Abstract
Background
This study aimed to explore the effect of long non-coding RNA (LncRNA) H19 on the proliferation and invasion of lung carcinoma cells A549, and to determine its molecular targets.
Methods
A549 cells were with either LncRNA H19 or LncRNA H19 shRNA, and the expression levels of LncRNA H19 were evaluated by quantitative real-time PCR (RT-PCR). We measured cell proliferation using the CCK-8 assay, cell counting assays, and colony formation assay in response to shLncRNA H19-2. Cell migration and invasion were assessed by wound healing assay and Transwell assay, respectively. The mRNA and protein expression levels of E-cadherin, N-cadherin, and vimentin were determined by RT-PCR and western blot, respectively.
Results
The three LncRNA H19 shRNAs used in our study significantly reduced the expression levels of LncRNA H19 in A549 cells (P<0.05). Moreover, LncRNA H19 shRNA 2 (shLncRNA-2) was the most potent inhibitor of LncRNA H19 expression, and was selected for further experimentation. Transfection with shLncRNA H19-2 significantly decreased the proliferation, migration, and invasion of A549 cells, while overexpression of LncRNA H19 had the opposite effect in these cells (P<0.05). In response to shLncRNA H19-2, the expression levels of E-cadherin were notably elevated (P<0.05), while the expression levels of N-cadherin and vimentin were decreased (P<0.05). In contrast, overexpression of LncRNA H19 induced the expression of E-cadherin, and blocked the expression of N-cadherin, and vimentin (P<0.05).
Conclusion
Our results suggest that LncRNA H19 mediates the proliferation and invasion of lung cancer cells via upregulation of N-cadherin and vimentin, and downregulation of E-cadherin.
Keywords: long non-coding RNA, lung cancer, E-cadherin, N-cadherin, vimentin
Introduction
Lung cancer is associated with an exceptionally high rate of mortality.1 The onset of lung cancer is linked not only to external factors, such as smoking, but also to the genetic background of the patient.2 The activation of proto-oncogenes and the inactivation of tumor suppressor genes may contribute to lung cancer tumorigenesis and metastasis.2
Long non-coding RNAs (LncRNAs) are RNA molecules composed of 200 or more nucleotides that are not translated into proteins. Because they do not have a reading frame, it first appears as if LncRNAs serve no biological function.3 However, they have been found to regulate multiple cellular processes, including histone modification, chromatin remodeling, and cell cycle progression.4 Furthermore, certain LncRNAs are now used as biomarkers in cancer.5 These LncRNAs are differentially expressed in malignant tumors, and can be used to diagnose cancer and to assess tumor origin when the primary site is unknown.6
LncRNA H19, located near the telomeric region of its chromosome, is 2.5 kb in length, which includes five exons and four introns, and has been linked to tumor development.7 LncRNA H19 is primarily localized to the cytoplasm and acts as a regulatory RNA or ribose nuclear factor to control cell growth. During embryonic development, its expression is concentrated in the endoderm- and mesoderm-derived tissues.8–10 After birth, LncRNA H19 is only expressed, to a small extent, in heart muscles, skeletal muscles, and the breasts.8–10 These observations suggest that LncRNA H19 plays a dual role in cell proliferation and differentiation.8–10 Additionally, it has been reported that LncRNA H19 can induce the expression of a variety of genes related to tumor invasion and migration.11
In this study, we aim to explore the effect of LncRNA H19 on the proliferation and invasion of lung carcinoma cells A549, and determine its molecular targets using gene silencing and overexpression of LncRNA H19. Our results provide insight into the role of LncRNA H19 in cancer progression, and suggest that LncRNA H19 is a novel therapeutic target in lung cancer.
Materials and methods
Reagents
Certified FBS was obtained from Biological Industries (04-001-1A; Beth Haemek, Israel). Complete RPMI-1640 medium containing FBS and penicillin-streptomycin was bought from KeyGEN Biotech (KGM31800S-500; Jiangsu, China). DL5000 DNA marker was purchased from Takara (A2001C; Tokyo, Japan). Horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG (H+L) was purchased from ZSGB-Bio (ZB-2301; Beijing, China). Luria-Bertani (LB) broth (HB0128) and LB agar (HB0129) were bought from Hopebiol (Shandong, China). Lipofectamine® 3000 was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Plasmid extraction kits (DP103-02) and gel extraction kits (DP209-02) were purchased from Tiangen Biotech (Beijing, China). The PLVX-Puro vector was obtained from Youbio (VT1465; Hunan, China). Primers were synthesized by Genscript (Jiangsu, China) and General Biosystems (Anhui, China). Rabbit anti-E-cadherin (bs-1519R) and anti-N-cadherin (bs-1172R) polyclonal antibodies were obtained from Bioss Antibodies (Beijing, China). Rabbit anti-vimentin polyclonal antibody was purchased from Abcam (ab1256; Cambridge, UK). The Trans 15K DNA marker was from Transgen Biotech (BM161-01; Beijing, China). Trypsin-EDTA buffer (0.25%) without phenol red (T1300) and ampicillin (NOA8180) was purchased from Solarbio (Beijing, China). Ultrapure RNA kits (CW0581M), HiFiScript cDNA synthesis kits (CW2569M), and Ultra-SYBR mixture (CW0957M) were purchased from CWBIO (Beijing, China). Human A549 lung carcinoma cells were provided by Stem Cell Bank, Chinese Academy of Sciences, Shanghai, China. Cells were cultured in RPMI-1640 medium supplemented with 10% (v/v) FBS.
Vector construction
The whole genome sequence of LncRNA H19 was identified from the NCBI database (https://www.ncbi.nlm.nih.gov/). Primers were designed and synthesized according to its whole genome sequence, and suitable restriction sites were added. shRNA constructs are shown in Table 1. The DNA of whole genome sequences was extracted, PCR amplified, and extracted from agarose gels to amplify and recover the constructs. We performed TA cloning of the LncRNA H19 and T vector constructs, and the products were transformed into Escherichia coli DH5α. The transformed cells were seeded into liquid LB media overnight, and plasmids were then extracted. DNA was obtained from positive clones via restriction enzyme digestion. The plasmids were then digested by endonucleases, linked by ligases, and transformed into E. coli DH5α. Finally, the recombinant plasmids were extracted from the bacteria.
Table 1.
Interference sequences
| Sequence name | Primer sequences (5′–3′) |
|---|---|
| shLncRNA H19-1 | F: GATCCTTAGCAAAGGTGACATCTTCTCTCTCGAGAGAAGATGTCACCTTTGCTAATTTTTG |
| R: AATTCAAAAAATTAGCAAAGGTGACATCTTCTCTCGAGAGAAGATGTCACCTTTGCTG | |
| shLncRNA H19-2 | F: GATCCTCCCAGAACCCACAACATGAACTCGAGTTCATGTTGTGGGTTCTGGGATTTTTG |
| R: AATTCAAAAAATCCCAGAACCCACAACATGAACTCGAGTTCATGTTGTGGGTTCTGGGAG | |
| shLncRNA H19-3 | F: GATCCTGAATATGCTGCACTTTACAACTCGAGTTGTAAAGTGCAGCATATTCATTTTTG |
Abbreviations: R, reverse; F, forward.
Cell transfection
According to the different transfected genes, A549 cells were divided into five groups: blank control (Control), interference negative control (shLncRNA H19 NC), expression negative control (LncRNA H19 NC), interference (shLncRNA H19), and overexpression (LncRNA H19). Cells without any treatment served as blank control. Different genes were transfected into A549 cells using Lipofectamine® 3000 per manufacturer’s manual. After transfection, A549 cells were cultured in FBS-free media for 4 hours. The media was then replaced with FBS-containing media and cells were cultured for an additional 44 hours.
Measurement of cell proliferation
A549 cells in the logarithmic phase were seeded into 96-well plates at a density of 4×103 cells/well. Six wells were seeded per group, and cells were cultured for 24 hours. Forty-eight hours post-transfection, cell counts were performed. For the Cell Counting Kit-8 (CCK-8) assay, media was replaced with fresh CCK-8 solution (10 µL per well) for 4 hours, and the absorbance was measured at 450 nm. Each experiment was repeated in triplicate.
After transfection, cells were seeded into culture dishes at a density of 50 cells/dish and cultured for 14 days. The culture media was then discarded, and the cells were washed twice with PBS. Cells were fixed in 4% paraformaldehyde for 15 minutes and stained in Giemsa solution for 20 minutes. They were then washed with water to remove the staining solution and air dried. The dishes were inverted and overlaid with a mesh transparent film. Clones larger than ten cells were counted using a microscope.
Measurement of cell migration
After transfection, cells were seeded into 24-well plates at a density of 5×105 cells/well and cultured overnight. A scratch was made with a pipette tip across the cell monolayer. The cells were washed with PBS for three times, cultured in serum-free media for 48 hours, and visualized using a microscope.
Measurement of cell invasion
Matrigel was uniformly distributed onto the membranelle of transwell inserts and incubated at 37°C. After transfection for 48 hours, the media of A549 cells was replaced with FBS-free media containing 0.2% BSA for 12 hours. The cells were then trypsinized, washed with PBS once, and suspended in FBS-free media containing 0.2% BSA. Cell suspensions were counted, and cell concentrations were adjusted to 2×105 cells/mL. Complete medium (500 µL) and cell suspensions (100 µL) were added into the lower and upper chambers, respectively. Transwell inserts were then incubated for 24 hours. Cells were fixed in 4% paraformaldehyde for 30 minutes, washed with PBS twice, and stained in Giemsa buffer for 30 minutes. After washing with PBS twice, the cells in the upper chamber were discarded gently with cotton swabs. Cells in five fields of view of the lower chamber were counted using a microscope. Invasive capacity was expressed as the average number of cells in each field of view.
Reverse transcription PCR (RT-PCR)
After transfection for 48 hours, total RNA was collected with an ultrapure RNA kit, and RNA was reversely transcribed into cDNA using a HiFiScript cDNA synthesis kit. cDNA was amplified using fluorogenic quantitative PCR (CFX Connect™; Bio-Rad, Hercules CA, USA). Primers (listed in Table 2) were added to 25 µL of PCR mix (9.5 µL of RNase free dH2O, 1 µL of cDNA, 1 µL of forward primer, 1 µL of reverse primer, 12.5 µL of 2× UltraSYBR mix) and reaction parameters were as follows: pre-denaturation for 10 minutes at 95°C, denaturation for 10 seconds at 95°C, annealing for 30 seconds at 55°C, elongation for 30 seconds at 72°C. Dissociation curves were analyzed as follows: 15 seconds at 95°C, 1 minute at 55°C, 15 seconds at 95°C, 15 seconds at 55°C, 15 seconds at 55°C, and measured stepwise from 92°C, every 0.5°C. Quantitative analysis was performed using fluorogenic quantitative RT-PCR (CFX Connect™; Bio-Rad). Glyceraldehyde-phosphate dehydrogenase served as an internal control.
Table 2.
Primers for RT-PCR
| Gene | Primer (5′–3′) |
|---|---|
| LncRNA H19 | F: TCAGCTCTGGGATGATGTGGT |
| R: CTCAGGAATCGGCTCTGGAAG | |
| E-cadherin | F: CAAATCCAACAAAGACAAAGAAGGC |
| R: ACACAGCGTGAGAGAAGAGAGT | |
| N-cadherin | F: CATCATCATCCTGCTTATCCTTGT |
| R: GGTCTTCTTCTCCTCCACCTTCT | |
| Vimentin | F: TCGTGATGCTGAGAAGTTTCG |
| R: TCTGGATTCACTCCCTCTGGT | |
| GAPDH | F: GGACCTGACCTGCCGTCTAG |
| R: GTAGCCCAGGATGCCCTTGA |
Abbreviations: RT-PCR, reverse transcription PCR; R, reverse; F, forward.
Western blot
After transfection for 48 hours, A549 cells were collected and lysed in RIPA buffer at 4°C for 30 minutes. Cell lysates were centrifuged at 8000× g and 4°C for 10 minutes, and the supernatants were collected. Protein concentrations were measured using a BCA kit. Proteins were then denatured and separated using SDS-PAGE electrophoresis for 1–2 hours. Proteins were subsequently transferred to membranes and incubated with the primary antibody (1:1,000 for anti-E-cadherin, 1:200 for anti-N-cadherin, 1:2,000 for anti-vimentin) overnight at 4°C. The membranes were then washed and incubated with the secondary antibody (1:2,000) for 1–2 hours at room temperature. Bands were visualized with chemiluminescence solution, and imaged with a gel imaging system (ChemiDoc™ XRS+; Bio-Rad, Hercules CA, USA). Quantity one software (Bio-Rad) was used to determine the relative amount of protein per band.
Statistical analysis
All experiments were repeated in triplicate. The data were presented as mean ± SD. Statistical analyses were carried out using ANOVA followed by a post hoc test using SPSS 19.0 software. P<0.05 was considered to be statistically significant.
Results
shRNA construct screening
Figure 1 shows the expression levels of LncRNA H19 in A549 cells after transfection with various LncRNA H19 shRNA constructs. Compared with the control group, shLn-cRNA H19-1, shLncRNA H19-2, and shLncRNA H19-3 significantly reduced the expression levels of LncRNA H19 in these cells (P=0.0005, 0.0003, and 0.0007, respectively). Moreover, shLncRNA H19-2 showed the greatest effect on the inhibition of LncRNA H19 expression. Thus, we selected shLncRNA H19-2 for all further experimentation.
Figure 1.
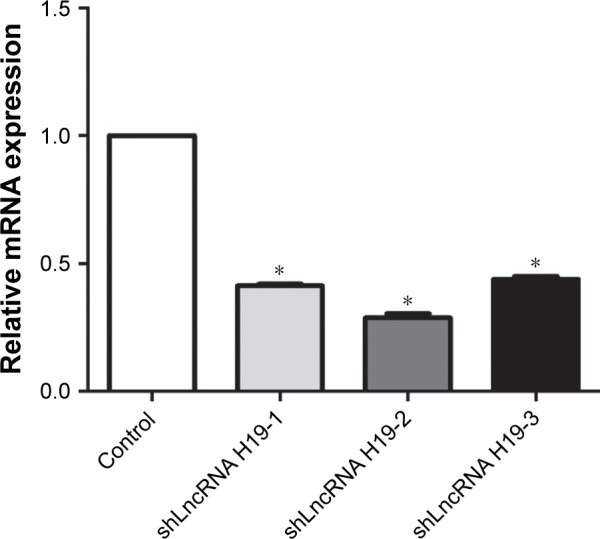
mRNA expression levels of LncRNA H19 in A549 cells after transfection with various LncRNA H19 silencing constructs, as measured by RT-PCR.
Notes: Treatment with shLncRNA H19-2 resulted in the lowest mRNA expression levels of LncRNA H19 compared with all other constructs. *P<0.05 vs the control group.
Abbreviations: RT-PCR, reverse transcription PCR; LncRNA, long non-coding RNA.
Screening of LncRNA H19 overexpression constructs
The expression levels of LncRNA H19 in A549 cells transfected with different LncRNA H19 gene vectors is depicted in Figure 2. We found that the shLncRNA H19 NC and LncRNA H19 NC groups had similar LncRNA H19 expression levels compared with the control group, confirming the reliability of our analyses. Interestingly, transfection with shLncRNA H19 remarkably downregulated LncRNA H19 expression compared with the control group (P=0.0000001), while transfection with LncRNA H19 increased the levels of LncRNA H19 (P=0.0000001).
Figure 2.
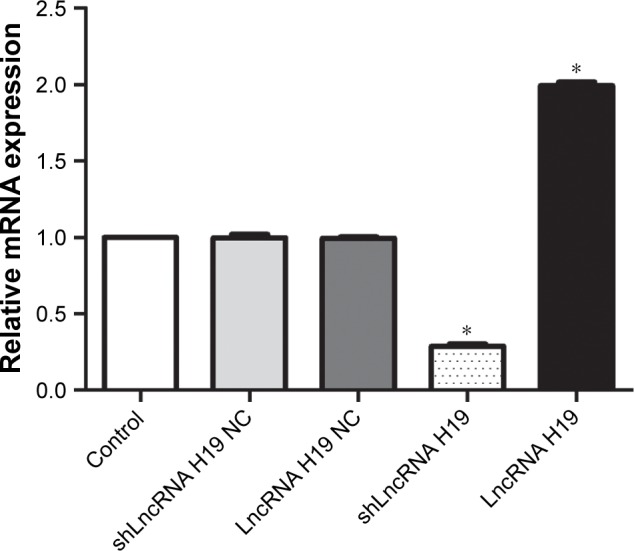
mRNA expression levels of LncRNA H19 in A549 cells transfected with different LncRNA H19 gene vectors, as measured by RT-PCR.
Notes: Transfection with shLncRNA H19 significantly reduced the mRNA expression levels of LncH19 in A549 cells, while LncRNA H19 significantly elevated the mRNA expression levels compared with the control group. *P<0.05 vs the control group.
Abbreviations: RT-PCR, reverse transcription PCR; LncRNA, long non-coding RNA.
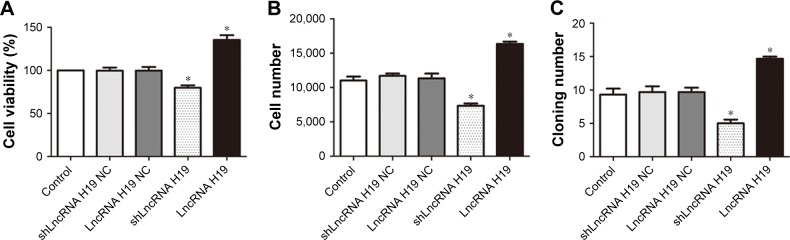
LncRNA H19 mediates cell proliferation in A549 cells
As shown in Figure 3, transfection with either shLncRNA H19 NC or LncRNA H19 NC exerted a negligible effect on the viability, cell number, and rate of colony formation of A549 cells. shLncRNA H19 significantly decreased the viability, cell number, and rate of colony formation of A549 cells compared with the control group (P=0.046, 0.042, and 0.039, respectively), while transfection with LncRNA H19 had the opposite effect (P=0.045, 0.043, and 0.044, respectively). These results suggest that inhibition of LncRNA H19 expression suppresses the proliferation of lung carcinoma cells, while its overexpression has an opposite effect.
Figure 3.
The proliferation of A549 cells transfected with different LncRNA H19 gene vectors, as assessed by CCK-8 assay (A), cell counting (B), and colony formation (C).
Notes: shLncRNA H19 decreased the proliferation of A549 cells, while LncRNA H19 increased their proliferation compared with the control group. *P<0.05 vs the control group.
Abbreviations: LncRNA, long non-coding RNA; CCK-8, Cell Counting Kit-8.
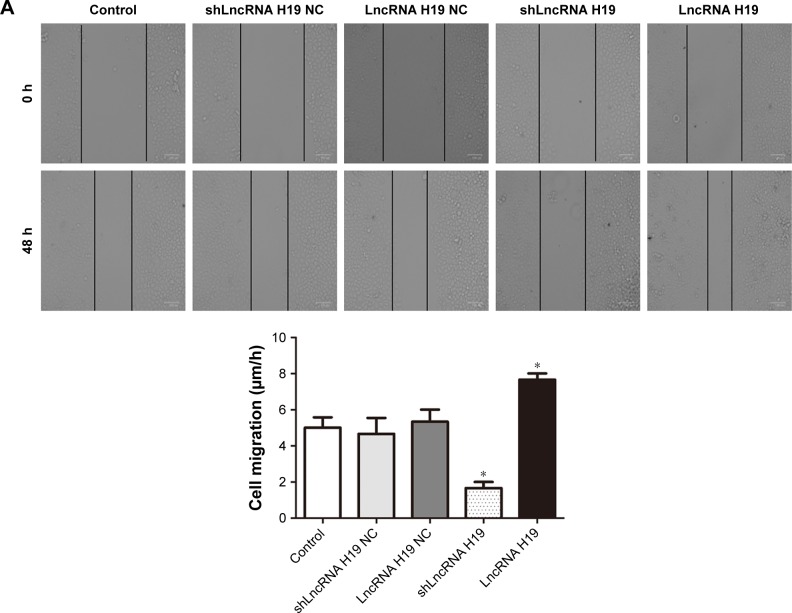
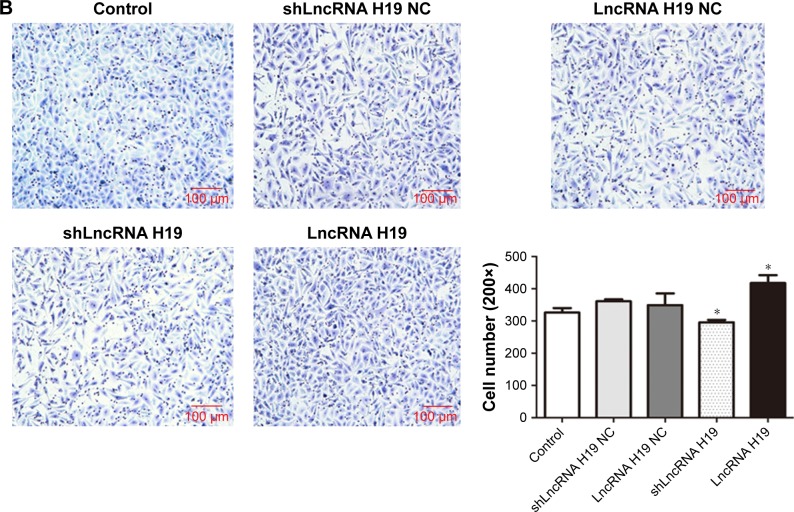
LncRNA H19 mediates the migration and invasion of A549 cells
As shown in Figure 4, transfection with either shLncRNA H19 NC or LncRNA H19 NC had no effect on the migration and invasion of A549 cells. However, compared with the control group, the migration and invasion of A549 cells was significantly blocked by shLncRNA H19 (P=0.025 and 0.038, respectively), while an opposite effect was observed with LncRNA H19 (P=0.039 and 0.041, respectively). These results suggest that shLncRNA H19 mediates the migratory and invasive potential of lung cancer cells.
Figure 4.
(A) The migration of A549 cells transfected with different LncRNA H19 gene vectors, as evaluated by wound healing assay. (B) Qualitative distribution and quantitative number of A549 cells transfected with different LncRNA H19 gene vectors in a typical visual field of the lower chamber, as evaluated by transwell assay. Compared with the control group, the migration and invasion of A549 cells was significantly reduced by shLncRNA H19 and was increased by LncRNA H19. *P<0.05 vs the control group.
Abbreviation: LncRNA, long non-coding RNA.
LncRNA H19 influences the expression of E-cadherin, N-cadherin, and vimentin
As shown in Figure 5, A549 cells transfected with either shLncRNA H19 NC or LncRNA H19 NC showed similar levels of E-cadherin, N-cadherin, and vimentin compared with the control group. After transfection with shLncRNA H19, the expression levels of E-cadherin were notably increased and those of N-cadherin and vimentin were reduced (all P=0.000001), while an opposite effect was observed with LncRNA H19 overexpression (all P=0.000001).
Figure 5.
mRNA and protein levels of E-cadherin, N-cadherin, and vimentin in A549 cells transfected with different LncRNA H19 gene vectors, as assessed by RT-PCR and Western blot, respectively.
Notes: In response to shLncRNA H19, the expression levels of E-cadherin were significantly increased, and the expression levels of N-cadherin and vimentin were reduced. An opposite effect with regards to the expression levels of E-cadherin, N-cadherin, and vimentin was observed with LncRNA H19 overexpression. *P<0.05 vs the control group.
Abbreviations: RT-PCR, reverse transcription PCR; LncRNA, long non-coding RNA.
Discussion
As the incidence of lung cancer has steadily increased worldwide, it is necessary to find effective novel therapeutics to treat this debilitating disease. LncRNA is currently of great interest to the therapeutics. Several LncRNAs play important regulatory roles in the development and progression of cancer.12 The abnormal expression of LncRNAs in multiple types of malignant tumors, such as liver cancer,13 prostate cancer,14 and breast cancer,15 can either inhibit or promote tumor growth, proliferation, and metastasis. A few LncRNAs have also been used as prognostic biomarkers.
Previous studies have shown that LncRNA H19 is associated with tumor proliferation and invasion,16 and promotes epithelial–mesenchymal transition (EMT) in alveolar epithelial cells.17 Zhou et al revealed that the expression levels of lncRNA H19 in lung cancer tissues were significantly higher compared with normal tissues and knockout of LncRNA H19 inhibited the proliferation of lung cancer cells in vitro (A549 cells) and in vivo.18 This study also found that overexpression of LncRNA H19 promoted the invasion and metastasis of lung cancer cells.18 These results are in agreement with our study, as the proliferative, migratory, and invasive potential of lung cancer cells were significantly decreased by knocking down LncRNA H19 expression, and overexpression of LncRNA H19 resulted in an opposite effect in these cells. Therefore, LncRNA H19 plays an important role in the development and progression of lung cancer, with significant effects on tumor proliferation and invasion.
However, Zhou et al do not report the involved mechanisms.18 Therefore, this study aims to further explore the involved mechanisms in vitro from the perspectives of EMT through the same A549 cell line. We investigate the expression of EMT biomarkers such as E-cadherin, N-cadherin, and vimentin. EMT refers to a change from epithelial cell morphology to mesenchymal morphology, including loss of cell polarity and increase of migratory capacity.19 E-cadherin, N-cadherin, and vimentin are classical biomarkers of EMT. E-cadherin is a calcium-dependent cell adhesion molecule that maintains the integrity of cell morphology and structure by mediating the connection between homotypic cells. Decreased expression of E-cadherin is a marker of EMT occurrence.20 N-cadherin exhibits an opposite expression pattern compared with E-cadherin, as it facilitates tumor invasion and metastasis.21 High expression levels of N-cadherin in epithelium-derived tumors are positively correlated with tumor growth, migration, invasion, lymph node metastasis, and EMT.22 Deficiency in E-cadherin expression in epithelium-derived cancer tissues induces the expression of N-cadherin, and the phenotypic transition from E-cadherin to N-cadherin is closely related to the enhancement of tumor metastasis and invasion.23 Vimentin is an intermediate filament protein that is expressed in mesenchymal cells and some ectodermal cells. Its primary biological function includes the maintenance of cell and organelle morphology, signal transduction, transplantation immunity, and apoptosis.24 The abnormal expression of vimentin can change the structure of cytoskeletal proteins and promote the transformation of cubic epithelial cells into spindle fibroblast-like cells, increasing their ability to migrate.25 In this study, we found that shLncRNA H19 reduced the expression levels of vimentin and N-cadherin and increased the levels of E-cadherin. The overexpression of LncRNA H19 had the opposite effect as it upregulated N-cadherin and vimentin, and downregulated E-cadherin. These results suggest that LncRNA H19 promotes EMT in lung cancer cells by modulating the expression of E-cadherin, N-cadherin, and vimentin. These results are consistent with previous reports,7,9 where LncRNA H19 promotes the expression of EMT genes, including vimentin, zinc-finger E-box binding homeobox 1 (ZEB1), and ZEB2 by regulating miR-138 and miR-200a.7 One study, which also used A549 cells, reports that LncRNA H19 promotes EMT by negatively regulating miR-484.26 LncRNA H19 also successively downregulates the expression of LET-7 and E-cadherin, and upregulates the expression of HMGA2 and Twist/snail to promote EMT.9
A549 cell line is a common lung cancer cell line used for in vitro studies.18,26 We recognize that a more rigorous experimental design, including more than one cell line, would be useful to further evaluate the effects of LncRNA H19. Further experimentation is required to confirm our findings.
Conclusion
LncRNA H19 mediates the proliferation, invasion, and metastasis of lung cancer cells, and modulates the expression of E-cadherin, N-cadherin, and vimentin, thereby promoting EMT. Silencing of LncRNA H19 inhibits the proliferation and invasion of lung cancer cells and results in the down-regulation of vimentin and N-cadherin, and the upregulation of E-cadherin. EMT is involved in the regulation of the invasion and metastasis of lung cancer cells by LncRNA H19. These results provide promising insight into the role of LncRNA H19 in tumor progression, and may allow for the development of novel therapeutics and diagnostic markers for patients with lung cancer.
Acknowledgments
This work was supported by the financial support from the National Natural Science Foundation of China (81260345).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372(18):1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 2.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Necsulea A, Soumillon M, Warnefors M, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505(7485):635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 4.Volders PJ, Helsens K, Wang X, et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41(Database issue):D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang JF, Yin QF, Chen T, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the Myc locus. Cell Res. 2014;24(5):513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X, Ye F, Yin C, et al. The interaction between miR-141 and lncRNA-H19 in regulating cell proliferation and migration in gastric cancer. Cell Physiol Biochem. 2015;36(4):1440–1452. doi: 10.1159/000430309. [DOI] [PubMed] [Google Scholar]
- 9.Matouk IJ, Halle D, Raveh E, et al. The role of the oncofetal H19 lncRNA in tumor metastasis: orchestrating the EMT-MET decision. Oncotarget. 2016;7(4):3748–3765. doi: 10.18632/oncotarget.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Hua Y, Jin J, et al. Association of genetic variants in lncRNA H19 with risk of colorectal cancer in a Chinese population. Oncotarget. 2016;7(18):25470–25477. doi: 10.18632/oncotarget.8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu T, Qu L, He G, et al. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7(10):11553–11566. doi: 10.18632/oncotarget.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua Q, Lv X, Gu X, et al. Genetic variants in lncRNA H19 are associated with the risk of bladder cancer in a Chinese population. Mutagenesis. 2016;31(5):531–538. doi: 10.1093/mutage/gew018. [DOI] [PubMed] [Google Scholar]
- 13.Li H, An J, Wu M, et al. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget. 2015;6(29):27847–27864. doi: 10.18632/oncotarget.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281(16):3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 15.Shi SJ, Wang LJ, Yu B, et al. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6(13):11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Li Y, Li Y, et al. Long non-coding RNA H19 promotes the proliferation and invasion of breast cancer through upregulating DNMT1 expression by sponging miR-152. J Biochem Mol Toxicol. 2017;31(9):e21933. doi: 10.1002/jbt.21933. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y, He R, An J, et al. The effect of H19-miR-29b interaction on bleomycin-induced mouse model of idiopathic pulmonary fibrosis. Biochem Biophys Res Commun. 2016;479(3):417–423. doi: 10.1016/j.bbrc.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Sheng B, Xia Q, Guan X, Zhang Y. Association of long non-coding RNA H19 and microRNA-21 expression with the biological features and prognosis of non-small cell lung cancer. Cancer Gene Ther. 2017;24(8):317–324. doi: 10.1038/cgt.2017.20. [DOI] [PubMed] [Google Scholar]
- 19.de Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 20.Yu JM, Sun W, Hua F, et al. BCL6 induces EMT by promoting the ZEB1-mediated transcription repression of E-cadherin in breast cancer cells. Cancer Lett. 2015;365(2):190–200. doi: 10.1016/j.canlet.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 21.Angadi PV, Patil PV, Angadi V, et al. Immunoexpression of epithelial mesenchymal transition proteins E-cadherin, b-catenin, and N-cadherin in oral squamous cell carcinoma. Int J Surg Pathol. 2016;24(8):696–703. doi: 10.1177/1066896916654763. [DOI] [PubMed] [Google Scholar]
- 22.Liu YA, Liang BY, Guan Y, et al. Loss of N-cadherin is associated with loss of E-cadherin expression and poor outcomes of liver resection in hepatocellular carcinoma. J Surg Res. 2015;194(1):167–176. doi: 10.1016/j.jss.2014.09.031. [DOI] [PubMed] [Google Scholar]
- 23.Song PP, Qian XY, Zhou H, et al. Expression of E-cadherin, N-cadherin, b-catenin and their clinical significance in laryngeal carcinoma. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2016;51(6):440–445. doi: 10.3760/cma.j.issn.1673-0860.2016.06.008. Chinese. [DOI] [PubMed] [Google Scholar]
- 24.Zhu MQ, de Broe ME, Nouwen EJ. Vimentin expression and distal tubular damage in the rat kidney. Exp Nephrol. 1996;4(3):172–183. [PubMed] [Google Scholar]
- 25.Dong Q, Zhu X, Dai C, et al. Osteopontin promotes epithelial-mesenchymal transition of hepatocellular carcinoma through regulating vimentin. Oncotarget. 2016;7(11):12997–13012. doi: 10.18632/oncotarget.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Li X, Li X, Li X, Chen Z. LncRNA H19 promotes epithelial-mesenchymal transition (EMT) by targeting miR-484 in human lung cancer cells. J Cell Biochem. 2018;119(6):4447–4457. doi: 10.1002/jcb.26537. [DOI] [PubMed] [Google Scholar]