Abstract
Accurate diagnosis of colorectal cancer (CRC) relies on the use of invasive tools such as colonoscopy and sigmoidoscopy. Non-invasive tools are less sensitive in detecting the disease, particularly in the early stage. A number of researchers have used metabolomics analyses on serum/plasma samples of patients with CRC compared with normal healthy individuals in an effort to identify biomarkers for CRC. The aim of the present review is to compare reported serum metabolomics profiles of CRC and to identify common metabolites affected among these studies. A literature search was performed to include any experimental studies on global metabolomics profile of CRC using serum/plasma samples published up to March 2018. The Quality Assessment of Diagnostic Accuracy Studies (QUADAS) tool was used to assess the quality of the studies reviewed. In total, nine studies were included. The studies used various analytical platforms and were performed on different populations. A pathway enrichment analysis was performed using the data from all the studies under review. The most affected pathways identified were protein biosynthesis, urea cycle, ammonia recycling, alanine metabolism, glutathione metabolism and citric acid cycle. The metabolomics analysis revealed levels of metabolites of glycolysis, tricarboxylic acid cycle, anaerobic respiration, protein, lipid and glutathione metabolism were significantly different between cancer and control samples. Although the majority of differentiating metabolites identified were different in the different studies, there were several metabolites that were common. These metabolites include pyruvic acid, glucose, lactic acid, malic acid, fumaric acid, 3-hydroxybutyric acid, tryptophan, phenylalanine, tyrosine, creatinine and ornithine. The consistent dysregulation of these metabolites among the different studies suggest the possibility of common diagnostic biomarkers for CRC.
Keywords: colorectal cancer, metabolomics, serum, biomarkers, pathways
1. Introduction
Colorectal cancer (CRC) is the third most common type of cancer worldwide and is one of the leading causes of cancer mortality (1). Mortality from CRC is primarily due to metastatic progression (2) and treatment in the early stage of the disease has been reported to have a high 5-year survival rate (80–90%) compared with cases when the tumor has metastasized (3). However, the majority of patients are diagnosed when they are already in the advanced stage of the disease. This is due to the fact that CRC may remain asymptomatic until the disease has advanced. Furthermore, diagnostic tests for CRC are invasive which may deter many patients from undergoing screening.
The gold standards for screening and detection of CRC are colonoscopy and sigmoidoscopy. These tests, apart from being expensive and requiring skilled operators, are also invasive and pose potential risks for complications particularly if performed on the elderly or seriously ill patients. Therefore, the non-invasive fecal occult blood test and fecal immunochemical test are the more popular screening methods. However, these tests lack sensitivity, particularly in detecting the early stage of the disease. Therefore, the need for screening methods that are non-invasive, specific and accurate for early identification of CRC has spurred a number of researchers to turn to the use of molecular techniques such as genomics, proteomics and, more recently, metabolomics, to identify serum biomarkers.
Metabolomics is the comprehensive study of low-molecular-mass metabolites in biological matrices. It is downstream of genomics, transcriptomic and proteomics. Therefore, alterations at the metabolomics level not only reflect the changes at the genomics and proteomics levels, but also are influenced by environmental factors. Differences in the levels of metabolites between a diseased and the normal states are used to identify altered metabolic pathways in that particular disease. In recent years, metabolomics studies have been used successfully to identify biomarkers and altered metabolic pathways in various cancer systems, including gastric (4), brain (5), breast (6) and lung (7) cancer.
Serum has been the sample of choice for the identification of biomarkers as it reflects the metabolite profile at the particular time when the sample is taken. Differences observed in the metabolites from normal profiles may serve as important indicators of the pathological states. Global serum metabolomics has been used to identify early CRC biomarkers. Since the altered metabolites may be influenced by biological and environmental factors, it is important to determine the common differentiating metabolites identified by the various studies which may serve as potential common biomarkers for CRC. Therefore, in the present systematic review, the results of the different studies on global serum metabolomics on CRC were compared to identify any common altered metabolites/biomarkers and the affected pathways.
2. Search strategy
A literature search on metabolomics profiling of colorectal cancer was conducted using EBSCO host, Web of Science and PubMed electronic databases up to and including March 2018. The search was conducted using the MeSH terms and Boolean operators: Metabolomic* (OR) metabonomic* (OR) metabolite* (OR) metabolome (OR) metabolic profiling in multiple combinations (AND) colorectal (OR) colon (OR) bowel (OR) rectal (AND) cancer* (OR) tumor* (OR) malignan* (OR) neoplas* (OR) benign* (OR) carcinoma* (OR) adenoma*. The search was limited to the English language. Study titles and abstracts were screened by four independent reviewers identified through the electronic search. Full texts of potentially relevant articles were then retrieved.
The inclusion criteria for the systematic review included global metabolomics profiling studies on CRC using serum or plasma. Exclusion criteria were studies on genomics, transcriptomic and proteomics, studies on polyp colon disease, animals and studies without a control group. Since the aim of the systematic review was to identify common altered metabolites reported, articles which did not include a report of the metabolites identified were excluded. Sample types other than serum such as tissue, urine and feces were excluded. Studies with interventions including surgery or chemotherapy were also excluded.
3. Study quality assessment
The Quality Assessment of Diagnostic Accuracy Studies (QUADAS) tool (8) was used to assess the quality of the included studies. It consists of 14 questions that evaluate spectrum, misclassification, disease progression, partial verification, differential verification, incorporation, review biases and bias associated with study withdrawals and uninterpretable results. Each question was answered with ‘yes’, ‘no’ or ‘unclear’. The answer ‘yes’ means that the risk of bias is low, whereas the answer ‘no’ or ‘unclear’ means the risk of bias is high.
4. Results
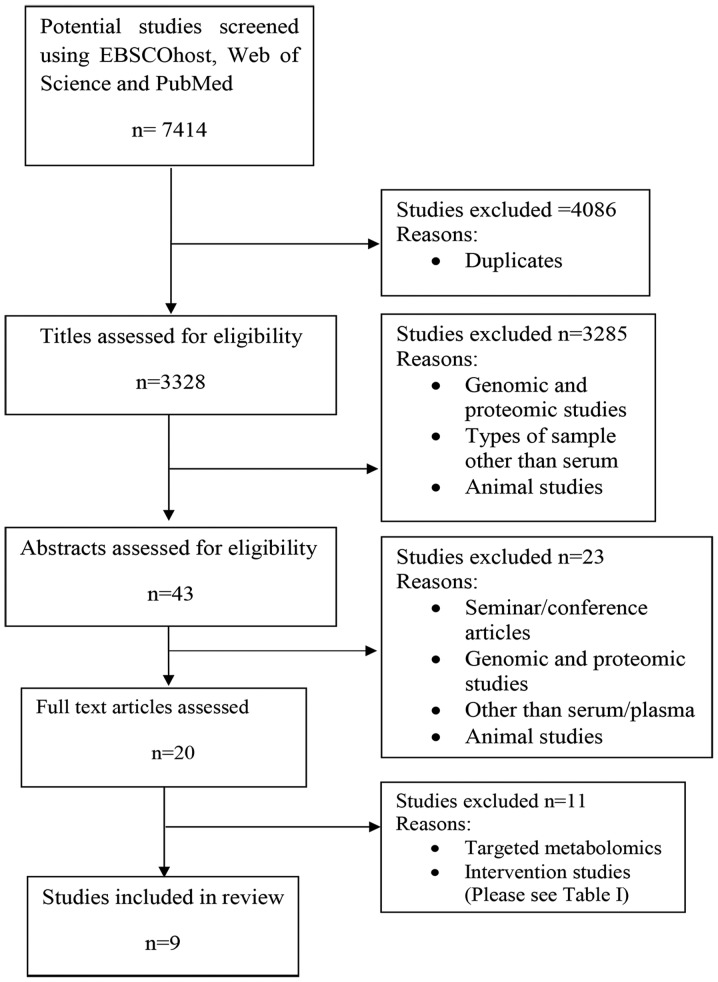
In total, 7,414 publications were identified during the electronic database search. However, 4,086 were excluded because they were duplicates. The remaining 3,328 titles were screened by four independent reviewers who removed 3,285 studies which did not meet the inclusion criteria. Abstracts of the remaining 43 publications were then screened and 23 studies were further excluded because they concerned genomics or proteomics, or the index test was not serum or plasma. Abstracts presented in seminars and conferences were also removed. This is because the details on affected metabolites are needed for analysis in the review. Only 20 publications were retrieved in full and reviewed in detail (Fig. 1). A further 11 were excluded for the reasons listed in Table I. Ultimately, nine articles met the inclusion criteria and were included in the present systematic review.
Figure 1.
Flow process of the systematic search and selection strategy.
Table I.
Excluded studies.
| No. | Study | Reason for exclusion | (Refs.) |
|---|---|---|---|
| 1 | Ma et al, 2010 | The study used subjects with surgical intervention | (9) |
| 2 | Bertini et al, 2012 | Patients with CRC with chemotherapy intervention | (10) |
| 3 | Zhu et al, 2014 | Targeted metabolomics | (11) |
| 4 | Zhu et al, 2015 | Targeted metabolomics | (2) |
| 5 | Guertin et al, 2015 | The study investigated the association between CRC and coffee consumption | (12) |
| 6 | Dowling et al, 2015 | Proteomic studies | (13) |
| 7 | Crotti et al, 2016 | Targeted metabolomics; focus was on lipid metabolism | (14) |
| 8 | Farshidfar et al, 2012 | Metabolite profile was not reported | (15) |
| 9 | Farshidfar et al, 2016 | Patients with CRC undergoing chemotherapy | (16) |
| 10 | Vahabi et al, 2017 | Comparing normal and stage I CRC | (17) |
| 11 | Shu et al, 2018 | Sample collection prior to cancer diagnosis | (18) |
CRC, colorectal cancer.
5. Characteristics of studies
There were nine studies that met the inclusion criteria in the present systematic review (19–27). All nine studies presented the demographic data of their patients and subjects. The index tests utilized for metabolomics analysis were serum and plasma. The analytical platforms used for metabolite detection included gas chromatography-mass spectrometry (GC-MS), gas chromatography-triple-quadrupole mass spectrometry (GC-QqQ-MS), gas chromatography-time-of-flight mass spectrometry (GC-TOF-MS), high-resolution magic angle spinning-nuclear magnetic resonance (1H-NMR), ultra-performance liquid chromatography-mass spectrometry (UPLC-MS), ultra-performance liquid chromatography quadrupole-time-of-flight mass spectrometry (UPLC-QTOF-MS) and capillary electrophoresis-time-of-flight mass spectrometry (CE-TOF-MS).
6. Summary of included studies
Table II presents a brief summary of the characteristics of the included studies.
Table II.
Characteristics of studies.
| Sample size | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Study | Country | Patients with CRC | Healthy controls | Polyps adenoma | Sample extraction | Analytical platform | Data analysis | Total metabolites detected | No. of metabolites significantly different | (Refs.) |
| Qiu et al, 2009 | China | 64 | 65 | – | Yes | GC-TOF-MS | MATLAB 7.0 | 22 | 21 | (19) |
| UPLC-TOF-MS | MarkerLynx 4.1 | 16 | 13 | |||||||
| Nishiumi et al, 2012 | Japan | 60 | 60 | – | Yes | GC-MS | MetAlign | 126 | 54 | (20) |
| Tan et al, 2013 | China | 101 | 102 | – | Yes | GC-TOF-MS | ChromaTOF | 107 | 34 | (21) |
| UPLC-TOF-MS | MarkerLynx 4.1 | 147 | 44 | |||||||
| Cross et al, 2014 | USA | 254 | 254 | – | Yes | UPLC-GC-MS | SAS 9.1.3 and R language 3.0.1 | 278 | 21 | (22) |
| Zamani et al, 2014 | Iran | 33 | 33 | – | No | NMR | Chenomix 6.4 | 14 | 14 | (23) |
| Deng et al, 2016 | USA | 28 | 55 | 44 | No | NMR | Bruker TopSpin 3.5 | 70 | 5 | (24) |
| Uchiyama et al, 2017 | Japan | 56 | 60 | 59 | Yes | CE-TOF-MS | PeakStat | 139 | 70 | (25) |
| Long et al, 2017 | USA | 30, 50 | 30, 50 | 30, 50 | Unknown | LC-MS | Stata version 10.1- | 404 | 50 | (26) |
| Nishiumi et al, 2017 | Japan | 282 | 291 | – | Yes | GC-QqQ-MS | JMP12 | 64 | 41 | (27) |
CRC, colorectal cancer; GC, gas chromatography; MS, mass spectrometry; TOF-MS, time-of-flight MS; UPLC, ultra-performance liquid chromatography; NMR, nuclear magnetic resonance; CE, capillary electrophoresis; LC, liquid chromatography; QqQ, triple-quadrupole.
Qiu et al, 2009 (19)
This study was conducted in China where the authors analyzed serum metabolomics profiles using GC-TOF-MS and UPLC-QTOF-MS. Fasting serum samples were collected from 64 patients with CRC and 65 healthy subjects in the morning. Samples were treated with methanol/chloroform (3:1) prior to metabolomics analysis. The data were then analyzed using multivariate analysis such as principal component analysis and orthogonal partial least-squares-discriminant analysis (OPLS-DA) coupled with univariate statistics. The authors were able to discriminate altered metabolites between CRC and normal samples, but failed to discriminate the different stages of the disease (stages I–IV). There were 21 metabolites reported as being significantly different between CRC and normal using GC-TOF-MS, and 13 metabolites identified using UPLC-QTOF-MS. Of these metabolites, five were detected by the two analytical platforms, including pyruvic acid, lactic acid, tyrosine, uridine and tryptophan.
Nishiumi et al, 2012 (20)
This Japanese study aimed to identify novel early biomarkers for CRC. Fasting blood samples were collected in the early morning and the serum was treated with a methanol/water/chloroform (2.5:1:1) mixture before analysis with GC-MS. The study consisted of two sets of tests: Training and validation sets. Serum samples from 60 patients with CRC and 60 healthy individuals were analyzed for the training set. In total, 54 metabolites were identified to be significantly different; four metabolites, i.e. 2-hydroxybutyrate, aspartic acid, kynurenine and cystamine, were subsequently used as their predictive model for CRC and tested during the validation test. The protocol of the study was well-described, including the stability studies of the analytical platform using quality control samples.
Tan et al, 2013 (21)
These authors replicated their earlier study (9) using the same analytical platforms, i.e. GC-TOF-MS and UPLC-QTOF-MS, to analyze serum samples from a new set of subjects which consisted of 62 patients with CRC and 62 healthy controls. The authors reported 107 altered metabolites using GC-TOF-MS and 147 metabolites when UPLC-TOF-MS was used. Metabolomics profile data obtained from the two platforms were then combined and analyzed using OPLS-DA; 249 differentiating metabolites were identified. A prediction model was then developed and validated on serum samples from 39 patients with CRC and 40 healthy controls. The authors provided a comprehensive protocol of their methodologies including statistical analysis.
Cross et al, 2014 (22)
This study was conducted in the USA. Serum samples were collected from 254 patients with CRC and 254 controls, and were analyzed for global metabolomics using UPLC-MS and GC-MS. Samples were first treated with methanol to precipitate the proteins prior to vacuum-drying. The samples for UPLC-MS analysis were reconstituted in 50 µl 0.1% formic acid in water, whereas for GC-MS analysis, aliquots were derivatized using equal parts bis(trimethylsilyl)trifluoroacetamide and solvent mixture of acetonitrile/dichloromethane/cyclohexane (5:4:1, by vol.) with 5% trimethylamine. The authors reported 278 altered metabolites, of which only 21 metabolites were identified to be significantly different between patients with CRC and normal controls. However, following Bonferroni correction for multiple comparisons, no association between the metabolites and CRC was identified.
Zamani et al, 2014 (23)
A total of 33 serum samples from Iranian patients with CRC on a liquid diet and 33 healthy controls were analyzed using 1H-NMR. The authors identified 14 differentiating metabolites which were statistically significant between patients with CRC and healthy individuals.
Deng et al, 2016 (24)
The authors performed global metabolomics on 28 patients with CRC, 44 patients with polyps and 55 healthy controls in the USA, using NMR. The results were then validated using targeted metabolomics with LC-tandem MS (MS/MS). In total, 70 differentiating metabolites were identified, of which five were significantly different. The authors also reported that the altered pathways in CRC were glycolysis, tricarboxylic acid (TCA) cycle, fatty acid metabolism, amino acid metabolism and glutaminolysis.
Uchiyama et al, 2017 (25)
Serum samples were obtained from 56 patients with CRC, 60 healthy controls and 59 patients with colonic adenoma in Japan. The samples were analyzed using CE-TOF-MS using the Advanced Scan package (Human Metabolome Technologies, Yamagata, Japan). In total, 139 metabolites were identified that were distinguishable from controls, of which 70 were statistically significant; 16 of these metabolites were reported to be correlated with CRC stages. Benzoic acid was identified as the best candidate for CRC biomarkers.
Long et al, 2017 (26)
This study was conducted in the USA and consisted of two phases: Discovery phase and validation phase. Blood samples were collected from 30 healthy controls, 30 patients with colorectal adenoma polyps and 30 patients with CRC for global metabolomics profile analysis using LC-MS. In the validation phase, targeted metabolomics was performed to validate the biomarkers identified earlier in the discovery phase using LC-MS/MS on a new set of samples comprising 50 healthy controls, 50 patients with polyps and 50 patients with CRC. Lower levels of xanthine and hypoxanthine, and higher levels of D-mannose were identified in polyps and CRC cases compared with controls.
Nishiumi et al, 2017 (27)
This study used GC-QqQ-MS to analyze plasma from 282 Japanese patients with CRC and 291 healthy volunteers. The study differed from the other studies in that CRC samples were collected from patients at stage I and II of the disease. Following statistical analysis, eight metabolites were used in their predictive model, i.e. pyruvic acid, glycolic acid, tryptophan, palmitoleic acid, fumaric acid, ornithine, lysine and 3-hydroxyisovaleric acid.
7. Methodological quality of included studies
The quality assessment results for the individual studies are presented in Table III. All studies described the demographic, clinical features and the inclusion and exclusion criteria of their subjects clearly. The diagnosis of study groups was confirmed by colonoscopy and biopsy which was the reference standard for all studies. This eliminated differential verification bias and partial verification bias. However, the majority of the studies reviewed did not indicate the duration of storage for samples before analyses. Overall, the strength of these studies was in the technical aspects of test description, but were limited in the amount of information regarding blinded samples during the experiment.
Table III.
Quality assessment of included studies.
| Study | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | Question | Qiu et al, 2009 (19) | Nishiumi et al, 2012 (20) | Tan et al, 2013 (21) | Cross et al, 2014 (22) | Zamani et al, 2014 (23) | Deng et al, 2014 (24) | Uchiyama et al, 2016 (25) | Long et al, 2017 (26) | Nishiumi et al, 2017 (27) |
| 1 | Was the spectrum of patients representative of the patients who will receive the test in practice? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 2 | Were selection criteria clearly described? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 3 | Is the reference standard likely to correctly classify the target condition? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 4 | Is the period between reference standard and index test short enough to be reasonably sure that the target condition did not alter between the two tests? | Unclear | Unclear | Unclear | Unclear | Unclear | Yes | Unclear | Unclear | Unclear |
| 5 | Did the whole sample, or a random selection of the sample, receive verification using a reference standard of diagnosis? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 6 | Did patients receive the same reference standard regardless of the index test results? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 7 | Was the reference standard independent of the index test (i.e. the index text did not form part of the reference standard)? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 8 | Was the execution of the index test described in sufficient detail to permit replication of the test? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 9 | Was the execution of the reference standard described in sufficient detail to permit its replication? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 10 | Were the index tests results interpreted without knowledge of the results of the reference standard? | No | No | No | No | No | No | No | No | No |
| 11 | Were the reference standard results interpreted without knowledge of the results of the index test? | No | No | No | No | No | No | No | No | No |
| 12 | Were the same clinical data available when test results were interpreted as would be available when the test is used in practice? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 13 | Were uninterpretable/intermediate test results reported? | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear |
| 14 | Were withdrawals from the study explained? | No | No | No | No | No | No | No | No | No |
Risk of bias summary: Review authors' judgments about each risk of bias item for each included study.
8. Altered metabolites
The numbers of differentiating metabolites that were significantly different between patients with CRC and normal controls identified by the studies differ. Furthermore, different studies detected different affected metabolites. The suggested biomarkers also differed in each study. However, there were some common altered metabolites detected among the studies. For the purpose of the present review, differentiating metabolites reported by more than four studies are identified (Table IV) and discussed.
Table IV.
Differentiating metabolites identified by four or more studies.
| Metabolite | GC-TOF-MS Qiu et al, 2009 (19) | UPLC-TOF-MS | GC-MS Nishiumi et al, 2012 (20) | GC-TOF-MS Tan et al, 2013 (21) | UPLC-TOF-MS | UPLC-GC-MS Cross et al, 2014 (22) | NMR Zamani et al, 2014 (23) | NMR Deng et al, 2016 (24) | CE-TOF-MS Uchiyama et al, 2017 (25) | LC-MS Long et al, 2017 (26) | GC-QqQ-MS Nishiumi et al, 2017 (27) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pyruvic acid | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | |||||
| Glucose | ↓ | ↑ | ↑ | ↓ | |||||||
| Lactic acid | ↑ | ↑ | ↓ | ↓ | ↑ | ||||||
| Malic acid | ↑ | ↑ | ↓ | ↑ | |||||||
| Fumaric acid | ↓ | ↑ | ↓ | ↑ | |||||||
| 3-Hydroxybutyric acid | ↑ | ↑ | ↑ | ↓ | ↑ | ||||||
| Ornithine | ↓ | ↑ | ↓ | ↓ | ↓ | ↑ | |||||
| Tryptophan | ↓ | ↓ | ↓ | ↓ | |||||||
| Phenylalanine | ↓ | ↑ | ↓ | ↓ | ↑ | ↓ | |||||
| Tyrosine | ↓ | ↓ | ↑ | ↓ | ↓ | ||||||
| Creatinine | ↓ | ↑ | ↑ | ↓ | ↑ |
GC, gas chromatography; MS, mass spectrometry; TOF-MS, time-of-flight MS; UPLC, ultra-performance liquid chromatography; NMR, nuclear magnetic resonance; CE, capillary electrophoresis; LC, liquid chromatography; QqQ, triple-quadrupole; ↑, increase; ↓, decrease.
There were 11 metabolites identified to be significantly different between CRC and normal individuals by four or more studies reviewed (Table IV). These metabolites included pyruvic acid, glucose, lactic acid, malic acid, fumaric acid, 3-hydroxybutyric acid, tryptophan, phenylalanine, tyrosine, creatinine and ornithine.
The data in Table IV indicate that three metabolites of the glycolytic pathway were altered in CRC. Pyruvic acid was reported to be upregulated in five studies (19–21,26,27). Glucose was reported to be downregulated in two studies (20,26) and upregulated in another two studies (24,26). Similarly, two studies reported downregulation of lactic acid (20,26), whereas two other studies identified it to be upregulated (19,27). Among the metabolites of the TCA cycle, upregulation of malic acid was reported by three studies (19,20,27), whereas one study reported it to be downregulated (26). Fumaric acid was reported to be upregulated by two studies (22,27) and downregulated in another two (21,26).
In general, there were fewer agreements in the reported metabolites of lipid metabolism among the studies. However, 3-hydroxybutyrate was reported by five studies to be altered; four studies noted the upregulation of this lipid (19–21,25), whereas one study identified it to be downregulated (23).
There were five altered metabolites of protein/amino acid metabolism reported by more than four studies: Ornithine, tryptophan, phenylalanine, tyrosine, and creatinine (Table IV). In total, six studies identified altered levels of ornithine; four of these studies reported downregulation of ornithine (19,21,25,26), whereas the other two noted its upregulation (20,26). Tryptophan was identified to be downregulated in four studies in CRC (19,21,25,27). Downregulation of phenylalanine was reported in three studies (19,21,25), whereas two studies identified that it was upregulated in CRC (20,24). With regard to tyrosine, three studies reported downregulation (19,25,27), whereas one study reported upregulation (20), in CRC. Creatinine was reported to be downregulated in two studies (20,24) and upregulated in two other studies (21,27).
9. Metabolic pathways
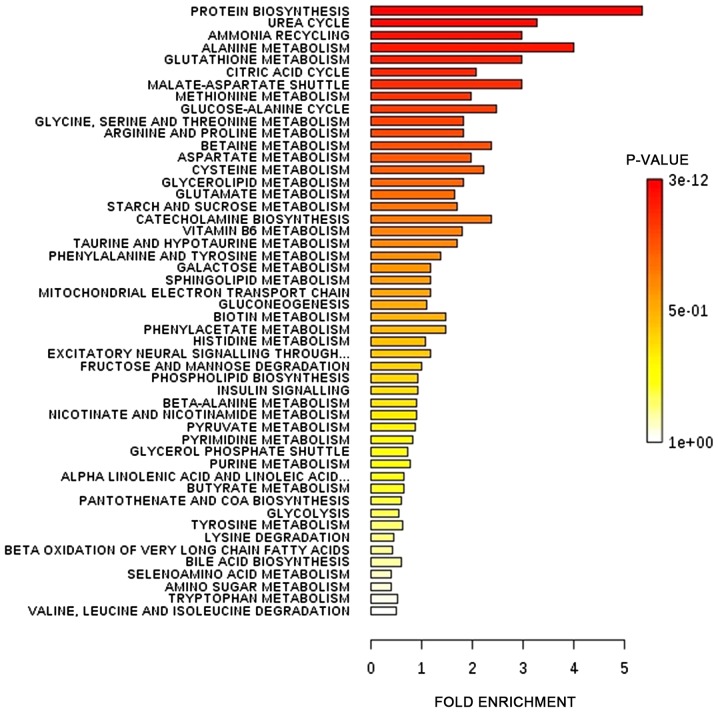
The differential metabolites identified by all studies under review were enriched into pathways. The enrichments were performed using MetaboAnalyst software (version 4.0; www.metaboanalyst.ca). Fig. 2 presents the affected pathways when data from all the studies were enriched into pathways. The six most affected pathways affected in CRC as presented in Fig. 2 were protein biosynthesis, urea cycle, ammonia recycling, alanine metabolism, glutathione metabolism and citric acid cycle.
Figure 2.
Affected metabolic pathways in colorectal cancer. The differential metabolites identified by all studies under review were enriched into pathways using MetaboAnalyst software (version 4.0; www.metaboanalyst.ca).
10. Discussion
Serum metabolomics profiling has been aided by the development of techniques which separate, detect, characterize and quantify the metabolomes. The studies investigated in the present systematic review used six different analytical platforms, which were UPLC-QTOF-MS, LC-MS, GC-MS, GC-TOF-MS, CE-TOF-MS, GC-QqQ-MS and NMR. The study populations were Chinese (19,21), Japanese (20,25,27), Iranian (23) and American (22,24,26). To interpret the complex metabolomics data, each study used different software as presented in Table II. Qiu et al (19) and Tan et al (21) analyzed their data using ChromaTOF and MarkerLynx Application Manager software. Nishiumi et al (20) used MetAlign and SIMCA software, whereas Nishiumi et al (27) used GCMS Solution Software and JMP12 software for statistical analysis. Zamani et al (23) used Chenomix 6.4 software. Uchiyama et al (25) used HMT Advanced Scan package for metabolome analysis and PeakStat for statistical analysis. Long et al (26) used Stata software in their statistical analysis.
The results of the present systematic review confirmed an earlier review by Kim et al (28) that different analytical platforms detect different altered metabolomes. The studies by Qiu et al (19) and Tan et al (21) on two different analytical platforms illustrated this fact despite conducting the analysis on the same samples. The differences in metabolites identified may be due to differences in the techniques of separation and detection of the different platforms. Furthermore, sample preparations were also different. GC-MS detects low-molecular-mass (between 18 and 350 Da) metabolites, including amino acids and organic acids, fatty acids, carbohydrates and cholesterol (19), whereas UPLC-MS detects higher-molecular-mass compounds of medium-to-high lipophilicity, including lipids (29). Amino acids with a molecular mass >147 Da can be detected by UPLC-MS. NMR has lower sensitivity and therefore detects compounds that are present in high amounts. Sample preparation also differs according to the analytical platform used. Analysis using GC-MS involves sample derivatization, whereas LC-MS requires the metabolites to be extracted from the serum; however, little or no sample preparation is required for NMR (30). In addition, there were also differences in the study design. Certain authors reported the metabolomics profiles following global analysis, whereas others went on to validate their results using targeted metabolomics analysis.
Another notable result is the difference in the altered metabolomes detected in the different populations such as Chinese (19,21), Japanese (20,25,27), Iranian (23) and American (22,24,26). This is not surprising as metabolomes have been identified to be affected by genetics as well as environmental factors such as nutrition and lifestyle (31). Furthermore, the cancer screening policies and levels of medical intervention differ in different countries. In certain countries, there are more patients in the early stage, whereas others diagnosed more patients at the metastatic stage. This may account for the differing numbers of patients at each stage of the disease and may affect the metabolomics profiles obtained. The same concern can also be applied to the different time of the study conducted. Since the present systematic review included only nine studies which used five different analytical platforms on four different populations, it is suggested that more studies are required to confirm the effect of populations on the metabolomics profiles. This is important as it may signify differences in the sensitivity and specificity of biomarkers for the detection of CRC in different populations.
For the purpose of the present review, the metabolites whose levels were altered in CRC compared with normal individuals identified by four or more studies are considered common altered metabolites. Therefore, the present review identified that the metabolites most affected in CRC are the metabolites of energy pathways, protein/amino acid and lipid metabolism (Table IV). However, different studies reported different expression of the metabolites, with the exception of pyruvic acid and tryptophan. The reason for this difference is unclear. The CRC subjects included patients at different stages of the disease and each study differed in the percentage of subjects at the different stages. Since the metabolism may differ at different stages of cancer progression, this may account for the stated observation. Furthermore, serum metabolomics profiling provides a snapshot of metabolism at that instant within the cells.
Pyruvic acid, glucose and lactic acid were the most commonly reported metabolites of glycolysis altered in CRC. Malic acid and fumaric acid, intermediates of the TCA cycle, were also commonly reported in the studies reviewed. High cellular glucose metabolism in cancer has been suggested to provide energy and precursors for the increased rate of anabolic processes in cancer cells (32). This is achieved by reprogramming of glucose metabolism which involves the activation of phosphoinositide 3-kinase and its downstream protein kinase B and mammalian target of rapamycin (mTOR) signaling pathways. The events of glucose reprogramming in cancer has been well-reviewed by Hay (33). The increased glycolysis is not met with an increased rate of oxidative phosphorylation in cancer cells because their proliferation rate often exceeds the rate of angiogenesis (34). Therefore, increased production of pyruvate from glycolysis leads to lactic acid production. The metabolic adaptation to hypoxia is coordinated by hypoxia-inducible factor 1 (35) through a variety of mechanisms, including hyperactivation of mTOR complex 1, accumulation of reactive oxygen species (ROS) and accumulation of the citric acid cycle metabolites (36).
Amino acids are important as building materials and as nutrient signals that regulate important signaling pathways (37). The amino acids ornithine, tryptophan, phenylalanine, tyrosine and creatinine were reported to be altered in CRC by the majority of the studies reviewed (Table IV), suggesting the potential of using these amino acids as biomarkers. Tryptophan metabolism is an important mechanism exploited by cancer to evade immune surveillance (38). Tryptophan is an essential amino acid and 95% of dietary tryptophan is metabolized along the kynurenine pathway (39). Decrease tryptophan levels are associated with catabolism of tryptophan into kynurenine via the kynurenine pathway (40). Mezrich et al (41) suggested that kynurenine binds to aryl hydrocarbon receptor to promote the generation of immune-suppressive T cells that support cancer development.
Tyrosine is phosphorylated by tyrosine kinases, and phosphorylated tyrosine acts in signal transduction and the regulation of enzyme activity. One such enzyme is pyruvate dehydrogenase kinase 1 which has been suggested to be involved in the promotion of the Warburg effect and cell proliferation (42).
It is interesting to note that the ornithine level was identified to be altered in CRC by six of the studies reviewed. Ornithine is catabolized by ornithine decarboxylase (ODC) in the pathway for polyamine synthesis. Polyamines are important molecules for normal cell proliferation and are highly expressed in various malignancies (43). Hoshino et al (44) reported that the ODC level is increased in CRC tumor cells compared with normal tissues.
Increased levels of lipid metabolic intermediates are primarily used for energy storage (45). Menendez and Lupu (46) suggested that lipid synthesis serves an important function in tumor pathogenesis (46). In the present review, five studies reported that 3-hydroxybutyric acid level was altered in CRC. 3-Hydroxybutyric acid is among the components of ketone bodies and the altered levels may indicate increased protein catabolism particularly involving the ketogenic branched-chain amino acids (BCAAs) and increased fatty acid oxidation.
The significance of the altered metabolites is reflected in the pathways affected (Fig. 2). Tumors are associated with increased protein synthesis required for cell proliferation. The increase in urea cycle, ammonia recycling and alanine metabolism may indicate increased muscle proteolysis generating BCAAs which are subsequently oxidized via the citric acid cycle, to meet the increased energy demand of a tumor cell. The data in Fig. 2 also indicated that glutathione metabolism is affected in CRC. The increased metabolic activity of cancer cells leads to increase production of ROS. ROS serve a significant function in the pathogenesis of cancer by activating signaling pathways that support cell proliferation, survival and metabolic adaptation. However, high levels of ROS may cause cell damage. Therefore, tumour cells react by producing glutathione, an antioxidant, to prevent ROS from reaching toxic levels (47).
11. Limitations
The number of published papers on global serum metabolomics that were included in the present systematic review is small, indicating an inadequate amount of research in this area. Different metabolite markers were identified by the different studies which were influenced by the analytical platforms used, the study design, stage of CRC, statistical analysis and the populations studied. Although certain common metabolites were identified, further studies are required to verify and identify distinguishable metabolites in patients with CRC from those in normal individuals in different populations, perhaps with standardized sample preparation and measurement and analysis of data.
12. Conclusion
Enrichment pathway analysis using data from all studies included indicated that protein biosynthesis, urea cycle, ammonia recycling, alanine metabolism, glutathione metabolism and citric acid cycle were among the most affected pathways. However, the altered metabolites in patients with CRC compared with in healthy individuals were identified to be different among different studies owing to differences in sample analysis and populations. Tryptophan, phenylalanine, tyrosine, ornithine, creatinine, pyruvic acid, glucose, lactic acid, malic acid, fumaric acid and 3-hydroxybutyrate were the altered metabolites identified by more than four of the studies reviewed. Further studies are required to determine whether metabolites can serve as common biomarkers for CRC in different populations.
Acknowledgements
The authors would like to thank the Ministry of Education Malaysia for providing the research grant. The authors would also like to thank Professor Datuk Abdul Rahman A. Jamal who secured funding for the present study.
Funding
The present study was funded by a Long Term Research Grant of the Ministry of Higher Education, Malaysia (grant no. LRGS/2014/UKM-UiTM/K/03).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article [and its supplementary information files].
Authors' contributions
NAAH searched for the published articles, performed the analysis and wrote the manuscript. SAR, LSS, MSAS and MM were involved in evaluating and reviewing the titles, abstracts and full papers and determining the quality of papers (using QUADAS) and that the papers included in the review fulfilled the inclusion and exclusion criteria. They were also involved in generating ideas in the content of the manuscript as well as editing the manuscript prior to submission. MM conceptualized the idea for the review, arranged for the writing and co-wrote and edited the manuscript prior to submission.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Djukovic D, Deng L, Gu H, Himmati F, Abu Zaid M, Chiorean EG, Raftery D. Targeted serum metabolite profiling and sequential metabolite ratio analysis for colorectal cancer progression monitoring. Anal Bioanal Chem. 2015;407:7857–7863. doi: 10.1007/s00216-015-8984-8. [DOI] [PubMed] [Google Scholar]
- 3.American Cancer Society. Cancer facts and figures 2013. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2013/cancer-facts-and-figures-2013.pdf. Accessed Month day, year.
- 4.Wang D, Li W, Zou Q, Yin L, Du Y, Gu J, Suo J. Serum metabolomic profiling of human gastric cancer and its relationship with the prognosis. Oncotarget. 2017;8:110000–110015. doi: 10.18632/oncotarget.21314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinnaiyan P, Kensicki E, Bloom G, Prabhu A, Sarcar B, Kahali S, Eschrich S, Qu X, Forsyth P, Gillies R. The metabolomic signature of malignant glioma reflects accelerated anabolic metabolism. Cancer Res. 2012;72:5878–5888. doi: 10.1158/0008-5472.CAN-12-1572-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadi NI, Jamal Q, Iqbal A, Shaikh F, Somroo S, Musharraf SG. Serum metabolomic profiles for breast cancer diagnosis, grading and staging by gas chromatography-mass spectrometry. Sci Rep. 2017;7:1715. doi: 10.1038/s41598-017-01924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar N, Shahjaman M, Mollah MNH, Islam SMS, Hoque MA. Serum and plasma metabolomic biomarkers for lung cancer. Bioinformation. 2017;13:202–208. doi: 10.6026/97320630013202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J. The development of QUADAS: A tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol. 2003;3:25. doi: 10.1186/1471-2288-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y, Liu W, Peng J, Huang L, Zhang P, Zhao X, Cheng Y, Qin H. A pilot study of gas chromatograph/mass spectrometry-based serum metabolic profiling of colorectal cancer after operation. Mol Biol Rep. 2010;37:1403–1411. doi: 10.1007/s11033-009-9524-4. [DOI] [PubMed] [Google Scholar]
- 10.Bertini I, Cacciatore S, Jensen BV, Schou JV, Johansen JS, Kruhøffer M, Luchinat C, Nielsen DL, Turano P. Metabolomic NMR fingerprinting to identify and predict survival of patients with metastatic colorectal cancer. Cancer Res. 2012;72:356–364. doi: 10.1158/0008-5472.CAN-11-1543. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J, Djukovic D, Deng L, Gu H, Himmati F, Chiorean EG, Raftery D. Colorectal cancer detection using targeted serum metabolic profiling. J Proteome Res. 2014;13:4120–4130. doi: 10.1021/pr500494u. [DOI] [PubMed] [Google Scholar]
- 12.Guertin KA, Loftfield E, Boca SM, Sampson JN, Moore SC, Xiao Q, Huang WY, Xiong X, Freedman ND, Cross AJ, Sinha R. Serum biomarkers of habitual coffee consumption may provide insight into the mechanism underlying the association between coffee consumption and colorectal cancer. Am J Clin Nutr. 2015;101:1000–1011. doi: 10.3945/ajcn.114.096099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowling P, Hughes DJ, Larkin AM, Meiller J, Henry M, Meleady P, Lynch V, Pardini B, Naccarati A, Levy M, et al. Elevated levels of 14-3-3 proteins, serotonin, gamma enolase and pyruvate kinase identified in clinical samples from patients diagnosed with colorectal cancer. Clin Chim Acta. 2015;441:133–141. doi: 10.1016/j.cca.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Crotti S, Agnoletto E, Cancemi G, Di Marco V, Traldi P, Pucciarelli S, Nitti D, Agostini M. Altered plasma levels of decanoic acid in colorectal cancer as a new diagnostic biomarker. Anal Bioanal Chem. 2016;408:6321–6328. doi: 10.1007/s00216-016-9743-1. [DOI] [PubMed] [Google Scholar]
- 15.Farshidfar F, Weljie AM, Kopciuk K, Buie WD, Maclean A, Dixon E, Sutherland FR, Molckovsky A, Vogel HJ, Bathe OF. Serum metabolomic profile as a means to distinguish stage of colorectal cancer. Genome Med. 2012;4:42. doi: 10.1186/gm341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farshidfar F, Weljie AM, Kopciuk KA, Hilsden R, McGregor SE, Buie WD, MacLean A, Vogel HJ, Bathe OF. A validated metabolomic signature for colorectal cancer: Exploration of the clinical value of metabolomics. Br J Cancer. 2016;115:848–857. doi: 10.1038/bjc.2016.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vahabi F, Sadeghi S, Arjmand M, Mirkhani F, Hosseini E, Mehrabanfar M, Hajhosseini R, Iravani A, Bayat P, Zamani Z. Staging of colorectal cancer using serum metabolomics with 1HNMR Spectroscopy. Iran J Basic Med Sci. 2017;20:835–840. doi: 10.22038/IJBMS.2017.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu X, Xiang YB, Rothman N, Yu D, Li HL, Yang G, Cai H, Ma X, Lan Q, Gao YT, et al. Prospective study of blood metabolites associated with colorectal cancer risk. Int J Cancer. 2018;143:527–534. doi: 10.1002/ijc.31574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, Ni Y, Zhao A, Xu LX, Cai S, Jia W. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J Proteome Res. 2009;8:4844–4850. doi: 10.1021/pr9004162. [DOI] [PubMed] [Google Scholar]
- 20.Nishiumi S, Kobayashi T, Ikeda A, Yoshie T, Kibi M, Izumi Y, Okuno T, Hayashi N, Kawano S, Takenawa T, et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS One. 2012;7:e40459. doi: 10.1371/journal.pone.0040459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan B, Qiu Y, Zou X, Chen T, Xie G, Cheng Y, Dong T, Zhao L, Feng B, Hu X, et al. Metabonomics identifies serum metabolite markers of colorectal cancer. J Proteome Res. 2013;12:3000–3009. doi: 10.1021/pr400337b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cross A, Moore SC, Boca S, Huang WY, Xiong X, Stolzenberg-Solomon R, Sinha R, Sampson JN. A prospective study of serum metabolites and colorectal cancer risk. Cancer. 2014;120:3049–3057. doi: 10.1002/cncr.28799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamani Z, Arjmand M, Vahabi F, Eshaq Hosseini SM, Fazeli SM, Iravani A, Bayat P, Oghalayee A, Mehrabanfar M, Haj Hosseini R, et al. A metabolic study on colon cancer using (1)h nuclear magnetic resonance spectroscopy. Biochem Res Int. 2014;2014:348712. doi: 10.1155/2014/348712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng L, Gu H, Zhu J, Nagana Gowda GA, Djukovic D, Chiorean EG, Raftery D. Combining NMR and LC/MS using backward variable elimination: Metabolomics analysis of colorectal cancer, polyps, and healthy controls. Anal Chem. 2016;88:7975–7983. doi: 10.1021/acs.analchem.6b00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchiyama K, Yagi N, Mizushima K, Higashimura Y, Hirai Y, Okayama T, Yoshida N, Katada K, Kamada K, Handa O, et al. Serum metabolomics analysis for early detection of colorectal cancer. J Gastroenterol. 2017;52:677–694. doi: 10.1007/s00535-016-1261-6. [DOI] [PubMed] [Google Scholar]
- 26.Long Y, Sanchez-Espiridion B, Lin M, White L, Mishra L, Raju GS, Kopetz S, Eng C, Hildebrandt MAT, Chang DW, et al. Global and targeted serum metabolic profiling of colorectal cancer progression. Cancer. 2017;123:4066–4074. doi: 10.1002/cncr.30829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishiumi S, Kobayashi T, Kawana S, Unno Y, Sakai T, Okamoto K, Yamada Y, Sudo K, Yamaji T, Saito Y, et al. Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple-quadrupole mass spectrometry. Oncotarget. 2017;8:17115–17126. doi: 10.18632/oncotarget.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SJ, Kim SH, Kim JH, Hwang S, Yoo HJ. Understanding metabolomics in biomedical research. Endocrinol Metab (Seoul) 2016;31:7–16. doi: 10.3803/EnM.2016.31.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 30.Serkova NJ, Standiford TJ, Stringer KA. The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am J Respir Crit Care Med. 2011;184:647–655. doi: 10.1164/rccm.201103-0474CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kastenmüller G, Raffler J, Gieger C, Suhre K. Genetics of human metabolism: An update. Hum Mol Genet. 2015;24:R93–R101. doi: 10.1093/hmg/ddv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn CS, Metallo CM. Mitochondria as biosynthetic factories for cancer proliferation. Cancer Metab. 2015;3:1. doi: 10.1186/s40170-015-0128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hay N. Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat Rev Cancer. 2016;16:635–649. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain RK, Munn LL, Fukumura D. Dissecting tumour pathophysiology using intravital microscopy. Nat Rev Cancer. 2002;2:266–276. doi: 10.1038/nrc778. [DOI] [PubMed] [Google Scholar]
- 35.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Ananieva E. Targeting amino acid metabolism in cancer growth and anti-tumor immune response. World J Biol Chem. 2015;6:281–289. doi: 10.4331/wjbc.v6.i4.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prendergast GC, Smith C, Thomas S, Mandik-Nayak L, Laury-Kleintop L, Metz R, Muller AJ. Indoleamine 2,3-dioxygenase pathways of pathogenic inflammation and immune escape in cancer. Cancer Immunol Immunother. 2014;63:721–735. doi: 10.1007/s00262-014-1549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters JC. Tryptophan nutrition and metabolism: An overview. Adv Exp Med Biol. 1991;294:345–358. doi: 10.1007/978-1-4684-5952-4_32. [DOI] [PubMed] [Google Scholar]
- 40.Grohmann U, Bronte V. Control of immune response by amino acid metabolism. Immunol Rev. 2010;236:243–264. doi: 10.1111/j.1600-065X.2010.00915.x. [DOI] [PubMed] [Google Scholar]
- 41.Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, Ge Q, Gu TL, Polakiewicz RD, Roesel JL, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell. 2011;44:864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pegg AE. Functions of polyamines in mammals. J Biol Chem. 2016;291:14904–14912. doi: 10.1074/jbc.R116.731661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoshino Y, Terashima S, Teranishi Y, Terashima M, Kogure M, Saitoh T, Osuka F, Kashimura S, Saze Z, Gotoh M. Ornithine decarboxylase activity as a prognostic marker for colorectal cancer. Fukushima J Med Sci. 2007;53:1–9. doi: 10.5387/fms.53.1. [DOI] [PubMed] [Google Scholar]
- 45.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 46.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 47.Benlloch M, Ortega A, Ferrer P, Segarra R, Obrador E, Asensi M, Carretero J, Estrela JM. Acceleration of glutathione efflux and inhibition of gamma-glutamyltranspeptidase sensitize metastatic B16 melanoma cells to endothelium-induced cytotoxicity. J Biol Chem. 2005;280:6950–6959. doi: 10.1074/jbc.M408531200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article [and its supplementary information files].