Abstract
Objective:
Leucyl-tRNA synthetase (LRS) is an aminoacyl-tRNA synthetase catalyzing ligation of leucine to its cognate tRNA and is involved in the activation of mTORC1 by sensing cytoplasmic leucine. In this study, the usefulness of LRS as a therapeutic target of non-small cell lung cancer (NSCLC) and the anticancer effect of the LRS inhibitor, BC-LI-0186, was evaluated.
Methods:
LRS expression and the antitumor effect of BC-LI-0186 were evaluated by immunohistochemical staining, immunoblotting, and live cell imaging. The in vivo antitumor effect of BC-LI-0186 was evaluated using Lox-Stop-Lox (LSL) K-ras G12D mice.
Results:
LRS was frequently overexpressed in NSCLC tissues, and its expression was positively correlated with mTORC1 activity. The guanosine-5’-triphosphate (GTP) binding status of RagB was related to the expression of LRS and the S6K phosphorylation. siRNA against LRS inhibited leucine-mediated mTORC1 activation and cell growth. BC-LI-0186 selectively inhibited phosphorylation of S6K without affecting phosphorylation of AKT and leucine-mediated co-localization of Raptor and LAMP2 in the lysosome. BC-LI-0186 induced cleaved poly (ADP-ribose) polymerase (PARP) and caspase-3 and increase of p62 expression, showing that it has the autophagy-inducing property. BC-LI-0186 has the cytotoxic effect at nanomolar concentration and its GI50 value was negatively correlated with the degree of LRS expression. BC-LI-0186 showed the antitumor effect, which was comparable with that of cisplatin, and mTORC1 inhibitory effect in a lung cancer model.
Conclusions:
BC-LI-0186 inhibits the noncanonical mTORC1-activating function of LRS. These results provide a new therapeutic strategy for NSCLC and warrant future clinical development by targeting LRS.
Keywords: aminoacyl-tRNA synthetase (ARS), BC-LI-0186, leucyl-tRNA synthetase (LRS), mTORC1, non-small cell lung cancer (NSCLC)
Introduction
Lung cancer is the third most common malignancy and still the leading cause of cancer death in the world. In the United States, it is projected that there may be approximately 222,500 new cases and 155,870 deaths from lung cancer in 2017.1 The development of target agents and immunotherapy has improved treatment outcomes, but ultimate resistance to these therapies requires new approaches to combat carcinogenesis and resistance to current therapies.2
Aminoacyl-tRNA synthetases (ARSs) are housekeeping enzymes that contribute to protein synthesis by catalyzing the ligation of specific amino acids to their corresponding tRNAs.3,4 ARSs bind ATP and an amino acid to form an aminoacyl adenylate and deliver the activated amino acid to the 3’-end of a tRNA.5 ARSs are divided into two classes: Class I synthetases have a nucleotide-binding Rossmann fold,6 whereas class II synthetases share a different catalytic domain.7 In addition to their role in protein synthesis, mammalian ARSs have other domains that enable them to perform noncanonical functions. For example, tryptophanyl- and tyrosyl-tRNA synthetases control angiogenesis and immune responses in the tumor microenvironment.8–10 Other ARSs play noncanonical roles by forming complexes with ARS-interacting multifunctional proteins.5 For example, lysyl-tRNA synthetase is involved in the development of melanoma through binding to the microphthalmia-associated transcription factor, and glutamyl-prolyl-tRNA synthetase is a translational silencer that suppresses vascular endothelial growth factor A.11,12
Leucyl-tRNA synthetase (LRS), which catalyzes ligation of L-Leu to its cognate tRNA, is a class I ARS with a large insertion connective peptide 1 (CP1) domain, a tRNA anticodon-binding domain, and a C-terminal extension domain.13 Recent studies have shown that LRS possesses another important noncanonical function: it is required for leucine-mediated mTORC1 activation. LRS binds and activates Rag GTPase in an amino-acid-dependent manner; Rag GTPase then activates mTORC1.14–17 These findings suggest that LRS overexpression can impact cancer progression and that LRS inhibitors are thus potential anticancer therapeutics. The compound BC-LI-0186 specifically inhibits the GTPase activating the function of LRS through binding to the RagD interacting site of LRS and inhibits lysosomal localization of LRS and mTORC1 activity. Furthermore, BC-LI-0186 showed the growth inhibiting activity in the cancer cells that express drug-resistant mTOR mutations.18
Missense mutations of KRAS, which introduce an amino acid substitution at position 12 or 13, results in constitutive activation of KRAS signaling and is one of the most common driver mutations in lung adenocarcinoma.19,20 However, appropriate therapeutic modality has not yet been developed for these subtypes of lung cancer in the metastatic setting and it is still one of the representative area of unmet need in the medical area. Activating mutation of KRAS induces cellular proliferation through constitutive activation of RAS → RAF → MEK → ERK signaling and activated ERK activates mTORC1 through inhibition of TSC. Meanwhile, activated KRAS can also induce mTORC1 activation through activation of the PI3K–AKT pathway.21 Therefore, the mTORC1 signaling pathway is one of the key targets in lung adenocarcinoma with KRAS activation mutation, although conventional rapalogs targeting FKBP-12 have limited effect as a single therapeutic agent. Therefore, we investigated the effect of novel mTORC1 inhibitor, BC-LI-0186, on the K-ras mouse lung cancer model.
In this study, we measured the expression of LRS and pS6, a marker of mTORC1 signaling, in non-small cell lung cancer (NSCLC) tissues. Using NSCLC cells, we assessed the effects of BC-LI-0186 on LRS and mTORC1 activity and confirmed its cytotoxic activity. The potential utility of BC-LI-0186 as a lung cancer therapeutic was tested using a K-ras mouse lung cancer model. This study provides evidence that BC-LI-0186 inhibits the noncanonical, mTORC1-activating function of LRS and could be useful as a therapeutic for NSCLC. Additional studies are needed to identify noncanonical functions of ARSs and to assess their potential as targets for cancer therapeutics.
Materials and methods
Study materials
Anti-actin (I-19) and anti-β-tubulin were purchased Santa Cruz Biotechnology (Dallas, TX, USA), rabbit polyclonal anti-RRAGD from Bethyl Laboratories (Montgomery, TX, USA), anti-Leucyl-tRNA synthetase (LRS) from Neomics (Suwon, Korea). Unless otherwise noted, antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). Guanosine-5’-triphosphate (GTP) mutant of RagB (Flag pLJM1 RagB Q99L, Plasmid #19315) and GDP mutant of RagD (Flag pLJM1 RagD S77L Plasmid #19317) were obtained from addgene.22 The leucine free (Cat #ML011-96) and amino acid (Cat #ML011-82) free cell culture media were purchased from Welgene (Gyeongsan-si, Korea). The 11 NSCLC cells used in this study were H1703, H1299, H2009, H460, A549, H1650, H596, HCC2228, SNU1330, H2279, and H358 obtained from ATCC (https://www.atcc.org/). A set of 117 NSCLC tissues from patients who underwent pulmonary resection between 2000 and 2008 and had submitted written consent providing the residual samples were randomly selected from the institutional tissue archive. To evaluate LRS expression, a formalin-fixed paraffin-embedded lung cancer tissue slide from each patient was analyzed by immunohistochemistry (IHC). The relationship between the expression of pS6 and LRS was evaluated using the serial section slides from the same formalin-fixed paraffin-embedded tissue block. This study was approved by the Institutional Review Board (IRB) of Gangnam Severance Hospital (IRB #3-2014-0299) and was carried out in compliance with the Declaration of Helsinki and Korean Good Clinical Practice guidelines. Lox-Stop-Lox (LSL) K-ras G12D mice were obtained from the NCI Mouse Repository, bred, and genotyped according to the supplier’s guidelines (https://ncifrederick.cancer.gov/Lasp/MouseRepository/Default.aspx). A total of 26 K-ras G12D mice, including five vehicles, eight cisplatin, seven BC-LI-0186, and six combination treatment mice were used for the experiment. This animal study was approved by our Institutional Animal Care and Use Committee (#2013-0278-1), following guidelines from the American Association for the Assessment and Accreditation of Laboratory Animal Care. AdCre virus was obtained from the Gene Transfer Vector Core of the University of Iowa (Iowa City, IA, USA).
Immunoblotting
Cells were harvested using 2×LSB lysis buffer containing protease and phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO, USA) on ice. After sonication, 30 mg of lysate was separated by gel electrophoresis on a 7.5–12% polyacrylamide gel and transferred onto a nitrocellulose membrane (Bio-Rad Laboratories Inc., Richmond, CA, USA). The expression level of each protein was measured using ImageJ (http://rsbweb.nih.gov/ij/) and quantified relative to β-actin.
RagB, RagD GTP-agarose bead pull-down assay
Tissue lysates were incubated with 100 µl GTP-agarose beads (Sigma-Aldrich, cat no. G9768) in 0.5 ml of GTP-binding buffer for 30 min at 4°C. The beads were washed with GTP-binding buffer, and the supernatant was retained. Then retained supernatant was incubated with beads for another 30 min. The beads were washed again, then incubated with the retained supernatant overnight at 4°C. After washing five times with GTP-binding buffer, GTP-bound protein extracts were eluted with 2× sample buffer and GTP-bound protein was visualized by immunoblotting.
Cell growth, viability assays, and flow cytometer
NSCLC cells were plated in 96-well plates, incubated for 24 h, and then treated with the indicated dose of BC-LI-0186 diluted in the media containing CellTox Green Dye (Promega, G8731). The cell growth and death were measured by photographing the confluency and green fluorescence of the well every 2 h from drug treatment for 24 h using the incuCyte Zoom System (Essen Bioscience). Briefly, cell growth was estimated from the cell confluency of each well. Values denoting cell growth in each reagent-treated well was the relative cell confluency to those obtained from the vehicle-treated wells. For the measurement of cell death, the green fluorescence spot was measured after adding CellTox green (promega, G8731, protocol attached) onto the media, and the values obtained from each treated well were normalized from those from the vehicle-treated wells. The growth and inhibition curves, half maximal inhibitory concentration (IC50), and half maximal effective concentration (EC50) were calculated using Graph Pad Prism (Version 5.01). To measure apoptotic cell death, cells were treated with the indicated dose of BC-LI-0186 or rapamycin for 48 h, stained with annexin V and propidium iodide (PI), and analyzed using a FACSCanto II flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA).
Immunocytochemistry and IHC
Cells were plated on the cover glasses in a 24-well plate and, at 50% confluency, cells were incubated in leucine-free RPMI media and treated with 10 μM of 0186 for 16 h. The next day, cells were stimulated with 0.8 mM of leucine for 10 min, fixed and permeabilized with cold methanol. After incubation in the primary antibody mixture in 1% bovine serum albumin (BSA) in PBS-T (10× concentrated phosphate buffered saline [PBS] supplemented with Tween 20) for 1 h, cells were visualized with the mixture of secondary antibodies and incubated in 4’,6-diamidino-2-phenylindole (DAPI) in PBS. Cell imaging was performed with a confocal laser-scanning microscope (Zeiss, LSM780) and images were captured with the ZEN2012 software (Zeiss) and stored as CZI image format. IHC staining was performed according to the manufacturer’s protocol using the LABS2 system (Dako, Carpentaria, CA, USA). Briefly, sections were deparaffinized, rehydrated, immersed in H2O2–methanol solution, and incubated overnight with primary antibodies against activated caspase-3, pS6, pAKT, pGSK-3β, or LRS at 1:100 dilution in antibody diluent (Dako). Sections were incubated for 10 min with a biotinylated linker and processed using avidin/biotin IHC techniques. The chromogen 3,3’-diaminobenzidine (DAB) was used in conjunction with a Liquid DAB Substrate Kit (Novacastra Laboratories Ltd., Newcastle, UK). LRS expression was scored as the product of staining intensity and percentage of positive tumor cells. Staining intensity was classified as 0, 1, 2, or 3 and frequency as 0 (<10%), 1 (10–50%), 2 (51–80%), or 3 (>80%). Expression was scored as the product of intensity and frequency, and overexpression was defined as expression ⩾6. The scoring system used for pS6 was adapted from Shin et al.23 Briefly, staining intensity was classified as 0 (negative), 1 (weak), 2 (moderate), or 3 (strong), and frequency was classified as 0 (negative), 1 (1–25%), 2 (26–50%), 3 (51–75%), or 4 (>75%). Expression of pS6 was scored as the sum of staining intensity and frequency. The staining and quantification of p-AKT Ser473 was adapted from Kim et al.19 Staining intensity was classified as 0, 1, 2, and 3 where intensity two means equal as that of positive control, breast ductal cancer tissue. Frequency was classified as 0, 1 (trace <5%), 1 (–10%), 2 (−30%), and 3 (>30%). The score was obtained by the product of intensity and frequency.
RT-PCR analysis
A549 or H460 cells were treated with DMSO or inhibitor for 6 h or 12 h. For LRS knockdown experiment, Cells were transfected with control siRNA or LRS-targeting siRNA for 48hr. Total RNA was isolated from cells using MiniBEST Universal RNA Extraction Kit (TAKARA). Each RNA (1ug) was reverse-transcribed into cDNA using PrimeScript™ 1st strand cDNA Synthesis Kit (TAKARA). Reverse transcription polymerase chain reaction (RT-PCR) was performed using EmeraldAmp® GT PCR Master Mix (TAKARA), then PCR products were analyzed by 1% agarose gel electrophoresis. Gene expression levels were determined using ImageJ and normalized to the housekeeping gene, GAPDH. All RT-PCR data were analyzed by using Student’s t-test, and error bars in figures represent the standard deviation (SD) of three biological experiments. Primer sequences are as follows: human LRS forward, TAAATTTGGGAAGCGGTATAC; human LRS reverse, GCATTGGGAGAACATAGATCACC; human SREBP-2 forward, CTTTCAAGTCCTTCAGCCT; human SREBP-2 reverse, CAGGCATTGTGGTCAGAAT; human HMGCR forward, ATATTGCTCGTGGAATGGC; human HMGCR reverse, GAGAAGGATCAGCTATCCA.
Mouse imaging and CT image analysis
LSL K-ras G12D mice inhaled 5 × 107 plaque-forming units (PFU) of AdCre virus at 8 weeks after birth.24 At 24 ± 2 weeks after AdCre particle inhalation, with mice under isoflurane anesthesia, a microcomputed tomography (µCT) scan was taken using an eXplore Locus Micro CT Scanner (GE Healthcare, Little Chalfont, UK; 45 μm resolution, 80 kV, 450 μA). Mice were randomized according to tumor burden and received either vehicle (10% dimethylacetamide and 10% Tween 80 in phosphate-buffered saline), BC-LI-0186 (20 mg/kg bid, 5 days per week, i.p.), cisplatin (5 mg/kg, weekly, i.p.), or a combination of both drugs for 2 weeks. Mice were sacrificed after the second µCT scan, and expression of activated caspase-3 and other proteins was analyzed by IHC. Treatment response was evaluated by CT image analysis. To measure the area of a tumor, at least three representative axial CT images were selected from the upper, middle, and lower levels of the lung. Briefly, at least three axial images were selected after reviewing entire scanned images using following criteria: (1) the tumor was precisely separated from the surrounding normal structures; (2) it had a measurable form; (3) the section containing the longest diameter of the mass. To select axial images of same lesion of one tumor before and after treatment, we referred to the bronchial and pulmonary vascular structures. The pixel of tumor area was quantified using Adobe Photoshop (Adobe Systems, San Jose, CA) and Paint.Net (dotPDN LLC., Kirkland, WA, USA) and the change of the tumor area (%) before and after treatment was estimated by the equation (Pixelbefore – Pixelafter)/Pixelbefore × 100 and averaged over the three images.25
Statistical analysis
The paired t-test was used to compare protein expression in paired samples between tumor and normal-appearing tissue lysates. For the comparison of continuous variables of independent groups, independent sample t-tests were used. Differences in the change in tumor area between groups were analyzed using the Mann–Whitney U test. SPSS software (v23; SPSS, Chicago, IL, USA) was used for statistical analysis. All statistical analyses were two-tailed, and a p-value < 0.05 was interpreted to indicate statistical significance.
Results
LRS expression in NSCLC is related to mTORC1 activity
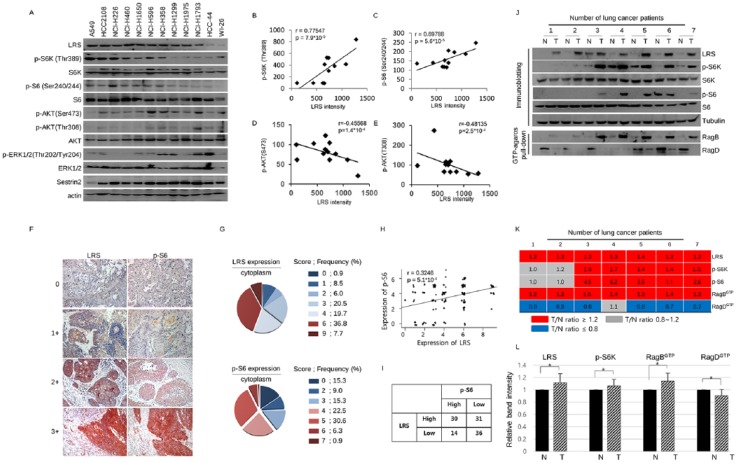
Because LRS plays a major role in activating the mTORC1 pathway by sensing leucine in the cytoplasmic nutrients,16 we first questioned whether there is a relationship between the level of LRS expression and the activity of mTORC1. To confirm this, immunoblotting was performed on the LRS and the component of mTOR signaling using 12 lung origin cell lines. The expression level of LRS, detected in the majority of lung cancer cells, was not correlated with the phosphorylation of ERK and showed inverse correlation with phosphorylation of AKT. However, it was clearly positively correlated with phosphorylation of S6K and S6, which represent mTORC1 activity (Figure 1a–e).
Figure 1.
Expression of leucyl-tRNA synthetase (LRS) has positive correlation with mTORC1 signal in non-small cell lung cancer (NSCLC). (a) Immunoblotting of LRS and molecules constituting mTOR signaling in various NSCLC cells and WI-26 cells and (b) the correlation plot between LRS and p-S6K, (c) that between LRS and p-S6, (d) that between LRS and p-AKT(Ser473), and (e) that between LRS and p-Akt(Thr308). p-values were obtained from Pearson’s correlation analysis and r denotes Pearson’s correlation coefficient. (f) Representative photographs of immunohistochemistry (IHC) staining for LRS and p-S6 from serial sections of 117 NSCLC tissues and (g) pie chart indicating their IHC staining scores. (h) A correlation plot between the LRS and p-S6 expression. There was a significant positive correlation between two molecules in NSCLC tissues (r = 0.3246, p-value = 5.0 × 10-4). p-values were obtained from Pearson’s correlation analysis and r denotes Pearson’s correlation coefficient. (i) A 2 × 2 table, denoting the relationship between the LRS expression and p-S6 expression. There was a significant relationship between LRS and p-S6 expression (p-value = 0.03801, χ2-test ). For detailed information on the scoring system using intensity and frequency of staining, refer to the ‘Immunocytochemistry and IHC’ section. (j) Immunoblotting and GTP-agarose pull-down assay for LRS and mTORC1 markers using paired lysate of adjacent normal appearing tissue and NSCLC tissue. (k) Heatmap indicating the protein expression ratio from paired tumor–normal tissue lysates. Representative results from 7 paired samples among 15 paired samples were shown. Red represents the protein intensity ratio of tumor/normal tissues is over 1.2 and gray represents the ratio between 0.8 and 1.2. Blue represents the ratio of tumor/normal tissues is below 0.8. (l) A histogram obtained by normalizing each protein band intensity obtained from tumor tissue to that from adjacent normal appearing lung tissue in 15 normal–tumor paired samples. N, adjacent normal-appearing lung tissue; T, lung cancer tissue. p-values were obtained from t-test. *p < 0.001.
In order to further confirm the relationship between the expression level of LRS and the activity of mTORC1, IHC staining was performed using serial tissues sections from 117 NSCLC cases (Figure 1f–i). The demographic characteristics of the study cases are noted in Table 1. There was significant positive correlation between cytoplasmic expression of LRS and that of pS6 in these study sets (Pearson’s r = 0.3246, p-value < 0.001; Figure 1h). To identify the relationship between the expression of LRS in NSCLC tissues and clinical and pathologic parameters, overexpression was set as a staining score 6 or more, and then analyzed. LRS was overexpressed in the cytoplasm of NSCLC cells in 52 (44.5%) out of 117 cases. The cases with cytoplasmic LRS overexpression were older than those without overexpression (58.46 ± 10.58 versus 62.79 ± 9.46 years, t-test p = 0.023); other clinical and pathological parameters including clinical outcome were not associated with LRS overexpression (Table 1 and Supplemental Data 1). The effect of LRS expression on clinical outcome of NSCLC was further evaluated using a publicly available dataset from The Cancer Genome Atlas. Of 520 cases of lung adenocarcinoma, 32 (6%) showed a high LRS mRNA level (Z score ⩾2). However, there was no significant difference in disease-free survival (DFS) or overall survival (OS) in these cases versus cases with a normal LRS mRNA level (Z score <2) (Supplemental Data 2).
Table 1.
Clinical characteristics of the patients according to leucyl-tRNA synthetase (LRS) expression.
| LRS expression | p-value | |||
|---|---|---|---|---|
| Low (0–5) | High (6–9) | |||
| n = 65 | n = 52 | |||
| Age | 58.46 ± 10.58 | 62.79 ± 9.46 | 0.023* | |
| Gender | Male | 51 | 35 | 0.208** |
| Female | 14 | 17 | ||
| Smoking history† | Never | 14 | 15 | 0.107 |
| Former | 23 | 8 | ||
| Current | 22 | 24 | ||
| Pack-year (years) | 26.98 ± 21.35 | 27.59 ± 23.13 | 0.890* | |
| Maximal tumor diameter | 3.92±1.71 | 4.47±2.39 | 0.155** | |
| Pathology | ||||
| Adenocarcinoma | 32 | 26 | 0.523 | |
| Squamous cell carcinoma | 33 | 25 | ||
| Other | 0 | 1 | ||
| pStage | I | 21 | 17 | 0.999 |
| II | 15 | 12 | ||
| III | 28 | 22 | ||
| IV | 1 | 1 | ||
Independent sample t-test.
χ2-test.
Smoking history of nine cases is missing.
The relationship between LRS expression and mTORC1 activity was additionally evaluated using paired lung cancer and adjacent normal-appearing lung tissue lysates by immunoblotting and GTP-agarose pull-down assay (Figure 1j–l). Of 15 paired of cases, 12 (80%) showed increased LRS expression in tumor tissue lysate than normal-appearing lung tissue lysates. Similarly, phosphorylation of S6K was higher in 8 (53.3%) cases of tumor tissue lysates when compared with matched normal-appearing lung tissue lysates and showed that there is a clear positive relationship between S6K phosphorylation and LRS expression. Because RagB/RagD heterodimer has higher affinity for LRS than RagA/RagD heterodimer and heterodimer of RagB-GTP/RagD-GDP activates mTORC1, GTP-agarose pull-down assays performed using these paired tissue lysates.16,22 The GTP-agarose pull-down of RagB was significantly increased in the tumor tissues lysates whereas that of RagD was decreased when compared with the normal-appearing adjacent tissues lysates, suggesting that the GTP-bound status of RagB/RagD is significantly related to the expression of LRS and mTORC1 activity. In summary, LRS is frequently overexpressed in NSCLC tissues, and its expression correlated with mTORC1 activity.
LRS plays an important role in leucine-mediated mTORC1 activation and cell growth
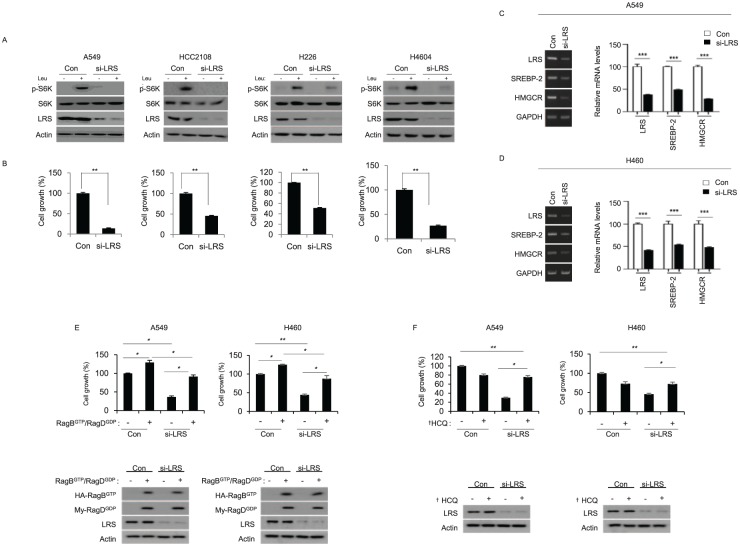
To confirm whether LRS mediates the activation of mTORC1 by leucine and participates in cell growth, siRNA against LRS was transfected to the NSCLC cells expressing high level of LRS. In the cells transfected with control siRNA, leucine induced phosphorylation of S6K, but phosphorylation of S6K by leucine was not observed in the cell line transfected with siRNA against LRS (Figure 2a). In addition, when cell growth was observed for 48 h after LRS silencing using live cell imaging system, significant inhibition of cell growth was observed in the cells transfected with siRNA against LRS comparing those transfected with control siRNA (Figure 2b). SREBP-2 and HMGCR, which are part of the cholesterol biosynthetic gene, are well-known downstream target genes for mTORC1.26 To demonstrate that LRS is involved in the activity of mTORC1, mRNA expression of SREBP-2 and MHGCR was evaluated after LRS knockdown. Indeed, silencing of LRS induced the suppression of mRNA expression of SREBP-2 and MHGCR. (Figure 2c and d). To confirm the interaction between LRS and RagB / RagD in cell growth and proliferation, A549 and H460 cells were co-transfected with GTP mutant of RagB (Q99L) and GDP mutant of RagD (S77L) in the presence of LRS siRNA and were observed using live cell imaging system. In 48 h, inhibition of cell growth by siRNA against LRS was significantly restored by co-transfection of RagBGTP/ RagDGDP, indicating that overexpression of active RagB/D could overcome inhibition of cell growth by LRS knockdown (Figure 2e). To observe the underlying mechanism of cell growth inhibition by LRS knockdown, hydroxychloroquine, an autophagy inhibitor, was treated in LRS knockdown cells and cell growth was monitored. Hydrox-ychloroquine inhibited cell growth in the control cells whereas it did not inhibit cell growth in LRS silenced cells, suggesting that suppression of LRS inhibit cell growth through induction of autophagy (Figure 2f). Taken together, LRS plays an important role in the leucine-mediated mTORC1 activation and could be an important therapeutic target in NSCLC.
Figure 2.
Leucyl-tRNA synthetase (LRS) plays an important role for mTORC1 activation and cell growth in response to leucine. (a) Lung cancer cells, which have relatively high LRS expression, were transfected with either control siRNA (Con) or siRNA against LRS (si-LRS). For 48 h, cells were starved for leucine for 90 min, and then re-stimulated with leucine for 20 min and subjected to immunoblotting. (b) At the same time, cell growth was evaluated by live cell imaging. p-values were obtained from t-test. (c) A549 cells and (d) H460 cells were transfected with Con or si-LRS for 48 h, and the mRNA level of SREBP-2 and HMGCR was analyzed by reverse transcription polymerase chain reaction (RT-PCR) (left). Relative mRNA level was quantified (right). (e) A549 and H460 cells were co-transfected with GTP mutant of RagB (Q99L) and GDP mutant of RagD (S77L) either in the presence or absence of si-LRS. 48 h later, cell growth was measured by live-cell imaging device. Active RagB/D overexpression overcomes the inhibitory effect of LRS knockdown on cell growth. In the bottom, the immunoblotting on the expression of RagB/RagD and LRS knockdown. (f) A549 and H460 NSCLC cells were transfected with si-LRS in the presence of DMSO (-) or hydroxychloroquine (+), which is an autophagy inhibitor. After 48 h, cell growth was monitored by live-cell imaging device. The immunoblotting on the expression of LRS knockdown by si-LRS was shown in the bottom of histogram. si-LRS denotes siRNA against LRS and Con denotes control siRNA. *p < 0.05; **p < 0.01; ***p < 0.001; †HCQ, hydroxychloroquine.
BC-LI-0186 inhibits the leucine-sensing function of LRS in mTORC1 signaling
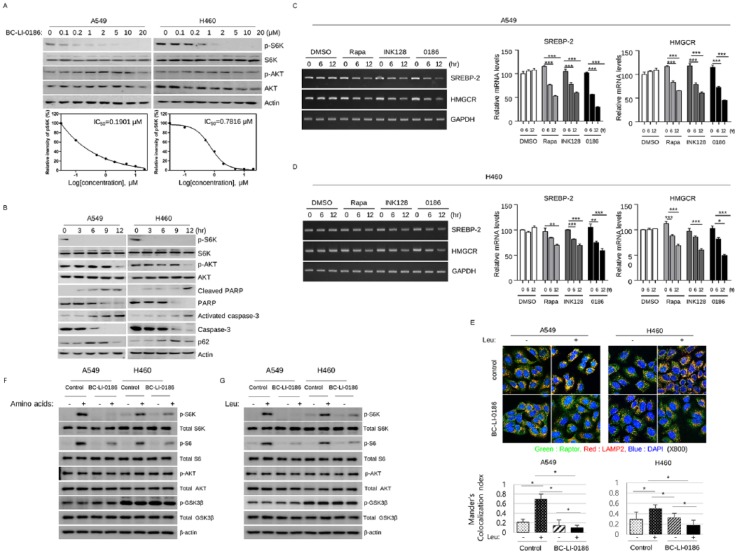
BC-LI-0186 binds to the RagD-interacting site of LRS and further specifically inhibits the GTPase activating protein (GAP) activity of LRS.18 To confirm that BC-LI-0186 is an LRS inhibitor, we first tested its effect on mTOR signaling in NSCLC cells (Figure 3a and b). Treatment of cells with BC-LI-0186 inhibited phosphorylation of S6K in a dose- and time-dependent manner. In the dose-dependent experiments, that harvested cells 15 min after treatment, inhibition of phosphorylation of S6K was observed from 0.2 μM, but the effect on phosphorylation of AKT (S473) was not observed. Treatment of BC-LI-0186 induced cleaved poly (ADP-ribose) polymerase (PARP) and caspase-3 and an increase of p62 from 6 h, suggesting that it has the property of inducing the activity of autophagy, which is a common characteristic of the mTORC1 inhibitor. Decrease of p-AKT was prominent at longer exposure to the BC-LI-0186 and these time points correspond to the emergence of cleaved PARP and activated caspase-3, suggesting that decrease of p-AKT related to the early cell death. For the clarification of mTORC1 inhibitory effect of BC-LI-0186, its effect on the SREBP-2 and MHGCR mRNA expression was compared in the cells treated with rapamycin and INK128, mTORC1 and mTORC1/2 inhibitor respectively. In the BC-LI-0186 treated cells, suppression of SREBP-2 and MHGCR mRNA expression was comparable with the rapamycin and INK128 (Figure 3c and d).
Figure 3.
BC-LI-0186 suppresses leucine-mediated mTORC1 activation. (a) Dose- and (b) time-dependent effects of BC-LI-0186 on mTOR signaling were evaluated by immunoblot. To evaluate the dose-dependent effect of BC-LI-0186, cells were starved for 90 min in the leucine-free medium and then treated with the indicated dose of BC-LI-0186 in the serum-free media for 15 min. To identify the time-dependent effect, 20 μM of BC-LI-0186 was treated by the same method and harvested at the indicated time. (c) A549 cells and (d) H460 cells were treated with 50 nM rapamycin (Rapa), 1 mM INK128, or 10 mM BC-LI-0186 (0186) and mRNA level of SREBP-2 and HMGCR was evaluated by reverse transcription polymerase chain reaction (RT-PCR) (left). Relative mRNA levels were quantified (right) (*p < 0.05; **p < 0.01; ***p < 0.001). (e) BC-LI-0186 inhibits leucine-mediated colocalization of Raptor and LAMP2 in lysosomes. A549 and H460 cells were starved for leucine for 90 min and then incubated in the media with either DMSO or 10 μM of BC-LI-0186 for 2 h, and then challenged with 0.8 mM of leucine for 10 min. After fixation cells were incubated with anti-Raptor and anti-LAMP2 primary antibodies and then visualized with Alexa 488- and Alexa 558-conjugated secondary antibodies. Colocalization results are shown in yellow. In the bottom, quantification of the colocalization between Raptor and LAMP2 was performed by Coloc 2 function of ImageJ. Histogram was obtained by the mean ± standard deviation (SD) of Mander’s colocalization index obtained from more than 10 cells for each staining. BC-LI-0186 inhibits (f) amino-acid- and (g) leucine-mediated mTORC1 signaling. Cells were plated in fetal bovine serum (FBS) supplemented growth media. The next day, cells were incubated in amino-acid-free or leucine-free media containing 10 μM of BC-LI-0186 for 16 h and then stimulated with either amino acid containing media or that with 0.8 mM of leucine for 10 min.
When stimulated by nutrients and Rags, mTORC1 translocates to the lysosome and is activated by GTP-bound Rheb on the surface of the lysosome.27 To further evaluate the effect of BC-LI-0186 on mTORC1, we measured its effect on leucine-induced colocalization of Raptor, a component of mTORC1, and LAMP2, a lysosomal marker (Figure 3e). In untreated cells, leucine stimulation induced colocalization of Raptor and LAMP2, while colocalization was not observed in BC-LI-0186 pretreated cells. To test whether BC-LI-0186 inhibits mTORC1 signaling by repressing the leucine-sensing function of LRS, the effects of amino acids and leucine on mTOR signaling were evaluated in cells pretreated with BC-LI-0186. The addition of amino acids increased phosphorylation of S6K and S6 in untreated cells, while pretreatment with BC-LI-0186 inhibited phosphorylation of S6K and S6 (Figure 3f). There were subtle differences in the phosphorylation of AKT by nutrient supplementation and drug treatment between A549 and H460 cells, which may attribute to the different activating mutations between A549 and H460 cells. A549 cells have functional mutations in the KRAS and LKB genes whereas H460 cells have not only in the KRAS and LKB gene but also activating mutations in the PI3K (p.E545K) gene.28,29 Leucine supplementation increased phosphorylation of S6K and S6 specifically, and BC-LI-0186 pretreatment suppressed this leucine-mediated S6K and S6 phosphorylation (Figure 3g). Taken together, these results show that BC-LI-0186 inhibits mTORC1 activity through repression of LRS-mediated leucine sensing.
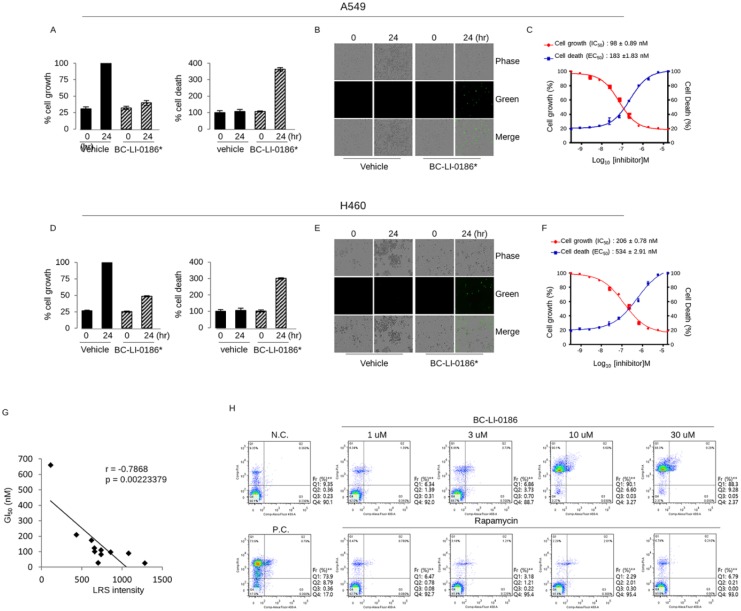
BC-LI-0186 has the cytotoxic effect at nanomolar concentration in NSCLC cells
To confirm that BC-LI-0186 could be developed as a therapeutic agent for lung cancer, the cytotoxicity and growth inhibitory effect were evaluated on several NSCLC cells. NSCLC cells were treated different dose of BC-LI-0186 and cell growth and death were monitored every 2 h by counting the cell confluency and green fluorescence under live cell imaging system, respectively (Figure 4a–f). The BC-LI-0186 IC50 and EC50 were 98 ± 0.89 nM and 183 ± 1.83 nM in A549 cells and 206 ± 0.78 nM and 534 ± 2.91 nM in H460 cells. The same experiment was carried out for the other nine NSCLC cells, and the IC50 and EC50 were shown in Table 2. There was a significant negative correlation between the degree of LRS expression shown in the Figure 1 and the GI50 value of BC-LI-0186 in the NSCLC cell lines (Figure 4g). Rapamycin, a prototypic drug of Rapalog such as temsirolimus, everolimus, and ridaforolimus, inhibits mTORC1 by binding to FK-binding protein 12 (FKBP-12). Because both Rapalog and BC-LI-0186 inhibit mTORC1 activity but target at different sites of signaling pathway, we compared the difference in the growth inhibitory effect and cell death between the two drugs. For this, several NSCLC cell lines were treated with 10 μM of BC-LI-0186 or rapamycin for 48 h, and apoptotic cell death was measured by flow cytometry using annexin V-PI staining (Supplemental Data 3). The representative data obtained from H460 cells by treating indicated dose of BC-LI-0186 for 48 h are shown in the Figure 4h. These findings suggested that BC-LI-0186 has highly effective cytotoxic effect comparing rapamycin in some cells and the cytotoxic effect of BC-LI-0186 did not correspond with that of rapamycin. Neither BC-LI-0186 nor rapamycin had a significant effect on cell cycle progression at the concentration used, and there was no difference in cell cycle progression between drug-sensitive and drug-resistant cell lines (data not shown).
Figure 4.
Cytotoxic effect of BC-LI-0186 on non-small cell lung cancer (NSCLC) cells. (a) A histogram and (b) representative phase contrast images on the cell growth and cell death obtained by incubating A549 cells for the indicated times with vehicle or 16.67 uM of BC-LI-0186. Black bar indicates vehicle whereas hatch bar indicates BC-LI-0186-treated cell. (c) Curved graph indicating the A549 cell growth and cell death at various BC-LI-0186 concentrations. IC50 and EC50 was estimated using a 12-drug concentration by serial dilution based on the maximum concentration of 50 uM. (d) A histogram and (e) representative phase contrast images on the cell growth and cell death obtained by incubating H460 cells for the indicated times with vehicle or 16.67 uM of BC-LI-0186. Black bar indicates vehicle whereas hatch bar indicates BC-LI-0186-treated cell. (f) Curved graph indicating the H460 cell growth and cell death at various BC-LI-0186 concentration. IC50 and EC50 was estimated using a 12-drug concentration by serial dilution based on the maximum concentration of 50 uM. For the measurement of cell growth and death, cells were incubated in the media containing CellToxTM GreenDye and cell confluence and green fluorescence were measured by IncuCyteTM Zoom assay. Note that a green spot indicates cell death. (g) Correlation plot between GI50 obtained from live cell imaging and leucyl-tRNA synthetase (LRS) expression from immunoblotting in 11 NSCLC cells. p-values were obtained from Pearson’s correlation analysis and r denotes Pearson’s correlation coefficient. (h) H460 cells were treated with indicated dose of BC-LI-0186 or rapamycin for 48 h, and cell death was measured by flow cytometry using annexin V and PI staining.
Table 2.
IC50 and EC50 of BC-LI-0186 in non-small cell lung cancer (NSCLC) cells.
| Lung cancer cells | IC50 (nM) | EC50 (nM) |
|---|---|---|
| H2228 | 55 ± 1.27 nM | 146 ± 2.01 nM |
| H1703 | 78 ± 3.18 nM | 158 ± 3.19 nM |
| SNU1330 | 83 ± 0.31 nM | 188 ± 2.07 nM |
| H1650 | 86 ± 1.01 nM | 106 ± 0.88 nM |
| A549 | 98 ± 2.13 nM | 183 ± 1.12 nM |
| H2009 | 102 ± 1.05 nM | 215 ± 0.89 nM |
| H358 | 109 ± 2.38 nM | 329 ± 0.67 nM |
| H2279 | 128 ± 1.99 nM | 289 ± 0.73 nM |
| H460 | 206 ± 0.78 nM | 534 ± 2.91 nM |
| H596 | 206 ± 0.89 nM | 534 ± 3.19 nM |
| H1299 | 1850 ± 0.19 nM | 2345 ± 1.91 nM |
Anticancer effect of BC-LI-0186 in the K-ras mouse lung cancer model
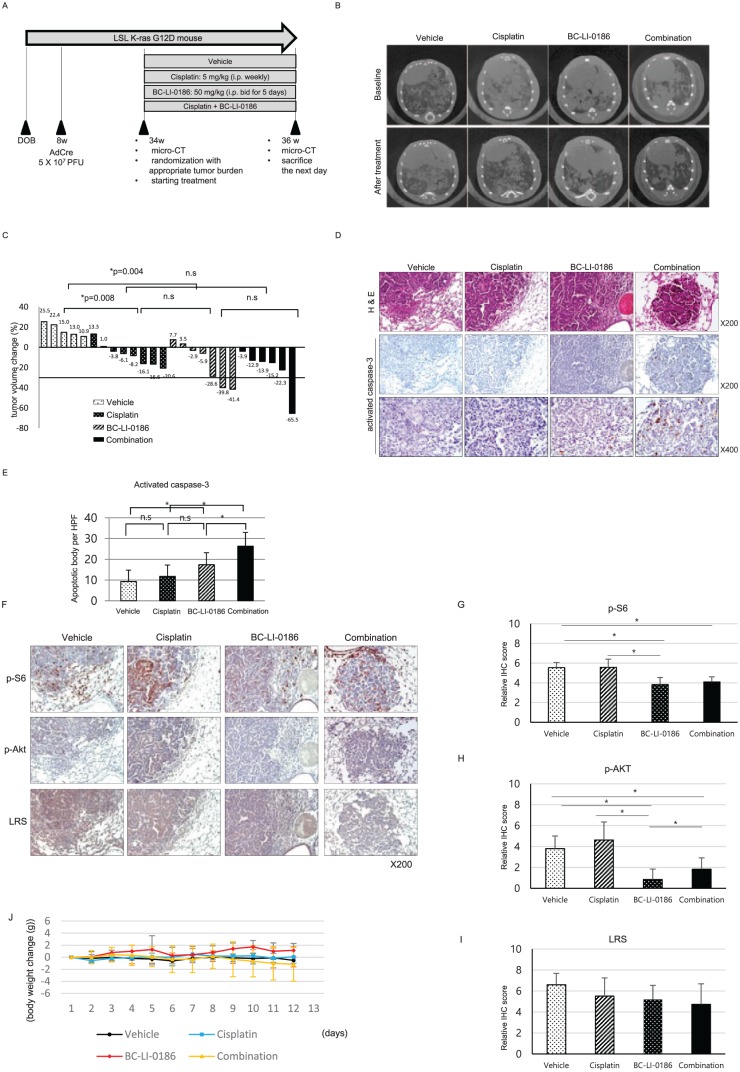
We then assessed whether BC-LI-0186 could be applied to the treatment of lung cancer using an LSL K-ras G12D lung cancer animal model. Tumor size was measured using µCT and the tumor size changes before and after treatment was compared among the groups treated with vehicle, cisplatin, BC-LI-0186, or a combination of BC-LI-0186 and cisplatin (Figure 5a). BC-LI-0186 was administered at 50 mg/kg bid (i.p.) for 5 days per week for 2 weeks. Treatment with BC-LI-0186, cisplatin, and a combination of both drugs significantly reduced tumor size compared with treatment with vehicle, and the antitumor effects of BC-LI-0186 and cisplatin were comparable (Figure 5b and c). To further evaluate the antitumor effect of BC-LI-0186, mice lungs were harvested after treatment, and hematoxylin and eosin (H&E) and IHC staining for activated caspase-3 was performed. The number of activated caspase-3-positive cells was significantly higher in the BC-LI-0186-treated group than in the vehicle or cisplatin-treated groups (Figure 5d and e). To confirm the effect of BC-LI-0186 on mTORC1 and mTORC2 signaling in vivo, IHC staining was performed for p-S6 and p-AKT, markers for mTORC1 and mTORC2 activity, respectively. Treatment with BC-LI-0186 reduced p-S6 and p-AKT level, whereas cisplatin alone has minimal effect on both p-S6 and p-AKT expression (Figure 5f and i). To assess how well the mice tolerated the treatment, change in body weight during the treatment period was compared among treatment groups. The BC-LI-0186-treated group showed a slight (not statistically significant) increase in body weight during the treatment period, whereas the combination-treatment group showed a slight decrease (Figure 5j). Taken together, these results indicate that treatment with BC-LI-0186 showed a significant antitumor effect in the K-ras mouse lung cancer model, comparable to treatment with cisplatin but with specific inhibition of mTORC1 and not mTORC2.
Figure 5.
Antitumor effect of BC-LI-0186 in a K-ras lung cancer mouse model. (a) Diagram of the treatment schedule in a Lox-Stop-Lox (LSL) K-ras G12D mouse lung cancer model. (b) Representative microcomputed tomography (µCT) images performed before and after 2-week treatment schedule and (c) waterfall plot for tumor size change comparing before and after treatment. p-values were obtained using the Mann–Whitney U test. (d) Representative photos of H&E and activated caspase-3 immunohistochemistry (IHC) staining on the lungs harvested after 2 weeks of treatment and (e) a histogram quantifying the activated caspase-3 spots in the tumor of each treatment group. A total of 10 tumor areas per mouse were selected and then photographed at a magnification of 400×. The histogram was obtained by counting the number of activated caspase-3 spots observed in each field and using the mean and standard error (SE) values. *p < 0.05. p-values were obtained by one-way analysis of variance (ANOVA) followed by Tukey’s post hoc multiple comparison tests. (f) Representative photograph of IHC staining of pS6, pAKT, and leucyl-tRNA synthetase (LRS) on the lung sections obtained after 2 weeks treatment and quantification of (g) p-S6, (h) p-AKT, and (i) LRS expression in the tumor of each treatment group. At least 10 tumor was photographed per mouse and scored as described in the ‘Materials and methods’ section. *p < 0.05. p-values were obtained by one-way ANOVA followed by Tukey’s post hoc multiple comparison tests. (j) Body weight change among treatment groups during treatment. There was no significant body weight change among the treatment groups.
Discussion
The findings of this study provide further evidence that LRS is involved in mTORC1 activation, and that an LRS inhibitor, BC-LI-0186, could be developed as a cancer therapeutic in line with recent reports.30 The classic mTOR inhibitors, rapalogs, are used in the coating of coronary stents, prevention of rejection after organ transplantation, treatment of tuberous sclerosis complex, and lymphangioleiomyomatosis.31–34 Everolimus, a rapalog, has indications for the treatment of neuroendocrine tumors, advanced breast cancer in postmenopausal women who failed letrozole or anastrozole, and advanced renal cell carcinoma that failed sunitinib or sorafenib.35–37 The main reason that rapalogs do not have wider indications in cancer is that their antitumor effect is modest whereas they have various adverse effects.
When comparing BC-LI-0186 with rapamycin in terms of cytotoxicity, there was no correlation between rapamycin and BC-LI-0186 (Supplemental Data 3). It also was encouraging that BC-LI-0186 showed a significant antitumor effect whereas the 2-week treatment of rapamycin did not show antitumor effect in the same experimental model used in this study (data not shown). This might be due to difference in the inhibitory mechanism: rapamycin inhibits mTORC1 by binding FKBP-12, whereas BC-LI-0186 inhibits mTORC1 by inhibiting binding of LRS to Rag GTPase. Moving the targeting site to the downstream of the signaling pathway could change the potency and efficacy of the drug.
However, a change in the targeting site of a drug may cause new problems. Long-term use of rapalogs has various side effects ranging from mild stomatitis, rash, increased susceptibility to infectious disease, diabetes, to interstitial pneumonitis leading drug discontinuation.38 Because this study had a primary goal of confirming the efficacy of the drug, investigation on the adverse effects was partially done. There were no discernible changes in the appearance, behavior, or body weight in the experimental animals during the 2 weeks of treatment, but biochemical, hematologic, and other additional careful investigations are needed to clarify the short-term and long-term adverse effect of BC-LI-0186.
Along with the studies on the side effects, additional investigation need to be preceded for therapeutic application of this drug. This study confirmed efficacy by measuring changes in tumor size after 2 weeks of short-term treatment. For effective long-term cancer management, studies on the treatment cycle, frequency and duration of therapy are needed and once these are determined, the additional observation is needed to identify overall survival is significantly different to the control group.
The direct effect of LRS and its inhibitor, BC-LI-0186, on AKT signaling was not been predicted. However, some interesting findings between BC-LI-0186 and AKT have been observed in this study; among other things, the inverse correlation between p-AKT and LRS expression in unstimulated NSCLC. This finding suggested that LRS might be involved in negative feedback loop on the AKT signaling loop composed of mTORC1–pS6K–IRS. On the other hand, inhibition of p-AKT by increased concentration and exposure time to BC-LI-0186 in the cancer cells, and decreased expression of p-AKT in the tumor tissues after 2 weeks of BC-LI-0186 treatment, may be attributed to the cell death rather than its direct effect on AKT.
The TP53 mutation, one of the most frequently observed variations in various cancers, confers resistance to chemotherapeutic agents and reduces their therapeutic effects.39 In a small pilot study using K-ras:p53fl/fl mice, BC-LI-0186 did not show significant tumor reduction (Supplemental Data 4). This finding suggests that loss of TP53 function can affect the response of mice to BC-LI-0186. However, subsequent experiments using NSCLC cell lines, which lack functional TP53, did not support this finding, because several cell lines showed high or moderate sensitivity to BC-LI-0186 in terms of cytotoxicity, whereas the remaining cells were not sensitive. To clarify the antitumor effect of LRS inhibitor on the cancer with TP53 mutation, confirmation is required with various models.
In conclusion, LRS is frequently overexpressed in NSCLC tissues, and its expression correlates with that of pS6, a marker of mTORC1 activity. A novel LRS inhibitor, BC-LI-0186, inhibits mTORC1 signaling and has cytotoxic effects in NSCLC cells and antitumor effects in a K-ras mouse lung cancer model. This study suggests that BC-LI-0186 inhibits the noncanonical, mTORC1-activating function of LRS and provides a novel therapeutic strategy for NSCLC.
Supplemental Material
Supplemental material, Supp_data_1 for Therapeutic effects of the novel Leucyl-tRNA synthetase inhibitor BC-LI-0186 in non-small cell lung cancer by Eun Young Kim, Jin Gu Lee, Jung Mo Lee, Arum Kim, Hee Chan Yoo, Kibum Kim, Minji Lee, Chulho Lee, Gyoonhee Han, Jung Min Han and Yoon Soo Chang in Therapeutic Advances in Medical Oncology
Supplemental Material
Supplemental material, Supp_data_2 for Therapeutic effects of the novel Leucyl-tRNA synthetase inhibitor BC-LI-0186 in non-small cell lung cancer by Eun Young Kim, Jin Gu Lee, Jung Mo Lee, Arum Kim, Hee Chan Yoo, Kibum Kim, Minji Lee, Chulho Lee, Gyoonhee Han, Jung Min Han and Yoon Soo Chang in Therapeutic Advances in Medical Oncology
Supplemental Material
Supplemental material, Supp_data_3 for Therapeutic effects of the novel Leucyl-tRNA synthetase inhibitor BC-LI-0186 in non-small cell lung cancer by Eun Young Kim, Jin Gu Lee, Jung Mo Lee, Arum Kim, Hee Chan Yoo, Kibum Kim, Minji Lee, Chulho Lee, Gyoonhee Han, Jung Min Han and Yoon Soo Chang in Therapeutic Advances in Medical Oncology
Supplemental Material
Supplemental material, Supp_data_4 for Therapeutic effects of the novel Leucyl-tRNA synthetase inhibitor BC-LI-0186 in non-small cell lung cancer by Eun Young Kim, Jin Gu Lee, Jung Mo Lee, Arum Kim, Hee Chan Yoo, Kibum Kim, Minji Lee, Chulho Lee, Gyoonhee Han, Jung Min Han and Yoon Soo Chang in Therapeutic Advances in Medical Oncology
Footnotes
Funding: This work was supported by the Global Frontier Project (grant numbers NRF-2013M3A6A4072536 and NRF-2014M3A6A4074817) and the Basic Science Research Program (grant number 2018R1A6A1A03023718) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education.
Conflict of interest statement: The authors declare that there are no conflicts of interest.
Supplemental material: Supplemental material for this article is available online.
ORCID iD: Yoon Soo Chang  https://orcid.org/0000-0003-3340-4223
https://orcid.org/0000-0003-3340-4223
Contributor Information
Eun Young Kim, Department of Internal Medicine, Yonsei University College of Medicine, Yonsei University, Seoul, South Korea.
Jin Gu Lee, Department of Thoracic and Cardiovascular Surgery, Yonsei University College of Medicine, Yonsei University, Seoul, South Korea.
Jung Mo Lee, Department of Internal Medicine, Yonsei University College of Medicine, Yonsei University, Seoul, South Korea.
Arum Kim, Department of Internal Medicine, Yonsei University College of Medicine, Yonsei University, Seoul, South Korea.
Hee Chan Yoo, College of Pharmacy, Yonsei Institute of Pharmaceutical Sciences, Yonsei University, Seoul, South Korea.
Kibum Kim, College of Pharmacy, Yonsei Institute of Pharmaceutical Sciences, Yonsei University, Seoul, South Korea.
Minji Lee, College of Pharmacy, Yonsei Institute of Pharmaceutical Sciences, Yonsei University, Seoul, South Korea.
Chulho Lee, Translational Research Center for Protein Function Control, Department of Biotechnology, Yonsei University, Seoul, South Korea.
Gyoonhee Han, Translational Research Center for Protein Function Control, Department of Biotechnology, Yonsei University, Seoul, South Korea.
Jung Min Han, College of Pharmacy, Yonsei Institute of Pharmaceutical Sciences, Yonsei University, 85 Songdogwahak-ro, Yeonsu-gu, Incheon, 21983, South Korea.
Yoon Soo Chang, Department of Internal Medicine, Yonsei University College of Medicine, Yonsei University, 4th Floor, Research Center for Future Medicine, 20, Eonju-ro 63-gil, Gangnam-gu, Seoul, 06229, South Korea.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017; 67: 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Chang YS, Choi CM, Lee JC. Mechanisms of epidermal growth factor receptor tyrosine kinase inhibitor resistance and strategies to overcome resistance in lung adenocarcinoma. Tuberc Respir Dis 2016; 79: 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Park SG, Choi EC, Kim S. Aminoacyl-tRNA synthetase-interacting multifunctional proteins (AIMPs): a triad for cellular homeostasis. IUBMB Life. 2010; 62: 296–302. [DOI] [PubMed] [Google Scholar]
- 4. Lee SW, Cho BH, Park SG, et al. Aminoacyl-tRNA synthetase complexes: beyond translation. J Cell Sci 2004; 117(Pt 17): 3725–3734. [DOI] [PubMed] [Google Scholar]
- 5. Kim S, You S, Hwang D. Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat Rev Cancer 2011; 11: 708–718. [DOI] [PubMed] [Google Scholar]
- 6. Arnez JG, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem Sci 1997; 22: 211–216. [DOI] [PubMed] [Google Scholar]
- 7. Cusack S, Hartlein M, Leberman R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res 1991; 19: 3489–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghanipour A, Jirstrom K, Ponten F, et al. The prognostic significance of tryptophanyl-tRNA synthetase in colorectal cancer. Cancer Epidemiol Biomarkers Prev 2009; 18: 2949–2956. [DOI] [PubMed] [Google Scholar]
- 9. Wakasugi K, Slike BM, Hood J, et al. Induction of angiogenesis by a fragment of human tyrosyl-tRNA synthetase. J Biol Chem 2002; 277: 20124–20126. [DOI] [PubMed] [Google Scholar]
- 10. Greenberg Y, King M, Kiosses WB, et al. The novel fragment of tyrosyl tRNA synthetase, mini-TyrRS, is secreted to induce an angiogenic response in endothelial cells. FASEB J 2008; 22: 1597–1605. [DOI] [PubMed] [Google Scholar]
- 11. Levy C, Fisher DE. Dual roles of lineage restricted transcription factors: the case of MITF in melanocytes. Transcription 2011; 2: 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ray PS, Jia J, Yao P, et al. A stress-responsive RNA switch regulates VEGFA expression. Nature 2009; 457: 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cusack S, Yaremchuk A, Tukalo M. The 2 A crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J 2000; 19: 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 2013; 14: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bonfils G, Jaquenoud M, Bontron S, et al. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell 2012; 46: 105–110. [DOI] [PubMed] [Google Scholar]
- 16. Han JM, Jeong SJ, Park MC, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012; 149: 410–424. [DOI] [PubMed] [Google Scholar]
- 17. Avruch J, Long X, Ortiz-Vega S, et al. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab 2009; 296: E592–E602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim JH, Lee C, Lee M, et al. Control of leucine-dependent mTORC1 pathway through chemical intervention of leucyl-tRNA synthetase and RagD interaction. Nat Commun 2017; 8: 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim EY, Kim A, Kim SK, et al. KRAS oncogene substitutions in Korean NSCLC patients: clinical implication and relationship with pAKT and RalGTPases expression. Lung Cancer (Amsterdam, Netherlands) 2014; 85: 299–305. [DOI] [PubMed] [Google Scholar]
- 20. Karachaliou N, Mayo C, Costa C, et al. KRAS mutations in lung cancer. Clin Lung Cancer 2013; 14: 205–214. [DOI] [PubMed] [Google Scholar]
- 21. Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 2018; 15: 273–291. [DOI] [PubMed] [Google Scholar]
- 22. Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science (New York, NY) 2008; 320: 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shin E, Choi CM, Kim HR, et al. Immunohistochemical characterization of the mTOR pathway in stage-I non-small-cell lung carcinoma. Lung Cancer (Amsterdam, Netherlands) 2015; 89: 13–18. [DOI] [PubMed] [Google Scholar]
- 24. Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001; 15: 3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim EY, Kim A, Kim SK, et al. AZD6244 inhibits cisplatin-induced ERK1/2 activation and potentiates cisplatin-associated cytotoxicity in K-ras G12D preclinical models. Cancer Lett 2015; 358: 85–91. [DOI] [PubMed] [Google Scholar]
- 26. Wang BT, Ducker GS, Barczak AJ, et al. The mammalian target of rapamycin regulates cholesterol biosynthetic gene expression and exhibits a rapamycin-resistant transcriptional profile. Proc Natl Acad Sci USA 2011; 108: 15201–15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Betz C, Hall MN. Where is mTOR and what is it doing there? J Cell Biol 2013; 203: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blanco R, Iwakawa R, Tang M, et al. A gene-alteration profile of human lung cancer cell lines. Hum Mutat 2009; 30: 1199–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mahoney CL, Choudhury B, Davies H, et al. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. Br J Cancer 2009; 100: 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao G, Yao Y, Li K, et al. A human leucyl-tRNA synthetase as an anticancer target. Onco Targets Ther 2015; 8: 2933–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lozano I, Serrador A, Lopez-Palop R, et al. Immediate and long-term results of drug-eluting stents in mammary artery grafts. Am J Cardiol 2015; 116: 1695–1699. [DOI] [PubMed] [Google Scholar]
- 32. Waldner M, Fantus D, Solari M, et al. New perspectives on mTOR inhibitors (rapamycin, rapalogs and TORKinibs) in transplantation. Br J Clin Pharmacol 2016; 82: 1158–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brakemeier S, Bachmann F, Budde K. Treatment of renal angiomyolipoma in tuberous sclerosis complex (TSC) patients. Pediatr Nephrol (Berlin, Germany) 2017; 32: 1137–1144. [DOI] [PubMed] [Google Scholar]
- 34. McCormack FX, Inoue Y, Moss J, et al. Efficacy and safety of sirolimus in lymphangioleiomyomatosis. N Engl J Med 2011; 364: 1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yao JC, Fazio N, Singh S, et al. Everolimus for the treatment of advanced, non-functional neuroendocrine tumours of the lung or gastrointestinal tract (RADIANT-4): a randomised, placebo-controlled, phase 3 study. Lancet (London, England) 2016; 387: 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baselga J, Campone M, Piccart M, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med 2012; 366: 520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Motzer RJ, Escudier B, Oudard S, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet (London, England) 2008; 372: 449–456. [DOI] [PubMed] [Google Scholar]
- 38. Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab 2014; 19: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aas T, Børresen A-L, Geisler S, et al. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med 1996; 2: 811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supp_data_1 for Therapeutic effects of the novel Leucyl-tRNA synthetase inhibitor BC-LI-0186 in non-small cell lung cancer by Eun Young Kim, Jin Gu Lee, Jung Mo Lee, Arum Kim, Hee Chan Yoo, Kibum Kim, Minji Lee, Chulho Lee, Gyoonhee Han, Jung Min Han and Yoon Soo Chang in Therapeutic Advances in Medical Oncology
Supplemental material, Supp_data_2 for Therapeutic effects of the novel Leucyl-tRNA synthetase inhibitor BC-LI-0186 in non-small cell lung cancer by Eun Young Kim, Jin Gu Lee, Jung Mo Lee, Arum Kim, Hee Chan Yoo, Kibum Kim, Minji Lee, Chulho Lee, Gyoonhee Han, Jung Min Han and Yoon Soo Chang in Therapeutic Advances in Medical Oncology
Supplemental material, Supp_data_3 for Therapeutic effects of the novel Leucyl-tRNA synthetase inhibitor BC-LI-0186 in non-small cell lung cancer by Eun Young Kim, Jin Gu Lee, Jung Mo Lee, Arum Kim, Hee Chan Yoo, Kibum Kim, Minji Lee, Chulho Lee, Gyoonhee Han, Jung Min Han and Yoon Soo Chang in Therapeutic Advances in Medical Oncology
Supplemental material, Supp_data_4 for Therapeutic effects of the novel Leucyl-tRNA synthetase inhibitor BC-LI-0186 in non-small cell lung cancer by Eun Young Kim, Jin Gu Lee, Jung Mo Lee, Arum Kim, Hee Chan Yoo, Kibum Kim, Minji Lee, Chulho Lee, Gyoonhee Han, Jung Min Han and Yoon Soo Chang in Therapeutic Advances in Medical Oncology