Abstract
The aim of the present study was to determine antimicrobial susceptibilities, biofilm production and, to discern a relationship between antimicrobial resistance, biofilm potential and virulence-related genes in strains of Yersinia entercocolitica biotype 1A. Thirty strains of Y. enterocolitica biotype 1A including clinical and non-clinical strains were investigated. Antimicrobial susceptibility for 15 antibiotics (representing different classes) was determined by disk-diffusion assay. Biofilm potential was determined on two different culture media using crystal violet assay. Also, a co-relation was studied between antimicrobial susceptibilities, biofilm production and virulence-related genes. All strains of biotype 1A produced biofilms and exhibited varied level of susceptibilities for different antibiotics. More than 60% of the strains were strong to moderate biofilm producers and, were exclusively associated with REP/ERIC clonal group B. Moderate and strong biofilm producers exhibited both sensitive and resistant phenotypes towards different antibiotics. Interestingly, weak biofilm producers were resistant to amoxicillin, amoxicillin-clavulanate and cefazolin. Analysis of antimicrobial susceptibilities, biofilm potential and virulence-related genes did not reveal any unequivocal relationships. The differential biofilm potential of Indian strains of Y. enterocolitica biotype 1A, suggests that biotype 1A strains are heterogeneous in nature.
Keywords: Microbiology, Molecular biology
1. Introduction
Bacteria which produce biofilms exhibit a significantly higher antimicrobial resistance and virulence than the planktonic forms [1, 2]. This suggests that antimicrobial resistance, biofilms and enhanced virulence might be related to each other. Although multiple mechanisms underlie the biofilm-associated enhancement of bacterial virulence and antimicrobial resistance the exact mechanisms are not understood well [3].
Yersinia enterocolitica is a gastrointestinal enteric pathogen which causes a variety of diseases in humans [4]. It can be classified into more than fifty serotypes and six biotypes, of which five (1B, 2, 3, 4, 5) have been considered pathogenic [5]. Due to the absence of pYV (plasmid for Yersinia virulence) and major chromosomal virulence genes, strains of biotype 1A were initially considered non-pathogenic [6]. However, strains of biotype 1A have been reported from clinical samples across the globe which indicates the pathogenic nature of these strains [7, 8]. Though, several studies have reported antimicrobial susceptibilities, virulence related genes and biofilm potential of many bacterial species, only a few have tried to discern the relationship between them in Y. enterocolitica. Further, information about a probable relationship between antimicrobial susceptibilities, virulence related genes and biofilm potential in Y. enterocolitica biotype 1A is scarce. Hence, in depth analysis of antimicrobial susceptibilities, virulence related genes and biofilm potential of Y. enterocolitica biotype 1A strains should be performed to understand the true pathogenic potential of these strains. Thus, the present study intended to study antimicrobial susceptibilities, virulence related genes and biofilm potential of Y. enterocolitica biotype 1A strains and, discern a relationship between them. Antimicrobial susceptibilities and biofilm potential of thirty strains of Y. enterocolitica biotype 1A were determined. The strains were isolated from various sources, like clinical samples, wastewater, pigs and pork. Though, the relationship between antimicrobials and biofilm formation has been studied in many members of the family Enterobacteriaceae, to the best of our knowledge, relationship between antimicrobials, biofilms and virulence has been reported in Y. enterocolitica biotype 1A strains for the first time.
2. Materials and methods
2.1. Bacterial strains
In the present study, 30 well-characterized strains of Y. enterocolitica biotype 1A were examined. These strains were authenticated and serotyped by the Yersinia National Reference Laboratory and WHO Collaborating Center, Pasteur Institute, Paris (France). These strains have also been deposited at the Pasteur Institute, Paris (France) and at the National Repository i.e. Microbial Type Culture Collection (MTCC) and Gene Bank located at Institute of Microbial Technology, Chandigarh, India. The strains were maintained in our laboratory at University of Delhi South Campus, New Delhi, India, on tryptone soy agar at 4°C. These strains have been isolated from various sources and genotyped using repetitive extragenic palindromic sequence (REP) and enterobacterial repetitive intergenic consensus sequence (ERIC) typing which revealed that these strains belonged to two clonal groups A or B [9]. The details of these strains viz. laboratory accession numbers, serotypes, source of isolation and clonal groups are mentioned in Table 2. Y. enterocolitica subsp. enterocolitica strain ATCC® 23715™ was included as a reference strain.
Table 2.
Biofilm formation, virulence-related genes and clonal groups based on REP/ERIC typing in Y. enterocolitica biotype 1A.
| Virulence-related genesc |
Biofilm Formation(OD values) Mean ± SEM (BF d) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. No. | Strain numbera | Serotype | Source of Isolation | Clonal groupb (based on REP-ERIC typing) | ystB, hreP, sat | myfA | fepA | MH medium |
TSB medium |
||
| 24 h | 48 h | 24 h | 48 h | ||||||||
| 1. | IP27359 | O:6,30–6,31 | Human stool | A | + | − | + | 0.76 ± 0.05(M) | 0.77 ± 0.01(M) | 0.35 ± 0.01(M) | 0.36 ± 0.00(M) |
| 2. | IP27360 | O:6,30–6,31 | Human stool | A | + | − | − | 0.66 ± 0.12(M) | 0.69 ± 0.10(M) | 0.26 ± 0.02(M) | 0.56 ± 0.16(S) |
| 3. | IP27362 | O:6,30–6,31 | Human stool | A | + | − | − | 0.60 ± 0.11(M) | 1.02 ± 0.00(S) | 0.15 ± 0.01(W) | 0.31 ± 0.04(M) |
| 4. | IP27363 | O:6,30–6,31 | Human stool | A | + | − | − | 0.38 ± 0.02(W) | 0.68 ± 0.02(M) | 0.14 ± 0.01(W) | 0.34 ± 0.07(M) |
| 5. | IP27364 | O:6,30–6,31 | Human stool | B | − | − | + | 0.79 ± 0.20(M) | 0.81 ± 0.10(M) | 0.34 ± 0.01(M) | 0.35 ± 0.01(M) |
| 6. | IP26311 | O:6,30–6,31 | Waste water | A | + | + | + | 0.39 ± 0.02(W) | 0.69 ± 0.02(M) | 0.36 ± 0.00(M) | 0.58 ± 0.09(S) |
| 7. | IP26144 | O:6,30–6,31 | Waste water | A | + | + | − | 0.38 ± 0.03(W) | 0.41 ± 0.02(W) | 0.28 ± 0.05(M) | 0.34 ± 0.05(M) |
| 8. | IP26145 | O:6,30–6,31 | Waste water | A | + | + | − | 0.39 ± 0.04(W) | 0.69 ± 0.04(M) | 0.14 ± 0.03(W) | 0.32 ± 0.09(M) |
| 9. | IP27425 | O:6,30 | Human stool | A | + | + | − | 0.62 ± 0.05(M) | 0.64 ± 0.05(M) | 0.10 ± 0.01(W) | 0.14 ± 0.01(W) |
| 10. | IP27431 | O:6,30 | Human stool | A | + | − | − | 1.05 ± 0.12(S) | 1.06 ± 0.01(S) | 0.30 ± 0.06(M) | 0.36 ± 0.02(M) |
| 11. | IP27433 | O:6,30 | Human stool | A | + | + | − | 0.40 ± 0.03(W) | 0.41 ± 0.01(W) | 0.30 ± 0.00(M) | 0.31 ± 0.00(M) |
| 12. | IP27434 | O:6,30 | Human stool | A | + | + | − | 0.51 ± 0.05(M) | 1.10 ± 0.03(S) | 0.12 ± 0.03(W) | 0.28 ± 0.03(M) |
| 13. | IP27481 | O:6,30 | Human stool | A | + | − | − | 1.14 ± 0.16(S) | 1.17 ± 0.10(S) | 0.35 ± 0.08(M) | 0.36 ± 0.09(M) |
| 14. | IP27404 | O:6,30 | Human stool | A | + | + | − | 1.23 ± 0.12(S) | 1.33 ± 0.10(S) | 0.15 ± 0.05(W) | 0.36 ± 0.05(M) |
| 15. | IP27408 | O:6,30 | Human stool | A | + | + | − | 0.74 ± 0.12(M) | 1.24 ± 0.10(S) | 0.15 ± 0.06(W) | 0.26 ± 0.02(M) |
| 16. | IP27426 | O:6,30 | Human stool | A | + | − | − | 0.80 ± 0.06(M) | 1.12 ± 0.05(S) | 0.33 ± 0.07(M) | 0.29 ± 0.11(M) |
| 17. | IP27428 | O:6,30 | Human stool | B | − | − | + | 1.14 ± 0.18(S) | 1.18 ± 0.08(S) | 0.31 ± 0.10(M) | 0.59 ± 0.11(S) |
| 18. | IP27430 | O:6,30 | Human stool | A | + | − | − | 0.54 ± 0.03(M) | 1.13 ± 0.01(S) | 0.18 ± 0.01(W) | 0.64 ± 0.14(S) |
| 19. | IP27432 | O:6,30 | Human stool | A | + | + | − | 0.50 ± 0.03(M) | 0.59 ± 0.03(M) | 0.28 ± 0.04(M) | 0.31 ± 0.10(M) |
| 20. | IP27484 | O:6,30 | Human stool | B | − | − | + | 0.58 ± 0.05(M) | 0.58 ± 0.05(M) | 0.33 ± 0.06(M) | 0.32 ± 0.03(M) |
| 21. | IP26261 | O:10-34 | Waste water | B | + | + | + | 0.80 ± 0.12(M) | 0.82 ± 0.10(M) | 0.59 ± 0.05(S) | 0.61 ± 0.04(S) |
| 22. | IP26147 | O:10-34 | Waste water | B | − | − | + | 0.73 ± 0.04(M) | 1.15 ± 0.04(S) | 0.32 ± 0.08(M) | 0.32 ± 0.03(M) |
| 23. | IP26316 | O:41,42 | Waste water | A | + | − | + | 0.30 ± 0.04(W) | 0.32 ± 0.04(W) | 0.11 ± 0.03(W) | 0.15 ± 0.01(W) |
| 24. | IP26151 | O:7,8-8-8,19 | Waste water | A | + | + | + | 1.02 ± 0.19(S) | 1.08 ± 0.09(S) | 0.36 ± 0.04(M) | 0.52 ± 0.00(S) |
| 25. | IP26153 | O:7,8-8-8,19 | Pork | B | − | − | + | 0.80 ± 0.12(M) | 1.07 ± 0.12(S) | 0.31 ± 0.01(M) | 0.35 ± 0.02(M) |
| 26. | - | ND | Pig throat | A | + | + | + | 0.81 ± 0.11(M) | 0.80 ± 0.11(M) | 0.34 ± 0.09(M) | 0.52 ± 0.00(S) |
| 27. | - | ND | Pig throat | A | + | + | − | 1.02 ± 0.08(S) | 1.07 ± 0.05(S) | 0.36 ± 0.05(M) | 0.73 ± 0.14(S) |
| 28. | IP27387 | NAG | Human stool | B | − | − | + | 0.74 ± 0.05(M) | 1.40 ± 0.03(S) | 0.36 ± 0.11(M) | 0.33 ± 0.03(M) |
| 29. | IP27388 | NAG | Human stool | A | + | + | + | 0.26 ± 0.02(W) | 0.80 ± 0.02(M) | 0.31 ± 0.09(M) | 0.35 ± 0.17(M) |
| 30. | IP27485 | NAG | Human stool | A | + | + | − | 0.70 ± 0.12(M) | 0.79 ± 0.12(M) | 0.31 ± 0.04(M) | 0.61 ± 0.20(S) |
ND – Non-determined, NAG – Non-agglutinable.
IP: Yersinia National Laboratory and WHO Collaboration Centre, Institute Pasteur, France.
The data on clonal groups based on REP/ERIC typing has been retrieved from Sachdeva and Virdi (2004).
The data on virulence-related genes has been retrieved from Bhagat and Virdi (2006).
BF – Biofilm formation; S – Strong biofilm producer; M – Moderate biofilm producer; W – Weak biofilm producer.
2.2. Antimicrobial susceptibility testing
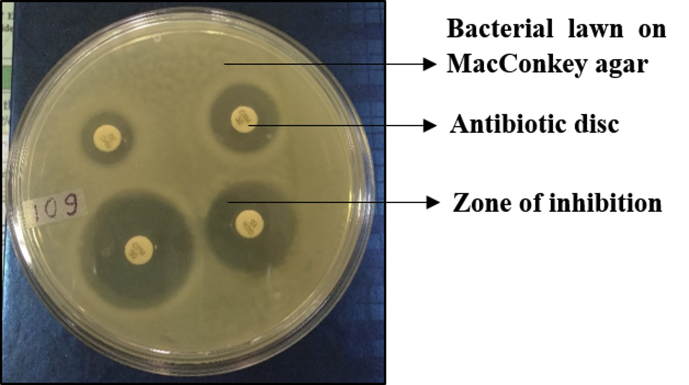
Antimicrobial susceptibilities of Y. enterocolitica biotype 1A strains were determined by disk diffusion test following the guidelines of Clinical and Laboratory Standards Institute [10]. Briefly, the bacteria were spread plated on Mac Conkey agar plate and the paper discs impregnated with different antibiotics were placed on the surface of agar plate. The plates were incubated at 37°C overnight and observed for the zone of inhibition (no bacterial growth) around the antibiotic disks. The diameter of the zone of inhibition around each antibiotic was measured and compared with a database of zone standards [10] and accordingly the bacterial strains were designated as antibiotic susceptible, intermediate susceptible or resistant (Fig. 1). The antibiotic disks (HiMedia, India) which were used in the present study represented major antibiotic classes, like β-lactam antibiotic - amoxicillin (AMX), β-lactam+β-lactamase inhibitor combination - amoxicillin-clavulanate (AMC), a first generation cephalosporin – cefazolin (CZ), a second generation cephalosporin - cefuroxime (CXM), third generation cephalosporins – cefoperazone (CPZ), cefixime (CFM), a fourth generation cephalosporin – cefepime (CPM) and a carbapenem – imipenem (IPM). The β-lactams and cephalosporins kill bacteria by inhibiting the synthesis of bacterial cell wall. Quinolones (inhibit bacterial DNA replication) were represented by ciprofloxacin (CIP) and aminoglycosides (inhibit bacterial protein synthesis) by tobramycin (TOB), gentamycin (GEM), and kanamycin (K). Erythromycin (E) represented the macrolides (inhibit bacterial protein synthesis), and furazolidone (FR) represented the nitrofurans (inhibit many bacterial enzyme systems). The results were interpreted as per the guidelines of Clinical Laboratory Standards Institute, CLSI [10]. The antibiotic susceptibility breakpoints suggested by CLSI in 2017 for most of the antibiotics are the same as in previous years, except for CPM, IPM and CZ [10].
Fig. 1.
Antimicrobial susceptibility testing of Y. enterocolitica biotype strains using disc diffusion test.
2.3. Assessment of biofilm formation
Assessment of biofilm formation by Y. enterocolitica biotype 1A strains was performed in two different broth media viz. Mueller-Hinton broth (MHB) and Tryptone Soya broth (TSB) following the published protocols, with slight modifications [11]. Briefly, 50 μl of overnight grown cultures at 28 °C with a cell density adjusted to 1 × 109 cells/ml were inoculated in 1.5 ml polypropylene microcentrifuge tubes (Tarsons, USA) containing 1 ml of MHB and TSB respectively, and incubated further at 28 °C for 24 h and 48 h each, without shaking. The medium was removed after 24 h and 48 h, and the microcentrifuge tubes were dried at 55 °C for 30 min. One ml of 0.1 % crystal violet (prepared in isopropanol: methanol: phosphate buffered saline in the ratio of 1:1:18, v/v) was added to all microcentrifuge tubes and incubated at room temperature for 30 min. After this, crystal violet was removed, followed by two washings with 1 ml of sterile distilled water. Microcentrifuge tubes were further dried at 55 °C for 30 min. The dye bound to the biofilm was dissolved in a 200 μl mixture of ethanol and acetone (4:1 v/v) and 100 μl of this mixture was added to a 96-well microtiter plate. Optical density was measured at 540 nm using ELISA plate reader (Thermo Scientific, USA). The strains were classified as non-adherent, weakly, moderately or strongly adherent based upon the ODs of bacterial biofilms. The cut-off OD (ODc) was defined as three standard deviations above the mean OD of the negative control (0.20 ± 0.00 for MHB and 0.09 ± 0.00 for TSB). Strains were classified using the following criteria:
| ≤ ODc or ODc < - ≥ 2x ODc: Non/weak biofilm producer |
| 2x ODc < - ≤ 4x ODc: Moderate biofilm producer |
| > 4x ODc: Strong biofilm producer |
Y. enterocolitica strain ATCC® 23715™ was included as a positive control. The assay was performed for each isolate in biological and technical triplicates and the average result was reported. The statistical significance was calculated using the Mann-Whitney U-test with R statistical package. A p-value<0.05 was considered as significant. A representative picture of the crystal violet assay has been presented as Fig. 2.
Fig. 2.
A representative picture showing crystal violet bound to biofilms formed by various bacterial strains.
2.4. Virulence-related genes
Information about the distribution of several virulence-related genes encoding adhesion (ail), transcriptional factor (virF), invasion (inv), enterotoxin (ystA, ystB, ystC), subtilisin/kexin-like protease (hreP), streptogramin acetyltransferase (sat), fimbriae (myfA), enterochelin receptor protein (fepA), insecticidal toxin complex-like protein (tccC), enterochelin ABC transporter (fepD), enterochelin esterase (fes) and Yersinia modulator (ymoA) was retrieved from an earlier study published from our laboratory [12].
3. Results and discussion
Results of antimicrobial susceptibility testing indicated that all the strains of biotype 1A were resistant to CZ - a first generation cephalosporin but sensitive to carbapenem - IPM, fluoroquinolone - CIP and an aminoglycoside-GEN (Table 1). The strains showed varied susceptibilities for other antibiotics. Among β-lactams, though only 8 (30%) strains were sensitive to AMX, a greater sensitivity (12 strains; 40%) was observed for β-lactam+β-lactamase inhibitor – AMC. Y. enterocolitica biotype 1A strains showed a good level of susceptibility level for the second generation cephalosporin – CXM, because 23 (76%) strains were sensitive to it. Biotype 1A strains showed different susceptibilities for the third generation cephalosporins – CPZ and CFX. Most of them exhibited intermediate susceptibility for CPZ (19 strains; 63%) while only 4 strains (13%) were sensitive to CFM. The susceptibility of biotype 1A strains for the fourth generation cephalosporin was better than that for other cephalosporins, as 15 (50%) strains were susceptible. Most of the biotype 1A strains (27 strains; 90%) were susceptible to TOB and KAN. Susceptibility of biotype 1A strains for E and FR was less, only 5 (16%) strains were susceptible to E and, 11 (36%) strains were susceptible to FR.
Table 1.
Antimicrobial susceptibilities of Y. enterocolitica biotype 1A strains.
|
Antimicrobial |
Antimicrobial susceptibility |
||
|---|---|---|---|
| Sensitive n | Intermediate n | Resistant n | |
| Amoxycillin | 8 | 3 | 19 |
| Amoxy-clavulanate | 12 | 6 | 12 |
| Cefazolin | - | - | 30 |
| Cefuroxime | 23 | 6 | 1 |
| Cefoperazone | 6 | 19 | 5 |
| Cefixime | 4 | 3 | 23 |
| Cefepime | 15 | 5 | 10 |
| Imipenem | 30 | - | - |
| Ciprofloxacin | 30 | - | - |
| Tobramycin | 27 | 1 | 2 |
| Gentamicin | 30 | - | - |
| Kanamycin | 28 | - | 2 |
| Erythromycin | 7 | 2 | 21 |
| Furazolidone | 11 | - | 19 |
n, denotes number of strains.
Results of the crystal violet assay indicated that all biotype 1A strains were capable of producing biofilms, even when cells were not exposed to any external stress, like depletion of iron, carbon, nitrogen or low concentration of oxygen. In MHB medium, after an incubation of 24 h, 11 (36.7%) strains were classified as strong biofilm producers, 12 (40%) strains as moderate biofilm producers, and 7 (23.3%) strains as weak biofilm producers. While after 48 h incubation in MHB medium, 14 (46.7%) strains were classified as strong biofilm producers, 13 (43.3%) strains as moderate biofilm producers, and 3 (10%) strains as weak biofilm producers (Table 2). In TSB medium, after an incubation of 24 h, only 1 (3.3%) strain was strong biofilm producer, 20 (66.7%) strains were moderate biofilm producers, and 9 (30%) strains were weak biofilm producers. After incubation of 48 h in TSB medium, 9 (30%) strains were found to be strong biofilm producers, 19 (63.3%) strains were moderate biofilm producers, and 2 (6.7%) strains were weak biofilm producers. A significant improvement in the biofilm forming capability of biotype 1A strains was observed after 48 h of incubation, in either type of culture medium (p < 0.05). The reference strain ATCC® 23715™ was found to be a moderate biofilm producer after 24 h incubation, but showed a strong biofilm forming potential after 48 h incubation, in both MHB and TSB. Thus, our results are in concordance with the results of earlier studies which reported that all strains of Y. enterocolitica produced biofilms [13, 14, 15]. A recent study however, has reported that strains of Y. enterocolitica biotype 1A isolated from meat samples either did not produce, or were weak producers of biofilms [8].
It was observed that those biotype 1A strains which were classified as weak biofilm producers were resistant to a β-lactam antibiotic - AMX, β-lactam and β-lactamase inhibitor combination – AMC, and the first generation cephalosporin – CZ. These strains however, displayed varied level of susceptibilities for other antibiotics. The strains classified as moderate or strong biofilm producers showed varied levels of susceptibility, exhibiting both sensitive and resistant phenotypes towards different antibiotics. A previous study reported also reported that biofilm forming Y. enterocolitica biotype 4 isolates were more resistant to antibiotics than the planktonic forms [15]. However, in the present study, it was observed that biotype 1A strains showed a range of susceptibilities to different antibiotics. Such associations of antimicrobial susceptibilities and biofilm formation in Y. enterocolitica biotype 1A strains emphasized an earlier suggestion that Y. enterocolitica biotype 1A represented a heterogeneous population of more than one subspecies [16].
Analysis of the biofilm forming potential of biotype 1A strains and genotyping using repetitive extragenic palindromic sequence (REP) and enterobacterial repetitive intergenic consensus sequence (ERIC) typing revealed an interesting observation. It was observed that, while REP/ERIC clonal group A was associated with weak, moderate and strong biofilm producers while strains strong and moderate biofilm producers belonged exclusively to the REP/ERIC clonal group B (Table 2). In an earlier study, it was reported that four virulence-associated genes viz. subtilisin/kexin-like protease (hreP), fimbriae (myfA), Yersinia stable toxin B (ystB), and streptogramin acetyltransferase (sat) were exclusively associated with strains of clonal group A [12]. Thus, clonal group appears to encompass strains exhibiting a diverse biofilm potential greater number of virulence associated genes. more No co-relation was observed in the biofilm forming potential of biotype 1A strains and source of isolation (clinical versus nonclinical). Thus, it might be inferred that the biofilm forming potential of biotype 1A strains might be related to the clonal groups, rather than the source from which they were isolated. Various studies have shown that Y. enterocolitica biotype 1A was genetically the most heterogeneous biotype of Y. enterocolitica, encompassing strains of numerous serotypes [17, 18]. Thus, it becomes reasonable to assume that biofilm formation in biotype 1A might be a strain-specific character which cannot be extrapolated to all strains of the same biotype, serotype, source of isolation.
An earlier study reported that Y. enterocolitica biotype 1A strains lacked both the pYV plasmid and some virulence genes located on chromosome encoding adhesion (ail), fimbriae (myfA), enterotoxin (ystA), TTSS and HPI [6]. However, biotype 1A strains have been associated with various food-borne outbreaks, indicating that these strains are pathogenic [7, 8]. An earlier study from our laboratory reported that certain virulence-related genes encoding adhesion (ail), transcriptional factor (virF), Yersinia stable toxins A and C (ystA, ystC) and, insecticidal toxin complex-like protein (tccC) were absent, while other virulence-related genes like Yersinia stable toxin B (ystB), subtilisin/kexin-like protease (hreP), streptogramin acetyltransferase (sat), fimbriae (myfA) and enterochelin receptor protein (fepA) were present in many strains of Y. enterocolitica biotype 1A [12]. Also, it was reported that four virulence-associated genes viz. subtilisin/kexin-like protease (hreP), fimbriae (myfA), Yersinia stable toxin B (ystB), and streptogramin acetyltransferase (sat) were exclusively associated with strains of clonal group A [12]. However, the results of the present study indicated that strong and moderate biofilm producers were associated with clonal group B.
The REP/ERIC-PCR based genotyping of Y. enterocolitica biotype 1A strains isolated from clinical and environmental sources revealed that despite the immense heterogeneity in the O-antigens, the strains clustered into limited clonal groups which suggest that there might be a limited genotypic diversity in biotype 1A strains studied [9]. Our study failed to reveal an unequivocal relationship between antimicrobial susceptibilities, biofilm production and virulence-related genes. However, our results indicated that there was a relationship between clonal groups and biofilm forming potential of Y. enterocolitica biotype 1A strains, with REP/ERIC clonal group B associated with strains exhibiting strong and moderate biofilm forming potential and REP/ERIC clonal group A with weak, moderate and strong biofilm producers. Also, it was observed that the biofilm potential of biotype 1A strains investigated in this study was different from biotype 1A strains isolated from other parts of the world [8]. These differences might be attributed to the heterogeneous nature of biotype 1A strains isolated from different parts of the world. However, further studies using serotypes of biotype 1A not represented in the present study are required to corroborate these findings and unravel the true virulence potential of biotype 1A strains.
Declarations
Author contribution statement
Neelja Singhal: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Anay Kumar Maurya: Performed the experiments; Analyzed and interpreted the data.
Nambram Somendro Singh: Performed the experiments.
Manish Kumar: Contributed reagents, materials, analysis tools or data.
Jugsharan Singh Virdi: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Neelja Singhal was supported by Science & Engineering Research Board (SERB) of Department of Science & Technology, New Delhi (SB/YS/LS-156/2014). Nambram Somendro Singh was supported by Indian Council of Medical Research, New Delhi (No. 80/919/2014-ECD-I).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Neelja Singhal, Email: neelja30@gmail.com.
Jugsharan Singh Virdi, Email: virdi_dusc@rediffmail.com.
References
- 1.Ito A., Taniuchi A., May T., Kawata K., Okabe S. Increased antibiotic resistance of Escherichia coli in mature biofilms. Appl. Environ. Microbiol. 2009;75(12):4093–4100. doi: 10.1128/AEM.02949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva A.J., Benitez J.A. Vibrio cholerae biofilms and Cholera pathogenesis. PLoS Neglected Trop. Dis. 2016;10(2) doi: 10.1371/journal.pntd.0004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giraud E., Rychlik I., Cloeckaert A. Antimicrobial resistance and virulence common mechanisms. Front. Microbiol. 2017;8:310. doi: 10.3389/fmicb.2017.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imoto A., Murano M., Hara A., Narabayashi K., Ogura T., Ishida K. Adult intussusception caused by Yersinia enterocolitica enterocolitis. Intern. Med. 2012;51:2545–2549. doi: 10.2169/internalmedicine.51.7890. [DOI] [PubMed] [Google Scholar]
- 5.Bottone E.J. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 1997;10(2):257–276. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tennant S.M., Grant T.H., Robins-Browne R.M. Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS Immunol. Med. Microbiol. 2003;38(2):127–137. doi: 10.1016/S0928-8244(03)00180-9. [DOI] [PubMed] [Google Scholar]
- 7.MacDonald E., Heier B.T., Nygård K., Stalheim T., Cudjoe K.S., Skjerdal T. Yersinia enterocolitica outbreak associated with ready-to-eat salad mix, Norway, 2011. Emerg. Infect. Dis. 2012;18(9):1496. doi: 10.3201/eid1809.120087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zadernowska A., Chajęcka-Wierzchowska W. Prevalence, biofilm formation and virulence markers of Salmonella sp. and Yersinia enterocolitica in food of animal origin in Poland. LWT - Food Sci. Technol. (Lebensmittel-Wissenschaft -Technol.) 2017;75:552–556. [Google Scholar]
- 9.Sachdeva P., Virdi J.S. Repetitive elements sequence (REP/ERIC)-PCR based genotyping of clinical and environmental strains of Yersinia enterocolitica biotype 1A reveal existence of limited number of clonal groups. FEMS Microbiol. Lett. 2004;240(2):193–201. doi: 10.1016/j.femsle.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 10.CLSI . 27th ed. Clinical and Laboratory Standards Institute; Wayne, PA: 2017. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. [Google Scholar]
- 11.Reeser R.J., Medler R.T., Billington S.J., Jost B.H., Joens L.A. Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl. Environ. Microbiol. 2007;73(6):1908–1913. doi: 10.1128/AEM.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhagat N., Virdi J.S. Distribution of virulence-associated genes in Yersinia enterocolitica biovar 1A correlates with clonal groups and not the source of isolation. FEMS Microbiol. Lett. 2007;266(2):177–183. doi: 10.1111/j.1574-6968.2006.00524.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim T.J., Young B.M., Young G.M. Effect of flagellar mutations on Yersinia enterocolitica biofilm formation. Appl. Environ. Microbiol. 2008;74(17):5466–5474. doi: 10.1128/AEM.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raczkowska A., Skorek K., Brzóstkowska M., Lasińska A., Brzostek K. Pleiotropic effects of a Yersinia enterocolitica ompR mutation on adherent-invasive abilities and biofilm formation. FEMS Microbiol. Lett. 2011;321(1):43–49. doi: 10.1111/j.1574-6968.2011.02308.x. [DOI] [PubMed] [Google Scholar]
- 15.Ioannidis A., Kyratsa A., Ioannidou V., Bersimis S., Chatzipanagiotou S. Detection of biofilm production of Yersinia enterocolitica strains isolated from infected children and comparative antimicrobial susceptibility of biofilm versus planktonic forms. Mol. Diagn. Ther. 2014;18(3):309–314. doi: 10.1007/s40291-013-0080-1. [DOI] [PubMed] [Google Scholar]
- 16.Sihvonen L.M., Jalkanen K., Huovinen E., Toivonen S., Corander J., Kuusi M. Clinical isolates of Yersinia enterocolitica biotype 1A represent two phylogenetic lineages with differing pathogenicity-related properties. BMC Microbiol. 2012;12(1):208. doi: 10.1186/1471-2180-12-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boghenbor K.K., On S.L., Kokotovic B., Baumgartner A., Wassenaar T.M., Wittwer M. Genotyping of human and porcine Yersinia enterocolitica, Yersinia intermedia, and Yersinia bercovieri strains from Switzerland by amplified fragment length polymorphism analysis. Appl. Environ. Microbiol. 2006;72(6):4061–4066. doi: 10.1128/AEM.01996-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard S.L., Gaunt M.W., Hinds J., Witney A.A., Stabler R., Wren B.W. Application of comparative phylogenomics to study the evolution of Yersinia enterocolitica and to identify genetic differences relating to pathogenicity. J. Bacteriol. 2006;188(10):3645–3653. doi: 10.1128/JB.188.10.3645-3653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]