Abstract
Telomeres play important roles in genome stability and cell proliferation. High risk neuroblastoma (HRNB), an aggressive childhood cancer, is especially reliant on telomere maintenance. Three recurrent genetic aberrations in HRNB (MYCN amplification, TERT re-arrangements, and ATRX mutations) are mutually exclusive and each capable of promoting telomere maintenance mechanisms (i.e., through telomerase or ALT). We analyzed a panel of 5 representative HRNB cell lines and 30 HRNB surgical samples using assays that assess average telomere lengths, length distribution patterns, single-stranded DNA on the G- and C-strand, as well as extra-chromosomal circular telomeres. Our analysis pointed to substantial and variable degrees of telomere DNA damage in HRNB, including pervasive oxidative lesions. Moreover, unlike other cancers, neuroblastoma consistently harbored high levels of C-strand ssDNA overhangs and t-circles, which are consistent with active “telomere trimming”. This feature is observed in both telomerase- and ALT-positive tumors and irrespective of telomere length distribution. Moreover, evidence for telomere trimming was detected in normal neural tissues, raising the possibility that TMMs in HRNB evolved in the face of a canonical developmental program of telomere shortening. Telomere trimming by itself appears to distinguish neuroectodermal derived tumors from other human cancers, a distinguishing characteristic with both biologic and therapeutic implications.
Abbreviations: HRNB, High risk neuroblastoma; ALT, Alternative lengthening of telomeres; TMM, telomere maintenance mechanisms
Introduction
Neuroblastoma is the most common extracranial solid tumor in children. It accounts for 6% of all pediatric tumor diagnosis, but 10% of mortality [1], making it one of the most aggressive and lethal childhood cancers. While many patients initially respond to front-line therapy, progression is frequent and leads to chemo-resistant disease and poor survival [2]. Genomic studies have revealed MYCN, TERT, and ATRX aberrations to be frequent and mutually exclusive drivers in high-risk disease. Because a common connection between these aberrations is their activation of telomere maintenance mechanisms (TMMs), telomeres and TMMs have emerged as pivotal attributes of high-risk neuroblastoma (HRNB) [3].
Telomeres are special nucleoprotein structures that cap chromosome ends to maintain genome stability, while concurrently playing a key role in controlling cellular proliferation [4]. Telomere DNA comprises numerous copies of a short repeat (5′-TTAGGG-3′/5′-CCCTAA-3′), which is G-rich on the 3′-end-containing strand (G-strand) and C-rich on the complementary, 5′-end-containing strand (C-strand). This telomere DNA nucleates the assembly of a special nucleoprotein structure at chromosome ends, which in turn allows the cells to recognize the normal ends (from abnormal double strand breaks) and to suppress inappropriate fusion and recombination reactions at telomeres [5], [6]. Moreover, an adequate amount of telomeric DNA is required to sustain cell proliferation, and a common hallmark of cancer cells is their acquisition of replicative immortality through up-regulation of TMMs [7]. In neuroblastoma, telomere DNAs are most often maintained by telomerase activation, which is strongly associated with MYCN amplification and TERT mutations [8], [9]. However, a significant percentage of neuroblastoma tumors (as high as 20% to 25% in some reports) utilizes alternative lengthening of telomeres (ALT, a recombination pathway) to replenish telomere DNA, and this pathway is tightly linked to ATRX mutations and deletions [10], [11]. (The frequency of ALT-positive tumors may be substantially lower in some cohorts diagnosed at a young age [8]). There is growing evidence that in addition to molecular biomarkers, telomere-specific features (length, heterogeneity, and extra-chromosomal telomeric DNA) are independently prognostic of high-risk disease [10], [12], [13]. This suggests that telomeres and TMMs are strongly connected to the underlying molecular pathogenesis of neuroblastoma and are of biological and clinical relevance. Indeed, a recent mechanistic classification of NB, derived from extensive profiling of numerous tumor samples, points to TMM as a key prognostic indicator [14].
While telomeres and TMMs have attracted considerable attention for their diagnostic and therapeutic potentials, previous characterization of telomere structures in this cancer is largely limited to measurements of average telomere lengths (by Southern analysis) and C-circle levels (marker of ALT activity) [12], [13]. However, there are other significant telomere features that impact on tumor biology. For example, the levels of single-stranded DNA on either the G- or C-strand could be indicative of abnormal telomere metabolism. While normal telomeres generally harbor a short (50 to 200 nt) 3′-overhang of the G-strand, “deprotected” telomeres can carry much longer overhangs due to excessive degradation of the C-strand [4]. 5′-overhangs of the C-strand have also been reported in selected settings, and been postulated to be a marker of telomere recombination and ALT [15], [16], [17]. In addition, the fraction of short telomeres in the cell instead of average telomere lengths, may be a better measure of cell proliferation, since even a few abnormally short telomeres are sufficient to trigger a senescence response [18]. Accordingly, assays such as STELA have been developed to specifically assess the levels of short telomeres [19], [20], [21]. These assays typically involve ligation of chromosome ends to one or two anchor oligonucleotides, followed by PCR amplification and Southern analysis to detect the amplified telomere fragments. To date, such assays (i.e., STELA or variants of STELA) have been applied mostly to primary cells (e.g., fibroblasts and lymphocytes) [20], [21], [22], and have not been used for comprehensive profiling of human cancers.
In this study, we broadened the characterization of telomeres in neuroblastoma by applying a series of assays that examined different aspects of telomere structures in cell lines and tumor samples derived from HRNB. Surprisingly, we found that HRNB harbors high levels of C-strand overhangs that are independent of ALT activation. The C-strand overhangs are also observed in normal neural tissues, and is likely due to active “telomere trimming”. The existence of telomere trimming in HRNB is further supported by the detection of substantial t-circles in multiple cell lines. In addition, in the course of analyzing telomere length distribution via STELA, we observed quite variable and often poor amplification of telomere DNA, indicative of substantial telomere damage. Further analysis suggests that the telomere DNA damages included 8-oxo-2′-deoxyguanine as well as other lesions. Our findings point to unique features of telomere dynamics in neuroblastoma that may have clinical implications.
Materials and methods
Tumor samples and cell lines
Use of neuroblastoma patient tissues for genetic studies was approved by the institutional review boards of Memorial Sloan Kettering Cancer Center (Clinicaltrials.gov NCT00588068). Written informed consent was obtained from patients or legal guardians at the time of surgical resection. All patients diagnosed with neuroblastoma at MSKCC had tumor samples archived in the tumor inventory. For this study, tumor samples were selected at random from the tumor registry to represent HRNB (n = 30). Only neuroblastomas from patients with metastatic disease were chosen. DNA was extracted and the integrity evaluated by gel electrophoresis. All tumors selected for the telomere studies passed pathology review for adequate tumor content (>50%) and adequate DNA quality. Primary acute myeloid leukemia (AML) specimens were obtained with informed consent and IRB approval at Weill Cornell Medical College. DNA was likewise extracted and the integrity evaluated by gel electrophoresis.
Neuroblastoma cell lines SK-N-MM, SK-N-BE(2)N, SK-N-F1 were originally established at MSKCC, while LAN-1 and LAN-6 were kindly provided by Dr. Robert Seeger at CHLA, Los Angeles. These cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and 100 units/ml penicillin–streptomycin. Leukemia cell lines MOLM13, K562 and KCL22 were obtained from the American Type Culture Collection (ATCC) or the DMSZ repository; these cell lines were authenticated and tested for mycoplasma. The leukemia cells were cultured in Iscove's Modified Dulbecco's Medium (IMDM; Life technologies) supplemented with 10%~20% fetal bovine serum (FBS) according to culture conditions specified by ATCC and 1% penicillin/streptomycin (Pen/Strep; Life Technologies). The human cervical cancer cell line HeLa and osteosarcoma cell line U2OS were also obtained from ATCC, and were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 100 units/ml penicillin–streptomycin.
Genomic DNA from tumors and cell lines were prepared using the All Prep kit (Qiagen), QiaAmp DNA Mini extraction kit (Qiagen), or the GeneJET genomic DNA purification kit (Thermo Fisher Scientific). The integrity of all preparations was verified by gel electrophoresis and concentrations determined by NanoDrop 2000 (Thermo Scientific).
Southern Analysis of Telomere Restriction Fragments
Standard Southern analysis of telomere restriction fragments (TRF) was performed using established protocols with minor modifications [23]. Briefly, chromosomal DNAs were digested with HinfI and RsaI, and fractionated in 0.8% agarose gels. Following transfer to nylon membranes, the blot was hybridized to a telomere repeat probe ((TTAGGG)82).
STELA
DNA from cell lines and tumor samples was extracted and purified using the GeneJET genomic DNA purification kit (Thermo Fisher Scientific). Ligation to telorette oligos [24] was performed in 15 μl reaction containing 10 ng genomic DNA, 0.01 μM telorette oligo, 1× CutSmart Buffer (NEB), 1 mM ATP, and 800 U T4 DNA ligase (NEB) at 35 °C for 20 hours [21]. PCR assays were carried out in 25 μl reactions containing 500 pg template DNA, 1 μM each of the subtelomeric forward primer and the teltail reverse primer, 1× Failsafe PCR PreMix H, and 2.5 U of Failsafe polymerase (Lucigen). Cycling reactions were performed in SimpliAmp (Applied Biosystems) with the following parameters: 30 sec at 94 °C, 30 sec at 60 °C and 12 min at 72 °C. Unless indicated otherwise, 33 cycles were utilized for these reactions. An alternative set of STELA amplification reactions (based on [25]) were carried out in 15 μl containing 1× Taq buffer (Thermo Fisher), 0.3 mM of each dNTP, 0.4 μM of forward and reverse primers, and 1 U of Pwo:Taq (10:1) polymerase mix. Twenty-nine cycles of 15 sec at 94 °C, 30 sec at 64 °C and 10 min at 68 °C were used for the alternative STELA. To ensure adequate coverage of the telomere size distribution, we performed 3 to 6 parallel PCR reactions for each ligated DNA sample. The reaction products were analyzed by electrophoresis in 0.8% agarose gels and subjected to Southern using the appropriate subtelomeric probes or a telomere repeat probe ((TTAGGG)82). Following PhosphorImager scanning (GE Healthcare), the sizes of individual telomeres were determined using TESLA software [21], and the results analyzed and plotted using Prism (GraphPad Software). In some of STELA assays, 50 ng of genomic DNA was digested with 5 U of Endonuclease VIII (NEB) and 4 U of Fpg (NEB) in 1× CutSmart buffer (NEB) at 37 °C for 2 h. Both glycosylases were inactivated at 75 °C for 20 min. Digested DNAs were applied to the ligation reaction for STELA analysis.
In-Gel Hybridization Analysis of ssDNA on Telomere G- and C-Strand
The standard in-gel hybridization analysis was performed using a combination of established protocols with minor modification [26]. For most assays, untreated genomic DNA was loaded directly onto a 0.8% agarose gel, and the current applied until the bromophenol blue dye has migrated about 4 cm into the gel. (This allows the signals to be concentrated into a tight band, thereby increasing the sensitivity of detection.) The gel was soaked in 2× SSC, and then dried for 30 minutes (into a thin layer) using a vacuum pressure of 550 mm Hg. The labeled G4 and C4 oligonucleotides, corresponding to four copies of the telomeric G-strand and C-strand repeats, were used as the probes, and hybridization was performed in the Church Mix at 42 °C. In some analysis, genomic DNA was digested with either Exonuclease I or RecJf before being subjected to the in-gel hybridization analysis.
C-Circle Assays
The C-circle assay was carried out as described [27] with minor modifications. Briefly, genomic DNAs were digested with EcoRI, and then subjected to rolling circle amplification in 20 μl reactions that contain 30 ng digested DNA, 0.2 mg/ml BSA, 0.1% Tween, 1 mM each dATP, dGTP and dTTP, 1× Φ29 Buffer and 5 U NxGen® phi29 DNA polymerase (Lucigen). The reactions were performed at 30 °C for 16 h, and then the DNA polymerase inactivated at 65 °C for 20 min. For quantification, the reaction products were dot-blotted onto a 2× SSC-soaked nylon membrane (Hybond-N, GE Healthcare, Inc.). Following UV-cross-linking, the G-strand products of C-circle assays were detected through hybridized at ~55 °C with 32P-labeled C8 probe ((CCCTAA)8).
Alkaline Gels
The alkaline gel electrophoresis was performed with modifications from a previously described protocol [28]. Genomic DNAs were digested with 135 U of Fpg (NEB) at 37 °C for 2 h and inactivation of Fpg was followed the addition of EDTA to 2 mM. After ethanol precipitation, DNAs were resuspended in 18 μl of alkaline loading buffer and denatured for 1.5 h at room temperature. The alkaline gel (0.8%) was prepared and pre-run at 1.5 V/cm for 1 h before the application of samples. After overnight electrophoresis at 1.5 V/cm, the gel was washed in 2× SSC for 30 min, stained for 45 min in 1 μg/mL of ethidium bromide, and destained in dH2O for 30 min. After the stained gel was photographed, it was soaked sequentially in 0.25 N HCl (30 min) and 0.5 N NaOH/1.5 M NaCl (45 min), and then rinsed with 10× SSC. The processed gel was blotted into nylon membrane (Hybond-N, GE Healthcare, Inc.) in 10× SSC and the blot was subjected to the standard telomere Southern using the 32P-labeled G8 ((TTAGGG)8) and C8 ((CCCTAA)8) probes. Following PhosphorImager scanning (GE Healthcare), the signals were quantified using ImageQuant software and plotted (Molecular Dynamics Inc.).
2D Gels
The two-dimensional gel analysis was performed as before [29] with slight modifications. Briefly, genomic DNA (5 μg) was digested with HinfI and AluI and subjected to two rounds of electrophoresis. The first dimension (0.5% agarose) was run at 0.5 V/cm for 16 h in the absence of ethidium bromide (EtBr). The gel was then stained with 0.3 μg/ml EtBr to visualize the size standards and bulk chromosomal DNA. Gel strips containing DNA in the 1.0 kb to ~15 kb size range are excised and impregnated in a 1.2% agarose gel containing 0.3 μg/ml EtBr. Electrophoresis was then performed in the orthogonal direction to the original gel lanes at 5 V/cm for 3.5 h. The DNAs in the gels were transferred to nylon membrane and probed with (TTAGGG)82.
Results
Telomeres in HRNB Cell Lines Contain Substantial C-Strand Overhangs Regardless of the Corresponding TMMs
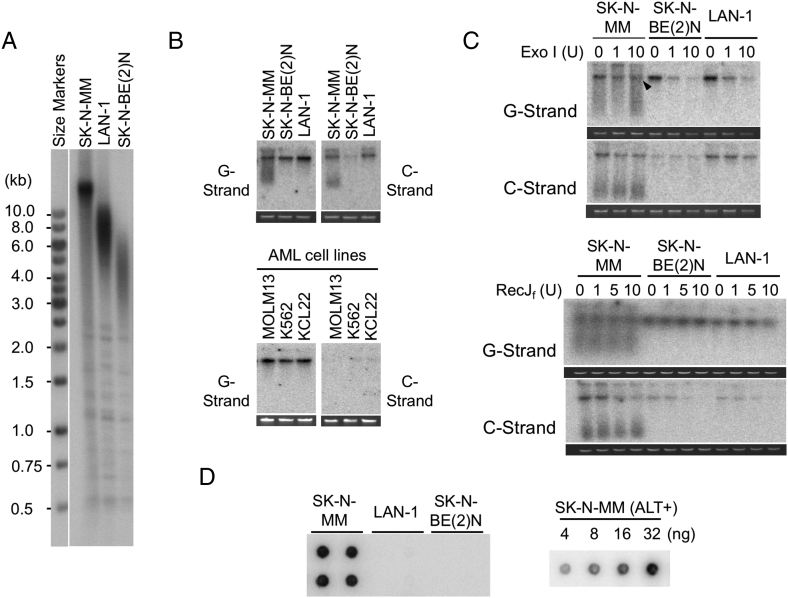
Our long-term objective is to comprehensively characterize the telomeres of HRNB tumors for diagnostic and therapeutic discoveries. As a benchmark for our telomere assays, we examined one ALT-positive cell line (SK-N-MM) and two telomerase-positive ones (LAN-1 and SK-N-BE(2)N) (Figure 1). First, we analyzed their overall telomere length distributions by standard TRF Southern. As expected for ALT-positive cancer cells, the telomere sizes of SK-N-MM were extremely heterogeneous and range from >10 kb to ~0.5 kb (Figure 1A). In contrast and also as expected, the telomeres of the two telomerase-positive cell lines were clustered in narrower size ranges, i.e., 5 to 10 kb for LAN-1 and 3 to 6 kb for SK-N-BE(2)N, respectively.
Figure 1.
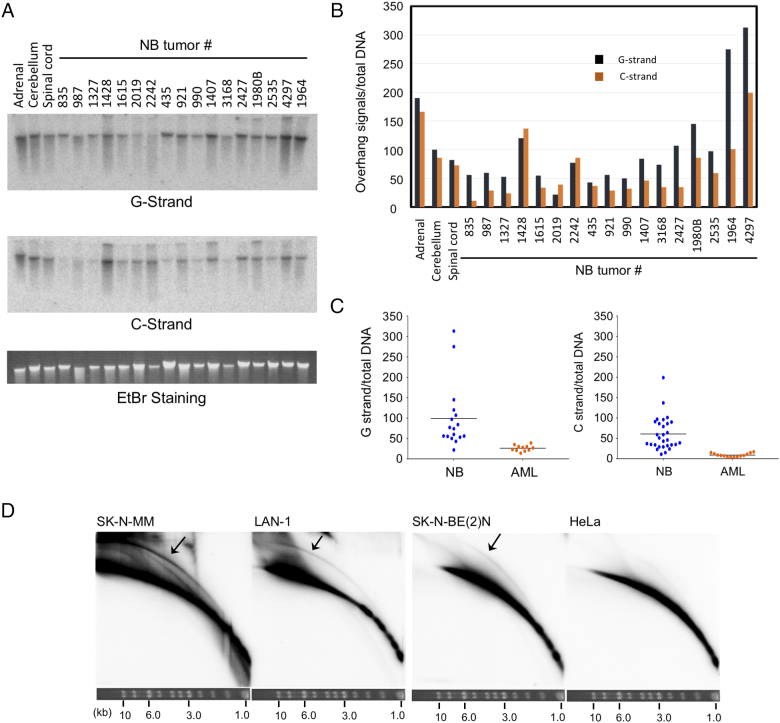
Analysis of telomere lengths and structures in neuroblastoma cell lines. A. Southern analysis for telomere restriction fragments (TRFs) was performed using DNA from one ALT positive cell line (SK-N-MM) and two telomerase positive cell lines (LAN-1 and SK-N-BE(2)N). B. Undigested genomic DNA from NB cell lines (Top) and AML cell lines (bottom) was subjected to in-gel hybridization assay to measure single-stranded DNA on the G- and C-strand. Ethidium bromide stained gels that were used for the analysis are shown below each blot. This applies to all in-gel hybridization assays presented in this paper. C. The G- and C-strand overhang signals were tested with respect to their sensitivity to Exonuclease I and RecJf. Different amounts of Exonuclease I (0 U, 1 U, and 10 U) and RecJf (0 U, 1 U, 5 U, 10 U) were used to digest 150 to 300 ng of genomic DNA at 37 °C 1 h and 2 h, respectively. Exonuclease I and RecJf degrade single-stranded 3′-overhang and 5′-overhang, respectively. Arrowhead indicates G-strand that appears to be partially resistant to Exonuclease I. D. C-circle assays were performed using 30 ng DNA from the indicated cell lines (left). All C-circle analysis of cell lines and tumor samples were performed in duplicates or quadruplets. A series of titration indicates that the assay was not saturated with up to 32 ng of SK-N-MM DNA, which exhibits the highest level of C-circles among all the analyzed samples (right).
Next, we assessed the levels of single-stranded telomere DNA on the G- and C-strand by in-gel hybridization. For this analysis, we applied undigested DNA directly to the gel such that the telomere ssDNA signal from the chromosomes was concentrated in a tight band, thus maximizing the sensitivity of detection. As noted before, normal chromosome ends typically possess short G-strand overhangs. Accordingly, we detected substantial signals for G-strand ssDNA in all three NB cell lines (Figure 1B, top-left panel). In addition to the signals from chromosome-sized DNA, SK-N-MM also exhibited lower molecular weight G-strand signals, which most likely derived from extrachromosomal telomere repeats (ECTR), known to be abundant in ALT-positive cells [15], [30]. Surprisingly, our assays revealed substantial C-strand ssDNA in all three NB cell lines (Figure 1B, top-right panel). While high levels of C-strand are often found in ALT-positive cells, they are rarely observed in telomerase-positive cells. The level of C-strand in LAN-1 was comparable to that in SK-N-MM, whereas the level in SK-N-BE(2)N was significantly lower. Consistent with the abundance of ECTR in ALT, only SK-N-MM exhibited substantially lower molecular weight C-strand signals. We confirmed that C-strand ssDNA was not a general feature of cancer cells as analyzed by our in-gel hybridization assay; none of the three AML cell lines that we analyzed in parallel contained detectable C-strand, although they did exhibit G-strand signals that were comparable to those in NB cell lines (Figure 1B, bottom, and data not shown).
To confirm that the ssDNA signal associated with chromosome-sized DNA was due to terminal overhangs, we digested the samples with E. coli Exo I or RecJf, which degraded single-stranded 3′-overhang and 5′-overhang, respectively (Figure 1C). As predicted, the G-strand signals were generally quite sensitive to Exo I, whereas the C-strand signals were sensitive to RecJf. Interestingly, in comparison to the telomerase-positive cell lines, a slightly higher fraction of SK-N-MM G-strand appeared to be resistant to Exo I (Figure 1C, marked by an arrowhead), suggesting that this ALT-positive cell line harbored more internal ssDNA on the G-strand (i.e., more gaps in the C-strand), possibly due to defective telomere replication. Notably, the low molecular weight G- and C-strands in SK-N-MM were refractory to both Exo I and RecJf, indicating that these ECTRs did not contain overhangs and were most likely circular DNAs.
Next, we examined the levels of C-circles, which were circular ECTRs that consisted of continuous C-strand and nicks or gaps on the G-strand. C-circles are known to be a specific and quantifiable marker of ALT TMM [27], and as expected, they were readily detected in SK-N-MM, but not in the two telomerase-positive cell lines (Figure 1D).
The Telomeres in NB Cell Lines Exhibited Reduced and Variable STELA Efficiency
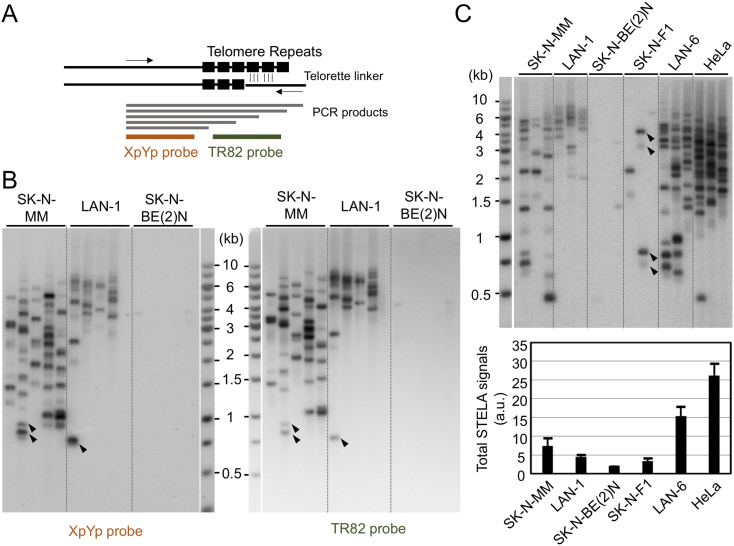
In addition to analyzing broad telomere size distributions, we sought to characterize at higher resolution the sub-population of relatively short telomeres because they are believed to have stronger impacts on cell proliferation [18]. For this, we utilized STELA, a strategy that entails the ligation of the 5′-end of C-strand to a linker (teltail), followed by PCR-based amplification of the ligated fragment and detection via Southern analysis (Figure 2A). Unlike Southern analysis, which preferentially detects longer telomeres (because they hybridize to more telomere repeat probes), STELA can efficiently identify even very short telomeres when a subtelomeric fragment is utilized as the probe (Figure 2A). We first applied STELA to the three NB cell lines and found that for two of these lines, the distributions of STELA fragments were largely consistent with those defined by Southern analysis. Specifically, the SK-N-MM STELA fragments were quite heterogeneous in size (from ~8 kb to <1 kb), whereas the LAN-1 fragments were more tightly clustered (from 8 to 3 kb) (Figure 2B). As predicted, the great majority of fragments in these assays hybridized to both the subtelomeric probe (XpYp) and the telomere repeat probe (TR82). Also as expected, the shorter STELA fragments generated stronger signals with the subtelomeric probe (Figure 2B, the fragments marked by arrowheads). Notably, for both SK-N-MM and LAN-1 samples, there were very few >8 kb fragments even though these were clearly visible in the Southern analysis. The absence of such long telomeres in the STELA analysis is consistent with a previous report and most likely due to inefficient PCR amplification [20].
Figure 2.
STELA analysis of telomere length distributions in neuroblastoma cell lines. A. A schematic diagram for STELA analysis was displayed. The telorette linker is annealed to telomere G-strand overhang and ligated to the 5′ end of the C-strand. The ligated DNA is then amplified by PCR (using primers that correspond to the linker sequence and a subtelomeric sequence), and detected by Southern. B-C. STELA analysis of five different NB cell lines and HeLa. To ensure adequate coverage of the telomere size distribution, 3 to 5 parallel PCR reactions for each ligated DNA sample were performed in all STELA assays in this paper. To confirm specificity of the PCR reactions, two different probes (subtelomere-specific XpYp and telomere-specific TR82) were used. Arrowheads in B indicate several very short telomeres. Arrowheads in C highlight a few STELA fragments of very different intensity in the same reaction. Total STELA signals from each cell line analyzed in C were quantified and plotted below the blot image.
Most unexpectedly, we found the efficiency of STELA to be very poor for the SK-N-BE(2)N DNA (Figure 2B). This was not due to the amount or quality of DNA used in the assay given that the same DNA generated quite robust Southern signals (Figure 1A). To further explore the variability in STELA efficiency, we examined two additional NB cell lines (SK-N-F1 and LAN-6) as well as HeLa, a common reference cell line from cervical carcinoma (Figure 2C, top). LAN-6 exhibited slightly higher STELA efficiency than the other NB cell lines, whereas SK-N-F1 generated very few fragments, just like SK-N-BE(2)N. Notably, in comparison to all five NB cell lines, the HeLa DNA manifested much higher overall STELA efficiency (Figure 2C, top and bottom). In addition, the intensities of STELA fragments derived from HeLa appeared to be uniformly strong, unlike the quite variable intensities found in individual NB samples (Figure 2C, some examples are marked by arrowheads). Together, these observations suggest that NB telomeres had features that substantially and variably reduced PCR efficiency.
The Telomeres in NB Cell Lines Harbor Substantial and Variable DNA Damage
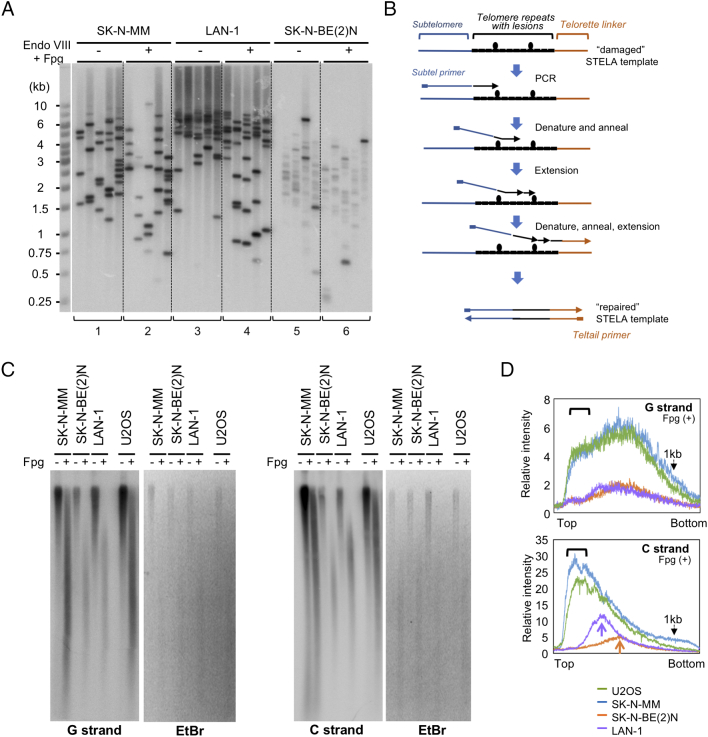
We reasoned that if the poor STELA efficiency of SK-N-BE(2)N and SK-N-F1 telomeres is due to impaired PCR amplification, this may be compensated by increasing the PCR cycles. Indeed, when the cycle number was increased from 33 to 36, we can readily detect STELA fragments in the SK-N-BE(2)N samples, even though the intensities of these fragments were ~20 fold lower than those from LAN-1 and SK-N-MM (Figure 3A, compare sample sets 1, 3, and 5). Thus, the inefficient STELA of selected NB samples was probably due to poor PCR amplification rather than poor ligation. Because DNA damages in cancer cells have been shown to cause sequencing errors by interfering with polymerase function [31], we hypothesized that NB tumors such as SK-N-BE(2)N may harbored more telomere damages that blocked the efficient elongation of the PCR polymerase (Failsafe PCR enzyme in our standard protocol). One model (shown in Figure 3B) for how such damaged telomeres might nevertheless be successfully amplified is as follows. During the first round of copying the ligated C-strand, the DNA polymerase could pause or terminate at a lesion. However, because of the repetitive nature of the telomere sequence, during the next round of PCR, the 3′-end of the partial product could anneal beyond the lesion site, allowing the DNA polymerase to carry out further extension until it reached the next lesion. By repeating this process in multiple cycles, a fully “repaired” STELA template (i.e., harboring both the subtelomeric primer sequence and the teltail linker sequence) can be generated, and subsequently amplified just like an undamaged STELA template. Notably, according to this model, the repaired STELA template is likely to be shorter than the original damaged template. Indeed, we found that the lengths of the SK-N-BE(2)N STELA fragments (1.5 to 4 kb, sample set 5 in Figure 3A) were substantially shorter than those of the corresponding TRFs (3 to 6 kb, Figure 1A).
Figure 3.
Analysis of the effect of glycosylases on STELA and DNA nicks in neuroblastoma samples. A. NB cell line DNA samples (with and without Fpg and Endonuclease VIII treatment) were subjected to STELA analysis. To enable detection of weak STELA fragments from the SK-N-BE(2)N samples (sample sets 5 and 6), the PCR cycle number was increased from 33 to 36 for these assays. For the glycosylase-treated samples, 50 ng of each genomic DNA was digested with 5 U of Endonuclease VIII and 4 U of Fpg at 37 °C for 2 h. B. A possible model to for the low STELA efficiency of SK-N-BE(2)N sample is presented. See the main text for a detailed discussion. C. DNA from the indicated cell lines with or without Fpg treatment were analyzed by alkaline denaturing gel electrophoresis followed by Southern analysis using 32P-labeled C8 ((CCCTAA)8) probe in the G-strand assay panel and G8 ((TTAGGG)8) probe in the C-strand panel. The ethidium bromide stained gels (EtBr) for this analysis are shown to the right of the Southern images. To facilitate visual comparison, the ethidium bromide stained gel images are inverted. D. The traces for the Fpg-treated samples from all cell lines are plotted together for easy comparison. The data for G- and C-strand are presented separately. Thick black brackets indicate longer ssDNA found only in ALT-positive cells. The purple and orange arrows highlight the different distributions of the LAN-1 and SK-N-BE(2)N C-strand fragments following Fpg treatment.
Because PCR polymerases vary in fidelity and tolerance to modified nucleotides, we wonder if the use of alternative polymerases might allow for more efficient amplification of damaged DNA. Indeed, when we employed an alternative STELA protocol that entailed amplification by Pwo and Taq polymerases [25], the SK-N-BE(2)N DNA became quite efficient at generating PCR products (Supp Figure 1). Moreover, the size distribution of the SK-N-BE(2)N STELA products from this alternative protocol (2 to 6 kb) corresponds well that in Southern analysis. Because Pwo is known to incorporate nucleotides with bulky side-chains, it may also be less sensitive to modified nucleotides in the telomere DNA template, thus allowing damaged DNA to be copied more efficiently.
One potential source of DNA damage is oxidized nucleotides [32]. To probe the existence of such nucleotides in telomeres, we analyzed the effects of glycosylase treatment on STELA. Both endonuclease VIII and Fpg are expected to induce nicks at oxidative DNA damage sites. Therefore, if there are many more such damaged sites in SK-N-BE(2)N, we expect that upon glycosylase treatment, STELA signals might be further reduced and the STELA fragments preferentially shortened in the SK-N-BE(2)N samples. However, this was not the case. Instead, we identified a new sub-population of slightly shorter telomeres in all three NB cell line samples following glycosylase digestion (Figure 3A, compare 1 vs 2, 3 vs 4, and 5 vs 6), suggesting that all three NB cell lines contained some endonuclease VIII- and Fpg-sensitive lesions in telomeres. Such lesions, however, were probably not responsible for the especially weak STELA of SK-N-BE(2)N DNA, given the lack of a preferential effect of the glycosylases on this DNA. Instead, SK-N-BE(2)N might contain other DNA polymerase-blocking lesions that impaired PCR efficiency.
To further investigate the extent of oxidative DNA damage in NB cells, we performed alkaline (denaturing) gel electrophoresis after treating the DNA with Fpg and/or endonuclease VIII. Remarkably, we observed substantial reduction in the size of ethidium bromide-strained ssDNA following Fpg but not endonuclease VIII treatment (data not shown), suggesting that the Fpg-sensitive lesions were quite prevalent throughout the genome of all three NB cell lines. We then specifically analyzed the fate of telomere-containing ssDNA following just Fpg treatment by alkaline gel electrophoresis and Southern analysis. In addition to the three NB cell lines, we analyzed in parallel U2OS, another ALT-positive cell line derived from osteosarcoma. Similar to the bulk genomic DNA, we found that both G-strand-containing ssDNA and C-strand-containing ssDNA were cleaved into smaller fragments by Fpg, consistent with the presence of Fpg-sensitive lesions in the telomere region (Figure 3, C and D). Following Fpg treatment, both ALT-positive cell lines (SK-N-MM and U2OS) harbored higher levels of longer ssDNA that hybridized to telomere repeats (Figure 3D, highlighted by thick black brackets), as expected from the presence of extra-long telomeres in these cells that might stochastically contain few lesions. For the two telomerase-positive cell lines, the sizes of the G-strand ssDNA were nearly identical after treatment, suggesting similar distribution of Fpg-sensitive lesions in the telomere regions (Figure 3D top diagram, orange and purple traces). In contrast, the sizes of the C-strand ssDNA were longer in LAN-1 than those in SK-N-BE(2)N (Figure 3D bottom diagram, marked by two arrows). This discrepancy mirrored the telomere lengths in these two cell lines and could possibly be explained by the absence of guanines on the C-strand repeat, making it unlikely for Fpg to cleave within the telomere repeats (although it could certainly cleave the subtelomeric region). We also attempted to correlate telomere DNA damage to DNA damage throughout the genome by comparing the pattern of ethidium bromide staining to those in the Southern analysis (Figure 3C). While the sizes of ssDNA in the stained gel following Fpg digestion appeared similar those in the Southern analysis, it was difficult to draw firm conclusions given the broad size distributions and the limited resolution of the gel. Taken altogether, our data suggested that oxidized purines (sensitive to Fpg) might be quite prevalent throughout the NB genome, and that because of its high purine content, the G-strand of telomeres might be enriched in such lesions.
Overall, the most notable findings from our analysis of the three NB cell line telomeres were (1) the detection of abundant C-strand overhangs in both ALT and non-ALT NB cells; and (2) evidence for relatively high levels of telomere DNA damage in NB. Therefore, we next proceeded to determine if these features are found among HRNB tumors.
C-Strand ssDNA Overhangs Were Consistently Detected in HRNB Tumors
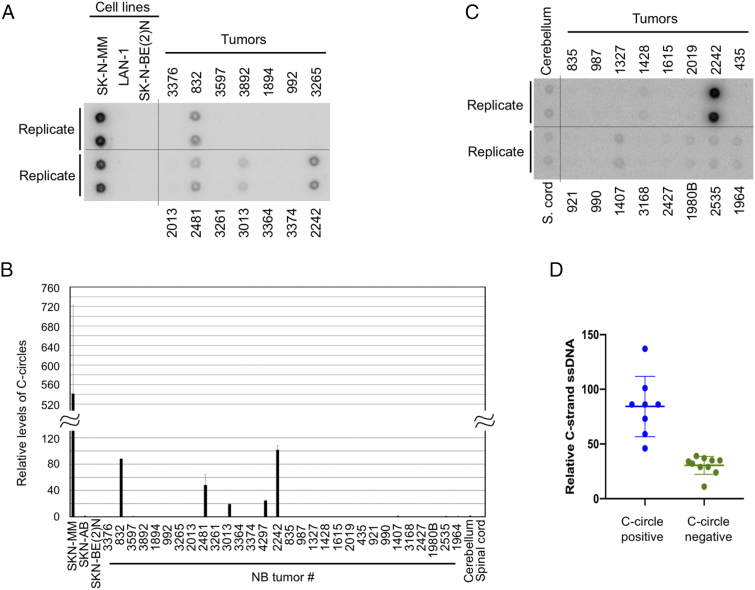
We proceeded to characterize single-stranded telomere DNA in 30 NB tumor samples using in-gel hybridization. However, before we analyzed the levels of G- and C-strand overhangs, the TMMs of these tumor samples were first assessed by C-circle analysis and their ATRX status, which was found to correlate strongly with ALT TMM. As shown in Figure 4, A and B, 4 of these NB tumors (832, 2481, 3013, and 2242) exhibited high levels of C-circles. Compared to the SK-N-MM cell line, the levels of C-circles in these tumors were lower (by ~5 to 27-fold), but still substantially elevated relative to other samples. All 4 tumors also carried deletions in ATRX, supporting a strong linkage between mutations in this gene and the ALT TMM. Interestingly, one other tumor (3374) that carried a point mutation in one ATRX allele (L407F) did not yield detectable C-circle, suggesting that this mutation did not affect TMM, and the HRNB was cured in this patient. We then analyzed the tumor DNA by in-gel hybridization, and in agreement with the results from cell lines, detected substantial and variable levels of G- and C-strand ssDNA in both ALT and non-ALT samples (Figure 5A). In general, there was a moderate but not tight correlation between G- and C-strand levels, with the former being slightly higher than the latter (Figure 5B). Unexpectedly, three normal neuroectoderm-derived tissues (adrenal, cerebellum, and spinal cord) also manifested significant G- and C-strand ssDNA (Figure 5, A and B). These results raise the intriguing possibility that the unusual ssDNA at NB telomeres was inherited from the developmental program of these cells. In comparison to NB, AML tumor samples consistently manifested lower levels of G-strand ssDNA, as well as undetectable C-strand, confirming that this aspect of telomere structure was distinct between neuroectodermal derived versus hematopoietic derived cancers (Figure 5C).
Figure 4.
Analysis of the levels of C-circles in NB tumors. A. C-circle assays were carried out using genomic DNA from the indicated NB cell lines and tumors. The cell line assays were done in quadruplet, and the tumor assays were done in duplicates. B. The levels of C-circles from the indicated cell lines, tumor samples, normal tissues were quantified and plotted. The signal from all samples were normalized to that from the spinal cord, which is set as 1 unit. C. C-circle assays were carried out using genomic DNA from the indicated normal tissues and tumor samples. After scanning, the ImageQuant scale is compressed to allow the visualization of weak signals (compare the signals for tumor 2422 in A and C). D. The C-strand ssDNA levels of the 7 samples with detectable C-circles and the 10 samples with undetectable C-circles were plotted as blue and green circles, respectively.
Figure 5.
Analysis of ssDNA on the G- and C-strand in NB tumors and the detection of t-circles in NB cell lines. A. The levels of single-stranded G- and C-strand signals were measured by in-gel hybridization assay. B. From each in-gel hybridization blot, the relative signals of the G- and C-strand overhangs to total DNA (quantified by ethidium bromide staining) were determined and plotted. G-strand signals are shown as dark blue bars and C-strand signals orange bars. C. The levels of G- and C-strand overhangs are compared between multiple NB and AML tumor samples. D. The levels of t-circles in the indicated cell lines were analyzed by 2D gel electrophoresis and hybridization to the TR82 probe. The arcs corresponding to t-circles in the NB samples are highlighted by arrows. The gel strip showing the migration of size standards after the first round of electrophoresis is displayed at the bottom of each blot. The diagram on the left illustrates the directions of electrophoresis and the expected paths for linear and circular DNAs.
C-Strand Overhangs in NB and Neural Cells Are Due To Telomere Trimming
While C-strand overhangs were initially thought to be related to ALT, more recent studies suggest that this structure is often a result of telomere trimming, a telomere shortening pathway that entails the recombinational excision of telomere repeats. Interestingly, low levels of C-circles (much lower than those found in ALT cells) were recently shown to be another marker of the telomere trimming pathway [17]. We re-analyzed our C-circle data to identify samples with low but detectable levels of this structure. Among the 17 non-ALT samples re-examined for this purpose (including 15 NB tumors), seven exhibited detectable C-circles (Figure 4C). Notably, these seven samples also consistently manifested high levels of C-strand ssDNA (Figure 4D, blue circles). This correlation lends additional support to the notion that the C-strand ssDNA in NB tumors may be a hallmark of telomere trimming activity. The most definitive marker of this activity is t-circle, which is thought to represent the product of recombinational excision and which can be unequivocally detected using 2D gels [17], [33]. Because the 2D gel assay requires substantial amount of DNA, we performed this analysis using samples from three NB cell lines and HeLa (Figure 5D). Indeed, we were able to detect clear t-circle arcs in all the NB DNA but not HeLa. Moreover, there is a modest quantitative correlation between the levels of t-circles and C-strand ssDNA in the different NB cell lines (i.e., more of both structures in the SK-N-MM and LAN-1 cells than in SK-N-BE(2)N). Altogether, the C-strand ssDNA, C-circle and t-circle analysis support the existence of active telomere trimming in HRNB. Therefore, in addition to incomplete end replication, NB telomeres may frequently suffer from another mechanism of telomere shortening.
NB Tumor DNA Samples Manifest Variable STELA Efficiency and Distinct Telomere Size Distributions
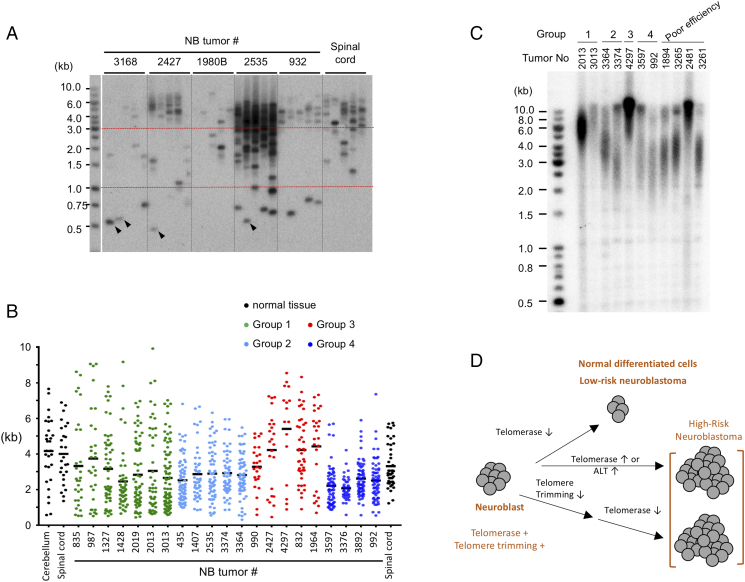
To determine if variable STELA efficiency is also a characteristic of NB tumors, we applied the assay to a total of 30 NB tumor samples (Figure 6). Interestingly, 9 of the 30 samples showed poor STELA efficiency, generating very few and weak fragments, similar to what was observed for SK-N-BE(2)N (Figure 6A, samples 3168 and 1980B, and data not shown). For the other 21 tumors and 2 normal neural tissues, we analyzed the STELA size distributions using TESLA software and plotted the results in Figure 6B. Based on the patterns of distribution, we classified the tumors into 4 groups. Group 1 (7/30 tumors, shown in green) exhibits broad distribution of telomeres, with some very long and some quite short fragments. Even though this pattern superficially resembled that from the ALT-positive cell line SK-N-MM, only one of 7 tumors in this group (3013) was ALT-positive based on C-circle assays. Group 2 (5/30 tumors, shown in light blue) yielded well defined telomere clusters in the 1.5 to 6 kb range (e.g., tumor 2535 in Figure 6A). Group 3 (5/30 tumors, shown in red) harbored mostly telomeres that were >3 kb, but with some containing a minor population of short telomeres (e.g., tumor 2427 in Figure 6A). Finally, Group 4 (4/30 tumors, shown in dark blue) harbored tightly clustered, short telomeres in the 1 to 3 kb range. For tumors from which sufficient DNA is available, we also analyzed the telomeres by TRF Southern (Figure 4C). All the samples, including those that were inefficient in STELA assays (Figure 4C, last 4 lanes), manifested sizable telomeres, indicating that the absence of STELA products was not due to absence of telomeres or degradation of DNA. In addition, similar to results for cell line DNA, the comparison of TRF and STELA analysis revealed under-representation of long telomeres in the STELA assays. For example, both of the group 2 samples manifested 8 to 10 kb telomeres by TRF Southern but generated few such fragments in STELA. Overall, our survey of NB tumor telomere distributions supported the existence of substantial telomere DNA damage in some samples, as well as heterogeneity of telomere size distributions across tumors.
Figure 6.
STELA analysis of telomere length distributions in neuroblastoma tumors; model for the roles of telomere trimming and TMMs in NB proliferation. A. Representative STELA analysis results for the indicated NB tumors are shown. A few especially short STELA fragments that correspond to the previously described T-stumps are designated by arrowheads. B. STELA signals were analyzed by TESLA software and plotted with prism software. Based on distributions of STELA signals, NB tumors are divided into 4 groups: broad distribution (group 1), well defined telomere cluster of 1.5 to 6 kb (group 2), mostly long telomeres (>3 kb) with a minor population of short telomeres (group 3), and tightly clustered short telomeres of 1 to 3 kb (group 4). NB tumors that showed poor STELA efficiency are not included in this classification. C. Selected NB tumor DNAs were subjected to TRF Southern analysis. Ethidium bromide staining of the gel indicate that all samples have comparable amounts of DNA except for tumor 3013, which contained two- to threefold less DNA. D. Model for how telomere trimming and TMMs may influence the proliferative capacity of neuroblast-derived normal and tumor cells. The neuroblast progenitor cells are proposed to harbor both telomere trimming and telomerase activity. Normal neural tissues and low-risk neuroblastoma have limited capacity for cell division due to the retention of telomere trimming and repression of TMMs (top branch). In contrast, high-risk neuroblastoma can sustain proliferation by either activating TMMs to compensate for the telomere trimming-mediated shortening (middle branch) or by repressing telomere trimming completely prior to shutting off telomerase (middle branch).
A previous STELA analysis of cancer cell lines indicate that some cancers may harbor a significant population of extra-short telomeres with very few telomere repeats [34]. Such telomeres are referred to as T-stumps and correspond to STELA fragments that are ~500 bp or shorter. T-stumps were also occasionally detected in our NB cell lines and tumor samples (see e.g., fragments marked by arrowheads in Figure 6A), but they represented a very minor fraction of total STELA fragments. In addition, our preliminary analysis did not reveal any association of T-stumps with other telomere features such as overall telomere distributions and telomere ssDNA.
Discussion
In this study, we expanded previous characterization of neuroblastoma telomeres by investigating for the first time single-stranded G- and C-overhangs at telomeres in multiple cell lines and a collection of HRNB tumor samples. We also characterized telomere length distribution using STELA, a technique especially well-suited to the identification of short telomeres. Our findings revealed surprising and unique features of neuroblastoma telomeres that may have both biological and therapeutic implications. These implications are discussed below.
C-Strand Overhangs and C-Circles in Neuroblastoma Cell Lines and Tumor Samples
Perhaps the most unexpected among our findings is the detection of high and variable levels of C-strand overhangs in neuroblastoma that is unrelated to ALT TMM. This has not been reported for any other cancer, and we confirmed that C-strand overhangs are rarely detected in e.g., acute myeloid leukemia. The co-detection of such C-strand overhangs with t-circles as well as low levels of C-circles suggests that they all reflect active “telomere trimming” [17]. This pathway for rapid telomere shortening is believed to be due to recombinational excision of telomere repeats. While telomere trimming has not been previously linked to a specific cancer, it was reported to be active in sperms, stem cells (both embryonic and induced pluripotent stem cells), and activated lymphocyte [17], [33]. These precedents suggest that that the variable levels of the telomere trimming activity in neuroblastoma may be due to the retention of an inherent activity in neural crest cells [35], from which neuroblastoma originates (Figure 6C). Even more surprising, we detected similar C-strand overhangs and C-circles in spinal cord and cerebellum, suggesting that this pathway persists in post mitotic neurons. Indeed, a recent analysis of telomeres in mouse brains during aging revealed telomere erosion in non-replicative neural cells [36], which is consistent with active telomere trimming. How this trimming activity and the resulting telomere C-strand overhangs impact on the biology of normal and cancerous neural cells is currently unknown. Notably, telomere trimming was hypothesized to be a homeostatic mechanism that counteract over-elongation of telomeres by telomerase, and therefore preferentially activated in cells with long telomeres [17], [33]. However, this is apparently not the case for neuroblastoma tumors; we found quite high levels of C-strand overhangs in many samples with short telomeres, suggesting that this trimming activity is widespread in this cancer, resulting in the continuous production of short telomeres, and a compensatory TMM.
The substantial and variable levels of C-overhangs in neuroblastoma suggest tumor heterogeneity in telomere dynamics. High levels of telomere trimming activity may continually generate relatively short telomeres that require TMM for maintaining tumor cell proliferation (Figure 6C). Conversely, low trimming activity may reduce or obviate the requirement for TMM. In this regard, it is worth noting that two neuroblastoma cell lines capable of extensive proliferation in vitro were recently found to possess no TMM [11]. Based on the level of t-circles, Dagg et al. proposed that the repression of trimming activity in these cell lines may have resulted in extremely elongated telomeres, which in turn sustained extensive proliferation (Figure 6C) [11]. Together, the prior and current findings support the further examination of the molecular basis and regulation of telomere trimming in neuroblastoma and other cancers. Insights on telomere trimming mechanisms remain quite limited; to date only XRCC3, Nbs1 (two recombination proteins) and TZAP/ZBTB48 (a telomere-binding protein) have been strongly implicated in this pathway [17], [33], [37], [38]. Whether these factors as well as other recombination proteins are responsible for the disparate levels of telomere trimming in different cancers is an interesting question for future investigation.
Telomere DNA Damage in Neuroblastoma Cell Lines and Tumor Samples
In the course of analyzing telomere length distribution via STELA, we observed substantial variability in both the abundance and intensity of PCR fragments across neuroblastoma cell lines and tumor samples. This has not been reported in previous applications of STELA to other cancers [34]. Although we do not have a full understanding of the molecular basis that underlies neuroblastoma STELA heterogeneity, one likely explanation is high levels of telomere DNA damage, including oxidative damage. In particular, we showed that treatment of the neuroblastoma DNA with Fpg induced high levels of nicks, causing substantial reductions in the sizes of telomere-bearing ssDNA upon alkaline gel electrophoresis (Figure 4). Notably, Fpg-sensitive lesions were probably not the only ones responsible for poor STELA efficiency. The SK-N-B(E)2 N and LAN-1 cell lines, which manifested very different STELA efficiencies, evidently harbored similar levels of Fpg-sensitive lesions. Previous RNA-seq profiling of neuroblastoma cell lines revealed significant variabilities in the expression of glycosylases and endonucleases involved in the repair of various DNA lesions [39], providing one explanation for the varying levels of such aberrations in our samples.
The DNA size reduction in alkaline gels caused by Fpg treatment was similar for telomere-bearing DNA (detected in Southern analysis) and the bulk of genomic DNA (visualized by ethidium bromide). Thus, the levels of 8-oxo-dG and other Fpg-sensitive lesions appeared comparable at telomeric and non-telomeric locations. In addition, the level of Fpg-sensitive lesions was unaffected by the TMM of the cell line given that all samples exhibited similar sized DNA following Fpg treatment in alkaline gels. While the DNA lesions in neuroblastoma were not telomere or TMM-specific, they might nevertheless have implications for telomere-targeting strategies. In particular, high levels of baseline telomere DNA damage may present greater challenges to the telomere replication machinery and increase the frequency of telomere truncation. In addition, 8-oxo-dG near telomere 3′ ends have been shown to inhibit telomerase activity in vitro, and this may account for the continuous telomere shortening observed in cells deficient in PRDX1 and MTH1, two factors that minimize oxidative DNA damage [40], [41], [42]. Moreover, the combined deficiency in telomerase and the DNA glycosylase NTH1 causes severe telomere loss [43]. Together, these considerations suggest that tumors with high levels of DNA damage may be more susceptible to telomerase inhibition or other telomere-targeting strategies. The connection between oxidative damage and telomeres is supported by another study, which investigated the potential of targeting telomeres through 6-thio-dG [44]. This nucleoside analog is a substrate for telomerase, and its incorporation into telomeres triggers a telomere DNA damage response. In a recent report, 6-thio-dG was shown to work synergistically with the mitochondrial inhibitor Gamitrinib (which increases reactive oxygen species and could cause substantial DNA damage) in suppressing the growth of NRAS-mutated melanoma [44]. A better understanding of the relationships between telomere DNA damage, telomere maintenance mechanisms, and telomere shortening pathways (both incomplete replication and telomere trimming) in specific cancers will be necessary to fully exploit the potential of telomeres in cancer diagnosis and therapies.
In summary, we have shown that NB telomeres (analogous to normal neural tissues) may be subjected to significant telomere trimming, and may harbor high levels of telomere DNA damage. Both features could render neuroblastoma more reliant on TMMs, thus accounting for the especially strong prognostic significance of TMMs for this cancer [14]. Our results also suggest that telomere trimming and DNA damage could serve as useful biomarkers for HRNB. Whether other cancers present similar opportunities for telomere-based diagnostics and therapies is an interesting question for future investigation.
The following is the supplementary data related to this article.
STELA analysis using alternative PCR polymerases. LAN-1 and SK-N-BE(2)N DNAs were subjected to STELA using a combination of Pwo and Taq polymerases (10:1). Six amplification reactions for each DNA were performed.
Funding
This work was supported by a pre-R01 seed grant from the Sandra and Edward Meyer Cancer Center (NFL and MLG) and the Bohmfalk Charitable Trust (NFL). NFL is a William Randolph Hearst Endowed Faculty Fellow in Microbiology. NKC was supported by Enid Haupt Endowed Chair, Robert Steel foundation and the Catie Hoch Foundation. MLG also received supports from Irma T. Hirschl/Monique Weill-Caulier Charitable Fund and R01CA172546.
Acknowledgments
We thank Yie Liu (NIA) for suggestions on the analysis of DNA damage at telomeres, and members of our laboratories for discussion.
References
- 1.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64:83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 2.Canete A, Gerrard M, Rubie H, Castel V, Di Cataldo A, Munzer C, Ladenstein R, Brichard B, Bermudez JD, Couturier J. Poor survival for infants with MYCN-amplified metastatic neuroblastoma despite intensified treatment: the International Society of Paediatric Oncology European Neuroblastoma Experience. J Clin Oncol. 2009;27:1014–1019. doi: 10.1200/JCO.2007.14.5839. [DOI] [PubMed] [Google Scholar]
- 3.Hertwig F, Peifer M, Fischer M. Telomere maintenance is pivotal for high-risk neuroblastoma. Cell Cycle. 2016;15:311–312. doi: 10.1080/15384101.2015.1125243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lange T. Shelterin-Mediated Telomere Protection. Annu Rev Genet. 2018;52:223–247. doi: 10.1146/annurev-genet-032918-021921. [DOI] [PubMed] [Google Scholar]
- 5.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnoult N, Karlseder J. Complex interactions between the DNA-damage response and mammalian telomeres. Nat Struct Mol Biol. 2015;22:859–866. doi: 10.1038/nsmb.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Hiyama E, Hiyama K, Yokoyama T, Matsuura Y, Piatyszek MA, Shay JW. Correlating telomerase activity levels with human neuroblastoma outcomes. Nat Med. 1995;1:249–255. doi: 10.1038/nm0395-249. [DOI] [PubMed] [Google Scholar]
- 9.Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Kramer A, Roncaioli JL, Sand F, Heuckmann JM. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, Heguy A, Pappo AS, Federico S, Dalton J. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307:1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dagg RA, Pickett HA, Neumann AA, Napier CE, Henson JD, Teber ET, Arthur JW, Reynolds CP, Murray J, Haber M. Extensive Proliferation of Human Cancer Cells with Ever-Shorter Telomeres. Cell Rep. 2017;19:2544–2556. doi: 10.1016/j.celrep.2017.05.087. [DOI] [PubMed] [Google Scholar]
- 12.Farooqi AS, Dagg RA, Choi LM, Shay JW, Reynolds CP, Lau LM. Alternative lengthening of telomeres in neuroblastoma cell lines is associated with a lack of MYCN genomic amplification and with p53 pathway aberrations. J Neurooncol. 2014;119:17–26. doi: 10.1007/s11060-014-1456-8. [DOI] [PubMed] [Google Scholar]
- 13.Ohali A, Avigad S, Ash S, Goshen Y, Luria D, Feinmesser M, Zaizov R, Yaniv I. Telomere length is a prognostic factor in neuroblastoma. Cancer. 2006;107:1391–1399. doi: 10.1002/cncr.22132. [DOI] [PubMed] [Google Scholar]
- 14.Ackermann S, Cartolano M, Hero B, Welte A, Kahlert Y, Roderwieser A, Bartenhagen C, Walter E, Gecht J, Kerschke L. A mechanistic classification of clinical phenotypes in neuroblastoma. Science. 2018;362:1165–1170. doi: 10.1126/science.aat6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oganesian L, Karlseder J. Mammalian 5' C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol Cell. 2011;42:224–236. doi: 10.1016/j.molcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oganesian L, Karlseder J. 5' C-rich telomeric overhangs are an outcome of rapid telomere truncation events. DNA Repair (Amst) 2013;12:238–245. doi: 10.1016/j.dnarep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera T, Haggblom C, Cosconati S, Karlseder J. A balance between elongation and trimming regulates telomere stability in stem cells. Nat Struct Mol Biol. 2017;24:30–39. doi: 10.1038/nsmb.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaul Z, Cesare AJ, Huschtscha LI, Neumann AA, Reddel RR. Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep. 2011;13:52–59. doi: 10.1038/embor.2011.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 20.Bendix L, Horn PB, Jensen UB, Rubelj I, Kolvraa S. The load of short telomeres, estimated by a new method, Universal STELA, correlates with number of senescent cells. Aging Cell. 2010;9:383–397. doi: 10.1111/j.1474-9726.2010.00568.x. [DOI] [PubMed] [Google Scholar]
- 21.Lai TP, Zhang N, Noh J, Mender I, Tedone E, Huang E, Wright WE, Danuser G, Shay JW. A method for measuring the distribution of the shortest telomeres in cells and tissues. Nat Commun. 2017;8:1356. doi: 10.1038/s41467-017-01291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang EE, Tedone E, O'Hara R, Cornelius C, Lai TP, Ludlow A, Wright WE, Shay JW. The maintenance of telomere length in CD28+ T cells during T lymphocyte stimulation. Sci Rep. 2017;7:6785. doi: 10.1038/s41598-017-05174-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sfeir A, Chai W, Shay J, Wright W. Telomere-end processing the terminal nucleotides of human chromosomes. Mol Cell. 2005;18:131–138. doi: 10.1016/j.molcel.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 25.Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu EY, Perez-Martin J, Holloman WK, Lue NF. Mre11 and Blm-dependent formation of ALT-like telomeres in Ku-deficient Ustilago maydis. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 28.Sutherland BM, Bennett PV, Sutherland JC. DNA damage quantitation by alkaline gel electrophoresis. Methods Mol Biol. 2006;314:251–273. doi: 10.1385/1-59259-973-7:251. [DOI] [PubMed] [Google Scholar]
- 29.Yu EY, Yen WF, Steinberg-Neifach O, Lue NF. Rap1 in Candida albicans: an unusual structural organization and a critical function in suppressing telomere recombination. Mol Cell Biol. 2010;30:1254–1268. doi: 10.1128/MCB.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Liu P, Evans TC, Jr., Ettwiller LM. DNA damage is a pervasive cause of sequencing errors, directly confounding variant identification. Science. 2017;355:752–756. doi: 10.1126/science.aai8690. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Rhee DB, Lu J, Bohr CT, Zhou F, Vallabhaneni H, de Souza-Pinto NC, Liu Y. Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickett HA, Henson JD, Au AY, Neumann AA, Reddel RR. Normal mammalian cells negatively regulate telomere length by telomere trimming. Hum Mol Genet. 2011;20:4684–4692. doi: 10.1093/hmg/ddr402. [DOI] [PubMed] [Google Scholar]
- 34.Xu L, Blackburn EH. Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol Cell. 2007;28:315–327. doi: 10.1016/j.molcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung NKV, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13:397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ain Q, Schmeer C, Penndorf D, Fischer M, Bondeva T, Forster M, Haenold R, Witte OW, Kretz A. Cell cycle-dependent and -independent telomere shortening accompanies murine brain aging. Aging (Albany NY) 2018;10:3397–3420. doi: 10.18632/aging.101655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jahn A, Rane G, Paszkowski-Rogacz M, Sayols S, Bluhm A, Han CT, Draskovic I, Londono-Vallejo JA, Kumar AP, Buchholz F. ZBTB48 is both a vertebrate telomere-binding protein and a transcriptional activator. EMBO Rep. 2017;18:929–946. doi: 10.15252/embr.201744095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li JS, Miralles Fuste J, Simavorian T, Bartocci C, Tsai J, Karlseder J, Lazzerini Denchi E. TZAP: A telomere-associated protein involved in telomere length control. Science. 2017;355:638–641. doi: 10.1126/science.aah6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harenza JL, Diamond MA, Adams RN, Song MM, Davidson HL, Hart LS, Dent MH, Fortina P, Reynolds CP, Maris JM. Transcriptomic profiling of 39 commonly-used neuroblastoma cell lines. Sci Data. 2017;4 doi: 10.1038/sdata.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aeby E, Ahmed W, Redon S, Simanis V, Lingner J. Peroxiredoxin 1 protects telomeres from oxidative damage and preserves telomeric DNA for extension by telomerase. Cell Rep. 2016;17:3107–3114. doi: 10.1016/j.celrep.2016.11.071. [DOI] [PubMed] [Google Scholar]
- 41.Ahmed W, Lingner J. PRDX1 and MTH1 cooperate to prevent ROS-mediated inhibition of telomerase. Genes Dev. 2018;32:658–669. doi: 10.1101/gad.313460.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fouquerel E, Lormand J, Bose A, Lee HT, Kim GS, Li J, Sobol RW, Freudenthal BD, Myong S, Opresko PL. Oxidative guanine base damage regulates human telomerase activity. Nat Struct Mol Biol. 2016;23:1092–1100. doi: 10.1038/nsmb.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallabhaneni H, O'Callaghan N, Sidorova J, Liu Y. Defective repair of oxidative base lesions by the DNA glycosylase Nth1 associates with multiple telomere defects. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reyes-Uribe P, Adrianzen-Ruesta MP, Deng Z, Echevarria-Vargas I, Mender I, Saheb S, Liu Q, Altieri DC, Murphy ME, Shay JW. Exploiting TERT dependency as a therapeutic strategy for NRAS-mutant melanoma. Oncogene. 2018;37:4058–4072. doi: 10.1038/s41388-018-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
STELA analysis using alternative PCR polymerases. LAN-1 and SK-N-BE(2)N DNAs were subjected to STELA using a combination of Pwo and Taq polymerases (10:1). Six amplification reactions for each DNA were performed.