Diarrheal illness is a major cause of morbidity and mortality throughout the world, yet the etiologic agent of many cases of gastrointestinal illness remains unspecified, often due to the lack of convenient, timely, and sensitive diagnostic testing. Although bulk fecal specimens remain the recommended specimen type for enteric culture, rectal swabs may be an option preferred by clinicians and patients due to the convenience and timing of collection.
KEYWORDS: diarrhea, enteric culture, fecal cup, gastrointestinal illness, rectal swab, stool culture
ABSTRACT
Diarrheal illness is a major cause of morbidity and mortality throughout the world, yet the etiologic agent of many cases of gastrointestinal illness remains unspecified, often due to the lack of convenient, timely, and sensitive diagnostic testing. Although bulk fecal specimens remain the recommended specimen type for enteric culture, rectal swabs may be an option preferred by clinicians and patients due to the convenience and timing of collection. However, the lack of data evaluating the sensitivity of rectal swabs compared to fecal specimens for detection of enteric pathogens precludes this specimen type from being recommended by national guidelines. In this study, we retrospectively reviewed 480 paired rectal swab and fecal specimens submitted for enteric culture to the Barnes-Jewish Hospital and St. Louis Children’s Hospital microbiology laboratories in St. Louis, MO, from 2002 to 2017. We report 32% positivity of paired specimens with an overall agreement of 93% and Cohen's κ of 0.84 (95% confidence interval, 0.78 to 0.89). Additionally, we evaluated the time to result from the time of patient presentation to the health care setting and demonstrate that rectal swabs have a significantly shorter time to an actionable result than bulk fecal specimens (median, 67.4 h versus 78.4 h, respectively; P < 0.001). These findings indicate that rectal swabs facilitate on-demand culture-based testing with a sensitivity comparable to that of fecal specimens and thus should be recommended for enteric bacterial culture when bulk fecal specimens are unavailable.
INTRODUCTION
Acute diarrheal illness is a significant cause of morbidity and mortality in children throughout the world. In 2015, 688 million illnesses and approximately 500,000 deaths worldwide were estimated to have occurred in children younger than 5 years of age, making acute diarrheal illness among the leading causes of death in this age group (1, 2). Annually in the United States, approximately 600,000 patients require hospitalization and nearly 5,000 individuals die due to acute gastroenteritis, with the estimated attributable cost being up to $145 billion (3). Although rotavirus is the leading cause of childhood diarrhea worldwide, Shigella and Shiga toxin-producing Escherichia coli (STEC) strains are among the major etiologies of moderate to severe diarrheal illness (1, 4). Complications related to the severe diarrhea caused by these agents, such as hemolytic-uremic syndrome (HUS), are medical emergencies necessitating a timely diagnosis in order to initiate appropriate management.
Currently, the Infectious Diseases Society of America (IDSA) guidelines recommend bulk fecal specimens (rather than rectal swabs) as the specimen of choice for diagnostic testing (5, 6); this recommendation is attributed to a perceived reduced sensitivity of swabs compared to bulk fecal specimens for enteric bacterial culture. Thus, some clinical microbiology laboratories reject swab specimens submitted for enteric culture, while other laboratories limit the use of rectal swabs to enteric culture in pediatric settings and/or when timely specimen submission is required (7). However, prompt initiation of diagnostic testing can be a challenge when a fresh bulk fecal specimen is not readily available. Difficulty in specimen collection likely contributes to reduced specimen submission in the setting of diarrhea, and transfer of the specimen into a sterile cup can also pose a substantial biohazard risk. Physicians have noted rectal swabs to be a preferable specimen type and suggest that the ability to routinely submit rectal swab specimens rather than bulk fecal specimens could potentially increase the rate of specimen submission (8). Further, the relative ease of collection and transport has also made rectal swabs a preferable specimen for outbreak investigations in the public health setting (9, 10). Recent developments in the design of the swabs have also made them more favorable for routine use. For example, flocked swabs and the widespread use of transport media have led to substantial improvements in culture recovery compared to that of traditional swab specimens (11, 12). There remains, however, a paucity of data evaluating the yield of rectal swab and fecal specimens for enteric culture.
Herein, our objective was to evaluate the yield of rectal swabs in comparison to that of bulk fecal specimens for enteric bacterial culture at an academic tertiary care medical center serving both adult and pediatric patients. The concordance and time to result for the two specimen types were evaluated.
MATERIALS AND METHODS
Study design.
We retrospectively reviewed the results for fecal and rectal swab specimens submitted for enteric bacterial cultures from Barnes-Jewish Hospital (BJH) and St. Louis Children's Hospital (SLCH) in St. Louis, MO. The laboratory information system (Cerner Millennium) was queried for enteric bacterial cultures for specimens submitted to the BJH and SLCH microbiology laboratories between January 2002 and November 2017. Patient location, encounter time, specimen type, specimen collection time, specimen laboratory receipt time, culture start time, and all preliminary and final culture results and times were captured. Nonpatient specimen cultures (proficiency specimens, test specimens, etc.), cultures of specimens other than feces or rectal swabs, and canceled cultures were removed from subsequent analyses. For paired specimen analysis, custom Python scripts were used to parse text data files and identify patients with rectal swab and bulk fecal specimens submitted for enteric bacterial culture within 72 h of each other.
Human studies approval.
This study was approved by the Human Research Protection Office of the Washington University in St. Louis School of Medicine Institutional Review Board (IRB; identification number 201712035).
Laboratory methods.
Rectal swabs and bulk fecal specimens (fresh or preserved in Cary-Blair transport medium) are routinely accepted for enteric bacterial culture. Samples were plated as soon as possible upon arrival in the laboratory on all shifts 7 days per week. Although the specific swab type and culture conditions did have modifications throughout the 15-year study period, rectal swab and bulk fecal specimens were evaluated using identical culture conditions throughout the study. From 2002 to 2011, rectal swab and fecal specimens from pediatric patients were cultured as previously described (13). From 2016 to the end of the study period, enteric cultures from adult and pediatric patients were routinely evaluated for Campylobacter, Escherichia coli O157, Salmonella, Shigella, Edwardsiella, Aeromonas, Plesiomonas, Yersinia, and Shiga-like toxin using blood agar, Hektoen enteric agar, MacConkey-sorbitol (SMAC) agar, Campylobacter blood agar, Yersinia selective agar, Gram-negative (GN) broth, and Shiga toxin immunoassay according to standard laboratory procedures. Throughout the entire study period, all isolates presumptively identified as E. coli O157 or aliquots of Shiga toxin-positive GN broths (if immunoassay results were positive and SMAC agar culture results were negative) were forwarded to the Missouri State Public Health Laboratory (Jefferson City, MO) for confirmation by PCR and serotyping.
Statistical analysis.
The chi-square test was used to compare the proportion of organisms identified in paired specimens. Cohen's κ statistic and 95% confidence intervals (CI) were calculated to assess the concordance of paired specimens. Mood's median test was used to compare the median age and the time to result between groups. All statistical analyses were performed with R (v.3.4.3) and RStudio (v.1.1.453) software.
RESULTS
Summary of cohort.
In total, 14,008 enteric bacterial cultures were performed for patients seen at BJH and SLCH between January 2002 and November 2017. The median (range) age of the patients and the percent positivity of each specimen type are summarized in Table 1. Overall, the median age of the patients was 31 years (range, 0.9 to >90 years) and 7.5% (1,055/14,008) of cultures were positive. Among all enteric cultures performed during the period of review, 94% were cultures of bulk fecal specimens, while only 6% of enteric bacterial cultures were cultures of rectal swabs. The median age of the patients who submitted rectal swab specimens was significantly lower than that of patients who submitted fecal specimens (15 versus 35 years, P < 0.001). During the study period, rectal swabs represented 15% of specimens submitted for enteric culture for pediatric patients but only 2.3% of specimens for adult patients, demonstrating that, in our experience, this specimen type is more commonly used for children. Notably, rectal swab specimens also had a higher positivity rate than bulk fecal specimens (13% versus 7.2%, P < 0.001).
TABLE 1.
Summary of characteristics of enteric cultures evaluated (2002 to 2017)
| Characteristic | All patients |
Adult patients (age ≥18 yr) |
Pediatric patients (age <18 yr) |
|||
|---|---|---|---|---|---|---|
| Bulk fecal | Swab | Bulk fecal | Swab | Bulk fecal | Swab | |
| No. (%) of cultures | ||||||
| Total | 13,142 (94) | 866 (6.2) | 9,355 (98) | 217 (2.3) | 3,787 (85) | 649 (15) |
| Positive | 945a (7.2) | 110a (13) | 646 (6.9) | 32 (15) | 299 (7.9) | 78 (12) |
| Negative | 12,197 (93) | 756 (87) | 8,709 (93) | 185 (85) | 3,488 (92) | 571 (88) |
| Median (range) age (yr) | 35b (0.9 to >90) | 15b (4 to >90) | 53 (18 to >90) | 24 (18 to >90) | 14 (0.9 to 17) | 14 (4 to 17) |
Chi-square test, P = 3.9 × 10−9.
Student’s t test, P < 2.2 × 10−16.
Analysis of paired specimens.
To assess the concordance between bulk fecal and rectal swab specimens, we identified patients with both specimen types submitted within 3 days of each other. During the 15-year review period, 480 individuals with 960 paired specimens were identified. Patients within the paired specimen cohort had a median age of 8 years (range, 0.5 to 55 years) and were primarily seen at SLCH (Table 2). For the patients in the paired specimen cohort, rectal swab specimens were submitted first in 430/480 instances (89.6%) and submission of rectal swab specimens preceded submission of bulk fecal specimens by a median of 9 h (interquartile range [IQR], 4 to 17 h), whereas submission of bulk specimens preceded submission of a swab by a median of 13 h (IQR, 3 to 32 h) in 22/480 (4.6%) instances.
TABLE 2.
Demographics of subjects with paired bulk fecal and rectal swab specimensa
| Characteristic | Value |
|---|---|
| Median (range) age (yr) | 8 (0.5–55) |
| No. of patients from the following hospital type: | |
| Academic, tertiary pediatric hospital | 477 |
| Academic, tertiary adult hospital | 3 |
| No. (%) of patients with the following encounter type: | |
| Emergency | 26 (5.4) |
| Inpatient | 260 (54) |
| Noninpatient | 194 (40) |
Paired specimens were defined as rectal swab and bulk fecal specimens collected from the same patient within 72 h.
The majority of cultures of both bulk fecal specimens (65%, 323/480) and rectal swabs (69%, 332/480) were negative for enteric bacterial pathogens. Culture results for positive paired specimens are summarized in Table 3. In positive enteric cultures, Shigella spp., Salmonella spp., Shiga toxin-producing E. coli, and Campylobacter spp. were the pathogens most commonly recovered from both bulk fecal specimens and rectal swabs, with comparable proportions of pathogens being detected between specimen types (chi-square test, P = 0.89). Culture results agreed for 447/480 paired cultures, demonstrating an overall percent agreement of 93.1% (Table 3). Of the 33 discordant pairs, an alternate pathogen was never identified; rather, cultures were positive for a pathogen in one specimen type and a pathogen was not detected in the other. More cultures were positive by bulk fecal specimen culture only than rectal swab only (21 versus 12). Negative percent agreement (NPA) and positive percent agreement (PPA) were 96.3% and 86.6%, respectively. As a measure of agreement, Cohen's κ statistic was calculated. κ was determined to be 0.84 (95% CI, 0.78 to 0.89), indicating very good agreement between specimen types. The distribution of pathogens reported for the paired cohort is representative of our overall epidemiology of enteric pathogens and was similar to previous reports from a similar study population which found Shigella species, Salmonella spp., Aeromonas spp., Campylobacter jejuni, and STEC to be the most common pathogens reported (13).
TABLE 3.
Overall and pathogen-specific agreement of paired bulk fecal and rectal swab specimens
| Pathogen and result obtained with rectal swab specimen | No. of bulk fecal specimens: |
|
|---|---|---|
| Positive | Negative | |
| Overall | ||
| Positive | 136 | 12 |
| Negative | 21 | 311 |
| κ (95% CI) | 0.84 (0.78–0.89) | |
| Shigella | ||
| Positive | 65 | 3 |
| Negative | 4 | 408 |
| κ (95% CI) | 0.94 (0.90–0.98) | |
| Salmonella | ||
| Positive | 37 | 3 |
| Negative | 5 | 435 |
| κ (95% CI) | 0.89 (0.82–0.97) | |
| Shiga toxin producing E. coli | ||
| Positive | 16 | 4 |
| Negative | 7 | 453 |
| κ (95% CI) | 0.73 (0.58–0.89) | |
| Campylobacter | ||
| Positive | 15 | 1 |
| Negative | 3 | 461 |
| κ (95% CI) | 0.88 (0.76–1.0) | |
| Aeromonas | ||
| Positive | 1 | 1 |
| Negative | 2 | 476 |
| κ (95% CI) | 0.40 (−0.28–1.1) | |
| Plesiomonas shigelloides | ||
| Positive | 1 | 0 |
| Negative | 0 | 479 |
| Yersinia | ||
| Positive | 1 | 0 |
| Negative | 0 | 479 |
Analysis by pathogen.
Of 960 cultures, 305 (32%) were positive for an enteric bacterial pathogen; 157 (51.5%) of these were from cultures of bulk fecal specimens and 148 (48.5%) were from cultures of rectal swabs. Shigella was the predominant pathogen identified, being identified in 55% (137/305) of all positive cultures. The high rates of Shigella among our cohort are consistent with our local epidemiology; several multiyear outbreaks were reported in our area during the study period (13). Shigella isolates were equally detected in both specimen types: 69 (43.9%) bulk fecal specimens and 68 (45.9%) rectal swabs. Likewise, the rates of detection among positive bulk fecal specimens versus rectal swab specimens were nearly equivalent for Salmonella (n = 42 [26.8%] versus n = 40 [27%]), Shiga toxin-producing E. coli (n = 23 [14.6%] versus n = 20 [13.5%]), and Campylobacter (n = 18 [11.5%] versus n = 16 [10.8%]). Aeromonas was detected in cultures of 5 specimens (3 [1.9%] bulk specimens and 2 [1.4%] rectal swab specimens), while Yersinia enterocolitica and Plesiomonas shigelloides were each detected in both the bulk specimen and the rectal swab of one paired set of specimens cultured. Fisher's exact test was performed to compare positivity rates between the specimen types, and no significant difference was observed for Shigella (P = 0.85), Salmonella (P = 0.82), Shiga toxin-producing E. coli (P = 0.64), and Campylobacter (P = 0.73). Pathogen-specific agreement was evaluated by the use of Cohen’s κ statistic for each pathogen detected (Table 3). Rectal swabs and bulk fecal specimens demonstrated excellent agreement for Shigella, Salmonella, and Campylobacter, with κ values of 0.94 (95% CI, 0.91 to 0.98), 0.89 (95% CI, 0.82 to 0.97), and 0.88 (95% CI, 0.76 to 1.0), respectively. Agreement was moderate to low for Shiga toxin-producing E. coli and Aeromonas, with κ values of 0.73 (95% CI, 0.58 to 0.89) and 0.4 (95% CI, −0.28 to 1.1), respectively. Among the paired cultures positive for Shiga toxin-producing E. coli, STEC was detected in both the rectal swab and bulk fecal specimens for 16/27 pairs (Table 3). Of the 11 pairs of specimens with discordant results, STEC was detected only in the rectal swabs in 4 cases and only in the bulk fecal specimens in 7 cases. Both the O157 and non-O157 E. coli serogroups were equally detected in either rectal swabs (n = 4) or bulk fecal specimens (n = 4). Interestingly, the three remaining pairs with discordant results in which Shiga toxin-producing E. coli was detected only in bulk fecal specimens were positive by the Shiga toxin assay (i.e., the specimens were negative using SMAC agar). Shiga toxin detection or recovery of a toxigenic E. coli isolate could not be confirmed by the state laboratory in 2/3 toxin-only reports, raising the possibility that the toxin-only detections could be false-positive results.
Time to result by specimen type.
Given the relative ease of collection and transport of rectal swab specimens compared to bulk fecal specimens, we hypothesized that collection of rectal swab specimens would result in a decrease in the time from patient presentation to a health care setting to reporting of the culture result. We defined the patient encounter time to result to be the time from patient admission to the time of a positive preliminary culture report or a final negative report. As expected, a significantly decreased time to result was found for positive cultures compared to negative cultures for both specimen types (for rectal swab specimens, 40.3 versus 72.1 h; for bulk fecal specimens, 46.5 versus 85.3 h; P < 0.001) (Fig. 1). The patient encounter time to result was significantly shorter for swab specimens than for bulk fecal specimens (67.4 versus 78.4 h; P = 0.005), though no difference in patient encounter time to result was observed between positive bulk fecal specimens and positive rectal swab specimens or negative bulk fecal specimens and negative rectal swab specimens (P = 0.22 and 0.58, respectively). There was no difference between specimen types in the total culture turnaround time, defined as the in-lab time from culture start to a positive preliminary result (data not shown). These data indicate that the differences in the time to a result from the time of the patient’s first encounter with the health care system were most likely due to a shorter time to collection.
FIG 1.
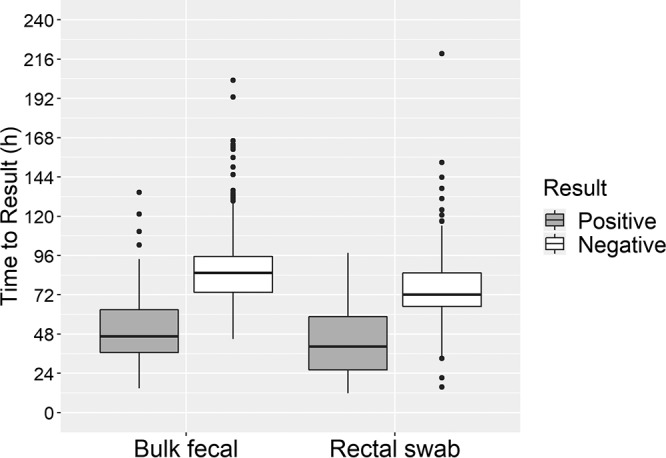
Time from patient encounter to a test result for rectal swab and bulk fecal specimens. The median and interquartile ranges for patient encounter time to result for paired enteric cultures are shown. Patient encounter time to result was defined as the time from admission to the time of a preliminary positive or final negative result. Median times were evaluated for statistical significance using Mood's median test. Rectal swabs had a significantly shorter time to result than bulk fecal specimens (67.4 versus 78.4 h, P = 0.005).
DISCUSSION
In this study, we evaluated the overall yield of rectal swabs compared to bulk fecal specimens for 480 paired specimens. We report that rectal swabs have a recovery of the most common bacterial pathogens comparable to that of bulk fecal specimens. Additionally, we found that rectal swab specimens had a decreased time from patient encounter with the health care system to the time of test result. While the high level of agreement between specimen types is consistent with recent reports in the literature, our study is the first, to our knowledge, to demonstrate a significant reduction in the time to a result with rectal swabs compared to bulk fecal specimens.
For more than 50 years, the convenience of rectal swabs for enteric culture has been noted, though early studies reported an inferiority of rectal swabs to bulk fecal specimens (14, 15). As such, rectal swab culture results are often viewed as presumptive, especially if they are negative, and testing with rectal swabs is frequently followed by testing with bulk fecal specimens. In their analysis of 513 prospectively collected paired rectal swab and bulk fecal specimens from symptomatic adult patients in the Philippines, Adkins and Santiago reported a positivity rate of 35.5%, and for nearly half of these positive cultures, a positive result was obtained only for either the bulk fecal specimen or the swab specimen (16). Our study demonstrated a similar positivity rate (32%) among the paired cohort of specimens reviewed; however, we reported a high overall concordance between the specimen types tested, indicating that either rectal swabs or bulk fecal specimens are adequate for enteric culture. Our increased rate of concordance is likely due to advances in the swab technologies used during the 15- to 33-year window between the two study periods as well as the detection of additional pathogens not assessed in our study, such as Vibrio species and rotavirus, which accounted for half of the organisms detected in positive cultures reported by Adkins and Santiago (16).
Similarly, Kotton and colleagues reported only moderate overall agreement between 155 paired rectal swab and bulk fecal specimens, with the sensitivity and the specificity of rectal swabs for the recovery of Salmonella from nine healthy volunteers administered a live-attenuated Salmonella enterica serovar Typhimurium vaccine reported to be 63.6% (95% CI, 0.54 to 0.71) and 90.0% (95% CI, 0.85 to 0.95), respectively (17). Interestingly, this study also reported that the sensitivity of rectal swabs compared to bulk fecal specimens was increased for specimens collected early in infection compared to those collected later in the infectious course (17).
Improvements in swab material and transport media have been reported to increase the sensitivity of rectal swab specimens. Rishmawi and colleagues demonstrated that rectal swabs transported with Copan M40 Transystem Amies medium had an excellent overall sensitivity of 98% (95% CI, 89.5 to 99.7%) compared to paired bulk fecal specimens for detecting the common enteric pathogens Salmonella, Shigella, and Campylobacter from symptomatic children (<10 years) (9). Likewise, Freedman and colleagues recently reported good agreement for paired cultures of rectal swab and bulk fecal specimens submitted from symptomatic children in an ambulatory care setting, with an overall Cohen’s κ value of 0.76 (95% CI, 0.71 to 0.80) (18). Despite the lower comparative yield of rectal swabs than bulk specimens, the authors reported that rectal swabs were submitted more often, resulting in a 10% increase in overall pathogen yield compared to that from bulk fecal specimens. Interestingly, this study also reported a greater concordance between specimen types for viral pathogens than for bacterial pathogens, with κ values of 0.82 (95% CI, 0.79 to 0.86) and 0.74 (95% CI, 0.68 to 0.80), respectively (18). While we did not evaluate the recovery of viral and parasitic pathogens in rectal swab and bulk fecal specimens in this study, others have also reported favorable sensitivity of flocked rectal swabs for the detection of norovirus, rotavirus, and Cryptosporidium using molecular methods (19, 20).
Given the need to rapidly diagnose E. coli O157 and other STEC infections to initiate measures that could prevent hemolytic-uremic syndrome, the low concordance between rectal swab and bulk fecal specimens reported in our study could be of concern. However, this low percent agreement is likely due to false-positive Shiga toxin immunoassay results, as Shiga toxin detection by PCR or recovery of an STEC isolate could not be confirmed by the state public health laboratory for 2/3 cultures positive for Shiga toxin only by immunoassay. Simultaneous testing for STEC via immunoassay or PCR and E. coli O157 culture on selective and differential media has been shown to increase the sensitivity of STEC detection and is recommended by the CDC (21). While the sensitivity and specificity of multiple tests have been reported to be >90%, false-positive immunoassay results have been reported, supporting the need for confirmatory testing (22). Our data reassuringly demonstrate that when likely false-positive Shiga toxin assay results were excluded, non-O157 and O157 STEC isolates were equally detected in both rectal swab and bulk fecal specimens.
Unlike previously published studies, where specimen collection, transport, and storage were tightly controlled, this study represents an analysis of real-world data, as the paired specimens evaluated by culture were collected during routine clinical care. Despite significant variability in organism burden, physician practice, stage of infection, and preanalytical variability, we report a sensitivity and a specificity of rectal swabs higher than those presented in previous reports, with rates of 86.6% (95% CI, 0.86 to 0.87) and 96.3% (95% CI, 0.96 to 0.96), respectively. Our study is also unique in that we assessed the impact of specimen type on patient encounter time to result. We report that rectal swab specimens had a significantly shorter time to result than bulk fecal specimens. This is likely due to the on-demand collection of rectal swab specimens immediately at the patient encounter, as no difference in laboratory analytical turnaround time was noted between specimen types. Since rectal swabs are also amenable to automated testing, it is likely that, when coupled with the on-demand specimen collection, the shorter incubation time due to improved culture conditions reported with automated culture workup will further reduce the time to culture results (23, 24). The reported length of the emergency department (ED) stay for children with gastroenteritis ranges from 125 to 372 min (25–27), and laboratory testing has been shown to delay patient discharge (27). These findings have the potential to decrease the length of stay in the ED, where 1.7 million children with gastroenteritis present each year (25), since the more timely specimen collection would likely lead to an earlier discharge or inpatient admission, freeing up limited ED resources.
Currently, rectal swab-based collection devices have not been cleared for use with any of the FDA-approved multiplex molecular platforms for gastrointestinal pathogens. Though some of these molecular tests can have an in-lab turnaround time of 1 to 5 h, the rapid turnaround time afforded by these methods remains subject to timely specimen collection. It is possible that future studies corroborating the sensitivity of rectal swabs for gastrointestinal molecular tests can facilitate the use of these tests in real-time patient management in the ED and outpatient settings.
The overall strengths of this study include an extensive study period spanning 15 years and identification of a large sample of paired specimens. Also, the culture results reviewed in this analysis were from clinical specimens collected during routine clinical care representing a spectrum of collection, transport, and storage conditions. Thirty-two percent of the paired cultures were positive for a bacterial pathogen, with broad representation of the most common bacterial causes of acute gastroenteritis in the United States, including Shigella, Salmonella, Campylobacter, and Shiga toxin-producing E. coli. The positivity rate for the paired cultures was much higher than that for all cultures performed during the study period (32% versus 7.5%). Thus, it is possible that the paired specimen cohort may be biased toward patients with a more severe clinical presentation, warranting continued observation and/or inpatient admission and submission of additional specimens. A limitation of our study is that very few or none of the cultures in this analysis were positive for less common enteric bacterial pathogens, such as Aeromonas, Plesiomonas shigelloides, Yersinia enterocolitica, and Vibrio, precluding comparison of the use of bulk fecal and rectal swab specimens for the detection of these pathogens. Additional studies with pathogens not well represented in our data set are warranted to explore the possibility of potential pathogen-specific differences in recovery with rectal swabs. This study was also limited to enteric bacterial culture and did not evaluate the recovery of viral and parasitic causes of gastroenteritis or the yield of molecular testing between bulk fecal and rectal swab specimens. Though the collection devices and culture conditions used varied throughout the 15-year study period, potentially impacting pathogen recovery (9, 28), this variance was accounted for, in that each rectal swab and bulk fecal specimen pair was processed under identical culture workup conditions. Since this study was also performed at an academic medical hospital, study findings may not extrapolate to community settings, and it remains uncertain how this will translate to the era of molecular diagnostics for stool pathogens.
In conclusion, we report that rectal swabs have recovery comparable to that of bulk fecal specimens for enteric bacterial culture. Despite variabilities in collection devices, culture conditions, and specimen quality, we demonstrate overall agreement of greater than 90% between specimen types with a Cohen's κ value of 0.84 (95% CI, 0.78 to 0.89). We also report a decreased patient encounter time to result with rectal swabs compared to bulk fecal specimens. These data suggest that rectal swabs have a performance equivalent to that of bulk fecal specimens and should not be rejected from clinical microbiology laboratories for enteric bacterial culture. Due to timely specimen collection, the routine use of rectal swabs for enteric culture can potentially improve clinical work flows and outcomes.
ACKNOWLEDGMENTS
We thank the staff of the Barnes-Jewish Hospital and former St. Louis Children's Hospital clinical microbiology laboratories for their efforts to provide timely and accurate results for patient care.
REFERENCES
- 1.Kotloff KL, Platts-Mills JA, Nasrin D, Roose A, Blackwelder WC, Levine MM. 2017. Global burden of diarrheal diseases among children in developing countries: incidence, etiology, and insights from new molecular diagnostic techniques. Vaccine 35:6783–6789. doi: 10.1016/j.vaccine.2017.07.036. [DOI] [PubMed] [Google Scholar]
- 2.GBD 2015 Mortality and Causes of Death Collaborators. 2016. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim HS, Rotundo L, Nasereddin T, Ike A, Song D, Babar A, Feurdean M, Demyen MF, Ahlawat SK. 2017. Time trends and predictors of acute gastroenteritis in the United States: results from National Health and Nutrition Examination Survey 2005-2014. J Clin Gastroenterol 51:693–700. doi: 10.1097/MCG.0000000000000907. [DOI] [PubMed] [Google Scholar]
- 4.GBD Diarrhoeal Diseases Collaborators. 2017. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 17:909–948. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller JM, Binnicker MJ, Campbell S, Carroll KC, Chapin KC, Gilligan PH, Gonzalez MD, Jerris RC, Kehl SC, Patel R, Pritt BS, Richter SS, Robinson-Dunn B, Schwartzman JD, Snyder JW, Telford S, Theel ES, Thomson RB, Weinstein MP, Yao JD. 2018. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2018 update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin Infect Dis 67:e1–e94. doi: 10.1093/cid/ciy381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shane AL, Mody RK, Crump JA, Tarr PI, Steiner TS, Kotloff K, Langley JM, Wanke C, Warren CA, Cheng AC, Cantey J, Pickering LK. 2017. 2017 Infectious Diseases Society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis 65:1963–1973. doi: 10.1093/cid/cix959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphries RM, Linscott AJ. 2015. Laboratory diagnosis of bacterial gastroenteritis. Clin Microbiol Rev 28:3–31. doi: 10.1128/CMR.00073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sperou AJ, Dickinson JA, Lee B, Louie M, Pang X-L, Chui L, Vanderkooi OG, Freedman SB. 2017. Physician perspectives on vaccination and diagnostic testing in children with gastroenteritis: a primary care physician survey. Paediatr Child Health 22:317–321. doi: 10.1093/pch/pxx078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rishmawi N, Ghneim R, Kattan R, Ghneim R, Zoughbi M, Abu-Diab A, Turkuman S, Dauodi R, Shomali I, Issa A-R, Siriani I, Marzouka H, Schmid I, Hindiyeh MY. 2007. Survival of fastidious and nonfastidious aerobic bacteria in three bacterial transport swab systems. J Clin Microbiol 45:1278–1283. doi: 10.1128/JCM.02110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arvelo W, Hall AJ, Estevez A, Lopez B, Gregoricus N, Vinjé J, Gentsch JR, Parashar U, Lindblade KA. 2013. Diagnostic performance of rectal swab versus bulk stool specimens for the detection of rotavirus and norovirus: implications for outbreak investigations. J Clin Virol 58:678–682. doi: 10.1016/j.jcv.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Horn KG, Audette CD, Tucker KA, Sebeck D. 2008. Comparison of 3 swab transport systems for direct release and recovery of aerobic and anaerobic bacteria. Diagn Microbiol Infect Dis 62:471–473. doi: 10.1016/j.diagmicrobio.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Van Horn KG, Audette CD, Sebeck D, Tucker KA. 2008. Comparison of the Copan ESwab system with two Amies agar swab transport systems for maintenance of microorganism viability. J Clin Microbiol 46:1655–1658. doi: 10.1128/JCM.02047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schindler EI, Sellenriek P, Storch GA, Tarr PI, Burnham C-A. 2014. Shiga toxin-producing Escherichia coli: a single-center, 11-year pediatric experience. J Clin Microbiol 52:3647–3653. doi: 10.1128/JCM.01231-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas M. 1954. Disadvantages of the rectal swab in diagnosis of diarrhoea. Br Med J 2:394–396. doi: 10.1136/bmj.2.4884.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaughnessy HJ, Friewer F, Snyder A. 1948. Comparative efficiency of rectal swabs and fecal specimens in detecting typhoid and salmonella cases and carriers. Am J Public Health Nations Health 38:670–675. doi: 10.2105/AJPH.38.5_Pt_1.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adkins HJ, Santiago LT. 1987. Increased recovery of enteric pathogens by use of both stool and rectal swab specimens. J Clin Microbiol 25:158–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotton CN, Lankowski AJ, Hohmann EL. 2006. Comparison of rectal swabs with fecal cultures for detection of Salmonella typhimurium in adult volunteers. Diagn Microbiol Infect Dis 56:123–126. doi: 10.1016/j.diagmicrobio.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Freedman SB, Xie J, Nettel-Aguirre A, Lee B, Chui L, Pang X-L, Zhuo R, Parsons B, Dickinson JA, Vanderkooi OG, Ali S, Osterreicher L, Lowerison K, Tarr PI, Chuck A, Currie G, Eltorki M, Graham T, Jiang J, Johnson D, Kellner J, Lavoie M, Louie M, MacDonald J, MacDonald S, Simmonds K, Svenson L, Tellier R, Drews S, Talbot J. 2017. Enteropathogen detection in children with diarrhoea, or vomiting, or both, comparing rectal flocked swabs with stool specimens: an outpatient cohort study. Lancet Gastroenterol Hepatol 2:662–669. doi: 10.1016/S2468-1253(17)30160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidler JA, Käch R, Noppen C, Dangel M, Battegay M, Bingisser R, Dubuis O, Widmer AF. 2014. Rectal swab for detection of norovirus by real-time PCR: similar sensitivity compared to faecal specimens. Clin Microbiol Infect 20:O1017–O1019. doi: 10.1111/1469-0691.12723. [DOI] [PubMed] [Google Scholar]
- 20.Goldfarb DM, Steenhoff AP, Pernica JM, Chong S, Luinstra K, Mokomane M, Mazhani L, Quaye I, Goercke I, Mahony J, Smieja M. 2014. Evaluation of anatomically designed flocked rectal swabs for molecular detection of enteric pathogens in children admitted to hospital with severe gastroenteritis in Botswana. J Clin Microbiol 52:3922–3927. doi: 10.1128/JCM.01894-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gould LH, Bopp C, Strockbine N, Atkinson R, Baselski V, Body B, Carey R, Crandall C, Hurd S, Kaplan R, Neill M, Shea S, Somsel P, Tobin-D'Angelo M, Griffin PM, Gerner-Smidt P, Centers for Disease Control and Prevention (CDC). 2009. Recommendations for diagnosis of Shiga toxin-producing Escherichia coli infections by clinical laboratories. MMWR Recommend Rep 58(RR-12):1–14. [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention (CDC). 2006. Importance of culture confirmation of Shiga toxin-producing Escherichia coli infection as illustrated by outbreaks of gastroenteritis—New York and North Carolina, 2005. MMWR Morb Mortal Wkly Rep 55:1042–1045. [PubMed] [Google Scholar]
- 23.Graham M, Tilson L, Streitberg R, Hamblin J, Korman TM. 2016. Improved standardization and potential for shortened time to results with BD KiestraTM total laboratory automation of early urine cultures: a prospective comparison with manual processing. Diagn Microbiol Infect Dis 86:1–4. doi: 10.1016/j.diagmicrobio.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Mutters NT, Hodiamont CJ, de Jong MD, Overmeijer HPJ, van den Boogaard M, Visser CE. 2014. Performance of Kiestra total laboratory automation combined with MS in clinical microbiology practice. Ann Lab Med 34:111–117. doi: 10.3343/alm.2014.34.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carson RA, Mudd SS, Madati PJ. 2017. Evaluation of a nurse-initiated acute gastroenteritis pathway in the pediatric emergency department. J Emerg Nurs 43:406–412. doi: 10.1016/j.jen.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Hendrickson MA, Zaremba J, Wey AR, Gaillard PR, Kharbanda AB. 2018. The use of a triage-based protocol for oral rehydration in a pediatric emergency department. Pediatr Emerg Care 34:227–232. doi: 10.1097/PEC.0000000000001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James C, Harper M, Johnston P, Sanders B, Shannon M. 2009. Effect of trainees on length of stay in the pediatric emergency department. Acad Emerg Med 16:859–865. doi: 10.1111/j.1553-2712.2009.00480.x. [DOI] [PubMed] [Google Scholar]
- 28.Bourbeau P. 2005. Just a swab you say? Balderdash! Clin Microbiol Newsl 27:19–23. doi: 10.1016/j.clinmicnews.2005.01.003. [DOI] [Google Scholar]


