Reports of methicillin-resistant Staphylococcus aureus (MRSA) harboring the mecC gene have increased in the UK since first being described. To our diagnostic S. aureus multiplex PCR, a mecC primer set was designed and implemented, and then the prevalence in our patient population was investigated.
KEYWORDS: MRSA, PCR, mecC
ABSTRACT
Reports of methicillin-resistant Staphylococcus aureus (MRSA) harboring the mecC gene have increased in the UK since first being described. To our diagnostic S. aureus multiplex PCR, a mecC primer set was designed and implemented, and then the prevalence in our patient population was investigated. Fewer than 1% of the clinical isolates possessed the mecC gene, confirming that mecA remains the dominant genetic determinant of MRSA in East London.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is such a significant health care issue that routine screening of all hospital admissions in the United Kingdom has been in place since 2009 (1). Screening for MRSA typically involves subculture of the patient sample to a selective chromogenic agar, followed by cefoxitin susceptibility testing and, if available, penicillin-binding protein 2′ (PBP2′) detection; PBP2′ is the protein encoded by mecA, which confers methicillin resistance (2–4). However, 2011 saw the emergence of a mecA homologue, now known as mecC, which defied these traditional screening methods and has since been reported in farm animals, animal products, and humans (5–8). Despite these reports, there have been few cases of mecC detection described in the UK patient population (9, 10).
Therefore, we sought to determine the mecC prevalence in our patient population in East London by modifying our existing S. aureus multiplex PCR and incorporating a mecC target.
MATERIALS AND METHODS
We are a district general hospital responsible for a patient population in East London of approximately 2.5 million, with specialist centers in trauma, cancer, cardiac, and emergency care.
S. aureus screening.
Any S. aureus isolates recovered in the laboratory undergo cefoxitin (30 µg) susceptibility testing, by disk diffusion, according to EUCAST guidelines (11). Isolates that are cefoxitin resistant are submitted for extended antibiotic susceptibility testing using the MicroScan (Beckman Coulter, UK). Where disk diffusion and automated susceptibility results disagree, latex agglutination for PBP2′ (Oxoid, UK) is performed. On the rare occasion that the latex agglutination and susceptibility results also disagree, and for suspect Panton-Valentine leucocidin (PVL) toxin producers, S. aureus isolates are investigated using the in-house S. aureus PCR.
S. aureus multiplex PCR.
The diagnostic S. aureus multiplex PCR includes primers for femB, the PVL toxin, and mecA (Table 1). PCR was performed using HotStarTaq Plus mastermix (Qiagen, Germany), with primers at a final concentration of 0.2 µM (PVL) and 0.48 µM (femB and mecA ), under the following conditions: initial denaturation at 95°C for 5 min; 35 cycles of 95°C for 45 s, 50°C for 45 s, and 72°C for 1 min; and a final elongation at 72°C for 2 min. PCR products were resolved and sized by gel electrophoresis.
TABLE 1.
S. aureus PCR primers used in this study
| Primer | Sequence | Size (bp) | Reference |
|---|---|---|---|
| FemB1 | TTACAGAGTTAACTGTTACC | 651 | 21 |
| FemB2 | ATACAAATCCAGCACGCTCT | ||
| PVL1 | ATCATTAGGTAAAAGTCGGAC | 433 | 22 |
| PVL2 | GCATCAAGTGTATTGGATAGCAA | ||
| MecA1 | GTAGAAATGACTGAACGTCCGAT | 310 | |
| MecA2 | CCAATTCCACATTGTTTCGGTCT | ||
| MecC Design_1 (F) | TGAACGAAGCAACAGTACACC | 238 | This study |
| MecC Design_1 (R) | AGATCTTTTCCGTTTTCAGCCT | ||
| MecC Design_2 (F) | CCCGAATTATTGGTAAATCTGGC | 163 | |
| MecC Design_2 (R) | GCATTATAGCTGGCCATCCC |
Addition of mecC to the S. aureus PCR.
As we already had an established S. aureus PCR as part of our diagnostic service, it was important to source mecC primers which were compatible with this assay. Therefore, the literature was reviewed for a mecC primer set that would anneal to target DNA at 50°C and produce a product that was either >651 bp or <310 bp. To maximize the primer choice, alternative mecC primers were also designed using Primer3 and the S. aureus M10/0061 mecC gene sequence (GenBank accession no. NG_047955).
mecC in silico analysis.
The NCBI database (https://www.ncbi.nlm.nih.gov/) was interrogated for all S. aureus strains with identified complete coding sequences for the mecC gene. The chosen mecC primer set was then aligned to each sequence in turn to determine the likely accuracy of detection, using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). In addition, the mecC primers were aligned to S. aureus strains harboring mecA to determine if there would be any cross-amplification.
Study isolates.
The following were used as control strains in this study: S. aureus M10/0061 (mecC positive), S. aureus LGA251 (mecC positive), S. aureus NCTC 12493 (mecA positive), and S. aureus NCTC 6571 (mecA and mecC negative). All mecA- and mecC-harboring control strains were methicillin resistant, as determined by cefoxitin disk susceptibility testing, with the exception of M10/0061, which was cefoxitin susceptible.
The number of isolates needed to determine the prevalence of mecC in our S. aureus population was calculated using the equation described by Jones et al., using a 95% confidence level, a 5% precision level, and an expected proportion of 6.2% based on the recently reported mecA prevalence (10, 12). Based on this equation, at least 89 isolates would need to be tested.
Therefore, 78 MRSA and 64 methicillin-sensitive S. aureus (MSSA) nonduplicate isolates, collected sequentially in the laboratory between July 2018 and February 2019, were used to evaluate the modified PCR. These isolates included 127 recovered from patient samples (Table 2), 7 external quality assessment (EQA) isolates, and 8 internal quality assessment (IQA) isolates. These isolates represented approximately 6% of the total annual S. aureus isolates recovered in the laboratory from MRSA and MSSA patient screens.
TABLE 2.
Patient demographics for the 127 clinical S. aureus isolates recovered from patient samples
| Demographic | No. (%) |
|---|---|
| Sex | |
| Male | 76 (60) |
| Female | 51 (40) |
| Age (yr) | |
| 0–18 | 39 (31) |
| 19–65 | 64 (50) |
| >65 | 24 (19) |
| Health care setting | |
| Inpatient | 41 (32) |
| Outpatient | 80 (63) |
| Unknown | 6 (5) |
| Sample/isolate origin | |
| Surveillance screen | 34 (27) |
| Skin and soft tissue infection | 65 (51) |
| Protected site | 5 (4) |
| Sepsis | 12 (9) |
| Respiratory infection/cystic fibrosis | 8 (6) |
| Unknown | 3 (3) |
RESULTS
Primer selection.
Based on our S. aureus PCR cycling conditions, the mecC primer sets from two studies were compatible with our assay (5, 13) and evaluated alongside the two primer sets designed as part of this study (Table 1).
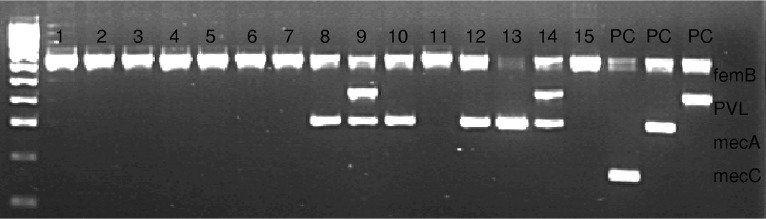
All four potential mecC primer sets were run within the S. aureus multiplex PCR using the PCR control strains. The two published primer sets failed to amplify the mecC target, whereas both designed primer sets successfully amplified the gene in the M10/0061 and LGA251 control strains (data not shown). For optimal gel resolution, the Design_2 primer set (here referred to as the mecC primer set) was selected for further testing, as it amplified a smaller DNA fragment compatible with the sizing of the other S. aureus targets (Fig. 1).
FIG 1.
Gel electrophoresis results for S. aureus isolates screened using our diagnostic assay. The ladder is composed of 100-bp increments. Lanes 1 to 15, clinical S. aureus isolates. PC, positive control. There are 3 PCs, one for mecC (163 bp), one for mecA (310 bp), and one for PVL (433 bp). All 3 controls are also positive for femB (651 bp).
In silico analysis.
A comparison of the chosen mecC primer set to complete mecC coding sequences in the GenBank database revealed the primer sequences and gene sequences to be identical for all mecC-harboring strains. The mecC primer set was also compared to complete MRSA mecA coding sequences to determine if there would be any cross-amplification. In silico analysis revealed the mecA sequence to differ in 9/23 positions in the mecC forward primer and 3/20 positions of the mecC reverse primer.
mecC PCR results.
Following the addition of the mecC primers to the S. aureus PCR, 142 S. aureus isolates were analyzed using this assay. Of these, mecC was detected in just one MRSA isolate, which was recovered from an EQA sample. All other MRSA isolates were positive for the mecA gene. The MSSA isolates were negative for both the mecC and mecA genes.
DISCUSSION
Despite the limited reports of mecC in human isolates, a number of commercial S. aureus PCR assays, available in the UK, now include a mecC target (9, 14, 15). Similarly, various research groups have designed mecC primers for their S. aureus PCR assays, which we also found to be the most appropriate method when selecting a mecC target for our established S. aureus PCR (16–18).
When designing primers for an assay, the concern for nonspecific amplification or cross-amplification of closely related genes is high. Given that mecC is a homologue of mecA, we not only confirmed that our mecC primers would specifically anneal to mecC genes only, but we also ensured there would be no cross-amplification of mecA genes. Lefever et al. reported that >5 mismatches between primer sequences and target sequences will result in PCR inhibition, and as the number of mismatches increases, the annealing temperature becomes affected (19). Our mecC primer sequences differed from the mecA primer binding regions in 9 and 3 positions, respectively, preventing cross-amplification of mecA by our mecC primers. This was clearly demonstrated by our study, with all S. aureus targets correctly amplified in their respective PCR control and the MRSA isolates possessing mecA or mecC, but not both (Fig. 1).
Though the specificity of our mecC primers was high, screening of our clinical S. aureus isolates identified just one mecC-harboring MRSA strain, which mirrors the low prevalence (0.45%) reported elsewhere in the UK (9, 10). However, this is also likely to be an artifact of our testing criteria, which predominantly focus on suspected PVL producers in recurrent skin infections, regardless of methicillin resistance, with fewer MRSA isolates analyzed for mecA and mecC detection and confirmation. In addition, as a hospital-based laboratory, we only test human isolates, which are reported to have a lower mecC prevalence than that of animal isolates, with livestock being the purported mecC reservoir (7, 9, 20). With such a lower number of mecC-positive isolates included in our study, it is important that more mecC strains are tested in the future to confirm the validity of our modified assay.
Our study demonstrates that mecA remains the dominant genetic determinant in human MRSA isolates in East London, and the multiplex PCR described here provides an additional appealing assay for the investigation of recurrent S. aureus skin infections.
ACKNOWLEDGMENTS
S. aureus LGA251 was kindly provided by Jodi Lindsay (St George’s, University of London), and S. aureus M10/0061 was kindly provided by David Coleman (Trinity College Dublin).
REFERENCES
- 1.Department of Health Expert Advisory Committee on Antimicrobial Resistance and Healthcare Associated Infection (ARHAI). 2014. Implementation of modified admission MRSA screening guidance for NHS (2014). Department of Health, London, United Kingdom: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/345144/Implementation_of_modified_admission_MRSA_screening_guidance_for_NHS.pdf. [Google Scholar]
- 2.Public Health England. 2014. UK standards for microbiology investigations: investigation of specimens for screening for MRSA. B29. Public Health England, London, United Kingdom: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/674688/B_29i6_under_review.pdf. [Google Scholar]
- 3.Tsang STJ, McHugh MP, Guerendiain D, Gwynne PJ, Boyd J, Simpson A, Walsh TS, Laurenson IF, Templeton KE. 2018. Underestimation of Staphylococcus aureus (MRSA and MSSA) carriage associated with standard culturing techniques: one third of carriers missed. Bone Joint Res 7:79–84. doi: 10.1302/2046-3758.71.BJR-2017-0175.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.French GL. 2009. Methods for screening for methicillin-resistant Staphylococcus aureus carriage. Clin Microbiol Infect 15:10–16. doi: 10.1111/j.1469-0691.2009.03092.x. [DOI] [PubMed] [Google Scholar]
- 5.Shore AC, Deasy EC, Slickers P, Brennan G, O’Connell B, Monecke S, Ehricht R, Coleman DC. 2011. Detection of Staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and crr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 55:3765–3773. doi: 10.1128/AAC.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuny C, Layer F, Strommenger B, Witte W. 2011. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS One 6:e24360. doi: 10.1371/journal.pone.0024360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison E, Paterson G, Holden M, Larsen J, Stegger M, Larsen AR, Petersen A, Skov R, Christensen JM, Bak Zeuthen A, Heltberg O, Harris S, Zadoks R, Parkhill J, Peacock S, Holmes M. 2013. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol Med 5:509–515. doi: 10.1002/emmm.201202413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paterson G, Morgan F, Harrison E, Peacock S, Parkhill J, Zadoks R, Holmes M. 2014. Prevalence and properties of mecC methicillin-resistant Staphylococcus aureus (MRSA) in bovine bulk tank milk in Great Britain. J Antimicrob Chemother 69:598–602. doi: 10.1093/jac/dkt417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paterson G, Morgan F, Harrison E, Cartwright E, Török M, Zadoks R, Parkhill J, Peacock S, Holmes M. 2014. Prevalence and characterisation of human mecC methicillin-resistant Staphylococcus aureus isolates in England. J Antimicrob Chemother 69:907–910. doi: 10.1093/jac/dkt462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horner C, Utsi L, Coole L, Denton M. 2017. Epidemiology and microbiological characterization of clinical isolates of Staphylococcus aureus in a single healthcare region of the UK, 2015. Epidemiol Infect 145:386–396. doi: 10.1017/S0950268816002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Committee on Antimicrobial Susceptibility Testing. 2019. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf. [Google Scholar]
- 12.Jones SR, Carley S, Harrison M. 2003. An introduction to power and sample size estimation. Emerg Med J 20:453–458. doi: 10.1136/emj.20.5.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kriegeskorte A, Ballhausen B, Idelevich E, Köck R, Friedrich A, Karch H, Peters G, Becker K. 2012. Human MRSA isolates with novel genetic homolog, Germany. Emerg Infect Dis 18:1016–1018. doi: 10.3201/eid1806.110910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker K, Denis O, Roisin S, Mellmann A, Idelevich E, Knaack D, van Alen S, Kriegeskorte A, Köck R, Schaumburg F, Peters G, Ballhausen B. 2016. Detection of mecA- and mecC-positive methicillin-resistant Staphylococcus aureus (MRSA) isolates by the new Xpert MRSA gen 3 PCR assay. J Clin Microbiol 54:180–184. doi: 10.1128/JCM.02081-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silbert S, Kubasek C, Galambo F, Vendrone E, Widen R. 2015. Evaluation of BD Max StaphSR and BD Max MRSAXT assays using ESwab-collected specimens. J Clin Microbiol 53:2525–2529. doi: 10.1128/JCM.00970-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pichon B, Hill R, Laurent F, Larsen AR, Skov RL, Holmes M, Edwards GF, Teale C, Kearns AM. 2012. Development of a real-time quadruplex PCR assay for simultaneous detection of nuc, Panton-Valentine leucocidin (PVL), mecA and homologue mecALGA251. J Antimicrob Chemother 67:2338–2341. doi: 10.1093/jac/dks221. [DOI] [PubMed] [Google Scholar]
- 17.Khairalla A, Wasfi R, Ashour H. 2017. Carriage frequency, phenotypic and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care personnel, patients, and environment. Sci Rep 7:7390. doi: 10.1038/s41598-017-07713-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersdorf S, Herma M, Rosenblatt M, Layer F, Henrich B. 2015. A novel staphylococcal cassette chromosome mec type XI primer for detection of mecC-harboring methicillin-resistant Staphylococcus aureus directly from screening specimens. J Clin Microbiol 53:3938–3941. doi: 10.1128/JCM.02328-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefever S, Pattyn F, Hellemans J, Vandesompele J. 2013. Single-nucleotide polymorphisms and other mismatches reduce performance of quantitative PCR assays. Clin Chem 59:1470–1480. doi: 10.1373/clinchem.2013.203653. [DOI] [PubMed] [Google Scholar]
- 20.Aires-de-Sousa M. 2017. Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin Microbiol Infect 23:373–380. doi: 10.1016/j.cmi.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi N, Wu H, Kojima K, Taniguchi K, Urasawa S, Uehara N, Omizu Y, Kishi Y, Yagihashi A, Kurokawa I. 1994. Detection of mecA, femA, and femB genes in clinical strains of Staphylococci using polymerase chain reaction. Epidemiol Infect 113:259–266. doi: 10.1017/S0950268800051682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClure J, Conly J, Lau V, Elsayed S, Louie T, Hutchins W, Zhang K. 2006. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible and -resistant staphylococci. J Clin Microbiol 44:1141–1144. doi: 10.1128/JCM.44.3.1141-1144.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]