Chlorhexidine gluconate (CHG) is a topical antiseptic widely used in health care settings. In Staphylococcus spp., the pump QacA effluxes CHG, while the closely related QacB cannot due to a single amino acid substitution.
KEYWORDS: CHG, Staphylococcus, Staphylococcus epidermidis, chlorhexidine, chlorhexidine gluconate, coagulase-negative staphylococci, qacA, qacB, qacA/B
ABSTRACT
Chlorhexidine gluconate (CHG) is a topical antiseptic widely used in health care settings. In Staphylococcus spp., the pump QacA effluxes CHG, while the closely related QacB cannot due to a single amino acid substitution. We characterized 1,050 cutaneous Staphylococcus isolates obtained from 173 pediatric oncology patients enrolled in a multicenter CHG bathing trial. CHG susceptibility testing revealed that 63 (6%) of these isolates had elevated CHG MICs (≥4 μg/ml). Screening of all 1,050 isolates for the qacA/B gene (the same qac gene with A or B allele) by restriction fragment length polymorphism (RFLP) yielded 56 isolates with a novel qacA/B RFLP pattern, qacA/B273. The CHG MIC was significantly higher for qacA/B273-positive isolates (MIC50, 4 μg/ml; MIC range, 0.5 to 4 μg/ml) than for other qac groups: qacA-positive isolates (n = 559; MIC50, 1 μg/ml; MIC range, 0.5 to 4 μg/ml), qacB-positive isolates (n = 17; MIC50, 1 μg/ml; MIC range, 0.25 to 2 μg/ml), and qacA/B-negative isolates (n = 418, MIC50, 1 μg/ml; MIC range, 0.125 to 2 μg/ml) (P = 0.001). A high proportion of the qacA/B273-positive isolates also displayed methicillin resistance (96.4%) compared to the other qac groups (24.9 to 61.7%) (P = 0.001). Whole-genome sequencing revealed that qacA/B273-positive isolates encoded a variant of QacA with 2 amino acid substitutions. This new allele, named qacA4, was carried on the novel plasmid pAQZ1. The qacA4-carrying isolates belonged to the highly resistant Staphylococcus epidermidis sequence type 2 clone. By searching available sequence data sets, we identified 39 additional qacA4-carrying S. epidermidis strains from 5 countries. Curing an isolate of qacA4 resulted in a 4-fold decrease in the CHG MIC, confirming the role of qacA4 in the elevated CHG MIC. Our results highlight the importance of further studying qacA4 and its functional role in clinical staphylococci.
INTRODUCTION
Staphylococcus epidermidis is a typical resident of the skin flora and an important cause of device-associated infections, especially central line-associated bloodstream infections (1). The success of S. epidermidis as an opportunistic pathogen derives from its ability to bind indwelling devices through the formation of a biofilm (2–4) and the high rate of antimicrobial resistance within the population (5, 6).
With a favorable safety profile and broad-spectrum and residual activity (7), chlorhexidine gluconate (CHG) is a promising option for skin cleansing and antisepsis for the prevention of device-associated infections. Bathing with CHG has been demonstrated to reduce the rates of central line-associated bloodstream infections (8, 9), acquisition of multidrug-resistant organisms (10), and blood culture contamination, which is frequently caused by S. epidermidis (11, 12). Furthermore, topical applications of CHG have been demonstrated to significantly reduce the cutaneous microbial burden (13, 14). However, increasing usage of CHG may select for organisms with decreased susceptibility to CHG and increased resistance to commonly prescribed antimicrobials (13, 15–17).
In Staphylococcus spp., qacA encodes a 514-amino-acid, 14-transmembrane-segment pump with the capacity to efflux CHG (18–20). The pump encoded by qacB (a qacA allelic variant) differs from that encoded by qacA by only 7 to 9 nucleotides but does not have the ability to efflux CHG (18, 21). A single nucleotide variant (SNV) (968C>A) resulting in a substitution, Ala323Asp, in transmembrane segment 10 accounts for the different substrate specificities of QacA and QacB (18). Currently, three alleles of qacA have been described; however, no functional differences between the pumps encoded by these three alleles have been reported (21).
Beyond its capacity to efflux CHG, QacA is responsible for the efflux of a broad range of mono- and divalent cations, including dyes and quaternary ammonium compounds (20). In S. epidermidis, qacA is most frequently carried by the plasmid pSK105, which also carries an aacA/aphD bifunctional aminoglycoside resistance gene (22). Other plasmids carrying qacA may contain the trimethoprim resistance gene dfrA, the blaZ β-lactamase, or genes encoding heavy metal efflux pumps (22).
In addition to QacA, the 107-amino-acid, 4-transmembrane-segment efflux pump encoded by smr, also known as qacC, has been implicated in the efflux of CHG (23–25). While it is unrelated to QacA and QacB, Smr demonstrates the capacity to efflux a similar, yet narrower range of monovalent cations (24, 25).
In our study, cutaneous Staphylococcus isolates were obtained from pediatric oncology patients enrolled in a multicenter randomized controlled CHG bathing trial. We identified a subpopulation of isolates with an elevated CHG MIC, which we defined as an MIC of ≥4 μg/ml. To investigate the genetic basis of the elevated CHG MIC, we screened the isolates for qacA/B (the same qac gene with A or B allele) via PCR and restriction fragment length polymorphism (RFLP). From this screening, we identified a previously undescribed RFLP pattern, termed qacA/B273, in a subset of isolates. We then determined whether the qacA/B273 RFLP pattern was associated with a significantly higher CHG MIC compared to that for the qacA-positive, qacB-positive, and qacA/B-negative isolates. We also describe the sequence of the novel qacA allele, referred to as qacA4, producing the novel qacA/B273 RFLP pattern and characterized the isolates carrying qacA4. Furthermore, through curing experiments, we investigated the role of qacA4 in causing elevated CHG MICs in S. epidermidis.
RESULTS
Overview of study population.
In total, 1,050 cutaneous Staphylococcus isolates were obtained from 173 patients. The study isolates primarily consisted of coagulase-negative Staphylococcus species, with S. epidermidis being the most frequently recovered species (53.1%), while S. aureus accounted for just 2.9% of the study population (Table 1). In addition to S. epidermidis, 17 other coagulase-negative Staphylococcus species were identified in the study population. Of note, the species of four coagulase-negative Staphylococcus isolates could not be determined by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS).
TABLE 1.
Overview of the cutaneous Staphylococcus isolates included in this studya
| Species | Total no. of isolates | % of isolates |
|---|---|---|
| S. aureus | 30 | 2.9 |
| Coagulase-negative staphylococci | 1,020 | 97.1 |
| S. epidermidis | 558 | 53.1 |
| S. hominis | 267 | 25.4 |
| S. capitis | 62 | 5.9 |
| S. warneri | 47 | 4.5 |
| S. haemolyticus | 22 | 2.1 |
| S. pasteuri | 16 | 1.5 |
| S. saprophyticus | 9 | 0.9 |
| S. lugdunensis | 8 | 0.8 |
| S. cohnii | 4 | 0.4 |
| S. caprae | 4 | 0.4 |
| S. pettenkoferi | 4 | 0.4 |
| S. condiment | 3 | 0.3 |
| S. schleiferi | 3 | 0.3 |
| S. simulans | 3 | 0.3 |
| S. auricularis | 2 | 0.2 |
| S. equorum | 2 | 0.2 |
| S. sciuri | 1 | 0.1 |
| S. xylosus | 1 | 0.1 |
| Staphylococcus spp. | 4 | 0.4 |
Data are for 1,050 cutaneous Staphylococcus isolates identified by MALDI-TOF MS.
A subset of Staphylococcus isolates has an elevated CHG MIC.
Measuring the CHG MICs across all 1,050 isolates yielded 63 isolates with elevated CHG MICs, defined as an MIC of ≥4 μg/ml (Fig. 1a). All of these isolates were identified as S. epidermidis.
FIG 1.

CHG MIC distribution of the 1,050 cutaneous Staphylococcus isolates included in our study, grouped by species (a), qacA/B PCR and RFLP patterns (b), and smr PCR results (c). The dashed line indicates the concentration that we defined as an elevated CHG MIC (≥4 μg/ml). MALDI ID, species identified by MALDI-TOF MS.
Identification of a novel qacA/B RFLP pattern.
Isolates were screened for the qacA/B genes to explore the genetic basis of the elevated CHG MICs. PCR amplification of the qacA/B genes resulted in an 864-bp product. Digestion of the qacA/B PCR product with AluI resulted in the presence of a characteristic 198-bp fragment for qacA-positive isolates and a characteristic 165-bp fragment for qacB-positive isolates. A third subpopulation of isolates was distinguished by the appearance of a 273-bp fragment (Fig. 2), and this subpopulation is hereafter referred to as qacA/B273-positive isolates.
FIG 2.
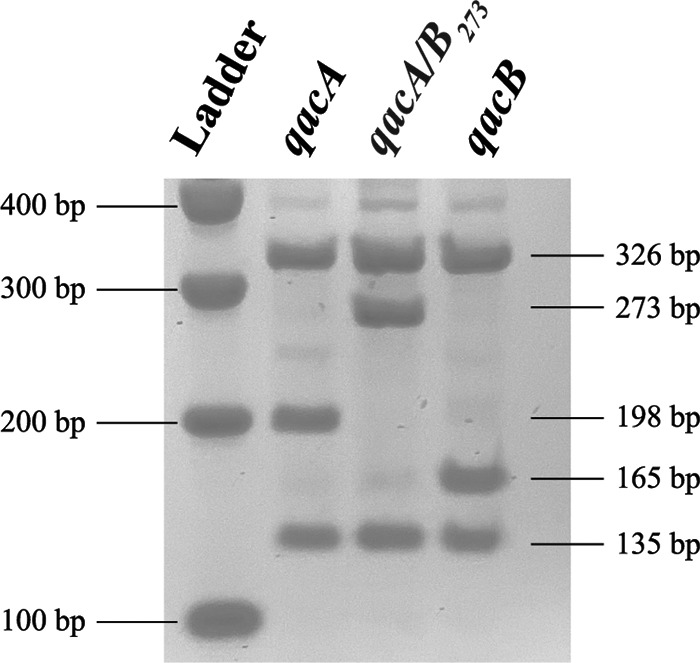
RFLP patterns observed from the AluI restriction digest of the qacA/B PCR amplicon. Isolates were classified as qacA positive, qacA/B273 positive, or qacB positive based on the presence of a 198-bp, 273-bp, or 165-bp fragment, respectively (21). Ladder, 100-bp markers (Promega).
Of the 1,050 isolates, 632 contained qacA/B, as identified by PCR. Based on the results of the RFLP analysis, 559 were classified as qacA positive, 17 as qacB positive, and 56 as qacA/B273 positive (Table 2). The qacA/B genes were detected in 8 different coagulase-negative Staphylococcus species. When screened for the carriage of smr, 279 of the 1,050 isolates were classified as smr positive (Table 2). In total, 12 unique coagulase-negative Staphylococcus species carried smr. Notably, the qacA/B genes and smr were not detected in any of the S. aureus isolates.
TABLE 2.
Comparison of CHG MIC distributions of the qacA-positive, qacB-positive, qacA/B273-positive, and qacA/B-negative isolates and CHG MIC distributions of smr-positive and smr-negative isolates
| Genotype | Total no. of isolates | CHG MIC (μg/ml) |
||
|---|---|---|---|---|
| 50% | 90% | Range | ||
| qacA positive | 559 | 1 | 2 | 0.5–4 |
| qacB positive | 17 | 1 | 1 | 0.25–2 |
| qacA/B273 positive | 56 | 4 | 4 | 0.5–4 |
| qacA/B negative | 418 | 1 | 1 | 0.125–2 |
| smr positive | 279 | 1 | 2 | 0.25–4 |
| smr negative | 771 | 1 | 1 | 0.125–4 |
The qacA/B273 RFLP pattern is associated with an elevated CHG MIC.
Next, the relationship between elevated CHG MICs and detection of the qacA, qacB, and smr genes was examined. qacA/B was detected in each of the 63 isolates with an elevated CHG MIC: 54 were classified as qacA/B273 positive and 9 were classified as qacA positive (Fig. 1b). None of the isolates with an elevated CHG were classified as qacB positive. Furthermore, 51 of the 63 isolates with an elevated CHG MIC were categorized as smr positive, and the remaining 12 were classified as smr negative (Fig. 1c).
To further investigate if the qacA/B273 RFLP pattern was associated with an elevated CHG MIC, differences in the CHG MIC distributions of the qacA/B-containing isolates were assessed. The CHG MIC was significantly higher for the qacA/B273-positive isolates than for the qacA-positive, qacB-positive, and qacA/B-negative isolates (P = 0.001); the results did not change when restricting the analyses to one randomly chosen isolate per patient per qacA/B group (Table 2). In addition, the CHG MIC distributions of the smr-positive and smr-negative isolates were compared. The CHG MIC was significantly higher for the smr-positive isolates than for the smr-negative isolates (P = 0.02); however, this comparison was no longer significant when the analyses were restricted to one randomly chosen isolate per patient per smr group, as one individual accounted for 20% of the smr-positive isolates with elevated MICs (P = 0.11) (Table 2).
Additionally, the CHG MIC distributions associated with the qacA/B and smr resistance gene combinations among all isolates were assessed to determine if a particular resistance gene combination was associated with elevated CHG MICs. This comparison revealed that qacA/B273 rather than a particular resistance gene combination was associated with elevated CHG MICs (P = 0.001); the results did not change when restricting the analyses to one randomly chosen isolate per patient per gene combination (Table 3).
TABLE 3.
Comparison of the CHG MIC distributions associated with the eight CHG resistance gene combinations detected in our isolates
| Genotype | Total no. of isolates | CHG MIC (μg/ml) |
||
|---|---|---|---|---|
| 50% | 90% | Range | ||
| qacA positive and smr positive | 101 | 1 | 2 | 0.5–4 |
| qacA positive and smr negative | 458 | 1 | 2 | 0.5–4 |
| qacB positive and smr positive | 5 | 1 | 1.6 | 0.5–2 |
| qacB positive and smr negative | 12 | 0.75 | 1 | 0.25–1 |
| qacA/B273 positive and smr positive | 50 | 4 | 4 | 4–4 |
| qacA/B273 positive and smr negative | 6 | 4 | 4 | 0.5–4 |
| qacA/B negative, smr positive | 123 | 0.5 | 1 | 0.25–1 |
| qacA/B negative and smr negative | 295 | 1 | 1 | 0.125–2 |
The qacA/B273-positive isolates exhibited higher rates of resistance to methicillin (96.4%) and other commonly prescribed antimicrobials, including erythromycin (ERY; 92.9%), ciprofloxacin (CIP; 96.4%), gentamicin (GEN; 89.3%), and sulfamethoxazole-trimethoprim (SXT; 98.2%), than the qacA-positive, qacB-positive, and qacA/B-negative isolates (P < 0.001 for all comparisons); the results did not change when restricting the analyses to one randomly chosen isolate per patient per qacA/B group (Table 4). All qacA/B genotypes exhibited rates of resistance of <1% to linezolid (LZD), rifampin (RIF), and vancomycin (VAN).
TABLE 4.
Comparison of proportion of qacA-positive, qacB-positive, qacA/B273-positive, and qacA/B-negative isolates resistant to commonly prescribed antimicrobials
| Antimicrobiala | % of isolates: |
|||
|---|---|---|---|---|
| qacA positive | qacB positive | qacA/B273 positive | qacA/B negative | |
| FOXb | 61.7 | 52.9 | 96.4 | 22.7 |
| ERYb | 76.0 | 88.2 | 92.9 | 30.1 |
| CIPb | 25.0 | 5.9 | 96.4 | 8.4 |
| GENb | 15.0 | 0 | 89.3 | 2.6 |
| SXTb | 60.1 | 35.3 | 98.2 | 22.7 |
| LZD | 0 | 0 | 0 | 0 |
| RIF | 0.9 | 0 | 0 | 0 |
| VAN | 0 | 0 | 0 | 0 |
CIP, ciprofloxacin; SXT, sulfamethoxazole-trimethoprim; FOX, cefoxitin; ERY, erythromycin; GEN, gentamicin; LZD, linezolid; RIF, rifampin;VAN, vancomycin.
P < 0.001 for all comparisons. Results were generated using all isolates; the results did not change when restricting the analyses to one randomly chosen isolate per patient per qacA/B group.
Whole-genome sequencing of qacA/B-positive isolates yields novel qacA alleles.
To further investigate the qacA/B genes in the qacA/B273-positive isolates, the genomes of 9 qacA/B273-positive S. epidermidis isolates were compared to the genomes of 10 qacA-positive and 4 qacB-positive S. epidermidis isolates (see Table S1 in the supplemental material). All 9 of the qacA/B273-positive isolates had elevated CHG MICs, while none of the 10 qacA-positive and 4 qacB-positive isolates had elevated CHG MICs.
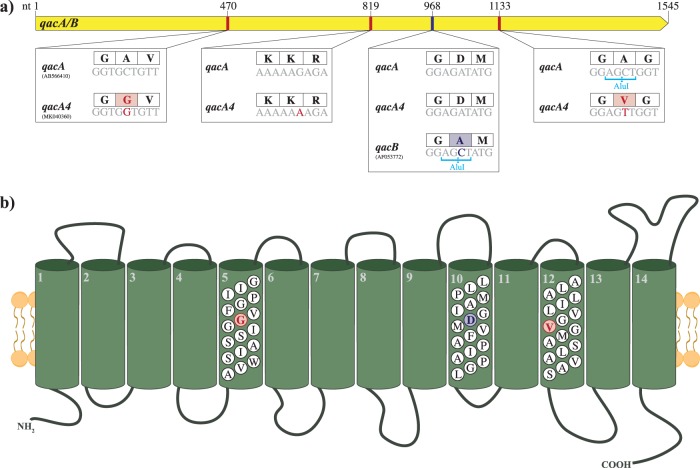
The sequences of the qacA/B genes were highly conserved in the 9 qacA/B273-positive isolates with elevated CHG MICs. As the qacA/B genes of the qacA/B273-positive isolates contained the distinguishing qacA nucleotide 968A, the gene was classified as a novel allele of qacA. As shown in Fig. 3a, this allele contained three SNVs (470C>G, 819G>A, and 1133C>T) compared to the sequence of the qacA gene (GenBank accession number AB566410) of a reference S. epidermidis sequence type 2 (ST2) strain (GenBank accession number GCA_900086615.1) (26) and is henceforth referred to as qacA4 (GenBank accession number MK040360). The SNV at position 1133 in qacA4 resulted in the loss of an AluI digestion site, explaining the novel RFLP pattern observed in Fig. 2. Two of the SNVs resulted in amino acid substitutions Ala157Gly and Ala378Val in transmembrane segments 5 and 12, respectively (Fig. 3b).
FIG 3.
(a) Comparison of the sequences of qacA4 (GenBank accession number MK040360), a reference qacA sequence (GenBank accession number AB566410), and a reference qacB sequence (GenBank accession number AF053772). The associated AluI restriction sites are shown below the nucleotide (nt) sequences, and the corresponding amino acid sequences are displayed in the boxes above the nucleotide sequences. (b) Structure of the predicted efflux pump encoded by qacA4 (adapted from reference 20 with permission). The residues which distinguish QacA4 from the reference QacA sequence are highlighted in red. Those which distinguish QacA from QacB are displayed in blue.
Compared to the three previously characterized alleles of qacA (qacA1 [GenBank accession number GU565967], qacA2 [21], and qacA3 [GenBank accession number MK040360]) and the qacA alleles of the 10 qacA-positive S. epidermidis isolates, qacA4 differed from these sequences by at least three SNVs, including all three that distinguished qacA4 from the reference qacA sequence (Fig. S1). Notably, from the 10 qacA-positive isolates that we sequenced, we identified 5 additional novel qacA alleles: qacA7 (GenBank accession number MK040363), qacA8 (GenBank accession number MK040364), qacA9 (GenBank accession number MK040365), qacA10 (GenBank accession number MK040366), and qacA11 (GenBank accession number MK040367) (Table S2; Fig. S1). Similarly, comparing the sequence of qacA4 to the other sequences of qacA deposited in the NCBI GenBank database further confirmed that the SNVs at the three positions described above were unique to qacA4.
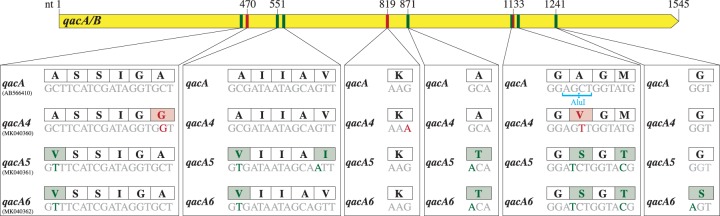
We also sequenced the genomes of the two qacA/B273-positive isolates that did not have elevated CHG MICs (MIC < 4 μg/ml) to investigate their discordant genotypic-phenotypic relationship (Table S1). Neither of the qacA/B genes in these two isolates contained the three identifying qacA4 mutations, and their sequences differed from the reference qacA sequence by six SNVs (Fig. 4). These SNVs resulted in six and seven amino acid substitutions compared to the reference qacA and qacA4 sequences, respectively (Fig. 4). The qacA/B genes in these isolates were classified as two additional alleles of qacA, referred to as qacA5 (GenBank accession number MK040361) and qacA6 (GenBank accession number MK040362). Due to an SNV at position 1132, which resulted in the loss of an AluI digestion site, qacA5 and qacA6 displayed digestion patterns identical to the pattern for qacA4.
FIG 4.
Comparison of the sequences of qacA4 (GenBank accession number MK040360), qacA5 (GenBank accession number MK040361), and qacA6 (GenBank accession number MK040362) and the reference qacA sequence (GenBank accession number AB566410). The associated AluI restriction sites are shown below the nucleotide sequences, and the corresponding amino acid sequences are displayed in the boxes above the nucleotide sequences. The nucleotides which distinguish the qacA4 sequence from the reference qacA, qacA5, and qacA6 sequences are highlighted in red. Those which distinguish the qacA5 and/or qacA6 sequence from the qacA4 and the reference qacA sequences are displayed in green.
We next sequenced the genomes from the nine qacA-positive isolates that had elevated CHG MICs.
From these isolates, we identified 4 qacA alleles: qacA10, qacA12, qacA13, and qacA14 (Table S2; Fig. S2). The sequences of the qacA genes in these isolates differed from the reference qacA sequence by 1 to 5 amino acid substitutions and from the qacA4 sequence by 2 to 8 amino acid substitutions (Fig. S2). The allele qacA14, identified in isolate 96.5, contained one of the distinguishing coding changes of qacA4, Ala157Gly, but not the other coding change. This allele encoded a unique amino acid substitution Pro328Leu, which distinguished the allele from the reference qacA and qacA4 sequences. Another isolate, 86.4, with an elevated CHG MIC carried the same qacA10 allele as isolate 110.3, which did not have an elevated CHG MIC. The amino acid substitutions in these novel qacA alleles occurred in transmembrane segments 5, 6, 9, 10, 12, and 13 and in the extracellular loop between transmembrane segments 5 and 6.
Identification of the novel resistance plasmid pAQZ1 containing the qacA4 allele.
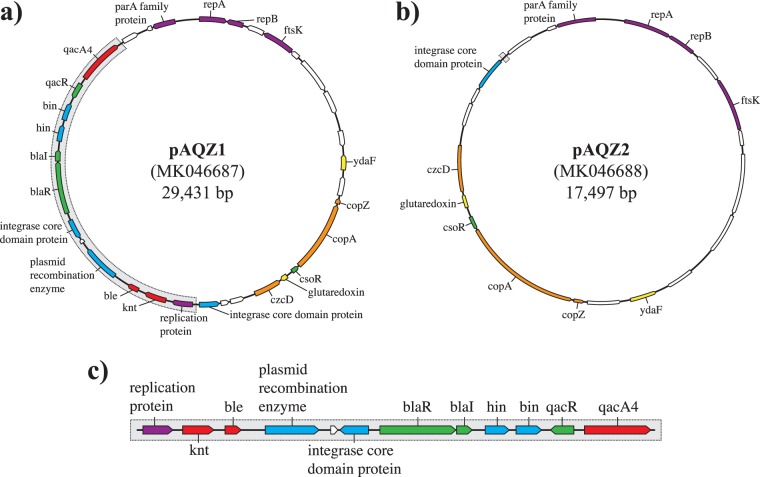
The genomic context of the qacA4 allele in de novo assemblies of two separate qacA/B273-positive isolates with high coverage, isolates 91.2 and 107.2, was examined to understand whether qacA4 was encoded on the chromosome or on a plasmid. Both isolates carried qacA4 on a 29,431-bp circular contig with coverage that was 2.8 times higher than the average chromosomal coverage, consistent with it being carried by a plasmid (Fig. 5a). The circular nature of the contig was verified by conducting PCR across the predicted junction site (data not shown). The plasmid, henceforth designated pAQZ1 (GenBank accession number MK046687), carrying qacA4 contained the RepA replication initiation protein with a RepA_N domain (pfam06970). Similar to other RepA_N family plasmids (27, 28), the origin of replication of pAQZ1 is likely contained within repA. The plasmid also carried several genes involved in heavy metal efflux, including copZ, copA, czcD, the knt kanamycin resistance gene, the ble bleomycin resistance gene, and an incomplete β-lactamase operon.
FIG 5.
Schematic representations of plasmid pAQZ1 containing qacA4 obtained from the de novo assemblies of isolates 91.2 and 107.2 (a), plasmid pAQZ2 retained in isolate 107.2cured (b), and the 11,934-bp segment of pAQZ1 eliminated through the curing experiments (c). Open reading frames (ORFs) shown in red depict resistance genes, ORFs in orange describe heavy metal efflux genes, ORFs in green represent transcriptional regulator genes, ORFs in blue depict recombinase genes, ORFs in purple describe replication genes, ORFs in white represent hypothetical proteins, and ORFs in yellow depict genes with other functions.
When the sequence of pAQZ1 was compared to plasmid sequences deposited in GenBank, several regions of pAQZ1 showed high sequence similarity (>99%) with previously characterized S. aureus and coagulase-negative Staphylococcus plasmids (GenBank accession numbers CP017465 and CP023967). The complete sequence of pAQZ1, however, did not fully align with any single, previously characterized S. aureus or coagulase-negative Staphylococcus plasmid. When queried against the sequences in the NCBI WGS database, the sequence of pAQZ1 showed high sequence similarity and a query coverage of 68% and 86%, respectively, to two previously sequenced contigs from two coagulase-negative Staphylococcus isolates (GenBank accession numbers JZUM01000030.1 and QSTD01000014.1).
Curing analysis in vitro confirms that qacA4 is responsible for the elevated CHG MICs.
Transformation of the pan-susceptible type strain S. epidermidis TÜ1457 with pAQZ1 was attempted but was unsuccessful (data not shown). Thus, to confirm the observed association between qacA4 and the elevated CHG MICs, we attempted to cure qacA4-carrying, smr-negative S. epidermidis isolate 107.2 of the pAQZ1 plasmid. We took advantage of the ability of QacA to efflux ethidium bromide (18) to screen for colonies which lost qacA4. Cells without qacA4 accumulate ethidium bromide in their cytoplasm, and the resulting colonies fluoresce under UV radiation. Those retaining qacA4 do not accumulate ethidium bromide, and thus, the resulting colonies do not fluoresce.
After 11 successive passages in Trypticase soy broth without selection, an isolate cured of qacA4, referred to as isolate 107.2cured, was identified. The CHG MIC of 107.2cured was 4-fold lower than that of 107.2 (Table 5). The 8-agent antimicrobial susceptibility profile of 107.2cured was identical to that of the parental strain (Table 5). Sequencing of 107.2cured (Table S1) revealed that recombination, presumably catalyzed by the recombinases on the plasmid, led to pAQZ1 eliminating an 11,934-bp segment and resulted in the formation of a new 17,497-bp plasmid. This new plasmid, pAQZ2 (GenBank accession number MK046688; Fig. 5b), retained the RepA protein of pAQZ1. The 11.9-kb segment lost in 107.2cured contained not only qacA4 but also the knt kanamycin resistance gene, the ble bleomycin resistance gene, the partial β-lactamase operon, and several recombinases (Fig. 5c). PCR testing further confirmed that isolate 107.2cured lost the 11.9-kb segment, distinguishing pAQZ1 from pAQZ2 (data not shown). Isolate 107.2cured contained one coding change in its chromosome compared to the sequence of the 107.2 parental strain. This coding change occurred in a GCN5-related N-acetyltransferase family protein (Gly225Glu).
TABLE 5.
Comparison of CHG resistance gene combinations and resistance patterns of isolates 107.2 and 107.2cured
| Isolatea | Presence of: |
CHG MIC (μg/ml) | Susceptibility tob
: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| qacA4 | smr | FOX | ERY | CIP | GEN | SXT | LZD | RIF | VAN | ||
| 107.2 | Yes | No | 4 | R | R | R | R | R | S | S | S |
| 107.2cured | No | No | 1 | R | R | R | R | R | S | S | S |
The isolates were determined to be S. epidermidis by MALDI-TOF MS.
CIP, ciprofloxacin; SXT, sulfamethoxazole-trimethoprim; FOX, cefoxitin; ERY, erythromycin; GEN, gentamicin; LZD, linezolid; RIF, rifampin; VAN, vancomycin; R, the isolate is resistant to the specified antimicrobial; S, the isolate is susceptible to the specified antimicrobial.
With the exception of qacA4, each of the genes present on the segment lost in isolate 107.2cured was identified in at least one of the qacA-positive control isolates without elevated CHG MICs. One of these qacA-positive isolates, 110.3, contained all of these other 11 genes present on the eliminated segment of pAQZ1.
Isolates carrying qacA4 belong to highly resistant and virulent S. epidermidis ST2.
The isolates carrying qacA4 harbored genes and mutations which confer resistance to several classes of commonly prescribed antimicrobials (Fig. 6). Additionally, all of the sequenced qacA4-carrying isolates contained the biofilm formation operon, icaADBC. When classified by multilocus sequence typing (MLST), all these isolates belonged to S. epidermidis ST2.
FIG 6.
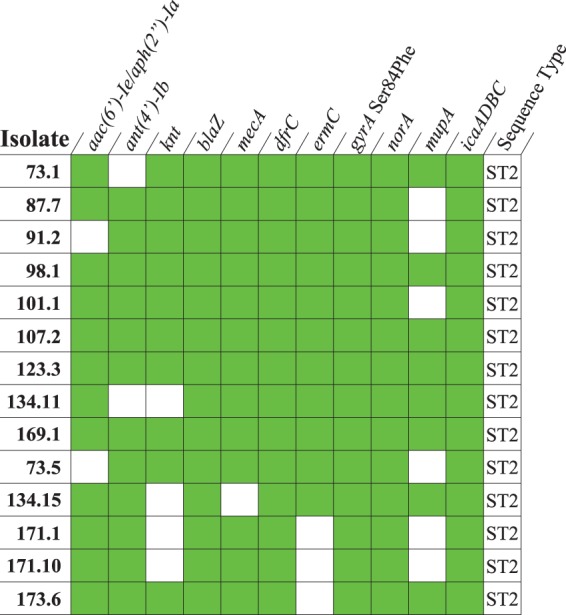
Presence-absence matrix displaying the antimicrobial resistance genes, resistance-associated mutations, and virulence genes identified in the isolates carrying qacA4. A green shaded box indicates that the resistant or virulence determinant was identified in a given isolate. The sequence type of each isolate, as determined by MLST, is shown.
Next, we performed a core genome analysis with 9 qacA4-carrying and 9 non-qacA4-carrying S. epidermidis ST2 isolates using the 2,793,003-bp chromosome of an S. epidermidis ST2 isolate from Australia (Genbank accession number GCA_900086615.1) as the reference genome (26). Our isolates differed from the reference strain by 1,904 to 2,186 SNVs (median, 2,148 SNVs). There was no apparent clustering based on the isolates’ collection location or date (Fig. S3). The qacA4-carrying isolates were more closely related to one another rather than the other non-qacA4-carrying ST2 isolates, with the exception of isolate 96.5. We included multiple isolates (isolates 36.3 and 36.4 and isolates 125.1, 125.3, and 125.8) collected at different time points from two patients in our core genome analysis, as these isolates had unusual qacA/B genotypes and CHG MICs, as described in Materials and Methods. These isolates differed from their corresponding coisolates by 0 to 3 SNVs (median, 2 SNVs) (Table S3).
The whole genomes of five additional qacA/B273-positive isolates that had elevated CHG MICs but that displayed discordant susceptibility patterns (susceptible to methicillin, gentamicin, or erythromycin) were sequenced (Table S1). Each of the isolates carried qacA4 and belonged to ST2. The divergent susceptibility patterns were explained by the absence of one or more resistance genes (Fig. 6).
qacA4-containing S. epidermidis isolates are globally distributed.
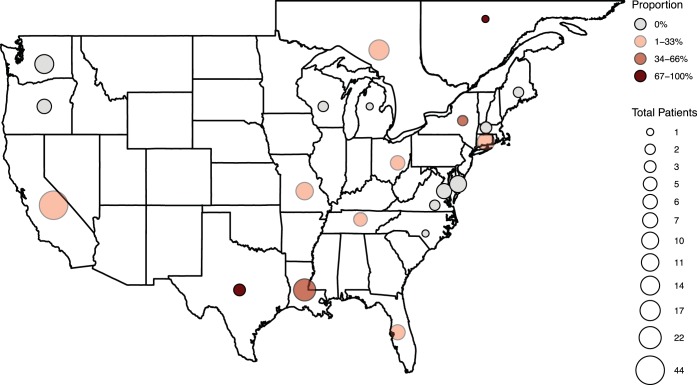
In total, 22 patients carried at least one cutaneous S. epidermidis isolate containing qacA4, as confirmed by sequencing or as presumed through the isolate’s qacA/B273-positive RFLP pattern and elevated CHG MIC. These 22 patients were enrolled at 14 study centers in 9 U.S. states and 2 Canadian provinces (Fig. 7). There was no obvious geographical clustering of the qacA4-carrying isolates.
FIG 7.
Geographic distribution of patients enrolled with the proportion of qacA4-carrying isolates within each geographic location. The size of the circles represents the total number of patients enrolled in each state or province, and the shade of the circle represents the proportion of the total patients with at least one qacA4-carrying S. epidermidis isolate.
We also identified isolates containing qacA4 that existed outside of our study (Fig. S4). A cutaneous S. epidermidis isolate containing qacA4 (Table S4) was identified from a CHG bathing pilot study (13) conducted at Seattle Children’s Hospital. In addition, we searched through the sequence reads of 922 isolates deposited in the NCBI Sequence Read Archive as part of several large S. epidermidis sequencing projects (BioProject accession numbers PRJEB12090, PRJNA239883, PRJNA382527, PRJNA433155, PRJNA434275, PRJNA470534, and PRJNA470752) (29–33). We further queried the NCBI WGS database and all of the bacterial whole-genome sequencing data sets in the European Nucleotide Archive through December 2016 (34). Including the isolate from the CHG bathing pilot study (13), we identified qacA4 in 39 additional S. epidermidis isolates collected at 10 sites in 5 different countries, including Australia, Belgium, Brazil, the United Kingdom, and the United States (Table S4). We obtained four local clinical S. epidermidis isolates containing qacA4 (isolates 6_SEPI, 642_SEPI, 872_SEPI, and 1295_SEPI) from a previous study (35) conducted at the University of Washington Medical Center that were identified during our search through publicly available sequencing data. Susceptibility testing of these isolates revealed that all four isolates had an elevated CHG MIC (4 μg/ml).
One of the 39 qacA4-carrying isolates (isolate MB591) contained the two coding changes but not the synonymous mutation (819G>A) of qacA4. Another qacA4-carrying isolate (isolate MB2095) appeared to contain qacA4 as well as an additional qacA allele.
Thirty-three of the 39 isolates contained reads that mapped to pAQZ1. Of these, 27 isolates contained reads that mapped to the entirety of pAQZ1. Five other isolates (isolates AUS16, BEL14, MB1595, 795_SEPI, and 798_SEPI) lacked reads corresponding to a 4.6-kb segment of pAQZ1 that contains the knt kanamycin resistance gene and the ble bleomycin resistance gene. One additional isolate (isolate 386_SEPI) lacked reads corresponding to a 10.4-kb segment of pAQZ1 that contains the copA and czcD heavy metal export genes, the knt kanamycin resistance gene, and the ble bleomycin resistance gene. Of note, we were unable to de novo assemble a single circular contig containing qacA4 in any these 33 isolates as a result of the short read length or low coverage. Furthermore, due to the lack of availability of sequencing reads and the quality of the genome assemblies, we were unable to determine if the remaining 6 isolates carried qacA4 on pAQZ1.
Of the 39 qacA4-carrying isolates from other sequencing projects, 35 belonged to ST2. The other 4 isolates (isolates 795_SEPI, 798_SEPI, 802_SEPI, and 803_SEPI) belonged to the novel ST847. ST847 differs from ST2 by a single SNV in the arcC loci (12A>T) and is therefore likely a descendant of ST2 and may share many characteristics of ST2.
DISCUSSION
In this study, we identified a novel qacA allele, termed qacA4, associated with an elevated CHG MIC in cutaneous S. epidermidis isolates and determined that qacA4 was contained on the novel pAQZ1 plasmid. We demonstrated that qacA4 is the determinant for the elevated CHG MIC by curing an isolate of the gene. Additionally, our analyses revealed that isolates carrying qacA4 displayed high rates of resistance to methicillin and other commonly prescribed antimicrobials, including erythromycin, ciprofloxacin, gentamicin, and sulfamethoxazole-trimethoprim. Whole-genome sequencing revealed that the isolates harbored several antimicrobial resistance determinants and resistance-associated mutations. Our analyses further demonstrated that these isolates contained the chromosomally encoded biofilm formation operon, icaADBC (2), and belonged to the highly resistant and pathogenic ST2 clone (36). We identified isolates proven or presumed to carry qacA4 from 22 patients enrolled in a multicenter, randomized controlled CHG bathing trial at 14 participating study centers across the United States and Canada. Furthermore, we identified qacA4 in clinical S. epidermidis isolates collected in prior studies at centers without patients participating in our CHG bathing trial (29, 33, 35, 37, 38).
Previous studies have characterized three alleles of qacA: qacA1, qacA2, and qacA3 (18, 21). Additional studies, however, have suggested that clinical and environmental Staphylococcus isolates may carry novel alleles of qacA (39, 40). Our identification of multiple novel qacA alleles supports this suggestion that the qacA allelic variation that exists within human staphylococcal populations is considerably greater than that previously appreciated.
The efflux pumps encoded by qacA1, qacA2, and qacA3 do not exhibit any functional differences (21). However, the efflux potential of QacA has been shown to vary with its amino acid sequence (41–44). Despite this recognition, no previous studies have examined the functional differences associated with the sequence variation of qacA observed in clinical and environmental Staphylococcus isolates. As the CHG MIC of the qacA4-carrying isolates was significantly higher than the CHG MICs of the isolates carrying other alleles of qacA, our results suggest that different alleles of qacA encode pumps with various CHG efflux potentials. This is further supported by our identification of four novel qacA alleles from the nine S. epidermidis isolates with prototypical qacA restriction patterns and elevated CHG MICs. These novel alleles indicate that other unique mutations may result in elevated CHG MICs and further underscore the importance of exploring the allelic variation of qacA in clinical and environmental Staphylococcus isolates.
It is tempting to speculate which of the two amino acid substitutions in QacA4, Ala157Gly and Ala378Val, is causal for the elevated CHG MIC observed in qacA4-carrying isolates. The Ala378Val substitution is particularly intriguing, as this mutation occurs in transmembrane segment 12. Transmembrane segment 12 is noteworthy when discussing the CHG efflux potential of QacA, since a previous study demonstrated that this segment lines the CHG binding pocket (41). The Ala157Gly substitution was identified in an S. epidermidis isolate with the prototypical qacA RFLP pattern and an elevated CHG MIC. This may indicate that the Ala157Gly substitution has a more causal role in the elevated CHG MICs. Future in vitro characterizations examining the structure-function relationship of the amino acid substitutions are merited.
Several studies have provided contradicting results as to whether the carriage of qacA influences the CHG MIC of Staphylococcus isolates (16, 19, 24, 45–49). These studies, however, did not distinguish between the qacA alleles carried by the isolates. Our findings highlight the importance of specifying the qacA allele carried by isolates when examining associations with CHG MICs: carriage of qacA4, as demonstrated by our curing analysis, results in a 4-fold increase in the CHG MIC of an isolate, while carriage of other alleles may not increase the CHG MIC.
Screening for qacA/B has been used as a proxy for determining whether an isolate exhibits reduced susceptibility or tolerance to CHG (48, 50–52), typically defined as a CHG MIC of ≥4 μg/ml (16, 48). All isolates carrying qacA4 but just 10.0% of all qacA/B-positive isolates had a CHG MIC of 4 μg/ml. Thus, screening for qacA4, rather than indiscriminately screening for qacA/B, may serve as a better indicator for reduced susceptibility to CHG in Staphylococcus spp.
Similar to previously described plasmids carrying qacA/B (22), pAQZ1 carries several genes involved in heavy metal efflux and a partial β-lactamase operon. As pAQZ1 contains only the β-lactamase transcriptional regulators blaI and blaR (53), it is unclear if carriage of the plasmid influences β-lactam resistance. The kanamycin nucleotidyltransferase encoded by knt on pAQZ1 showed high sequence similarity to the kanamycin nucleotidyltransferase of S. aureus (GenBank accession number X03408.1) and may contribute to aminoglycoside resistance (54). Since the cured isolate also contained other aminoglycoside resistance determinants, including aac(6′)-Ie plus aph(2ʺ)-Ia and ant(4′)-Ib, we were unable to assess the contribution of knt to aminoglycoside resistance.
All of the qacA4-carrying isolates that we sequenced belonged to ST2, an S. epidermidis clone frequently implicated in device-associated infections (5, 36, 55–58). Consistent with the findings of previous studies (36, 55–57), our ST2 isolates contained genes and mutations which confer resistance to several classes of commonly prescribed antimicrobials. Additionally, our isolates contained genes associated with binding to foreign materials (5, 36), including the biofilm formation operon, icaADBC. These results suggest that qacA4 may allow the highly resistant S. epidermidis ST2 clone to better persist following topical application of CHG and, thus, further succeed as an opportunistic pathogen. However, as the concentration of CHG used in clinical settings (8, 10) is much higher than that tested in vitro (2,000 μg/ml versus 4 μg/ml), further study is required to fully understand the clinical implications of carriage of qacA4 by the ST2 clone.
Our results suggest that qacA4 is distributed in pediatric oncology populations at centers across the United States and Canada. In addition, we identified 39 isolates carrying qacA4 collected at institutions in 5 countries (29, 33, 35, 37, 38). This suggests that qacA4 follows the wide distribution of both the S. epidermidis ST2 clone and qacA (36, 48) and is likely disseminated throughout health care settings globally.
Our study was limited by the nature of the RFLP screening analysis. While our method of screening for qacA/B allowed us to identify all the isolates with a mutation at positions 1131 to 1134, we were unable to easily detect the other novel qacA alleles that may have been present in our study population. From just the 35 qacA-positive and qacA/B273-positive isolates that we sequenced, we identified 11 novel qacA alleles, and, as we demonstrated, at least one of these alleles exhibits a functional difference with respect to CHG efflux. This emphasizes the necessity of using sequencing to screen for allelic variation in resistance determinants, especially in those determinants in which allelic variation has been underappreciated. Furthermore, reflecting the difficulty of performing transformations in Staphylococcus spp. (59), we were unable to perform a gain-of-function analysis for qacA4, despite trying three different methods of preparing electrocompetent cells and two separate electroporation conditions for each cell preparation. Despite this limitation, we were able to perform a loss-of-function analysis to confirm the role of qacA4 in the elevated CHG MIC. It is remarkable that the loss of function was achieved by recombination and that the cured isolate retained more than half of pAQZ1. Beyond qacA4, each of the 11 other genes contained on the segment lost in the cured isolate may explain the 4-fold decrease in the CHG MIC exhibited by this cured isolate. Many of these genes, however, have well-described functions unrelated to CHG efflux (53, 54, 60–62). Additionally, we identified each of the other 11 genes in an isolate without an elevated CHG MIC. Thus, the decrease in the CHG MIC observed in the cured isolate is most consistent with the loss of qacA4.
Our results highlight the importance of screening for allelic variation in qacA. Just as a single SNV between qacA and qacB accounts for the different substrate specificities of the resulting efflux pumps (18), the three SNVs of qacA4 are associated with a 4-fold increase in the CHG MIC. Further study should focus on understanding the functional differences of the various qacA alleles identified in clinical and environmental Staphylococcus isolates. Moreover, our results indicate the qacA4 is carried by the highly resistant S. epidermidis ST2 clone and related strains. Future study is required to understand if the use of CHG selects for qacA4 and this pathogenic clone of S. epidermidis.
MATERIALS AND METHODS
Collection and identification of cutaneous Staphylococcus isolates.
Skin swab specimens were obtained from patients between 2 months and 21 years of age who were undergoing allogeneic hematopoietic cell transplantation for any reason or treatment for a cancer diagnosis that required at least 3 months of additional chemotherapy from the time of enrollment and who were enrolled in a randomized double-blind placebo-controlled trial of CHG bathing versus control bathing conducted at 37 centers in the United States and Canada from January 2014 to April 2017 (Children’s Oncology Group ACCL1034; ClinicalTrials.gov identifier NCT01817075). The study was approved by the National Cancer Institute’s Pediatric Central Institution Review Board as well as the local review boards at the participating institutions, if required.
After enrollment, patients were followed for 90 days both during hospitalizations and while they were outpatients. Cutaneous swab samples were obtained by swabbing a 3- by 3-cm area on the side or back of the neck and axilla regions with a sterile nylon swab (Copan Diagnostics) for 20 s and transported in 1 ml of the accompanying liquid Amies medium. The swab and Amies medium were vigorously vortexed, and the medium was plated on the following agar plates: tryptic soy agar with 5% sheep’s blood (Remel), chocolate agar (Remel), Sabouraud dextrose agar (Remel), MacConkey agar (Remel), and mannitol salt agar (Remel). The plates were incubated at 35°C for 48 h. Staphylococcus isolates were identified via MALDI-TOF MS.
Isolates were prepared for MALDI-TOF MS according to the manufacturer’s direct transfer sample preparation procedure (63). A MicroFlex LT mass spectrometer (Bruker Daltonics, Inc.) operated in the positive linear mode with FlexControl software (version 3.4; Bruker) was used to obtain spectra. The resulting spectra were processed and classified using Biotyper software (version 3.2; Bruker). Identification results were interpreted according to the manufacturer’s guidelines. The isolates and the corresponding phenotypic information included in the study are presented in Data Set S1 in the supplemental material.
Five additional qacA4-carrying isolates were obtained for phenotypic testing: one was collected during a pilot study conducted at Seattle Children’s Hospital (13), and the other four were collected in a previous study at the University of Washington Medical Center (35).
Antimicrobial susceptibility testing.
Following CLSI guidelines (64), susceptibility testing was performed by disk diffusion (Becton, Dickinson and Company) for the following antimicrobials: ERY (15 μg), CIP (5 μg), GEN (10 μg), LZD (30 μg), cefoxitin (FOX; 30 μg), RIF (5 μg), and SXT (23.75/1.25 μg). Additionally, VAN (0.016 to 256 μg/ml) susceptibility testing was performed using the Etest (bioMérieux) MIC method. FOX was used as a surrogate for determination of methicillin susceptibility per CLSI guidelines (64). Isolates were classified as resistant, intermediate, or susceptible to a given agent using the breakpoints specified by CLSI (65).
CHG MICs (0.0625 to 64 μg/ml) were determined via the broth microdilution method (65, 66). For the CHG MICs, the following strains were included as controls: Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 25923, Staphylococcus epidermidis (a laboratory control strain) (13), and Escherichia coli ATCC 25922.
Detection of qacA/B and smr.
Isolates were tested for the presence of qacA/B via PCR using AmpliTaq DNA polymerase (Applied Biosystems) following the manufacturer’s instruction for the reaction mixture (67) with the previously described primers and reaction conditions (21). To distinguish between qacA and qacB, the PCR products were digested with AluI, and the resulting fragments were visualized by agarose gel electrophoresis (21). Control qacA-positive and qacB-positive strains were obtained from Nobumichi Kobayashi.
Isolates were additionally screened for the presence of smr via PCR with the following primers: forward primer 5′-AAAACAATGCAACACCTACCAC-3′ and reverse primer 5′-ATGCGATGTTCCGAAAATGT-3′. The following reaction conditions were used: an initial denaturation for 3 min at 95°C, followed by 30 cycles of denaturation for 1 min at 95°C, primer annealing for 30 s at 55°C, and elongation for 1 min at 72°C, with completion with a final elongation for 10 min at 72°C. A control smr-positive strain was obtained from Arnold Bayer.
Statistical testing.
Kruskal-Wallis or Wilcoxon rank-sum tests were used to assess differences in the distribution of CHG MICs between qac groups. Fisher’s exact test was used to assess the proportion of isolates resistant to commonly used antimicrobials by qac group. All analyses were first performed on all isolates and then repeated using one randomly chosen isolate per patient per qac group. Analyses were performed using STATA (version 14; College Station, TX) and R (version 3.3.2; R Core Team) software.
Whole-genome sequencing.
In total, the genomes of 40 Staphylococcus species isolates were sequenced. An initial group of 10 qacA-positive, 4 qacB-positive, and 10 qacA/B273-positive isolates was selected for whole-genome sequencing. The pool of qacA-positive and qacB-positive isolates was restricted to those identified as S. epidermidis since all of the qacA/B273-positive isolates with elevated CHG MICs were identified as S. epidermidis. The pool of isolates was then further restricted to the first qacA-positive isolate from 10 patients randomly chosen from the 91 patients with at least one qacA-positive isolate, the first qacA/B273-positive isolate obtained from 10 patients randomly chosen from the 24 patients with at least one qacA/B273-positive isolate, and the first qacB-positive isolate from all 4 patients with at least one qacB-positive isolate. One of the initially selected qacA/B273-positive isolates contained two alleles of qacA, with one being qacA4. As a result, this isolate was excluded from all subsequent analyses. Additional isolates were selected for whole-genome sequencing based on their antimicrobial susceptibility phenotypes: 2 qacA/B273-positive isolates were chosen as they did not have elevated CHG MICs, 9 qacA-positive isolates were selected as they had elevated CHG MICs, and 5 qacA/B273-positive isolates were chosen as they had discordant antimicrobial susceptibility patterns (susceptible to FOX, GEN, or ERY). Of note, selection of isolates with divergent qacA and qacB genotypes and antimicrobial susceptibility phenotypes was not restricted to the first S. epidermidis isolate per patient. As a result, 5 patients contributed multiple isolates.
Isolates were grown in brain heart infusion broth (Remel) for 24 h at 37°C at a constant shaking of 150 rpm. DNA was extracted from these isolates using a QIAamp DNA minikit (Qiagen) while following the manufacturer’s protocol for isolating DNA from Gram-positive bacteria (68). An initial cell lysis step was completed using a 200-μg/ml lysostaphin solution (Sigma-Aldrich).
Libraries were prepared with a Kapa Hyper preparation kit. Isolates were sequenced to at least 27× coverage using 2 × 300-bp Illumina MiSeq runs. De novo assemblies were constructed with the SPAdes program, annotated by use of the prokka program, and visualized using Geneious (version 10.2.3) software.
Multilocus sequence typing was completed by uploading the assemblies to PubMLST’s Staphylococcus epidermidis MLST website (https://pubmlst.org/sepidermidis/) (69). A core genome analysis was performed as previously described (70, 71) using BWA-MEM, SAMtools, and VCFtools (parameters, minDP 10, minQ 200, and minGQ 10), based on mapping to an S. epidermidis ST2 reference genome (GCA_900086615.1) (26). Resistance genes, limited to only those matching “perfect” and “strict” criteria, were detected with the Comprehensive Antibiotic Resistance Database’s Resistance Gene Identifier (https://card.mcmaster.ca/analyze/rgi) (72). The sequence of the DNA gyrase A protein in our isolates was compared with that of the DNA gyrase A protein of S. epidermidis ATCC 12228 (GenBank accession number AE015929), a ciprofloxacin-sensitive strain, to determine the identity of the residue at position 84. Isolates containing the Ser84Phe mutation were determined to contain a gyrA gene conferring resistance to fluoroquinolones (73). The assemblies were additionally screened for virulence genes, including icaADBC (GenBank accession number U43366), aap (GenBank accession number KJ920749), and bhp (GenBank accession number AY028618).
Curing of qacA4.
An isolate carrying qacA4, isolate 107.2, was selected for the curing analysis, as this isolate did not contain smr, which can efflux ethidium bromide. The isolate was successively passaged in tryptic soy broth (Remel) under four separate curing conditions: with no selection, in which it was incubated for 24 h at 37°C at a constant shaking of 150 rpm; at an increased temperature, in which it was incubated for 24 h at 42°C; in the presence of increasing subinhibitory concentrations of sodium dodecyl sulfate (Sigma-Aldrich), in which it was incubated for 24 h at 37°C at a constant shaking of 150 rpm with 0.001% to 0.01% sodium dodecyl sulfate; and in the presence of increasing subinhibitory concentrations of novobiocin (Sigma-Aldrich), in which it was incubated for 24 h at 37°C at a constant shaking of 150 rpm with 0.01 μg/ml to 0.1 μg/ml novobiocin.
After each passage, broth samples were plated onto tryptic soy agar plates (Remel) containing 0.375 μg/ml of filter-sterilized ethidium bromide (VWR). The plates were incubated at 35°C for 48 h. Screening for cured strains was completed with UV light as previously described (46). PCR was used to confirm that the cured strain eliminated qacA4. Whole-genome sequencing was used to confirm that the cured strain contained minimal chromosomal mutations compared to the sequence of the parental strain.
Three PCRs were conducted on the plasmids predicted from whole-genome sequencing of the isolates. To confirm the circular nature of the contig presumed to be a plasmid, PCR was conducted with the following primers: forward primer 5′-GGCTACTGTTGTTTTACCTACACCACC-3′ and reverse primer 5′-GCATACATAACCTTTGCGTCAGTTGTC-3′. To confirm that the curing resulted in the formation of a novel plasmid, PCR was conducted with the following primers: forward primer 5′-CCATTGTGGCGTCATTTCACGGC-3′ and reverse primer 5′-CGGCGAAATCCTTGAGCCATATCTG-3′ and forward primer 5′-GAAGAATCTGTAGTGGGCGCTG-3′ and reverse primer 5′-GATGAAAGTTGCTACTAGTGCTCC-3′. The following reactions conditions were used: an initial denaturation for 3 min at 95°C, followed by 30 cycles of denaturation for 1 min at 95°C; primer annealing for 30 s at 53°C, 57°C, or 52°C for the three sets of primers, respectively; and elongation for 1 min at 72°C, with completion with a final elongation for 10 min at 72°C.
Transformation of pAQZ1 into S. epidermidis TÜ1457.
In preparation for the extraction of the pAQZ1 plasmid, qacA4-carrying S. epidermidis isolate 107.2 was grown in tryptic soy broth (Remel) for 24 h at 37°C at a constant shaking of 150 rpm. The plasmid was extracted using a QIAprep Spin miniprep kit (Qiagen) following the manufacturer’s instructions (Qiagen, Hilden, Germany).
The pan-susceptible S. epidermidis TÜ1457 strain (74) was used for the transformations. Three previously described methods were used for preparing electrocompetent cells (59, 75, 76). For electroporation, 100 μl of the prepared cells was mixed with 100 ng of pAQZ1 DNA in a 1-mm electroporation cuvette (Bio-Rad). Two electroporation conditions were used for each preparation of electrocompetent cells: 21 kV/cm, 100 Ω, and 25 μF and 23 kV/cm, 100 Ω, and 25 μF. The pulsed cells were resuspended in 1,000 μl of broth, with the type of broth being selected based on the previously described methods, and incubated at 37°C at a constant shaking of 150 rpm for 1 h. The cells were plated onto tryptic soy agar plates (Remel) containing either 2 μg/ml CHG (Sigma-Aldrich), 15 μg/ml ethidium bromide (VWR), or 10 μg/ml of kanamycin (Sigma-Aldrich) and incubated overnight at 37°C.
Accession number(s).
The sequence of qacA4 was deposited in GenBank under accession number MK040360. The accession numbers for the additional 10 novel qacA alleles identified in this study are listed in Table S2. The sequences of pAQZ1 and pAQZ2 were deposited under accession numbers MK046687 and MK046688, respectively. Draft genome assemblies are available in GenBank under study BioProject accession number PRJNA415995. The accession numbers as well as the phenotypic data for the individual isolates sequenced are displayed in Table S1.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patients who agreed to participate in this study as well as the research teams at all the involved sites and the ACCL1034 committee. We also thank Stephen Salipante for sharing previously collected bacterial isolates, Paul Fey for providing the S. epidermidis TÜ1457 strain, Nobumichi Kobayashi for supplying the qacA/B-positive control strains, and Arnold Bayer for sharing the smr-positive control strain.
This work was supported by the National Cancer Institute at the National Institutes of Health (R01CA163394 and UG1CA189955). The research was also supported by St. Baldrick’s Foundation.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02607-18.
REFERENCES
- 1.Becker K, Heilmann C, Peters G. 2014. Coagulase-negative staphylococci. Clin Microbiol Rev 27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerke C, Kraft A, Süssmuth R, Schweitzer O, Götz F. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem 273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 3.Fey PD, Olson ME. 2010. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol 5:917–933. doi: 10.2217/fmb.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. 1997. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun 65:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otto M. 2009. Staphylococcus epidermidis—the “accidental” pathogen. Nat Rev Microbiol 7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M, SENTRY Participants Group. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific Region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis 32(Suppl 2):S114–S132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 7.Edmiston CE, Bruden B, Rucinski MC, Henen C, Graham MB, Lewis BL. 2013. Reducing the risk of surgical site infections: does chlorhexidine gluconate provide a risk reduction benefit? Am J Infect Control 41(Suppl):S49–S55. doi: 10.1016/j.ajic.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 8.Milstone AM, Elward A, Song X, Zerr DM, Orscheln R, Speck K, Obeng D, Reich NG, Coffin SE, Perl TM. 2013. Daily chlorhexidine bathing to reduce bacteraemia in critically ill children: a multicentre, cluster-randomised, crossover trial. Lancet 381:1099–1106. doi: 10.1016/S0140-6736(12)61687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bleasdale SC, Trick WE, Gonzalez IM, Lyles RD, Hayden MK, Weinstein RA. 2007. Effectiveness of chlorhexidine bathing to reduce catheter-associated bloodstream infections in medical intensive care unit patients. Arch Intern Med 167:2073–2079. doi: 10.1001/archinte.167.19.2073. [DOI] [PubMed] [Google Scholar]
- 10.Climo MW, Yokoe DS, Warren DK, Perl TM, Bolon M, Herwaldt LA, Weinstein RA, Sepkowitz KA, Jernigan JA, Sanogo K, Wong ES. 2013. Effect of daily chlorhexidine bathing on hospital-acquired infection. N Engl J Med 368:533–542. doi: 10.1056/NEJMoa1113849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mimoz O, Karim A, Mercat A, Cosseron M, Falissard B, Parker F, Richard C, Samii K, Nordmann P. 1999. Chlorhexidine compared with povidone-iodine as skin preparation before blood culture. A randomized, controlled trial. Ann Intern Med 131:834–837. doi: 10.7326/0003-4819-131-11-199912070-00006. [DOI] [PubMed] [Google Scholar]
- 12.Marlowe L, Mistry RD, Coffin S, Leckerman KH, McGowan KL, Dai D, Bell LM, Zaoutis T. 2010. Blood culture contamination rates after skin antisepsis with chlorhexidine gluconate versus povidone-iodine in a pediatric emergency department. Infect Control Hosp Epidemiol 31:171–176. doi: 10.1086/650201. [DOI] [PubMed] [Google Scholar]
- 13.Soma V, Qin X, Zhou C, Adler A, Berry J, Zerr D. 2012. The effects of daily chlorhexidine bathing on cutaneous bacterial isolates: a pilot study. Infect Drug Resist 5:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vernon MO, Hayden MK, Trick WE, Hayes RA, Blom DW, Weinstein RA. 2006. Chlorhexidine gluconate to cleanse patients in a medical intensive care unit: the effectiveness of source control to reduce the bioburden of vancomycin-resistant enterococci. Arch Intern Med 166:306–312. doi: 10.1001/archinte.166.3.306. [DOI] [PubMed] [Google Scholar]
- 15.Block C, Furman M. 2002. Association between intensity of chlorhexidine use and micro-organisms of reduced susceptibility in a hospital environment. J Hosp Infect 51:201–206. doi: 10.1053/jhin.2002.1246. [DOI] [PubMed] [Google Scholar]
- 16.Wang J-T, Sheng W-H, Wang J-L, Chen D, Chen M-L, Chen Y-C, Chang S-C. 2008. Longitudinal analysis of chlorhexidine susceptibilities of nosocomial methicillin-resistant Staphylococcus aureus isolates at a teaching hospital in Taiwan. J Antimicrob Chemother 62:514–517. doi: 10.1093/jac/dkn208. [DOI] [PubMed] [Google Scholar]
- 17.Vali L, Davies SE, Lai LLG, Dave J, Amyes S. 2008. Frequency of biocide resistance genes, antibiotic resistance and the effect of chlorhexidine exposure on clinical methicillin-resistant Staphylococcus aureus isolates. J Antimicrob Chemother 61:524–532. doi: 10.1093/jac/dkm520. [DOI] [PubMed] [Google Scholar]
- 18.Paulsen IT, Brown MH, Littlejohn TG, Mitchell BA, Skurray RA. 1996. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci U S A 93:3630–3635. doi: 10.1073/pnas.93.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell BA, Brown MH, Skurray RA. 1998. QacA multidrug efflux pump from Staphylococcus aureus: comparative analysis of resistance to diamidines, biguanidines, and guanylhydrazones. Antimicrob Agents Chemother 42:475–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown MH, Skurray RA. 2001. Staphylococcal multidrug efflux protein QacA. J Mol Microbiol Biotechnol 3:163–170. [PubMed] [Google Scholar]
- 21.Alam MM, Kobayashi N, Uehara N, Watanabe N. 2003. Analysis on distribution and genomic diversity of high-level antiseptic resistance genes qacA and qacB in human clinical isolates of Staphylococcus aureus. Microb Drug Resist 9:109–121. doi: 10.1089/107662903765826697. [DOI] [PubMed] [Google Scholar]
- 22.Leelaporn A, Paulsen IT, Tennent JM, Littlejohn TG, Skurray RA. 1994. Multidrug resistance to antiseptics and disinfectants in coagulase-negative staphylococci. J Med Microbiol 40:214–220. doi: 10.1099/00222615-40-3-214. [DOI] [PubMed] [Google Scholar]
- 23.Paulsen IT, Brown MH, Dunstan SJ, Skurray RA. 1995. Molecular characterization of the staphylococcal multidrug resistance export protein QacC. J Bacteriol 177:2827–2833. doi: 10.1128/jb.177.10.2827-2833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Littlejohn TG, Paulsen IT, Gillespie MT, Tennent JM, Midgley M, Jones IG, Purewal AS, Skurray RA. 1992. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol Lett 74:259–265. doi: 10.1111/j.1574-6968.1992.tb05376.x. [DOI] [PubMed] [Google Scholar]
- 25.Bjorland J, Sunde M, Waage S. 2001. Plasmid-borne smr gene causes resistance to quaternary ammonium compounds in bovine Staphylococcus aureus. J Clin Microbiol 39:3999–4004. doi: 10.1128/JCM.39.11.3999-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JYH, Monk IR, Pidot SJ, Singh S, Chua KYL, Seemann T, Stinear TP, Howden BP. 2016. Functional analysis of the first complete genome sequence of a multidrug resistant sequence type 2 Staphylococcus epidermidis. Microb Genom 2:e000077. doi: 10.1099/mgen.0.000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weaver KE, Kwong SM, Firth N, Francia MV. 2009. The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids. Plasmid 61:94–109. doi: 10.1016/j.plasmid.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwong SM, Ramsay JP, Jensen SO, Firth N. 2017. Replication of staphylococcal resistance plasmids. Front Microbiol 8:2279. doi: 10.3389/fmicb.2017.02279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JYH, Monk IR, Gonçalves da Silva A, Seemann T, Chua KYL, Kearns A, Hill R, Woodford N, Bartels MD, Strommenger B, Laurent F, Dodémont M, Deplano A, Patel R, Larsen AR, Korman TM, Stinear TP, Howden BP. 2018. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat Microbiol 3:1175–1185. doi: 10.1038/s41564-018-0230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Méric G, Mageiros L, Pensar J, Laabei M, Yahara K, Pascoe B, Kittiwan N, Tadee P, Post V, Lamble S, Bowden R, Bray JE, Morgenstern M, Jolley KA, Maiden MCJ, Feil EJ, Didelot X, Miragaia M, de Lencastre H, Moriarty TF, Rohde H, Massey R, Mack D, Corander J, Sheppard SK. 2018. Disease-associated genotypes of the commensal skin bacterium Staphylococcus epidermidis. Nat Commun 9:5034. doi: 10.1038/s41467-018-07368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Post V, Harris LG, Morgenstern M, Mageiros L, Hitchings MD, Méric G, Pascoe B, Sheppard SK, Richards RG, Moriarty TF. 2017. Comparative genomics study of Staphylococcus epidermidis isolates from orthopedic-device-related infections correlated with patient outcome. J Clin Microbiol 55:3089–3103. doi: 10.1128/JCM.00881-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tewhey R, Gu B, Kelesidis T, Charlton C, Bobenchik A, Hindler J, Schork NJ, Humphries RM. 2014. Mechanisms of linezolid resistance among coagulase-negative staphylococci determined by whole-genome sequencing. mBio 5:e00894-14. doi: 10.1128/mBio.00894-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Arias CA, Aitken SL, Galloway Peña J, Panesso D, Chang M, Diaz L, Rios R, Numan Y, Ghaoui S, DebRoy S, Bhatti MM, Simmons DE, Raad I, Hachem R, Folan SA, Sahasarabhojane P, Kalia A, Shelburne SA. 2018. Clonal emergence of invasive multidrug-resistant Staphylococcus epidermidis deconvoluted via a combination of whole-genome sequencing and microbiome analyses. Clin Infect Dis 67:398–406. doi: 10.1093/cid/ciy089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradley P, den Bakker HC, Rocha EPC, McVean G, Iqbal Z. 2019. Ultrafast search of all deposited bacterial and viral genomic data. Nat Biotechnol 37:152–159. doi: 10.1038/s41587-018-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roach DJ, Burton JN, Lee C, Stackhouse B, Butler-Wu SM, Cookson BT, Shendure J, Salipante SJ. 2015. A year of infection in the intensive care unit: prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet 11:e1005413. doi: 10.1371/journal.pgen.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoenfelder SMK, Lange C, Eckart M, Hennig S, Kozytska S, Ziebuhr W. 2010. Success through diversity—how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int J Med Microbiol 300:380–386. doi: 10.1016/j.ijmm.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 37.The Human Microbiome Jumpstart Reference Strains Consortium, Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, Chinwalla AT, Feldgarden M, Gevers D, Haas BJ, Madupu R, Ward DV, Birren BW, Gibbs RA, Methe B, Petrosino JF, Strausberg RL, Sutton GG, White OR, Wilson RK, Durkin S, Giglio MG, Gujja S, Howarth C, Kodira CD, Kyrpides N, Mehta T, Muzny DM, Pearson M, Pepin K, Pati A, Qin X, Yandava C, Zeng Q, Zhang L, Berlin AM, Chen L, Hepburn TA, Johnson J, McCorrison J, Miller J, Minx P, Nusbaum C, Russ C, Sykes SM, Tomlinson CM, et al. 2010. A catalog of reference genomes from the human microbiome. Science 328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh P, Bekaert M, Carroll J, Manning T, Kelly B, O’Driscoll A, Lu X, Smith C, Dickinson P, Templeton K, Ghazal P, Sleator RD. 2015. Draft genome sequences of six different Staphylococcus epidermidis clones, isolated individually from preterm neonates presenting with sepsis at Edinburgh’s Royal Infirmary. Genome Announc 3:e00471-15. doi: 10.1128/genomeA.00471-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wassenaar T, Ussery D, Nielsen L, Ingmer H. 2015. Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur J Microbiol Immunol (Bp) 5:44–61. doi: 10.1556/EUJMI-D-14-00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hijazi K, Mukhopadhya I, Abbott F, Milne K, Al-Jabri ZJ, Oggioni MR, Gould IM. 2016. Susceptibility to chlorhexidine amongst multidrug-resistant clinical isolates of Staphylococcus epidermidis from bloodstream infections. Int J Antimicrob Agents 48:86–90. doi: 10.1016/j.ijantimicag.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 41.Hassan KA, Skurray RA, Brown MH. 2007. Transmembrane helix 12 of the Staphylococcus aureus multidrug transporter QacA lines the bivalent cationic drug binding pocket. J Bacteriol 189:9131–9134. doi: 10.1128/JB.01492-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J, Hassan KA, Skurray RA, Brown MH. 2008. Functional analyses reveal an important role for tyrosine residues in the staphylococcal multidrug efflux protein QacA. BMC Microbiol 8:147. doi: 10.1186/1471-2180-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hassan KA, Galea M, Wu J, Mitchell BA, Skurray RA, Brown MH. 2006. Functional effects of intramembranous proline substitutions in the staphylococcal multidrug transporter QacA. FEMS Microbiol Lett 263:76–85. doi: 10.1111/j.1574-6968.2006.00411.x. [DOI] [PubMed] [Google Scholar]
- 44.Xu Z, O'Rourke BA, Skurray RA, Brown MH. 2006. Role of transmembrane segment 10 in efflux mediated by the staphylococcal multidrug transport protein QacA. J Biol Chem 281:792–799. doi: 10.1074/jbc.M508676200. [DOI] [PubMed] [Google Scholar]
- 45.Sheng W-H, Wang J-T, Lauderdale T-L, Weng C-M, Chen D, Chang S-C. 2009. Epidemiology and susceptibilities of methicillin-resistant Staphylococcus aureus in Taiwan: emphasis on chlorhexidine susceptibility. Diagn Microbiol Infect Dis 63:309–313. doi: 10.1016/j.diagmicrobio.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Costa SS, Ntokou E, Martins A, Viveiros M, Pournaras S, Couto I, Amaral L. 2010. Identification of the plasmid-encoded qacA efflux pump gene in meticillin-resistant Staphylococcus aureus (MRSA) strain HPV107, a representative of the MRSA Iberian clone. Int J Antimicrob Agents 36:557–561. doi: 10.1016/j.ijantimicag.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Skovgaard S, Larsen MH, Nielsen LN, Skov RL, Wong C, Westh H, Ingmer H. 2013. Recently introduced qacA/B genes in Staphylococcus epidermidis do not increase chlorhexidine MIC/MBC. J Antimicrob Chemother 10:2226–2233. doi: 10.1093/jac/dkt182. [DOI] [PubMed] [Google Scholar]
- 48.Horner C, Mawer D, Wilcox M. 2012. Reduced susceptibility to chlorhexidine in staphylococci: is it increasing and does it matter? J Antimicrob Chemother 67:2547–2559. doi: 10.1093/jac/dks284. [DOI] [PubMed] [Google Scholar]
- 49.Hayden MK, Lolans K, Haffenreffer K, Avery TR, Kleinman K, Li H, Kaganov RE, Lankiewicz J, Moody J, Septimus E, Weinstein RA, Hickok J, Jernigan J, Perlin JB, Platt R, Huang SS. 2016. Chlorhexidine and mupirocin susceptibility of methicillin-resistant Staphylococcus aureus isolates in the REDUCE-MRSA Trial. J Clin Microbiol 54:2735–2742. doi: 10.1128/JCM.01444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McClure J-A, Zaal DeLongchamp J, Conly JM, Zhang K. 2017. Novel multiplex PCR assay for detection of chlorhexidine-quaternary ammonium, mupirocin, and methicillin resistance genes, with simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J Clin Microbiol 55:1857–1864. doi: 10.1128/JCM.02488-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noguchi N, Suwa J, Narui K, Sasatsu M, Ito T, Hiramatsu K, Song J-H. 2005. Susceptibilities to antiseptic agents and distribution of antiseptic-resistance genes qacA/B and smr of methicillin-resistant Staphylococcus aureus isolated in Asia during 1998 and 1999. J Med Microbiol 54:557–565. doi: 10.1099/jmm.0.45902-0. [DOI] [PubMed] [Google Scholar]
- 52.Warren DK, Prager M, Munigala S, Wallace MA, Kennedy CR, Bommarito KM, Mazuski JE, Burnham C-A. 2016. Prevalence of qacA/B genes and mupirocin resistance among methicillin-resistant Staphylococcus aureus (MRSA) isolates in the setting of chlorhexidine bathing without mupirocin. Infect Control Hosp Epidemiol 37:590–597. doi: 10.1017/ice.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hackbarth CJ, Chambers HF. 1993. blaI and blaR1 regulate beta-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 37:1144–1149. doi: 10.1128/AAC.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedersen LC, Benning MM, Holden HM. 1995. Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. Biochemistry 34:13305–13311. doi: 10.1021/bi00041a005. [DOI] [PubMed] [Google Scholar]
- 55.Li M, Wang X, Gao Q, Lu Y. 2009. Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J Med Microbiol 58:456–461. doi: 10.1099/jmm.0.007567-0. [DOI] [PubMed] [Google Scholar]
- 56.Iorio NLP, Caboclo RF, Azevedo MB, Barcellos AG, Neves FPG, Domingues RMCP, dos Santos KRN. 2012. Characteristics related to antimicrobial resistance and biofilm formation of widespread methicillin-resistant Staphylococcus epidermidis ST2 and ST23 lineages in Rio de Janeiro hospitals, Brazil. Diagn Microbiol Infect Dis 72:32–40. doi: 10.1016/j.diagmicrobio.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 57.Hellmark B, Söderquist B, Unemo M, Nilsdotter-Augustinsson Å. 2013. Comparison of Staphylococcus epidermidis isolated from prosthetic joint infections and commensal isolates in regard to antibiotic susceptibility, agr type, biofilm production, and epidemiology. Int J Med Microbiol 303:32–39. doi: 10.1016/j.ijmm.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 58.Widerström M, McCullough CA, Coombs GW, Monsen T, Christiansen KJ. 2012. A multidrug-resistant Staphylococcus epidermidis clone (ST2) is an ongoing cause of hospital-acquired infection in a Western Australian hospital. J Clin Microbiol 50:2147–2151. doi: 10.1128/JCM.06456-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monk IR, Shah IM, Xu M, Tan M-W, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grkovic S, Brown MH, Roberts NJ, Paulsen IT, Skurray RA. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J Biol Chem 273:18665–18673. doi: 10.1074/jbc.273.29.18665. [DOI] [PubMed] [Google Scholar]
- 61.Novick RP. 1989. Staphylococcal plasmids and their replication. Annu Rev Microbiol 43:537–563. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- 62.Sugiyama M, Kumagai T, Matsuo H, Bhuiyan MZ, Ueda K, Mochizuki H, Nakamura N, Davies JE. 1995. Overproduction of the bleomycin-binding proteins from bleomycin-producing Streptomyces verticillus and a methicillin-resistant Staphylococcus aureus in Escherichia coli and their immunological characterisation. FEBS Lett 362:80–84. doi: 10.1016/0014-5793(95)00218-X. [DOI] [PubMed] [Google Scholar]
- 63.Bruker Daltonics. 2015. Instructions for use: Bruker Matrix HCCA. Bruker Daltonics, Billerica, MA. [Google Scholar]
- 64.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial disk susceptibility tests (M100). Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 65.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically (M07-A9). Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 66.Clinical and Laboratory Standards Institute. 1999. Methods for determining bactericidal activity of antimicrobial agents (M26-A). Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 67.Applied Biosystems. 2014. AmpliTaq DNA polymerase protocol. Applied Biosystems, Waltham, MA. [Google Scholar]
- 68.Qiagen. 2016. QIAamp® DNA mini and blood mini handbook. Qiagen, Hilden, Germany. [Google Scholar]
- 69.Jolley KA, Bray JE, Maiden M. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kozyreva VK, Jospin G, Greninger AL, Watt JP, Eisen JA, Chaturvedi V. 2016. Recent outbreaks of shigellosis in California caused by two distinct populations of Shigella sonnei with either increased virulence or fluoroquinolone resistance. mSphere 1:e00344-16. doi: 10.1128/mSphere.00344-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kozyreva VK, Truong C-L, Greninger AL, Crandall J, Mukhopadhyay R, Chaturvedi V. 2017. Validation and implementation of Clinical Laboratory Improvements Act-compliant whole-genome sequencing in the public health microbiology laboratory. J Clin Microbiol 55:2502–2520. doi: 10.1128/JCM.00361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sreedharan S, Peterson LR, Fisher LM. 1991. Ciprofloxacin resistance in coagulase-positive and -negative staphylococci: role of mutations at serine 84 in the DNA gyrase A protein of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother 35:2151–2154. doi: 10.1128/AAC.35.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Galac MR, Stam J, Maybank R, Hinkle M, Mack D, Rohde H, Roth AL, Fey PD. 2017. Complete genome sequence of Staphylococcus epidermidis 1457. Genome Announc 5:e00450-17. doi: 10.1128/genomeA.00450-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andreote FD, Gullo MJM, de Souza Lima AO, Júnior WM, Azevedo JL, Araújo WL. 2004. Impact of genetically modified Enterobacter cloacae on indigenous endophytic community of Citrus sinensis seedlings. J Microbiol 42:169–173. [PubMed] [Google Scholar]
- 76.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett 73:133–138. doi: 10.1111/j.1574-6968.1992.tb05302.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.