Haemophilus influenzae exclusively colonizes the human nasopharynx and can cause a variety of respiratory infections as well as invasive diseases, including meningitis and sepsis. A key virulence determinant of H. influenzae is the polysaccharide capsule, of which six serotypes are known, each encoded by a distinct variation of the capsule biosynthesis locus (cap-a to cap-f).
KEYWORDS: Haemophilus influenzae, capsule, genomics, serotyping, surveillance
ABSTRACT
Haemophilus influenzae exclusively colonizes the human nasopharynx and can cause a variety of respiratory infections as well as invasive diseases, including meningitis and sepsis. A key virulence determinant of H. influenzae is the polysaccharide capsule, of which six serotypes are known, each encoded by a distinct variation of the capsule biosynthesis locus (cap-a to cap-f). H. influenzae type b (Hib) was historically responsible for the majority of invasive H. influenzae disease, and its prevalence has been markedly reduced in countries that have implemented vaccination programs targeting this serotype. In the postvaccine era, nontypeable H. influenzae emerged as the most dominant group causing disease, but in recent years a resurgence of encapsulated H. influenzae strains has also been observed, most notably serotype a. Given the increasing incidence of encapsulated strains and the high frequency of Hib in countries without vaccination programs, there is growing interest in genomic epidemiology of H. influenzae. Here we present hicap, a software tool for rapid in silico serotype prediction from H. influenzae genome sequences. hicap is written using Python3 and is freely available at https://github.com/scwatts/hicap under the GNU General Public License v3 (GPL3). To demonstrate the utility of hicap, we used it to investigate the cap locus diversity and distribution in 691 high-quality H. influenzae genomes from GenBank. These analyses identified cap loci in 95 genomes and confirmed the general association of each serotype with a unique clonal lineage, and they also identified occasional recombination between lineages that gave rise to hybrid cap loci (2% of encapsulated strains).
INTRODUCTION
Haemophilus influenzae is a pleomorphic Gram-negative bacterium that is exclusive to humans, typically colonizing the upper respiratory tract and occasionally causing disease. It was the first free living organism to be completely sequenced and served as a stepping stone toward DNA sequencing technology development in preparation for the Human Genome Project (1). H. influenzae is often classified on the basis of the production and antigenicity of polysaccharide capsule. Strains that produce capsule are divided into six serotypes (H. influenzae a to f [Hia to Hif]), and nonencapsulated strains are designated nontypeable H. influenzae (NTHi) (2).
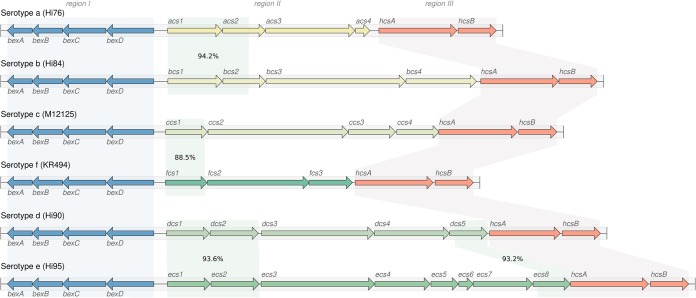
Biosynthesis of the polysaccharide capsule is controlled by the cap loci (cap-a to cap-f), each of which includes three contiguous but functionally distinct regions (I, II, and III) (Fig. 1). Regions I and III are common to all six cap loci and are associated with cellular transport (bex operon, region I) and posttranslational processing (hcs operon, region III) (3, 4). Region II encodes several genes involved in polysaccharide biosynthesis that are specific to each serotype (Fig. 1) (5–9). The cap locus is regularly subject to duplication, deletion, and interruption (10, 11). For example the cap-b locus is often duplicated, creating two tandem copies of the locus flanked by IS1016, and regularly coincides with a 1.2-kbp deletion of the terminal bexA-IS1016 copy (9). The arrangement and copy number of cap locus genes also have clinical relevance, as certain structural variants are associated with increased levels of virulence (12).
FIG 1.
Schematic representation of the six known H. influenzae cap loci. Capsule nucleotide sequences and annotations were collected from genome assemblies representing each of the six serotypes. Shading indicates homologous regions between reference loci as determined by BLAST identity values shown in region II. Regions I and III are homologous across the entire sequence for all loci, with nucleotide identities of ≥87% and ≥90%, respectively.
H. influenzae is capable of causing a variety of respiratory infections and invasive diseases. Prior to the introduction of capsular conjugate vaccines against Hib in the 1980s, this serotype was responsible for almost all H. influenzae-related morbidity and mortality (13). In the period subsequent to wide-spread adoption of childhood Hib vaccination programs, the incidence of Hib-related disease reduced markedly (14). However, following implementation of Hib vaccination programs, disease caused by NTHi has been increasing globally. The prevalence of disease caused by other encapsulated strains is also increasing at an alarming rate, and the Hia disease burden now exceeds that of Hib during the pre-Hib vaccination era in some regions and populations (15). A recent report of particular interest found that Hia constituted 50% of all H. influenzae cases between 2010 and 2015 in northwestern Ontario, Canada (16). Importantly Hib also remains an issue in countries that have not implemented a vaccination program (17).
Public health and clinical laboratories are now beginning to incorporate whole-genome sequencing (WGS) technologies into diagnostic, outbreak, and surveillance programs (18, 19). The departure from molecularly based diagnostics has been driven largely by the considerably higher resolution and accuracy afforded by WGS (20). Currently there are no dedicated tools for H. influenzae serotype prediction that seek to leverage WGS data for cap locus detection. The need for such a tool continues to grow with the resurgence of encapsulated H. influenzae and the increasingly routine use of WGS in the public health setting.
Here we describe hicap, a software tool specifically designed for rapid in silico serotype prediction from H. influenzae WGS data. hicap is an open source Python3 package and is freely available at https://github.com/scwatts/hicap under a GNU General Public License v3 (GPLv3). We further apply hicap to identify and extract cap loci from all H. influenzae genomes currently available in GenBank, and we explore the diversity and distribution of these loci in the H. influenzae population.
MATERIALS AND METHODS
hicap implementation and validation.
hicap uses a reference database to identify genes expected in the six cap loci (cap-a to cap-f). To this end, a curated nucleotide sequence database of cap locus genes was constructed by extracting the protein-coding sequences annotated from cap loci in publicly available sequences of well-defined H. influenzae serotypes (Table 1). The process adopted by hicap to perform serotype prediction from WGS assemblies by using this database is described in Fig. 2
TABLE 1.
cap locus sequences used to create the hicap reference database
| Gene or region | Strain | Accession no. | Reference |
|---|---|---|---|
| bexA | KR494 | GCA_000465255.1 | 43 |
| bexB | KR494 | GCA_000465255.1 | 43 |
| bexC | KR494 | GCA_000465255.1 | 43 |
| bexD | KR494 | GCA_000465255.1 | 43 |
| cap-a | Hi76 | ERX1834399 | 44 |
| cap-b | NCTC 8468 | ERX704106 | 45 |
| cap-c | Hi85 | ERX1834408 | 44 |
| cap-d | ATCC 9008 (cap locus) | HQ424464.1 | Haemophilus influenzae ATCC 9008 |
| cap-e | hi467 | GCA_001975845.1 | 46 |
| cap-f | KR494 | GCA_000465255.1 | 43 |
| hcsA | KR494 | GCA_000465255.1 | 43 |
| hcsB | KR494 | GCA_000465255.1 | 43 |
| IS1016 | Hae18 | X59756.1 | 47 |
FIG 2.
Summary of the hicap serotype prediction method. hicap takes an assembled genome in FASTA format as input and detects all open reading frames (ORFs) using Prodigal. Constituent cap genes and IS1016 copies are identified by performing alignments of either the ORF sequence or input assembly sequence against the reference database using BLAST+. The identified cap genes and IS1016 alignments are then used to inform structural composition of the locus. Serotype is predicted using the gene complement information of region II.
First, all open reading frames (ORFs) are identified in the query assembly using Prodigal (21). Each ORF nucleotide sequence then is queried against the hicap reference database using BLAST+ (22). The resulting alignments are filtered on the basis of subject coverage and nucleotide identity. The default parameters to designate an ORF a complete match to a cap locus gene are subject coverage of ≥80% and nucleotide identity of ≥70%. Often cap genes that are expected to be present lack a complete match to any ORF annotated by Prodigal. This typically occurs when an ORF in the Prodigal annotation has been truncated due to missense mutations, mobile elements, or incomplete assembly. hicap infers the number of genes missing from the predicted cap locus by examining the count of complete ORFs and comparing this to the expected count for the complete form of that locus.
Generally, hicap will attempt to find at least one copy of each gene expected in the cap locus. In the case that there are missing genes, hicap searches the remaining ORF database alignments for the expected gene fragment(s) using more relaxed filtering (defaults for this are alignment length of ≥60 bp and nucleotide identity of ≥80%). Failing this, hicap employs BLAST+ to identify regions of the input assembly that are homologous to missing genes proximal to the predicted cap locus (filtering alignments with a bit score of ≤200). An ORF or sequence is designated truncated if it is identified by either of these adjusted filters but does not meet the criteria for a complete match. Additionally, hicap searches for IS1016 in the cap locus and nearby regions by aligning the reference IS1016 sequence with the input assembly using BLAST+.
The resulting set of alignments and ORFs are used to predict serotype and various locus characteristics. Specifically, hicap predicts serotype by considering all complete and truncated alignments of region II genes. The predicted serotype is defined as the serotype observed to have the most complete set of region II genes. Where an ORF has multiple alignments to the hicap database, a single best alignment is selected on the basis of E value, with ties broken by bit score. ORFs identified as belonging to the cap locus and surrounding region are summarized in a tab-delimited report file, and the annotated cap locus sequence is output in GenBank format. A visualization of the locus annotation is also created using the graphics module in Biopython (23) and output in SVG format (examples are shown in Fig. 3).
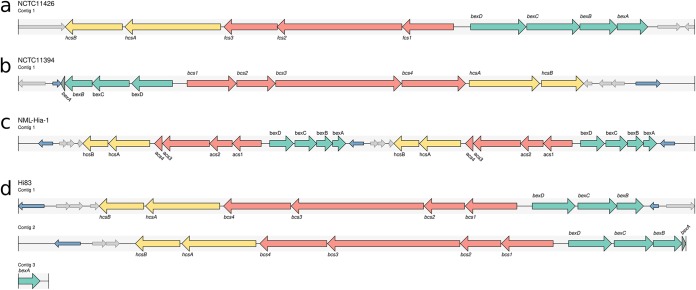
FIG 3.
Examples of hicap visualization for selected genomes. cap locus genes are annotated as large arrows with the direction representing the strand. Genes of the cap locus are colored to indicate region (region I, green; region II, red; region III, yellow). A truncated cap gene is given a darker shade of color for the respective region. Copies of IS1016 are denoted as small blue arrows, and open reading frames that do not generally belong to the cap locus are show as small gray arrows. (a) The complete and contiguous annotation of the NCTC 11426 cap-f locus. (b) The NCTC 11394 cap-b locus, which contains a truncated bexA gene and two copies of IS1016. (c) A duplication of the cap-a locus is observed in the assembly of NML-Hia-1. (d) The cap-b locus of Hi83 is also duplicated but is present across multiple contigs in the input assembly, as represented by multiple tracks.
To test the ability of hicap to predict serotypes, we reviewed the literature and identified publicly available WGS data for H. influenzae isolates with known serotypes (Table 2). The genome assemblies were downloaded and analyzed using hicap run with default parameters. For 26 isolates, only read data were available; hence, de novo assemblies were generated using SPAdes v3.12.0 (24) prior to analysis with hicap. All validation was performed using hicap v1.0.0. The full set of assemblies used for testing is available in FigShare (https://doi.org/10.26180/5c352c5110712). When discrepancies were observed between expected the serotype and hicap results, the output of nucleotide BLAST+ v2.7.1+ searches of genome assemblies against the hicap database was manually inspected.
TABLE 2.
Strains used in the validation of the hicap method, with predicted serotype and fragmentation status of the cap locus as determined by hicap
| Strain | Serotype |
Fragmented locus | Assembly identifiera | Read identifier | |
|---|---|---|---|---|---|
| Described | Predicted by hicap | ||||
| Hi75 | a | a | No | ERX1834398 | |
| Hi76 | a | a | No | ERX1834399 | |
| Hi77 | a | a | No | ERX1834400 | |
| Hi78 | a | a | No | ERX1834401 | |
| Hi79 | a | a | No | ERX1834402 | |
| Hi609 | a | a | No | GCA_003363335.1 | |
| Hi642 | a | a | No | GCA_003363355.1 | |
| NML-Hia-1 | a | a | No | GCA_001856725.1 | |
| 10810 | b | b | No | GCA_000210875.1 | |
| ATCC 10211 | b | b | Yes | GCA_001997355.1 | |
| Hi80 | b | b | No | ERX1834403 | |
| Hi81 | b | b | Yes | ERX1834404 | |
| Hi82 | b | b | Yes | ERX1834405 | |
| Hi83 | b | b | Yes | ERX1834406 | |
| Hi84 | b | b | No | ERX1834407 | |
| NCTC 13377 | b | b | No | GCA_900478275.1 | |
| NCTC 8468 | b | b | No | NCTC 8468 (Sanger FTP) | |
| Hi85 | c | c | No | ERX1834408 | |
| Hi86 | c | c | No | ERX1834409 | |
| Hi87 | c | c | No | ERX1834410 | |
| Hi88 | c | c | No | ERX1834411 | |
| M12125 | c | c | No | GCA_003351605.1 | |
| M17648 | c | c | No | GCA_003351465.1 | |
| Hi89 | d | d | No | ERX1834412 | |
| Hi90 | d | d | No | ERX1834413 | |
| hi467 | e | e | No | GCA_001975845.1 | |
| Hi91 | e | e | Yes | ERX1834414 | |
| Hi92 | e | e | No | ERX1834415 | |
| Hi93 | e | e | No | ERX1834416 | |
| Hi94 | e | e | No | ERX1834417 | |
| Hi95 | e | e | No | ERX1834418 | |
| NCTC 8455 | e | e | No | GCA_900478735.1 | |
| Hi100 | f | f | No | ERX1834168 | |
| Hi96 | f | f | No | ERX1834419 | |
| Hi97 | f | f | No | ERX1834420 | |
| Hi98 | f | f | Yes | ERX1834421 | |
| Hi99 | f | f | No | ERX1834422 | |
| KR494 | f | f | No | GCA_000465255.1 | |
| NCTC 11394 | f | b | No | NCTC 11394 (Sanger FTP) | |
| NCTC 11426 | f | f | No | GCA_900475755.1 | |
| WAPHL1 | f | f | Yes | GCA_002237715.1 | |
| 86-028NP | NTHi | No cap locus | GCA_000012185.1 | ||
| PittEE | NTHi | No cap locus | GCA_000016465.1 | ||
| Rd KW20 | NTHi | No cap locus | GCA_000027305.1 | ||
Assemblies that were available for each isolate were downloaded and screened. Assemblies obtained from the Sanger FTP (https://sanger.ac.uk/resources/downloads/bacteria/nctc) were additionally converted from GFF3 to FASTA format. Where an assembly was not available for an isolate, read sets were downloaded and assembled using SPAdes (as described in Materials and Methods) before screening. All assemblies used for testing are available through FigShare (https://doi.org/10.26180/5c352c5110712).
cap locus distribution, variation, and recombination.
To demonstrate a practical application of hicap, we investigated the distribution of capsular serotypes predicted by hicap among all H. influenzae genomes available in NCBI GenBank as of 8 October 2018 (n = 698, listed in Table S1 in the supplemental material). Whole-genome assemblies were downloaded via FTP, and a phylogeny was constructed using mashtree v0.33 (https://github.com/lskatz/mashtree) (25). Genomes were excluded from analysis where the assembly length was more than four standard deviations from the mean or the genomic content was sufficiently dissimilar to that of H. influenzae (n = 7). Specifically genomic content was assessed by simulating 50,000 error-free reads using wgsim v0.3.1-r13 (https://github.com/lh3/wgsim), which were taxonomically classified by centrifuge v1.0.4-beta (26) and samples with ≤80% H. influenzae reads excluded.
The sequence type (ST) for each assembly was determined via comparison to the multilocus sequence typing (MLST) database for H. influenzae (https://pubmlst.org/hinfluenzae) (27) using mlst v2.15 (https://github.com/tseemann/mlst). Capsular serotypes were inferred using hicap v1.0.0 with the default settings. Nucleotide sequence homology between hybrid cap loci was assessed by BLAST+ v2.7.1+ and visualized using genoPlotR v0.8.7 (28) in R v3.4.4 (29). The mashtree phylogeny was annotated with the ST and predicted capsular serotype in R v3.4.4 using ggtree v1.12.7 (30).
To establish the relationship between capsular serotype and allelic variants of genes encoded in regions I and III, we constructed individual gene trees. Nucleotide sequences were extracted for all complete region I and III genes that were detected by hicap during analysis of the H. influenzae GenBank data set. For each individual gene, nucleotide sequences were aligned using MAFFT v7.407 with default settings (31) and phylogenies inferred from the alignment using FastTree v2.1.10 with the general time-reversible substitution model (32). Nucleotide divergence was calculated using ape v5.2 (33) in R v3.4.4 from gene nucleotide alignments.
RESULTS AND DISCUSSION
hicap validation.
To validate hicap as a tool for in silico serotyping, we analyzed 41 publicly available H. influenzae genomes with reported serologically confirmed capsule types, including representatives for each of the six serotypes and three NTHi strains (Table 2). The results show that hicap robustly identifies the H. influenzae cap locus even in highly discontiguous assemblies. For each cap locus, the completeness, presence of truncated genes, duplication, contiguity, and serotype were correctly reported.
Capsule loci were detected by hicap in 41/41 genomes with reported serotypes and in 0/3 serologically determined NTHi genomes (Table 2). The predicted serotype matched the reported serotype in 40/41 cases (98%) (Table 2). We found that hicap yielded accurate predictions even from draft genomes where the cap locus was fragmented across multiple contigs (observed in 7 genomes from the validation set). Examples of the cap loci identified and visualized using hicap are shown in Fig. 3.
The single genome with a discrepancy between predicted and reported serotype, NCTC 11394, is described as Hif in the National Collection of Type Cultures (NCTC) but was confidently assigned Hib by hicap analysis of the completed PacBio genome assembly (Fig. 3b). Manual assessment of the cap locus in the NCTC 11394 genome assembly additionally confirmed the presence of a complete cap-b locus (uninterrupted, in a single contig) and the absence of any cap-f region II genes, with all expected cap-b protein-coding genes present at ≥95% coverage and ≥84% homology to those annotated in the cap-b reference sequence (excluding a truncated bexA gene). The standard slide agglutination test classically used for serological typing of H. influenzae has been shown to lack specificity and has been estimated to yield incorrect results at a rate of 17.5% (34). It therefore appears likely this discordance is due to inaccuracies in the described serotype rather than misidentification by hicap.
cap locus distribution and variation.
To demonstrate the utility of in silico serotype prediction with hicap, we used it to investigate all publicly available H. influenzae genomes in GenBank that passed quality filtering criteria (n = 691; see Table S1 in the supplemental material). hicap identified a complete cap locus in 95/691 (13.7%) genomes (8 cap-a, 54 cap-b, 4 cap-c, 1 cap-d, 20 cap-e, and 8 cap-f). All genomes contained either zero or one cap locus type, but duplication events were observed in 15/95 (15.8%) cap-positive genomes (14 cap-b and 1 cap-a).
Duplication of the cap-b locus has been frequently reported and is associated with enhanced virulence, conferred by an increased ability to produce capsule (12, 35). This duplication is thought to be driven by copies of IS1016 flanking the capsule locus in some isolates. A common variant of the duplicated cap-b locus involves the deletion of 1.2 kbp in one copy of region I, resulting in the truncation of bexA and IS1016. We observed this duplication deletion variant in 14/54 (26.0%) predicted Hib genomes. Complete cap-b duplication without truncation of bexA was not observed. In addition, hicap identified a single isolate (NML-Hia-1) containing a tandem duplication of the cap-a locus. Strains identified to be carrying cap loci were not assessed for capsule production; however, several of these strains are known to synthesize capsule (e.g., 10810 and NML-Hia-1).
To examine the distribution of cap loci in the H. influenzae population, we constructed a whole-genome phylogeny (Fig. 4) and inferred STs according to the H. influenzae MLST scheme (Table 3). We observed a high degree of exclusivity for STs in regard to predicted capsular serotypes, with each ST containing zero or one cap locus serotype.
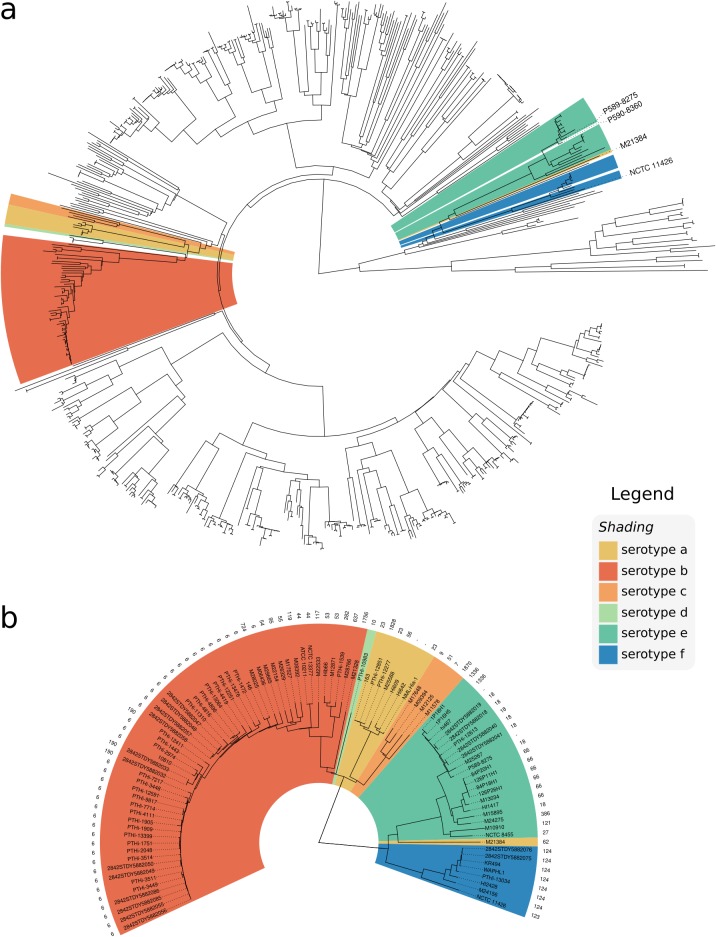
FIG 4.
Whole-genome neighbor-joining phylogeny inferred from MASH distances of assemblies in the GenBank data set. Isolates are annotated with the respective serotype as predicted by hicap. (a) Distribution of capsular serotypes in the complete data set. (b) The phylogeny subtree including only isolates that contained a cap locus, additionally annotated with the sequence type.
TABLE 3.
Sequence types associated with each serotype in the GenBank data set
| Serotype | ST | No. | Frequency (%) of serotype within ST |
|---|---|---|---|
| a | 23 | 3 | 100 |
| 2 | |||
| 1828 | 1 | 100 | |
| 56 | 1 | 100 | |
| 62 | 1 | 100 | |
| b | 6 | 38 | 100 |
| 190 | 3 | 100 | |
| 44 | 2 | 100 | |
| 53 | 2 | 100 | |
| 117 | 1 | 100 | |
| 119 | 1 | 100 | |
| 1756 | 1 | 100 | |
| 282 | 1 | 100 | |
| 54 | 1 | 100 | |
| 55 | 1 | 100 | |
| 637 | 1 | 100 | |
| 724 | 1 | 100 | |
| 95 | 1 | 100 | |
| c | 1870 | 1 | 100 |
| 51 | 1 | 100 | |
| 7 | 1 | 100 | |
| 9 | 1 | 100 | |
| d | 10 | 1 | 100 |
| e | 18 | 7 | 87.5 |
| 66 | 6 | 100 | |
| 2 | |||
| 1336 | 2 | 100 | |
| 121 | 1 | 100 | |
| 27 | 1 | 100 | |
| 386 | 1 | 100 | |
| f | 124 | 7 | 87.5 |
| 123 | 1 | 100 |
The whole-genome phylogeny confirmed that encapsulated strains are relatively clonal and are generally restricted to serotype-specific monophyletic clades (Fig. 4a), suggesting that each serotype emerged once within the H. influenzae population. This is consistent with earlier studies based on electrophoretic typing (36, 37), 16S rRNA (38), MLST (27), and WGS (39). Here, the whole-genome phylogeny resolves the monophyletic nature of each capsule locus with respect to phylogenetic lineage on a larger scale and in greater detail.
hicap did not detect cap loci in a small number of isolates within these serotype-specific clades, indicating occasional capsule loss. For example, P590-8360 clustered with the Hie clade (Fig. 4), but no cap locus was identified by hicap or by manual inspection of the assembly data. The high nucleotide identity between P590-8360 and the cap-e-positive strain P589-8275 along the rest of the genome (Fig. 4a) suggests that loss of the cap-e locus in the P590-8360 genome is the mostly likely explanation. Indeed, the loss of capacity to synthesize capsule has previously been observed to occur by partial or complete deletion of the cap locus (39), and the rate of spontaneous capsule loss is estimated to occur at a frequency of 0.1 to 0.3% (40). Our data are consistent with deletion of the cap locus being a cause of this phenomenon. Interestingly one cap-a genome (M21384) falls within the cap-e serotype-specific clade, suggesting possible recombination in this strain (Fig. 4) (further evidence for this is discussed below).
The serotype-specific clades cluster into two superclades within the H. influenzae phylogeny: one containing cap loci of Hia, Hib, Hic, and Hid and the other containing Hie and Hif cap loci (Fig. 4). Individual gene trees for the region I (bex) and III (hcs) genes show the same two-clade structure (Fig. 5) as the core genomes of their host strains. This observation is consistent with diversification of these cap locus regions in situ within their host chromosomes following introduction into two distinct H. influenzae superclade ancestors. While there is a general lack of homology between region II genes (Fig. 1), two of the three pairs that do show a measure of homology (cap-c/cap-f and cap-d/cap-e) span both superclades; hence, the evolutionary history of region II (and thus the distinct capsular serotypes) remains cryptic.
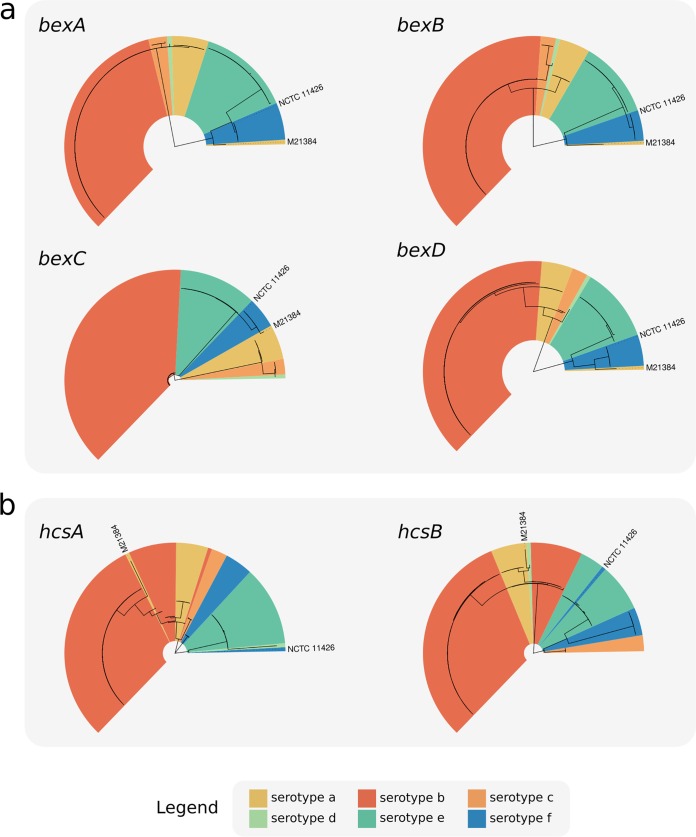
FIG 5.
Phylogenies of all complete cap locus region I (a) and III (b) genes identified in the GenBank data set. FastTree was used to recover phylogenies from MAFFT gene nucleotide sequence alignments, and isolates were annotated using the serotype as predicted by hicap.
Variation in each region I or III gene was associated with serotype, suggesting that the sequence of any could potentially be used to predict capsule type with a relatively high degree of certainty (Fig. 5). Indeed, both bexA and bexB have been proposed and used in single-gene PCR assays for the purpose of serotyping (41, 42). Here the gene bexB showed the greatest differentiation between serotype-specific alleles (0.63% to 17.71% median pairwise nucleotide divergence; see Fig. S1 in the supplemental material) and contained only serotype-specific monophyletic clades in the gene tree. These data suggest bexB to be the most suitable single marker gene for use in PCR or sequenced-based prediction of serotype. In contrast, bexA showed less differentiation then bexB, particularly between serotypes a, b, c, and d (0.17% to 0.85% median pairwise nucleotide divergence).
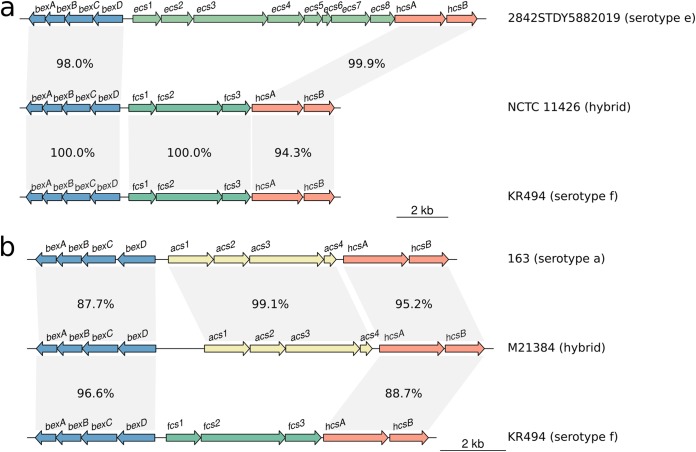
The exceptions to the general association between region I/III genes and predicted serotype were two isolates, M21384 and NCTC 11426 (labeled in Fig. 5). hicap predicted isolates M21384 and NCTC 11426 to be of serotype a and serotype f, respectively. However, both carry cap region I and/or III gene sequences distinct from other strains sharing the same cap II region type (and thus the same predicted serotype), indicative of recombination involving the cap locus within these isolates. Thus, there is evidence for occasional recombination within the cap locus between the different serotype-specific variants, which would limit the accuracy of any single marker gene-based approach to serotype prediction.
Recombination affecting the cap locus.
The isolate M21384 was the only exception to clonal clustering by serotype in the whole-genome phylogeny (Fig. 4). While this isolate is predicted to be Hia based on the presence of cap-a region II genes, the genome falls outside the Hia clade and within the Hie/Hif superclade (Fig. 4b). In all gene trees, M21384 also did not cluster in the expected Hia serotype clade, suggesting that there has been recombination within the cap locus of this isolate (see Fig. 5). Similarly in the hcsA and hcsB gene trees, the isolate NCTC 11426 did not cluster with the expected serotype Hif clade (Fig. 5b). Given the phylogenetic relation of M21384 and NCTC 11426 to other capsular serotypes, it was suspected that these two strains result from recombination events affecting the cap locus (representing a 2% recombination rate involving the cap locus).
To better understand the recombinant cap loci in isolates M21384 and NCTC 11426, we first examined their positions in the whole-genome phylogeny (Fig. 4) and the cap locus gene trees (Fig. 5) and then compared the full-length cap locus sequences of both isolates to reference cap locus sequences (Fig. 6). NCTC 11426 (predicted to be Hif) belongs to the Hif clade in the whole-genome tree and carries typical cap-f regions I and II but contains region III genes more similar to those from cap-e (Fig. 6a). Hence, it appears that the cap locus of NCTC 11426 has resulted from a small recombination event between a Hif clade strain and the cap locus from a Hie clade strain. M21384 clusters within the Hie/Hif superclade of the whole-genome phylogeny and carries cap-f-like region I genes (Fig. 5a and 6b). However, this isolate carries cap-a-like region II genes with a cap-b-like region III gene (hcsA) (Fig. 5b and 6b). The gene content of the M21384 cap locus suggests at least one recombination event involving import of foreign cap locus DNA into a Hif strain. It would be interesting to ascertain whether the isolates with recombinant cap loci described here do in fact express capsule and, if so, to then establish the serological phenotype. However, to our knowledge serotyping has not been performed for either strain, or the data are not publicly available.
FIG 6.
Homology plots generated using R and genoPlotR, showing the cap loci of two isolates which appear to have been subject to recombination. Different regions of the NCTC 11426 (a) and M21384 (b) cap loci show varying homology to different reference cap loci, suggesting a recombinogenic ancestry.
Conclusion.
The need for new tools and methods that leverage WGS continues to become increasingly pivotal with the adoption of WGS by public health laboratories. In this study, we validated and demonstrated the robustness of hicap for prediction of H. influenzae serotype and capsule locus structure. The application of hicap to WGS enables rapid and accurate acquisition of capsule information to aid genomic studies at both individual and population scales. We were also able to explore the diversity and distribution of cap loci in the H. influenzae population at unprecedented nucleotide resolution, identifying a likely misreported serotype in NCTC and describing two novel H. influenzae cap locus recombinants. The resurgence of disease caused by encapsulated H. influenzae and the potential for further antigenic diversification through recombination present a potential public health issue. An important question is whether the geographically disparate reports of increasing cases of infection with non-Hib encapsulated strains reflect the emergence and wide dissemination of a small number of highly successful disease-causing subclones (i.e., a rare but worrying event) or multiple independent events reflecting sporadic but localized outbreaks of non-Hib disease. hicap will facilitate extracting answers to these and other questions from genomic surveillance data.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Bill & Melinda Gates Foundation, Seattle, WA, and the by Australian Government Research Training Program. K.E.H. is supported by a Senior Medical Research Fellowship from the Viertel Foundation of Victoria.
We declare that there are no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00190-19.
REFERENCES
- 1.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Pittman M. 1931. Variation and type specificity in the bacterial species Hemophilus influenzae. J Exp Med 53:471–492. doi: 10.1084/jem.53.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kroll JS, Loynds B, Brophy LN, Moxon ER. 1990. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol Microbiol 4:1853–1862. doi: 10.1111/j.1365-2958.1990.tb02034.x. [DOI] [PubMed] [Google Scholar]
- 4.Sukupolvi-Petty S, Grass S, St Geme JW. 2006. The Haemophilus influenzae type b hcsA and hcsB gene products facilitate transport of capsular polysaccharide across the outer membrane and are essential for virulence. J Bacteriol 188:3870–3877. doi: 10.1128/JB.01968-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Follens A, Veiga-da-Cunha M, Merckx R, van Schaftingen E, van Eldere J. 1999. acs1 of Haemophilus influenzae type a capsulation locus region II encodes a bifunctional ribulose 5-phosphate reductase-CDP-ribitol pyrophosphorylase. J Bacteriol 181:2001–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Eldere J, Brophy L, Loynds B, Celis P, Hancock I, Carman S, Kroll JS, Moxon ER. 1995. Region II of the Haemophilus influenzae type be capsulation locus is involved in serotype-specific polysaccharide synthesis. Mol Microbiol 15:107–118. doi: 10.1111/j.1365-2958.1995.tb02225.x. [DOI] [PubMed] [Google Scholar]
- 7.Lâm T-T, Claus H, Frosch M, Vogel U. 2011. Sequence analysis of serotype-specific synthesis regions II of Haemophilus influenzae serotypes c and d: evidence for common ancestry of capsule synthesis in Pasteurellaceae and Neisseria meningitidis. Res Microbiol 162:483–487. doi: 10.1016/j.resmic.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Giufrè M, Cardines R, Mastrantonio P, Cerquetti M. 2010. Genetic characterization of the capsulation locus of Haemophilus influenzae serotype e. J Clin Microbiol 48:1404–1407. doi: 10.1128/JCM.01721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satola SW, Schirmer PL, Farley MM. 2003. Genetic analysis of the capsule locus of Haemophilus influenzae serotype f. Infect Immun 71:7202–7207. doi: 10.1128/IAI.71.12.7202-7207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corn PG, Anders J, Takala AK, Käyhty H, Hoiseth SK. 1993. Genes involved in Haemophilus influenzae type b capsule expression are frequently amplified. J Infect Dis 167:356–364. doi: 10.1093/infdis/167.2.356. [DOI] [PubMed] [Google Scholar]
- 11.Kroll JS, Hopkins I, Moxon ER. 1988. Capsule loss in H. influenzae type b occurs by recombination-mediated disruption of a gene essential for polysaccharide export. Cell 53:347–356. doi: 10.1016/0092-8674(88)90155-9. [DOI] [PubMed] [Google Scholar]
- 12.Kapogiannis BG, Satola S, Keyserling HL, Farley MM. 2005. Invasive infections with Haemophilus influenzae serotype a containing an IS1016-bexA partial deletion: possible association with virulence. Clin Infect Dis 41:e97–e103. doi: 10.1086/498028. [DOI] [PubMed] [Google Scholar]
- 13.Bijlmer HA. 1991. World-wide epidemiology of Haemophilus influenzae meningitis; industrialized versus non-industrialized countries. Vaccine 9:S5–S9. (Discussion, 9:S25. doi: 10.1016/0264-410X(91)90172-3. [DOI] [PubMed] [Google Scholar]
- 14.Peltola H. 2000. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin Microbiol Rev 13:302–317. doi: 10.1128/CMR.13.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulanova M. 2013. Global epidemiology of invasive Haemophilus influenzae type a disease: do we need a new vaccine? J Vaccines 2013:14. doi: 10.1155/2013/941461. [DOI] [Google Scholar]
- 16.Eton V, Schroeter A, Kelly L, Kirlew M, Tsang RSW, Ulanova M. 2017. Epidemiology of invasive pneumococcal and Haemophilus influenzae diseases in Northwestern Ontario, Canada, 2010-2015. Int J Infect Dis 65:27–33. doi: 10.1016/j.ijid.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 17.Puig C, Grau I, Marti S, Tubau F, Calatayud L, Pallares R, Liñares J, Ardanuy C. 2014. Clinical and molecular epidemiology of Haemophilus influenzae causing invasive disease in adult patients. PLoS One 9:e112711. doi: 10.1371/journal.pone.0112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Revez J, Espinosa L, Albiger B, Leitmeyer KC, Struelens MJ, ECDC National Microbiology Focal Points and Experts Group. 2017. Survey on the use of whole-genome sequencing for infectious diseases surveillance: rapid expansion of European national capacities, 2015-2016. Front Public Health 5:347. doi: 10.3389/fpubh.2017.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E. 2018. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin Microbiol Infect 24:335–341. doi: 10.1016/j.cmi.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grad YH, Lipsitch M. 2014. Epidemiologic data and pathogen genome sequences: a powerful synergy for public health. Genome Biol 15:538. doi: 10.1186/s13059-014-0538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon M. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D, Song L, Breitwieser FP, Salzberg SL. 2016. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res 26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, Popovic T, Spratt BG. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol 41:1623–1636. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guy L, Roat Kultima J, Andersson S. 2010. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 30.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 31.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 34.Satola SW, Collins JT, Napier R, Farley MM. 2007. Capsule gene analysis of invasive Haemophilus influenzae: accuracy of serotyping and prevalence of IS1016 among nontypeable isolates. J Clin Microbiol 45:3230–3238. doi: 10.1128/JCM.00794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kroll JS, Moxon ER, Loynds BM. 1993. An ancestral mutation enhancing the fitness and increasing the virulence of Haemophilus influenzae type b. J Infect Dis 168:172–176. doi: 10.1093/infdis/168.1.172. [DOI] [PubMed] [Google Scholar]
- 36.Musser JM, Granoff DM, Pattison PE, Selander RK. 1985. A population genetic framework for the study of invasive diseases caused by serotype b strains of Haemophilus influenzae. Proc Natl Acad Sci U S A 82:5078–5082. doi: 10.1073/pnas.82.15.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musser JM, Kroll JS, Granoff DM, Moxon ER, Brodeur BR, Campos J, Dabernat H, Frederiksen W, Hamel J, Hammond G. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev Infect Dis 12:75–111. doi: 10.1093/clinids/12.1.75. [DOI] [PubMed] [Google Scholar]
- 38.Sacchi CT, Alber D, Dull P, Mothershed EA, Whitney AM, Barnett GA, Popovic T, Mayer LW. 2005. High level of sequence diversity in the 16S rRNA genes of Haemophilus influenzae isolates is useful for molecular subtyping. J Clin Microbiol 43:3734–3742. doi: 10.1128/JCM.43.8.3734-3742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Chiara M, Hood D, Muzzi A, Pickard DJ, Perkins T, Pizza M, Dougan G, Rappuoli R, Moxon ER, Soriani M, Donati C. 2014. Genome sequencing of disease and carriage isolates of nontypeable Haemophilus influenzae identifies discrete population structure. Proc Natl Acad Sci U S A 111:5439–5444. doi: 10.1073/pnas.1403353111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoiseth SK, Connelly CJ, Moxon ER. 1985. Genetics of spontaneous, high-frequency loss of b capsule expression in Haemophilus influenzae. Infect Immun 49:389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis GS, Sandstedt SA, Patel M, Marrs CF, Gilsdorf JR. 2011. Use of bexB to detect the capsule locus in Haemophilus influenzae. J Clin Microbiol 49:2594–2601. doi: 10.1128/JCM.02509-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. 1994. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol 32:2382–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su Y-C, Hörhold F, Singh B, Riesbeck K. 2013. Complete genome sequence of encapsulated Haemophilus influenzae type f KR494, an invasive isolate that caused necrotizing myositis. Genome Announc 1:e00470-13. doi: 10.1128/genomeA.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staples M, Graham RMA, Jennison AV. 2017. Characterisation of invasive clinical Haemophilus influenzae isolates in Queensland, Australia using whole-genome sequencing. Epidemiol Infect 145:1727–1736. doi: 10.1017/S0950268817000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pittman M. 1942. Antibacterial action of several sulfonamide compounds on Hemophilus influenzae type B. Public Health Rep 57:1899–1910. [Google Scholar]
- 46.Giufrè M, Cardines R, Cerquetti M. 2017. First whole-genome sequence of a Haemophilus influenzae type e strain isolated from a patient with invasive disease in Italy. Genome Announc 5:e00059-17. doi: 10.1128/genomeA.00059-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobson SRM, Kroll JS, Moxon ER. 1992. Insertion sequence IS1016 and absence of Haemophilus capsulation genes in the Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius. Infect Immun 60:618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.