Here, we identified mcr-4.3 in Acinetobacter baumannii, which had not been previously observed to carry an mcr gene. The mcr-4.3-harboring A. baumannii strain AB18PR065 was isolated from pig feces from a slaughterhouse in Guangdong Province of China.
KEYWORDS: Acinetobacter baumannii, colistin resistance, mcr-4.3
ABSTRACT
Here, we identified mcr-4.3 in Acinetobacter baumannii, which had not been previously observed to carry an mcr gene. The mcr-4.3-harboring A. baumannii strain AB18PR065 was isolated from pig feces from a slaughterhouse in Guangdong Province of China. The mcr-4.3-carrying pAB18PR065 is 25,602 bp in size and could not be transferred in conjugation, transformation, and electroporation experiments, as we did not find any conjugation-related genes therein. pAB18PR065 harbors two copies of type II toxin-antitoxin systems, which are functional in plasmid stabilization and maintenance. pAB18PR065 shares similarity only with one recently identified plasmid, pAb-MCR4.3 (35,502 bp), from a clinical A. baumannii strain. It is likely that the emergence of pAb-MCR4.3 was due to the insertion of an 11,386-bp, ISAba19-based, composite transposon into pAB18PR065. These data indicate that mcr-4.3 was captured by an A. baumannii-original plasmid via horizontal gene transfer.
INTRODUCTION
Colistin is considered one of the last-resort treatments against human infections caused by multidrug-resistant Gram-negative bacteria (1). The first plasmid-mediated colistin resistance gene, mcr-1 (1,626 bp), was identified on an IncI2 plasmid from Escherichia coli and Klebsiella pneumoniae in China (2). Since then, mcr-1 has been proven to be disseminated ubiquitously among Enterobacteriaceae strains (3). While mcr-1 remains the predominant plasmid-mediated colistin resistance gene, mcr-2 (1,617 bp) (4), mcr-3 (1,626 bp) (5), mcr-4 (1,626 bp) (6), mcr-5 (1,644 bp) (7), mcr-6 (1,617 bp) (8), mcr-7 (1,620 bp) (9), and mcr-8 (1,698 bp) (10) have been identified in various species from humans and animals. There are 56 mcr variant sequences available in GenBank to date, including mcr-1.1 to mcr-1.15, mcr-2.1, mcr-2.2, mcr-3.1 to mcr-3.24, mcr-4.1 to mcr-4.6, mcr-5.1 to mcr-5.3, mcr-6.1, mcr-7.1, mcr-8.1, mcr-8.2, and mcr-8.4 (11).
Acinetobacter baumannii, which is a member of the ESKAPE group of pathogens (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), is an emerging pathogen and a leading cause of nosocomial infections (12). As an opportunistic pathogen, Acinetobacter primarily attacks immunocompromised patients and causes infections such as ventilator-associated pneumonia, catheter-related bacteremia, wound and soft tissue infections, urinary tract infections, postsurgical endocarditis, and meningitis (13).
The mcr-4 gene was first described to be located on an 8,749-bp ColE10 plasmid in pig-origin Salmonella enterica from Italy. Transformants with a mcr-4-carrying plasmid exhibited colistin MICs of 2 μg/ml, 8-fold higher than that of the untransformed recipient (6). mcr-4.3 (originally named mcr-4.2) differs from mcr-4 in two missense mutations, which lead to codon changes of V179G and V236F (11). mcr-4.3 was first identified on a ColE10-type plasmid in 6 sequence type 54 (ST54) Enterobacter cloacae strains isolated from humans in Singapore in 2014 (14). This gene was also found on an 8,639-bp ColE plasmid in a clinical ST84 E. cloacae strain from China (15). Here, we identified a novel A. baumannii-original plasmid, pAB18PR065, carrying mcr-4.3 in an A. baumannii strain recovered from pig feces in China. The transferability of pAB18PR065 was investigated using conjugation experiments, transformation, and electroporation. The birth and evolution of pAB18PR065, based on a comparative analysis of multiple plasmids, are also discussed.
RESULTS AND DISCUSSION
Identification of the mcr-4.3-harboring A. baumannii strain.
Among the samples collected, we identified 84 mcr-harboring isolates from pigs (n = 63), patients (n = 10), and healthy individuals (n = 11). These isolates included E. coli (mcr-1, n = 75; mcr-3, n = 1; mcr-1 and mcr-3, n = 3), Shewanella spp. (mcr-4.3, n = 4), and A. baumannii (mcr-4.3, n = 1) (see Table S1 in the supplemental material). The mcr-4.3-harboring Shewanella sp. strains did not show resistance to colistin, and mcr-4.3 was found to be located on ∼140- to 220 kb plasmids (Fig. S1). However, A. baumannii strain AB18PR065 exhibited resistance to both colistin and polymyxin B, with MICs of 8 μg/ml, while remaining susceptible to almost all other antimicrobial agents tested (Table S2). This strain was recovered from pig feces in a slaughterhouse in Guangdong province on 25 May 2018. Sequence typing of AB18PR065 determined that alleles of the seven housekeeping genes were cpn60-48, gdhB-49, gltA-51, gpi-25, gyrB-90, recA-11, and rpoD-4 in the Oxford scheme (newly assigned to ST1929) and cpn60-3, fusA-3, gltA-16, pyrG-1, recA-13, rplB-1, and rpoB-15 in the Pasteur scheme (newly assigned to ST1303), both of which indicated a new ST (Table S3). Except for mcr-4.3, this strain harbors the naturally occurring blaOXA-51-like (blaOXA-430 with additional H7Q and S90G mutations) and blaADC-like (blaADC-184 with an additional P113S mutation) genes in A. baumannii (16). The ISAba1 element can provide a promoter to upregulate the expression of blaADC or blaOXA, leading to resistance to cephalosporins or carbapenems in A. baumannii (17, 18). No ISAba1 element was found upstream of the blaOXA-51-like and blaADC-like genes in AB18PR065. A retrospective screening of mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 using a strain collection of 90 colistin-resistant A. baumannii clinical isolates identified in 2012 to 2015 by our group did not find a mcr-carrying strain.
A. baumannii is considered a serious threat to human health because it causes serious infections that are associated with high morbidity and mortality rates. The challenge in treating Acinetobacter infections is primarily related to its high intrinsic tolerance to most antibiotics. Compromise of antibiotics to treat A. baumannii infections is due to the exceptionally low permeability to antibiotics, constitutively expressed efflux pumps, resistance genes harbored on genetic islands, and high genetic plasticity of Acinetobacter (19, 20). Polymyxins have remained effective against Acinetobacter infections (13). Available approaches use novel combinations of drugs, such as daptomycin-colistin-teicoplanin and intensified meropenem-polymyxin B (21). The presence of mcr-4.3 and its location on a plasmid would facilitate the spread of mcr-4.3 among Acinetobacter strains.
mcr-4.3 was found on pAB18PR065 without a flanking mobile element.
S1-nuclease digestion–pulsed-field gel electrophoresis (S1-PFGE) and Southern blotting hybridization revealed that mcr-4.3 was located on an ∼25-kb plasmid within AB18PR065 (Fig. 1). Using whole-genome sequencing (WGS) data, we found a scaffold of 25,602 bp in the genome assembly that carries mcr-4.3. BLASTn was used to analyze the close plasmid sequences. Unexpectedly, this contig showed a high level of similarity (94% coverage, 99% identity) only to a 35,502-bp mcr-4.3-carrying plasmid (pAb-MCR4.3 [GenBank accession no. CP033872]) identified in A. baumannii (Fig. S2). pAb-MCR4.3 was found in an ST233 (Oxford scheme) or ST79 (Pasteur scheme) A. baumannii strain that had been collected from the cerebrospinal fluid of a patient in Brazil in 2008; the plasmid sequence had been released in GenBank by 26 November 2018. We used the pAb-MCR4.3 sequence as the reference to design primers to close the plasmid. The complete sequence of the 25,602-bp plasmid pAB18PR065 was found to encode 31 open reading frames (ORFs).
FIG 1.
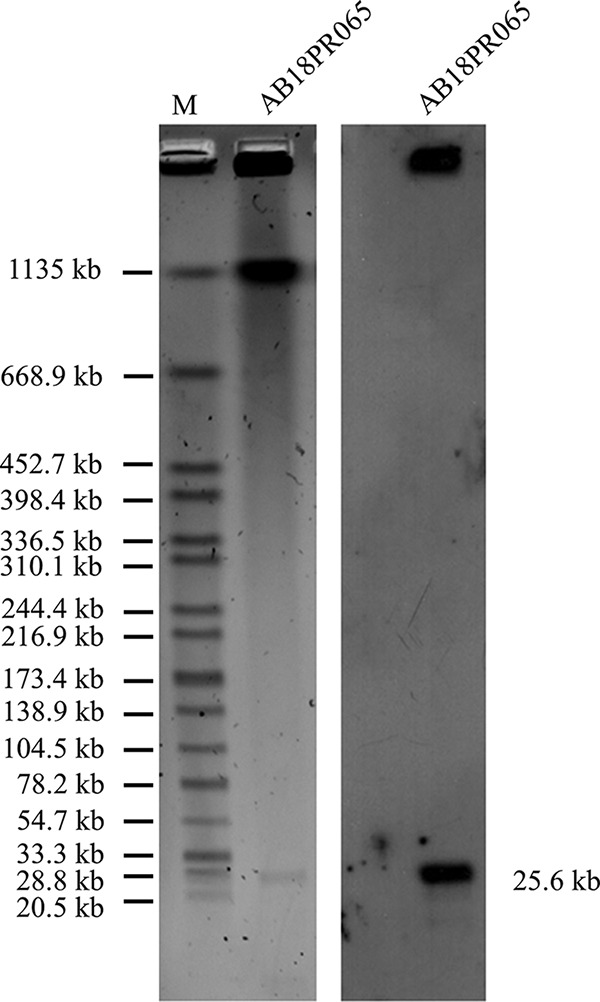
S1-PFGE and Southern hybridization with the mcr-4.3 probe. (Left) S1-PFGE map of the AB18PR065 strain. (Right) Southern blotting hybridization using the mcr-4.3 probe. M, Salmonella H9812 (New England BioLabs, Beverly, MA).
mcr-4.3 was found downstream of two ORFs that encode putative recombinase on pAB18PR065, whereas no mobile element was found adjacent to mcr-4.3. We identified a Tn3 element (3,087 bp) upstream of mcr-4.3 at a 1,528-bp distance. However, this transposon is more likely to mediate the location of type II toxin-antitoxin (TA) systems of parE and phd, since a 4,460-bp sequence including Tn3, parE, phd, and the two ORFs encoding recombinases has been found in various plasmids (Klebsiella oxytoca, pKOR-b08d [GenBank accession no. CP026279]; E. cloacae, pENT-2c5 [GenBank accession no. CP017992]; E. coli, pECO-dc1b [GenBank accession no. CP026207]). Of note, this sequence combined with the mcr-4.3 gene was found to be homologous to the genomic sequence of Shewanella frigidimarina NCIMB 400 (GenBank accession no. CP000447), and mcr-4.3 showed 100% nucleotide identity to the sequence in the genome. These data supported the conclusion that mcr-4 and its alleles originated from Shewanella frigidimarina (6), while the identification of mcr-4.3 in A. baumannii resonated with the indication of horizontal gene transfer of mcr-4.3, according to the other two reports (14, 15).
pAB18PR065 is a novel nonconjugative plasmid and carries plasmid stabilization elements.
pAB18PR065 encodes a replication initiation protein, RepB, that differs from the known Inc types. Phylogenetic analysis using replicon sequences revealed that repB within pAB18PR065 did not link with other plasmid types (Fig. S3). Conjugation experiments using AB18PR065 failed to transfer the plasmid into an E. coli recipient. Indeed, we did not find any conjugation-related ORFs on this plasmid. However, either heat shock transformation or electroporation using multiple recipient strains failed to yield pAB18PR065-harboring transformants. We could not determine whether transfer of pAB18PR065 could confer resistance to colistin. According to the other two studies, mcr-4.3 alone did not confer phenotypic resistance to colistin (14) and showed a lack of lipid A modification function (15). Recently, a study on the action and mechanism of MCR-4 showed that revertants of mcr-4.3 (either V179G or V236F) led to its expression and conferred colistin resistance of 8 μg/ml, together with the acquired function of lipid A modification (22).
Although pAB18PR065 is nonconjugative, it employs type II TA systems as a postsegregational killing operation to ensure plasmid stabilization and maintenance. Type II TA systems are generally composed of two genes, encoding a labile antitoxin and a stable toxin (23). These systems are thought to move from one genome to another by horizontal gene transfer, since TA loci were found in integrons in Vibrio cholerae and closely linked attC sites, which are regarded as the recombination sites of gene cassettes (24). pAB18PR065 has two TA gene families (parDE and phd/doc), and each has two copies. These systems preferentially guarantee the growth of pAB18PR065-carrying daughter cells in a bacterial population via killing of newborn bacteria that did not inherit a plasmid copy at cell division (25).
pAB18PR065 involvement in recombination in A. baumannii.
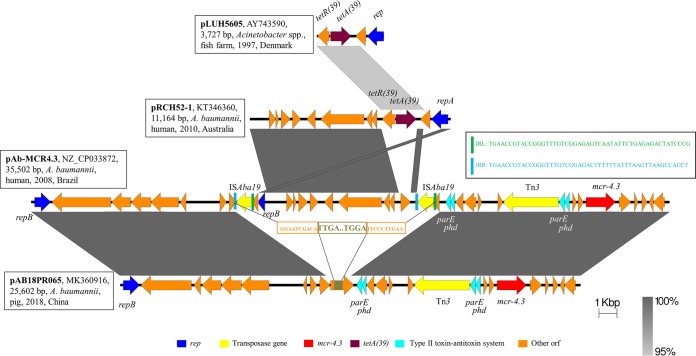
Given the fact that pAB18PR065 matched only pAb-MCR4.3, we analyzed their genetic contexts to understand the structural evolution (Fig. 2). The main difference between them was that pAB18PR065 lacked an 11,386-bp sequence that existed in pAb-MCR4.3, at the positions from 11,367 bp to 22,751 bp. Of note, this ∼11-kb sequence was flanked by two copies of ISAba19 in the same direction, and the sequences shared 99% nucleotide identity when ISAba19, with a unique 11,164-bp plasmid (pRCH52-1 [GenBank accession no. KT346360]) identified in clinical A. baumannii isolates recovered prior to 2010 in Australia (26), was excluded. ISAba19 is an A. baumannii-original insertion sequence with a length of 1,309 bp, which was found inserted into the blaOXA-78 gene in clinical A. baumannii isolates (27). Additionally, pRCH52-1 harbors a tetA(39)-tetR(39) unit (1,895 bp); the original study showed that tetA(39) was located on a plasmid from A. baumannii isolated in 1997, but only a 3,727-bp fragment was sequenced (28, 29). With all of this information, we hypothesized that the emergence of the mcr-4.3-carrying plasmids involved recombination in A. baumannii (Fig. 2). The process may be summarized as follows. The tetA(39)-tetR(39) unit was first inserted into a plasmid, causing the production of pRCH52-1. This unit was then transferred to other environments, which led to the appearance of a pRCH52-1-like plasmid without this unit. The transfer of two copies of intact ISAba19 constituted a composite transposon to translocate the pRCH52-1-like plasmid into a DNA polymerase-encoding ORF on pAB18PR065, leading to the formation of pAb-MCR4.3.
FIG 2.
Schematic presentation of the major structural features of pAB18PR065, in comparison with the reference plasmids pLUH5605, pRCH52-1, and pAb-MCR4.3. Areas shaded gray indicate homologous regions of ≥95% nucleotide sequence identity in the plasmid scaffold regions. ORFs are portrayed by arrows to indicate the direction of transcription and are colored based on their predicted gene functions. The figure is drawn to scale.
Conclusion.
We identified the colistin resistance gene mcr-4.3 on a novel nonconjugative plasmid, pAB18PR065, in A. baumannii from pig feces in China. There was no mobile element adjacent to mcr-4.3 on pAB18PR065, which suggests that mcr-4.3 might have been acquired in the A. baumannii community via horizontal gene transfer.
MATERIALS AND METHODS
Bacterial strains.
To detect mcr-carrying isolates in humans and animals, 185 pig fecal samples, 320 rectal swabs from healthy persons, and 170 rectal swabs from patients were collected from a pig slaughterhouse and hospitals in Guangdong Province, China, with one sample per individual, in April to June 2018. Samples were cultured by adding 3 ml of nutrient broth and incubated for 18 to 24 h at 37°C. Subsequently, the total DNA was extracted and screened for mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 using multiplex PCR, as described previously (30). Sanger sequencing using the positive amplicons was used to determine the mcr variants. Isolation of mcr-harboring strains from the PCR-positive samples was performed by inoculating the culture cells on LB agar containing colistin at 2 μg/ml, and different morphological colonies were selected to screen the mcr genes. Preliminary species identification was achieved by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonik GmbH, Bremen, Germany) and 16S rRNA sequencing, and the identification of A. baumannii species was confirmed by WGS.
Antimicrobial susceptibility testing.
MICs were determined for colistin, polymyxin B, tigecycline, ampicillin, amoxicillin-clavulanate, cefotaxime, ceftazidime, cefepime, gentamicin, amikacin, ertapenem, imipenem, meropenem, fosfomycin, and ciprofloxacin for all mcr-carrying isolates, using the agar dilution method except for colistin, for which used the broth dilution method was used, in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines (31). The results were interpreted according to CLSI instructions (31), and colistin resistance was defined according to EUCAST clinical breakpoints (32).
S1-PFGE and Southern blotting.
The plasmid and/or chromosomal locations of mcr-4.3 were determined by S1-PFGE (33), followed by Southern blotting hybridizations. Southern blotting hybridizations of plasmid DNA were performed with a digoxigenin-labeled mcr-4.3 probe, according to the manufacturer's instructions (Roche Diagnostics, Germany).
Plasmid conjugation, transformation, and electroporation.
Conjugation experiments were performed to test the transferability of the mcr-4.3-harboring plasmid, using streptomycin-resistant E. coli C600 as the recipient strain. Briefly, cultured cells of mcr-4.3-carrying isolates and E. coli C600 were mixed (at a ratio of 1:9) and subjected to overnight incubation (34). The mixture was then spread on LB agar plates containing sodium streptomycin (2,000 μg/ml) plus colistin (2 μg/ml) to select transconjugants, which were checked by PCR and Sanger sequencing. When the mcr-4.3-harboring plasmid could not be transferred by conjugation, we tried transformation and electroporation to harvest the transformants. Plasmid DNA was isolated from donor strains and transformed into E. coli DH5α cells or electroporated into E. coli DH10B, E. coli MG1655, E. coli C600, and A. baumannii ATCC 19606 cells, and transformants were selected on LB agar plates with 0.5, 1, and 2 μg/ml colistin.
Whole-genome sequencing.
The genomic DNA of the mcr-4.3-carrying isolates was extracted using the Qiagen Blood and Tissue kit (Qiagen, Hilden, Germany). DNA libraries were constructed with 350-bp paired-end fragments and sequenced using an Illumina HiSeq 2000 platform. Raw reads were assembled using SPAdes 3.10 (35). In silico multilocus sequence typing (MLST) was performed using MLST 1.8 (https://cge.cbs.dtu.dk/services/MLST). Antimicrobial resistance genes (ARGs) were identified by submitting the sequence to the Center for Genomic Epidemiology (https://cge.cbs.dtu.dk/services/ResFinder). The virulence factors of A. baumannii were identified using the VFDB database (36). Plasmid assembly was performed using plasmidSPAdes (37) and was confirmed by a PCR-based closure, using the primers listed in Table S4 in the supplemental material. Plasmid replicon types were detected using PlasmidFinder 1.3 (38). Insertion sequence elements were confirmed by searching in ISFinder (https://isfinder.biotoul.fr).
Accession number(s).
Genome assemblies of newly sequenced A. baumannii strain AB18PR065 (GenBank accession no. RZNI00000000) and the sequence of pAB18PR065 (GenBank accession no. MK360916) have been deposited in the NCBI database under BioProject no. PRJNA512224.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (grants 81830103 and 81722030), the Guangdong Natural Science Foundation (grant 2017A030306012), and the Science and Technology Planning Project of Guangdong (grant 2016A020219002).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00133-19.
REFERENCES
- 1.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 3.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 4.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 5.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 8.AbuOun M, Stubberfield EJ, Duggett NA, Kirchner M, Dormer L, Nunez-Garcia J, Randall LP, Lemma F, Crook DW, Teale C, Smith RP, Anjum MF. 2017. mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother 72:2745–2749. doi: 10.1093/jac/dkx286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. 2018. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother 73:1791–1795. doi: 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- 10.Wang XM, Wang Y, Zhou Y, Li JY, Yin WJ, Wang SL, Zhang SX, Shen JZ, Shen ZQ, Wang Y. 2018. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect 7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partridge SR, Di Pilato V, Doi Y, Feldgarden M, Haft DH, Klimke W, Kumar-Singh S, Liu J-H, Malhotra-Kumar S, Prasad A, Rossolini GM, Schwarz S, Shen J, Walsh T, Wang Y, Xavier BB. 2018. Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J Antimicrob Chemother 73:2625–2630. doi: 10.1093/jac/dky262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santajit S, Indrawattana N. 2016. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int 2016:2475067. doi: 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teo JWP, Kalisvar M, Venkatachalam I, Ng OT, Lin RTP, Octavia S. 2018. mcr-3 and mcr-4 variants in carbapenemase-producing clinical Enterobacteriaceae do not confer phenotypic polymyxin resistance. J Clin Microbiol 56:e01562-17. doi: 10.1128/JCM.01562-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chavda B, Lv J, Hou M, Chavda KD, Kreiswirth BN, Feng Y, Chen L, Yu F. 2018. Coidentification of mcr-4.3 and blaNDM-1 in a clinical Enterobacter cloacae isolate from China. Antimicrob Agents Chemother 62:e00649-18. doi: 10.1128/AAC.00649-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown S, Young HK, Amyes SG. 2005. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect 11:15–23. doi: 10.1111/j.1469-0691.2004.01016.x. [DOI] [PubMed] [Google Scholar]
- 17.Biglari S, Alfizah H, Ramliza R, Rahman MM. 2015. Molecular characterization of carbapenemase and cephalosporinase genes among clinical isolates of Acinetobacter baumannii in a tertiary medical centre in Malaysia. J Med Microbiol 64:53–58. doi: 10.1099/jmm.0.082263-0. [DOI] [PubMed] [Google Scholar]
- 18.Lopes BS, Amyes SG. 2012. Role of ISAba1 and ISAba125 in governing the expression of blaADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J Med Microbiol 61:1103–1108. doi: 10.1099/jmm.0.044156-0. [DOI] [PubMed] [Google Scholar]
- 19.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vila J, Marti S, Sanchez CJ. 2007. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J Antimicrob Chemother 59:1210–1215. doi: 10.1093/jac/dkl509. [DOI] [PubMed] [Google Scholar]
- 21.Lenhard JR, Bulitta JB, Connell TD, King-Lyons N, Landersdorfer CB, Cheah SE, Thamlikitkul V, Shin BS, Rao G, Holden PN, Walsh TJ, Forrest A, Nation RL, Li J, Tsuji BT. 2017. High-intensity meropenem combinations with polymyxin B: new strategies to overcome carbapenem resistance in Acinetobacter baumannii. J Antimicrob Chemother 72:153–165. doi: 10.1093/jac/dkw355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Hou M, Xu Y, Srinivas S, Huang M, Liu L, Feng Y. 2019. Action and mechanism of the colistin resistance enzyme MCR-4. Commun Biol 2:36. doi: 10.1038/s42003-018-0278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leplae R, Geeraerts D, Hallez R, Guglielmini J, Dreze P, Van Melderen L. 2011. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res 39:5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen RB, Gerdes K. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol 17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 26.Hamidian M, Holt KE, Pickard D, Hall RM. 2016. A small Acinetobacter plasmid carrying the tet39 tetracycline resistance determinant. J Antimicrob Chemother 71:269–271. doi: 10.1093/jac/dkv293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vali L, Dashti K, Opazo-Capurro AF, Dashti AA, Al Obaid K, Evans BA. 2015. Diversity of multi-drug resistant Acinetobacter baumannii population in a major hospital in Kuwait. Front Microbiol 6:743. doi: 10.3389/fmicb.2015.00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agerso Y, Guardabassi L. 2005. Identification of Tet 39, a novel class of tetracycline resistance determinant in Acinetobacter spp. of environmental and clinical origin. J Antimicrob Chemother 55:566–569. doi: 10.1093/jac/dki051. [DOI] [PubMed] [Google Scholar]
- 29.Agerso Y, Petersen A. 2007. The tetracycline resistance determinant Tet 39 and the sulphonamide resistance gene sulII are common among resistant Acinetobacter spp. isolated from integrated fish farms in Thailand. J Antimicrob Chemother 59:23–27. doi: 10.1093/jac/dkl419. [DOI] [PubMed] [Google Scholar]
- 30.Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, Guerra B, Malorny B, Borowiak M, Hammerl JA, Battisti A, Franco A, Alba P, Perrin-Guyomard A, Granier SA, De Frutos C, Escobar M-KS, Villa L, Carattoli A, Hendriksen RS. 2018. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill 23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; 28th informational supplement. M100-S28 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.European Committee on Antimicrobial Susceptibility Testing. 2019. Breakpoint tables for interpretation of MICs and zone diameters. EUCAST, Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf. [Google Scholar]
- 33.Sirichote P, Hasman H, Pulsrikarn C, Schonheyder HC, Samulioniene J, Pornruangmong S, Bangtrakulnonth A, Aarestrup FM, Hendriksen RS. 2010. Molecular characterization of extended-spectrum cephalosporinase-producing Salmonella enterica serovar Choleraesuis isolates from patients in Thailand and Denmark. J Clin Microbiol 48:883–888. doi: 10.1128/JCM.01792-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel L, Lagrutta E, Taylor P, Pham J, Nordmann P. 2010. Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob Agents Chemother 54:4914–4916. doi: 10.1128/AAC.00878-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Zheng D, Liu B, Yang J, Jin Q. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res 44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. 2016. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 38.Carattoli A, Zankari E, Garcia-Fernandez A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.