella pneumoniae NTUH-K2044 reveals a tight link between tyrosine phosphorylation and virulence. Mol Cell Proteomics 8:2613–2623. doi: 10.1074/mcp.M900276-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haeggman S, Löfdahl S, Paauw A, Verhoef J, Brisse S. 2004. Diversity and evolution of the class A chromosomal beta-lactamase gene in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:2400–2408. doi: 10.1128/AAC.48.7.2400-2408.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotsanas D, Wijesooriya WR, Korman TM, Gillespie EE, Wright L, Snook K, Williams N, Bell JM, Li HY, Stuart RL. 2013. “Down the drain”: carbapenem-resistant bacteria in intensive care unit patients and handwashing sinks. Med J Aust 198:267–269. doi: 10.5694/mja12.11757. [DOI] [PubMed] [Google Scholar]
- 20.De Geyter D, Blommaert L, Verbraeken N, Sevenois M, Huyghens L, Martini H, Covens L, Piérard D, Wybo I. 2017. The sink as a potential source of transmission of carbapenemase-producing Enterobacteriaceae in the intensive care unit. Antimicrob Resist Infect Control 6:24. doi: 10.1186/s13756-017-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Decker BK, Lau AF, Dekker JP, Spalding CD, Sinaii N, Conlan S, Henderson DK, Segre JA, Frank KM, Palmore TN. 2018. Healthcare personnel intestinal colonization with multidrug-resistant organisms. Clin Microbiol Infect 24:82.e1–82.e4. doi: 10.1016/j.cmi.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulrich N, Gastmeier P, Vonberg RP. 2018. Effectiveness of healthcare worker screening in hospital outbreaks with gram-negative pathogens: a systematic review. Antimicrob Resist Infect Control 7:36. doi: 10.1186/s13756-018-0330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopman J, Tostmann A, Wertheim H, Bos M, Kolwijck E, Akkermans R, Sturm P, Voss A, Pickkers P, Vd Hoeven H. 2017. Reduced rate of intensive care unit acquired gram-negative bacilli after removal of sinks and introduction of 'water-free' patient care. Antimicrob Resist Infect Control 6:59. doi: 10.1186/s13756-017-0213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gan HM, Rajasekaram G, Eng WWH, Kaniappan P, Dhanoa A. 2017. Whole-genome sequences of two carbapenem-resistant Klebsiella quasipneumoniae strains isolated from a tertiary hospital in Johor, Malaysia. Genome Announc 5:e00768-17. doi: 10.1128/genomeA.00768-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shankar C, Nabarro LEB, Muthuirulandi Sethuvel DP, Raj A, Devanga Ragupathi NK, Doss GP, Veeraraghavan B. 2017. Draft genome of a hypervirulent Klebsiella quasipneumoniae subsp. similipneumoniae with novel sequence type ST2320 isolated from a chronic liver disease patient. J Glob Antimicrob Resist 9:30–31. doi: 10.1016/j.jgar.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Martínez-Romero E, Rodríguez-Medina N, Beltrán-Rojel M, Silva-Sánchez J, Barrios-Camacho H, Pérez-Rueda E, Garza-Ramos U. 2018. Genome misclassification of Klebsiella variicola and Klebsiella quasipneumoniae isolated from plants, animals and humans. Salud Publica Mex 60:56–62. doi: 10.21149/8149. [DOI] [PubMed] [Google Scholar]
- 27.Gomi R, Matsuda T, Yamamoto M, Chou PH, Tanaka M, Ichiyama S, Yoneda M, Matsumura Y. 2018. Characteristics of carbapenemase-producing Enterobacteriaceae in wastewater revealed by genomic analysis. Antimicrob Agents Chemother 60:02501-17. doi: 10.1128/AAC.02501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wailan AM, Sartor AL, Zowawi HM, Perry JD, Paterson DL, Sidjabat HE. 2015. Genetic contexts of blaNDM-1 in patients carrying multiple NDM-producing strains. Antimicrob Agents Chemother 59:7405–7410. doi: 10.1128/AAC.01319-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snesrud E, Ong AC, Corey B, Kwak YI, Clifford R, Gleeson T, Wood S, Whitman TJ, Lesho EP, Hinkle M, McGann P. 2017. Analysis of serial isolates of mcr-1-positive Escherichia coli reveals a highly active ISApl1 transposon. Antimicrob Agents Chemother 61:e00056-17. doi: 10.1128/AAC.00056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabowski ME, Kang H, Wells KM, Sifri CD, Mathers AJ, Lobo JM. 2017. Provider role in transmission of carbapenem-resistant Enterobacteriaceae. Infect Control Hosp Epidemiol 38:1329–1334. doi: 10.1017/ice.2017.216. [DOI] [PubMed] [Google Scholar]
- 32.Mathers AJ, Poulter M, Dirks D, Carroll J, Sifri CD, Hazen KC. 2014. Clinical microbiology costs for methods of active surveillance for Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Infect Control Hosp Epidemiol 35:350–355. doi: 10.1086/675603. [DOI] [PubMed] [Google Scholar]
- 33.Mathers AJ, Stoesser N, Sheppard AE, Pankhurst L, Giess A, Yeh AJ, Didelot X, Turner SD, Sebra R, Kasarskis A, Peto T, Crook D, Sifri CD. 2015. Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae at a single institution: insights into endemicity from whole-genome sequencing. Antimicrob Agents Chemother 59:1656–1663. doi: 10.1128/AAC.04292-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoesser N, Sheppard AE, Peirano G, Anson LW, Pankhurst L, Sebra R, Phan HTT, Kasarskis A, Mathers AJ, Peto TEA, Bradford P, Motyl MR, Walker AS, Crook DW, Pitout JD. 2017. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci Rep 7:5917. doi: 10.1038/s41598-017-06256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheppard AE, Stoesser N, German-Mesner I, Vegesana K, Walker AS, Crook DW, Mathers AJ. 2018. TETyper: a bioinformatic pipeline for classifying variation and genetic contexts of transposable elements from short-read whole-genome sequencing data. Microb Genom 4. doi: 10.1099/mgen.0.000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Illumina paired-end sequence data can be accessed from NCBI BioProject identifier (ID) PRJNA411762. The accession numbers for completed closed genomes from hybrid assembly of PacBio and Illumina are GCA_003146655.1 (CAV2013), GCA_003146685.1 (CAV1947), GCA_003146635.1 (CAV2018), and GCA_003146705.1 (CAV1761). All other relevant data for the manuscript are within Data Set S1 in the supplemental material.