Mycoplasma pneumoniae is the leading cause of bacterial community-acquired pneumonia in persons of all ages. Due to the fastidious nature of this bacterium and the necessary specialized growth media, nucleic acid amplification testing is currently the most reliable means for patient diagnostics.
KEYWORDS: ELITe InGenius, Mycoplasma pneumoniae, PCR, molecular detection, respiratory
ABSTRACT
Mycoplasma pneumoniae is the leading cause of bacterial community-acquired pneumonia in persons of all ages. Due to the fastidious nature of this bacterium and the necessary specialized growth media, nucleic acid amplification testing is currently the most reliable means for patient diagnostics. Analytical sensitivity, specificity, reproducibility, and clinical performance of the ELITe InGenius automated PCR platform with its MGB Alert M. pneumoniae real-time PCR research use only reagents (ELITechGroup, Inc., Bothell, WA) were compared with those of a laboratory-developed real-time PCR assay targeting repMp1 for detection of M. pneumoniae. The ELITe InGenius PCR assay successfully detected 31 distinct M. pneumoniae clinical isolates and reference strains, and there was no cross-reactivity with other mollicutes, Gram-positive bacteria, or Gram-negative bacteria. In testing 223 clinical samples, the ELITe InGenius PCR showed 95.79% and 99.22% positive and negative agreement with the repMp1 assay, respectively. Additionally, the ELITech platform showed 98.91% positive and 96.95% negative predictive values, and there was no significant difference detected between the two assays (McNemar’s test, P = 0.375). The ELITe InGenius PCR assay limit of detection was 0.16 CFU/PCR test or 4.16 genome copies (GCs)/test. Accuracy, instrument ease-of-use, and decreased hands-on time make the ELITe InGenius platform suitable for detection of M. pneumoniae directly from clinical specimens.
INTRODUCTION
Mycoplasma pneumoniae is a highly genome-reduced human pathogen with a unique need for host-derived metabolites and cholesterol (1). It is a common cause of community-acquired respiratory infections in persons of all ages. An estimated 2 million cases of pneumonia due to M. pneumoniae occur annually. Even though most cases are relatively mild, there can be as many as 100,000 hospitalizations of adults in the United States each year (1–3). Clinical manifestations are rarely sufficiently distinct to allow precise diagnosis based on presentation alone. Laboratory diagnosis is most often sought when there is serious illness requiring hospitalization, failure to respond to antimicrobial treatment, presence of extrapulmonary manifestations, occurrence in an immunocompromised host, and/or an outbreak in the community (4). Due to its fastidious nature and slow growth in vitro paired with the specialized expertise required for culture, microbiologic diagnosis of persons suspected of having M. pneumoniae infection typically requires either demonstration of seroconversion or nucleic acid amplification testing (NAAT) (1). Even with analysis of acute-phase and convalescent-phase sera for IgG and IgM, results of commercial assays are often insensitive and/or nonspecific (1). Therefore, NAAT is now the preferred method for M. pneumoniae detection in clinical specimens. In the past 5 years, single and multiplex NAATs for detection of M. pneumoniae have received FDA clearance, and additional assays are undergoing commercial development (1).
The ELITechGroup MDx ELITe InGenius platform (ELITechGroup, Inc., Bothell, WA) is a fully automated sample-to-result PCR system, integrating nucleic acid extraction and real-time PCR amplification and reporting directly from primary patient samples in either single analyte or multiplex formats. The purpose of this study was to evaluate the analytical sensitivity, specificity, reproducibility, and clinical performance of the ELITe InGenius MGB Alert Mycoplasma pneumoniae research use only (RUO) detection reagent mix M4000033 real-time PCR (RT-PCR) assay targeting repMp4 compared to those of a laboratory-developed real-time PCR reference method targeting repMp1 (5, 6).
MATERIALS AND METHODS
ELITe InGenius detection of M. pneumoniae isolates and analytical specificity determination.
The analytical specificity of the ELITe InGenius automated RT-PCR platform with its MGB Alert M. pneumoniae RUO detection reagent mix M4000033 (ELITechGroup, Inc., Bothell, WA) was determined by testing a collection of 31 reference strains and clinical isolates of M. pneumoniae (see Table S1 in the supplemental material) and a panel of genomic DNA previously isolated from viruses, bacteria, yeasts, humans, and all other mollicutes known to colonize or infect humans (see Table S2 in the supplemental material). For genomic DNA analysis, all cycles with a cycle threshold (CT) of >45 were considered nonreactive and negative for detectable signal. M. pneumoniae isolates representing both genetically distinct P1 subtypes, 1 and 2, and obtained over a 30-year period from a broad geographic area (United States, Europe, Asia, and Australia) were included to analyze the ability of both assays to detect a diverse assortment of strains. Three strains were determined to be subtype P2 and two were subtype P1. P1 subtyping was not available for the remaining strains and clinical isolates. DNA for reference strains and clinical isolates of M. pneumoniae was isolated using the Roche MagNA Pure LC system using the large-volume DNA isolation kit per the manufacturer’s specifications (Roche, Indianapolis, IN).
Purified DNA was then stored at −80°C until PCR analysis. To be able to compare the real-time PCR components of the ELITech PCR assay and those of the reference method directly, we bypassed the automated DNA extraction protocol incorporated into the ELITe InGenius system utilizing the “PCR-only function” according to the manufacturer’s instructions. The same volume (15 µl) of DNA in Tris-EDTA (TE) buffer purified using the Roche system as described above was analyzed by both assays enabling direct comparison of RT-PCR analytic performance. M. pneumoniae (ATCC 29343) and Escherichia coli (ATCC 25922) were utilized as positive and negative controls.
ELITe InGenius analytical sensitivity determination.
Analyses were undertaken to determine the limit of detection (LoD) for the ELITe InGenius PCR platform and the repMp1 real-time PCR. Frozen aliquots of M. pneumoniae strain M129 (ATCC 29343) of known CFU per milliliter were thawed and serially diluted in 10-fold increments in SP4 broth to provide a broad range of concentrations (107 CFU/ml to 10−2 CFU/ml). For these analyses, the concentration of M129 frozen stock was 1.02 × 108 CFU/ml. For analysis of genome copy (GC) number in PCR LoDs, DNA stock concentration was at 1.2 × 107 GC/ml in the initial sample. The Qiagen DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) was used as per the manufacturer’s specifications to isolate DNA from samples (200 μl) to be analyzed by repMp1 PCR. Primers, probes, and PCR conditions were adapted for the Roche LC480 instrument (Roche, Indianapolis, IN), and the repMp1 PCR assay was performed as previously described (5, 6). For the ELITech PCR assay, total nucleic acid (NA) was extracted from each dilution using the ELITe InGenius total nucleic acid isolation protocol as per the manufacturer’s instructions. rRNA from spiked-in Torula sp. (Life Technologies, Carlsbad, CA, USA) was utilized as a NA extraction protocol control as per the manufacturer’s instructions. Purified NA samples and spiked internal controls were then subjected to the RT-PCR component of the ELITech assay as per the manufacturer’s instructions. The parameters for NA extraction followed by RT-PCR analysis were preprogrammed into the ELITe InGenius system enabling full automation subsequent to loading of the sample. The lowest dilution able to produce a positive result on repeat testing (2/2 independent repeat samples per PCR run) was designated as the LoD of each assay. Linear regression analysis of LoD data was used to determine R-squared values.
ELITe InGenius reproducibility determination.
Frozen aliquots of M. pneumoniae strain M129 (ATCC 29343) of known CFU per milliliter were thawed and diluted to concentrations of 105, 104, and 103 CFU/ml in SP4 broth and then placed in cryovials that were frozen at −80°C until use. Genomic DNA was extracted from each dilution, and PCR was run on the purified DNA samples as described above. Analyses were performed daily in duplicate on freshly thawed samples over a time course of 10 nonconsecutive days to ensure reproducible cycle threshold (CT) detection over time without loss of signal. Signal deviation of >2 to 3 cycles was considered unacceptable.
ELITe InGenius clinical performance evaluation.
A collection of 95 repMp1 PCR-positive and 128 PCR-negative previously analyzed respiratory specimens obtained from symptomatic pediatric and adult patients between the years 1997 and 2018 was analyzed with the ELITe InGenius assay by a technologist blinded to prior results. Specimens were centrifuged at 10,000 rpm for 30 min prior to DNA isolation and analysis by repMp1 PCR for clinical diagnostic purposes and then stored at −80°C in SP4 broth. A volume of 200 μl of each concentrated specimen was aliquoted and processed using the ELITe InGenius total nucleic acid extraction kit specifications with all parameters preprogrammed and fully automated. PCR analysis was performed in accordance with the manufacturer’s specifications with onboard automation. These data were used to determine the positive and negative percent agreement for the ELITech PCR assay and the reference PCR method. Test performance characteristics were determined by standard methods and include concordance, positive predictive value (PPV), negative predictive value (NPV), positive percent agreement (PPA), and negative percent agreement (NPA). McNemar’s test was used to evaluate statistically significant differences between the two PCRs. Statistical analyses were performed with GraphPad Prism software v.8.0 (GraphPad, San Diego, CA).
RESULTS
The ELITe InGenius PCR assay successfully detected DNA from all 31 M. pneumoniae clinical isolates and reference strains, and there was no cross-reactivity detected with DNA from other mollicutes, Gram-positive or Gram-negative bacteria, viruses, yeasts, or humans (see Table S2 in the supplemental material). Additionally, no appreciable cycle threshold deviations (>2 to 3 cycles) were detected with the ELITe InGenius PCR platform across sample concentrations (105, 104, and 103 CFU/ml) over a 10-nonconsecutive-day period of testing, indicating acceptable qualitative and quantitative assay precision/reproducibility over time.
A summary of the 223 clinical specimens stratified according to sample type is provided in Table 1. The ELITe InGenius PCR assay detected M. pneumoniae in 91 of 95 respiratory specimens that were previously positive by repMp1 PCR (see Table S3 in the supplemental material). However, there was one nasal/nasopharyngeal (NP) swab sample that was negative by repMp1 PCR that was positive by the ELITe InGenius assay. The remaining 127 repMp1 PCR-negative samples tested negative by the ELITe InGenius assay. Each of these discordant specimens was retested by both methods and results were unchanged. A summary of the comparative performances of the two PCRs is provided in Table 2. The repMp1 and ELITe InGenius assays had an overall concordance of 97.59% (κ = 0.954). The PPA was 95.79%, and the NPA was 99.22%. PPV and NPV were 98.91% and 96.95%, respectively. McNemar’s comparison showed no significant difference between the two assays (P = 0.375).
TABLE 1.
Clinical specimen summary
| Specimen type (no.) | No. of negative results |
No. of positive results |
||
|---|---|---|---|---|
| ELITe InGenius | repMp1 PCR | ELITe InGenius | repMp1 PCR | |
| Bronchoalveolar lavage/bronchial wash fluid (45) | 37 | 37 | 8 | 8 |
| Nasal/NP swab (13) | 6 | 7 | 7 | 6 |
| Sputum/endotracheal aspirate (52) | 40 | 38 | 12 | 14 |
| Throat (109) | 44 | 43 | 65 | 66 |
| Miscellaneous/unknowna (4) | 4 | 3 | 0 | 1 |
| Total (223) | 131 | 128 | 92 | 95 |
Miscellaneous/unknown sample types include blood (1), pericardial fluid (2), and unknown swab (1).
TABLE 2.
Performance comparison of ELITe InGenius and repMp1 PCRs
| Statistical outcome | Comparison value (%) (95% confidence interval) |
|---|---|
| Positive percent agreement | 95.79 (89.57–98.84) |
| Negative percent agreement | 99.22 (95.72–99.98) |
| Positive predictive value | 98.91 (92.81–99.84) |
| Negative predictive value | 96.95 (92.40–98.81) |
| Accuracy | 97.76 (94.85–99.27) |
| Concordance | 97.59 (κ = 0.954) |
| McNemar’s test | P = 0.375 (NSa ) |
NS, not significantly different.
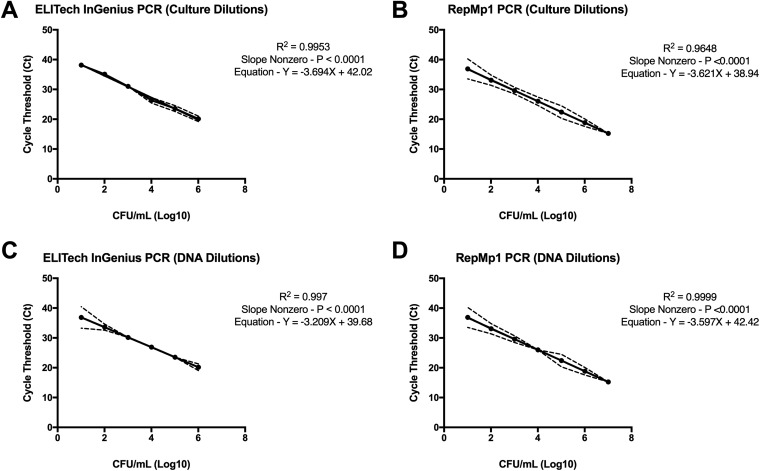
The LoD of the repMp1 PCR showed a high level of sensitivity at 0.41 CFU/PCR test or 1.66 GC/test. The ELITe InGenius assay LoD was found to be similar at 0.16 CFU/PCR test or 4.16 GC/test. Both the ELITe InGenius and repMp1 PCR assays exhibited dynamic detection ranges over 7 logs (101 to 107 CFU/ml of serially diluted M. pneumoniae stocks [Fig. 1]). R-squared values showed a statistically significant association between CFU per milliliter concentrations and CT values (0.995 for ELITe InGenius PCR and 0.965 for repMp1 PCR).
FIG 1.
Analytical sensitivity of M. pneumoniae detection by ELITe InGenius and repMp1 PCR assays. Ten-fold serial culture dilutions and subsequent detection of M. pneumoniae M129 (ATCC 29343) showing cycle threshold (CT) versus CFU per milliliter for ELITe InGenius PCR (A) and repMp1 PCR (B). Ten-fold serial DNA dilutions and subsequent detection of M. pneumoniae M129 (ATCC 29343) showing CT versus CFU per milliliter for ELITe InGenius PCR (C) and repMp1 PCR (D). Graphs depict group mean ± standard deviation. Each assay was run twice to ensure reproducibility with specimens tested in duplicate within each run. The ELITech PCR LoD was 0.16 CFU/PCR test or 4.16 genome copies (GC)/test. The repMp1 assay LoD was 0.41 CFU/PCR test or 1.66 GC/test. R-squared values for each dilution curve showed significant association between CFU per milliliter concentrations and CT values (0.995 for ELITe InGenius PCR and 0.965 for repMp1 PCR).
DISCUSSION
Commercial NAATs for detection of M. pneumoniae have been available in Europe for several years (1, 7–9). However, until the recent FDA clearance of four molecular-based assays, diagnostic testing for M. pneumoniae in the United States was limited primarily to serology and a few laboratory-developed PCRs performed in reference laboratories. The FDA-cleared illumigene Mycoplasma (Meridian Bioscience, Inc., Cincinnati, OH) assay is a loop-mediated isothermal amplification, single-analyte system in which M. pneumoniae is the only microbe detected (4). Currently three FDA-cleared multiplex PCR systems detect M. pneumoniae in addition to various bacterial and viral respiratory pathogens. They include the FilmArray (Biofire Diagnostics, Salt Lake City, UT), ePlex (GenMark Diagnostics, Inc., Carlsbad, CA), and NxTAG, (Luminex Molecular Diagnostics, Inc., Toronto, ON, Canada) platforms. Direct “sample-to-answer” assays, such as the Biofire FilmArray and GenMark ePlex are simple and convenient, enabling laboratories to perform syndromic panel testing. These closed systems minimize the likelihood for cross-contamination, allow decreased hands-on time, and have rapid turnaround times (∼2 to 3 h) (10). However, multiplex panels require a relatively large initial capital investment, exhibit high costs per test, and are poorly reimbursed given the lack of clinical actionability for most viral targets. In contrast, the ELITech MGB Alert assay is a singleplex assay yielding a clinically actionable result for M. pneumoniae. It is performed on the easy-to-use sample-to-answer ELITe InGenius platform with a turnaround time of approximately 3 h (setup to analysis). The ability to perform multiple assays for different targets on single or multiple specimens, simple reporting readouts, and the ability to be operated with and without preprogrammed control over reaction conditions are all valuable features of this platform (11, 12).
In the present study, the ELITe InGenius accurately detected a wide variety of M. pneumoniae strains and had no cross-reactivity with other mollicutes and respiratory pathogens. Additionally, it also showed satisfactory performance for detection of M. pneumoniae in a variety of upper and lower respiratory tract specimens compared with that of our laboratory-developed reference PCR assay targeting repMp1 with a 98.96% PPV, a 96.67% NPV, and no significant difference by the McNemar comparison. We have previously determined in internal laboratory validation procedures that the repMp1 PCR assay has a sensitivity of 100% and a specificity of 97.65% compared with M. pneumoniae culture in oropharyngeal swabs from children with respiratory disease.
Data from the current study also showed that LoD values for both PCR assays compared favorably with those for other M. pneumoniae gene targets, such as P1, ATPase, and community-acquired respiratory distress syndrome (CARDS) toxin (1). R-squared values showed high correlation between CT values and CFU dilutions, showing similar dynamic ranges and detection capacity. Differences between the LoDs for each system (∼1 to 5 estimated genome equivalents) are not significantly different and, most likely, cannot be explained by copy number variations between gene targets. repMp1 can be found in approximately 14 copies across the M. pneumoniae genome, while the InGenius PCR target (repMp4) is found in approximately 8 copies, showing negligible genocopy variation (5, 13–15).
Study limitations include a retrospective study design, lack of culture data, and lack of another molecular-based method for discordant sample adjudications. Currently, the Respiratory Bacterial ELITe MGB panel offered by ELITechGroup is only available in Europe for in vitro diagnostic use. Comparisons of this singleplex assay with the multiplex panel would be useful information if the panel is eventually offered in the United States, but this is beyond the designs of our current study. Overall, the ELITe InGenius PCR is a rapid, accurate, and easy-to-use method for detection of M. pneumoniae in clinical specimens from the respiratory tract in laboratories with varied test volumes.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Melanie Fecanin, Lynn Duffy, and Warren Simmons at UAB for technical support and assistance with study development and implementation.
Financial support was provided by ELITechGroup, Inc., Bothell, WA.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00287-19.
REFERENCES
- 1.Waites KB, Xiao L, Liu Y, Balish MF, Atkinson TP. 2017. Mycoplasma pneumoniae from the respiratory tract and beyond. Clin Microbiol Rev 30:747–809. doi: 10.1128/CMR.00114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marston BJ, Plouffe JF, File TM Jr, Hackman BA, Salstrom SJ, Lipman HB, Kolczak MS, Breiman RF. 1997. Incidence of community-acquired pneumonia requiring hospitalization. Results of a population-based active surveillance study in Ohio. Arch Intern Med 157:1709–1718. doi: 10.1001/archinte.1997.00440360129015. [DOI] [PubMed] [Google Scholar]
- 3.Winchell JM. 2013. Mycoplasma pneumoniae–a national public health perspective. Curr Pediatr Rev 9:324–333. doi: 10.2174/15733963113099990009. [DOI] [Google Scholar]
- 4.Ratliff AE, Duffy LB, Waites KB. 2014. Comparison of the illumigene Mycoplasma DNA amplification assay and culture for detection of Mycoplasma pneumoniae. J Clin Microbiol 52:1060–1063. doi: 10.1128/JCM.02913-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumke R, Schurwanz N, Lenz M, Schuppler M, Luck C, Jacobs E. 2007. Sensitive detection of Mycoplasma pneumoniae in human respiratory tract samples by optimized real-time PCR approach. J Clin Microbiol 45:2726–2730. doi: 10.1128/JCM.00321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waites KB, Xiao L, Paralanov V, Viscardi RM, Glass JI. 2012. Molecular methods for the detection of mycoplasma and ureaplasma infections in humans: a paper from the 2011 William Beaumont Hospital Symposium on molecular pathology. J Mol Diagn 14:437–450. doi: 10.1016/j.jmoldx.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Touati A, Benard A, Hassen AB, Bebear CM, Pereyre S. 2009. Evaluation of five commercial real-time PCR assays for detection of Mycoplasma pneumoniae in respiratory tract specimens. J Clin Microbiol 47:2269–2271. doi: 10.1128/JCM.00326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillet S, Lardeux M, Dina J, Grattard F, Verhoeven P, Le Goff J, Vabret A, Pozzetto B. 2013. Comparative evaluation of six commercialized multiplex PCR kits for the diagnosis of respiratory infections. PLoS One 8:e72174. doi: 10.1371/journal.pone.0072174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumke R, Jacobs E. 2014. Evaluation of five real-time PCR assays for detection of Mycoplasma pneumoniae. J Clin Microbiol 52:4078–4081. doi: 10.1128/JCM.02048-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babady NE, England MR, Jurcic Smith KL, He T, Wijetunge DS, Tang YW, Chamberland RR, Menegus M, Swierkosz EM, Jerris RC, Greene W. 2018. Multicenter evaluation of the ePlex respiratory pathogen panel for the detection of viral and bacterial respiratory tract pathogens in nasopharyngeal swabs. J Clin Microbiol 56:e01658-17. doi: 10.1128/JCM.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girlich D, Bernabeu S, Fortineau N, Dortet L, Naas T. 2018. Evaluation of the CRE and ESBL ELITe MGB kits for the accurate detection of carbapenemase- or CTX-M-producing bacteria. Diagn Microbiol Infect Dis 92:1–7. doi: 10.1016/j.diagmicrobio.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Robert-Gangneux F, Brenier-Pinchart MP, Yera H, Belaz S, Varlet-Marie E, Bastien P, Molecular Biology Study Group of the French National Reference Center for Toxoplasmosis. 2017. Evaluation of Toxoplasma ELITe MGB real-time PCR assay for diagnosis of toxoplasmosis. J Clin Microbiol 55:1369–1376. doi: 10.1128/JCM.02379-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wenzel R, Herrmann R. 1988. Repetitive DNA sequences in Mycoplasma pneumoniae. Nucleic Acids Res 16:8337–8350. doi: 10.1093/nar/16.17.8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruland K, Wenzel R, Herrmann R. 1990. Analysis of three different repeated DNA elements present in the P1 operon of Mycoplasma pneumoniae: size, number and distribution on the genome. Nucleic Acids Res 18:6311–6317. doi: 10.1093/nar/18.21.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spuesens EB, Oduber M, Hoogenboezem T, Sluijter M, Hartwig NG, van Rossum AM, Vink C. 2009. Sequence variations in RepMP2/3 and RepMP4 elements reveal intragenomic homologous DNA recombination events in Mycoplasma pneumoniae. Microbiology 155:2182–2196. doi: 10.1099/mic.0.028506-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.