Abstract
Oxidative stress is detrimental to animals and can depress the growth performance and regulate the gene expression of animals. However, it remains unclear how oxidative stress regulates the expression of long noncoding RNAs (lncRNAs) and mRNAs. Therefore, the purpose of this article was to explore the profiles of lncRNAs and mRNAs in the liver of piglets under oxidative stress. Here, we constructed a piglet oxidative stress model induced by diquat and evaluated the effects of oxidative stress on the growth performance and antioxidant enzyme activity of piglets. We also used RNA-Seq to examine the global expression of lncRNAs and mRNAs in piglets under oxidative stress. The targets of lncRNAs and mRNAs were enriched in gene ontology (GO) terms and signaling pathways. The results show that the growth performance and activities of antioxidant enzymes were decreased in piglets under oxidative stress. Moreover, eight lncRNAs (6 upregulated and 2 downregulated) and 30 mRNAs (8 upregulated and 22 downregulated) were differentially expressed in the oxidative stress group of piglets compared to the negative control group. According to biological processes in enriched GO terms, the oxoacid metabolic process, intramolecular oxidoreductase activity, and oxidation-reduction process play important roles in oxidative stress. Pathway analysis showed that the signaling pathways involved in insulin and glucose metabolism had a close relationship with oxidative stress. Further in vitro experiments showed that the expression of the upregulated gene GNMT was significantly increased in primary porcine hepatocytes after diquat stimulation. In contrast, the level of the downregulated gene GCK was significantly decreased at 12 h in primary porcine hepatocytes after diquat stimulation. Our results expand our knowledge of the lncRNAs and mRNAs transcribed in the livers of piglets under oxidative stress and provide a basis for future research on the molecular mechanisms mediating oxidative stress and tissue damage.
1. Introduction
In the process of metabolism, organisms can produce many kinds of reactive oxygen species (ROS). These ROS free radicals are maintained at certain physiological steady-state levels, and excessive ROS are generally eliminated by the antioxidant defense system, which includes antioxidant enzymes (superoxide dismutase (SOD) and glutathione peroxidase (GPx)) and nonenzymatic antioxidants (for example, glutathione, Se, vitamin E, and vitamin C) [1]. Therefore, the antioxidant system can protect tissues and cells from ROS damage. When the level of ROS produced by cells is higher than the antioxidant defense ability of cells, the redox state is unbalanced. Excessive ROS in tissues or cells will induce oxidative stress and lead to oxidative damage, such as tissue damage and the oxidation of proteins, lipids, and nucleic acids [2, 3]. In livestock production, numerous factors, such as environmental factors, oxidized diets, and weaning, can induce oxidative stress to damage cellular antioxidant defenses [4, 5]. Oxidative stress can result in suboptimal health conditions of livestock and a reduction in production efficiency and can also cause serious economic losses in animal husbandry systems.
Cells maintain homeostasis through transcription and posttranscriptional regulation that cause changes in gene expression [6, 7]. Long noncoding RNAs (lncRNAs), as important transcripts, play key roles in posttranscriptional mechanisms by regulating RNA stability and translation [8]. lncRNAs are a class of mRNA-like transcripts that are longer than 200 nucleotides (nt) and have no protein-coding potential [9–11]. lncRNAs are involved in numerous biological processes, including cell development and differentiation, immune responses, the cell cycle, and apoptosis, by regulating gene expression [12]. Recent evidence indicates that lncRNAs participate in the response to diverse stressful stimuli [13, 14]. Giannakakis et al. indicated that thousands of lncRNA transcripts were transiently induced by oxidative stress and linked with polysomes, a complex of the mRNA molecule and two or more ribosomes that act to form polypeptide chains during active translation [15]. However, very few oxidative stress-induced lncRNAs have been subjected to functional analysis.
The domestic pig (Sus scrofa) is an economically important farm animal worldwide. Pigs, especially weaned piglets on farms, suffer from various challenges, such as weaning, changes in the nutritional source, gastrointestinal disorders, infections, and diarrhea, and these challenges might produce excessive amounts of ROS and induce oxidative stress [4, 16], which may result in the growth retardation of piglets. Diquat (DQ) is a commercially available herbicide and is used extensively in China. It induces animal oxidative stress [5]. Previous research indicated that an ideal piglet model, which is useful for understanding the effects of oxidative stress, could be constructed by intraperitoneal injection of DQ [17, 18]. However, little is known about the effect of oxidative stress on genome-wide lncRNA expression in piglets. Therefore, in the present study, the growth performance of piglets was investigated, and the livers were collected to screen the differentially expressed lncRNAs and mRNAs and pathways using high-throughput sequencing. Furthermore, the key lncRNAs and genes were verified by real-time quantitative polymerase chain reaction (qRT-PCR) to explore the toxicity mechanism of oxidative damage induced by DQ. These results may provide information necessary to propose a new genetic strategy for oxidative stress resistance.
2. Materials and Methods
All experimental procedures were conducted in accordance with the protocol approved by the Animal Care and Use Committee of Shandong Agricultural University.
2.1. Animals, Housing, and Experimental Design
Landrace piglets weaned at 21 days were collected from 6 litters, and two male pigs were chosen from each litter. The two pigs were divided into an oxidative stress group and a negative control group. After 3 days of adaptation, piglets with an average initial body weight of 5.76 ± 0.22 kg were given free access to water and commercial feed and housed individually in wire cages in constant-temperature (25-27°C) animal rooms with a 12 h light-dark cycle.
At the beginning of the experiment, the oxidative stress group piglets received an intraperitoneal injection of DQ (Sigma-Aldrich, Saint Louis, USA) at 10 mg/kg of body weight, and the negative control group was injected with the same volume of isotonic saline. One piglet in the oxidative stress group died on the 3rd day. The trial lasted for 7 days, and then there were six piglets in the negative control group and five piglets in the oxidative stress group.
2.2. Rectal Temperature Measurement and Sample Collection
The rectal temperature of all the pigs was measured at 8:00 am on days 1, 3, 5, and 7. The body weight was also measured at the end of the experiment, and the average daily gain (ADG) was calculated.
All pigs were anesthetized with an intravenous injection of phenobarbital (0.25 mg/kg body weight) and slaughtered by exsanguinations according to protocols approved by the Shandong Agricultural University Animal Care and Use Committee. Fresh liver samples at the same location were collected and divided into two subsamples: one subsample was immediately frozen in liquid nitrogen and then stored at -80°C for extraction of total RNA, and the other subsample was kept in 10% formalin for H&E staining.
2.3. Measurement of the Oxidative Stress Index
Liver tissue (200 mg) was homogenized in a tissue lyser (ULTRA-TURRAX® T18 basic, IKA, Germany) for 2 min and then centrifuged for 15 min at 15,000 g at 4°C to remove the cellular debris; the supernatant was collected for oxidative stress index analysis. Total antioxidant capacity (T-AOC), superoxide dismutase (SOD) activity, glutathione peroxidase (GSH-Px) activity, and malondialdehyde (MDA) content in the liver were measured using spectrophotometric kits in accordance with the manufacturer's instructions (Nanjing Jiancheng Biotechnology Institute, Nanjing, China).
2.4. Liver Histological Examination
Liver tissue was fixed in 10% formalin for 24 h, followed by embedding in paraffin wax and then cut into 5 μm sections (Leica RM2135, Germany). After 3 h at 60°C, the slides were deparaffinized with xylene, rehydrated with a decreasing series of ethanol, and stained with hematoxylin and eosin (HE) according to standard histological protocols. Slides were photographed with an optical microscope (Olympus BX53).
2.5. Library Preparation for lncRNA Sequencing and Data Analysis
According to the rectal temperature, antioxidant enzyme activity, and HE staining of the liver in the oxidative stress piglets, three piglets with the most severe clinical manifestations were chosen from the oxidative stress group. At the same time, the three full sibs of the chosen piglets were also collected from the negative control group. Therefore, six piglets (three piglets in each group) were chosen for the lncRNA sequencing analysis. Total RNA was extracted from liver tissue using a total RNA extractor (Sangon, Shanghai, China) according to the manufacturer's instructions. The quantity and quality of isolated total RNAs were estimated by running on a 1% agarose gel and using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). Then, the rRNA was removed using the Ribo-off rRNA Depletion Kit (Vazyme Biotech, Nanjing, China). The lncRNA libraries were generated using the VAHTS™ stranded mRNA-seq library prep kit for Illumina® (Vazyme Biotech, Nanjing, China). The libraries were sequenced using an Illumina HiSeq X Ten platform, and 150 bp paired-end reads were generated.
Sequencing reads or raw reads were trimmed using Trimmomatic (version 0.36) [19] to remove the adaptor sequences, unknown nucleotides larger than 5%, or Q20 < 20%, followed by FastQC (version 0.11.2) quality control checks. Trimmed reads used for downstream analyses had a quality score of Q30. All clean reads were aligned to the reference genome of Sus scrofa (Sscrofa 10.2; ftp://ftp.ensembl.org/pub/release-75/fasta/sus_scrofa/) using the HISAT2 (version 2.1.0) software [20].
The transcript expression levels were calculated using the transcript per million (TPM) values generated by the StringTie (version 1.3.3b) software [21], and then the expression data was normalized using the R package RUVSeq (version 1.17.1) [22]. To obtain high-quality lncRNAs, transcripts with lengths greater than 200 nt were used for the prediction. The coding potential of these transcripts was predicted using CPC2 (version 2.0) [23], CNCI (version 2.0) [24], Pfam [25], and PLEK (version 1.2) [26]. The selected transcripts without coding potential from these software analysis results were considered lncRNAs. The lncRNAs and mRNAs (TPM > 0.1) that were detected commonly in all six samples were assigned as co-genes. The differentially expressed (DE) lncRNAs and mRNAs were analyzed using the R package DESeq [27]. The lncRNAs and mRNAs with fold changes (log2 ratio) ≥ 2 and qvalue significance scores < 0.05 were regarded as DE lncRNAs and mRNAs.
All the DE genes and the predicted target genes of DE lncRNAs were submitted to the databases of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) for enrichment analysis.
2.6. Isolation of Primary Porcine Hepatocytes and Oxidative Stress Treatment
The five three-day-old pig livers were perfused, and hepatocytes were isolated and purified using the two-step procedure described by Fang et al. [28]. The isolated hepatocytes were resuspended in RPMI 1640 medium supplemented with 10% fetal bovine serum, and the hepatocytes (5 × 106 cells/mL) were treated with 50 μM DQ for 6 and 12 h. Control cells were cultured under the same conditions except without DQ treatment. Total RNA of the primary porcine hepatocytes was extracted using TRIzol (Takara Biotechnology, Dalian, China) according to the manufacturer's instructions. The quantity and quality of the isolated RNA were determined via UV260/280 using a biophotometer (Eppendorf, Hamburg, Germany). The GNMT and GCK expression levels in primary porcine hepatocytes were determined by qRT-PCR.
2.7. Quantitative Real-Time RT-PCR (qPCR)
Eight DE lncRNAs and 13 DE mRNAs (Table S1) were selected to verify the expression changes observed in our RNA-Seq results. Primers of selected lncRNAs and mRNAs were designed with Primer3web software (version 4.1.0) [29] and then synthesized by Sangon Biotech (Shanghai, China). Real-time PCR was performed using the SYBR® Premix Ex TaqTM (Takara, Dalian, China) and a LightCycler® 96 Real-Time PCR System (Roche). Briefly, the amplifications were performed with a 20 μL reaction volume containing 10 μL of 2× SYBR Premix ExTaq, 0.5 μL of each primer, 2 μL of diluted cDNA, and sterile water to the volume to 20 μL. The PCR amplification was carried out as follows to calculate the melting curve: 95°C for 10 s; then 40 cycles of 95°C for 5 s, 58°C for 10 s, and 72°C for 15 s; followed by 1 cycle of 95°C for 1 min, 61°C for 30 s, and 95°C for 30 s. To exclude between-run variations, all samples were amplified in triplicate, and the mean was used for further analysis. A negative control reaction without template was performed for each primer to ensure that RNAs were free of genomic DNA. GAPDH and TOP2B genes were used as a control for expression normalization. Only primers showing an efficiency between 85 and 115% were used for qRT-PCR. The comparative cycle threshold (CT) method (2−ΔΔCT) was used to quantitate mRNA expression [30].
2.8. Statistical Analyses
Data were analyzed by using the statistical R package (http://www.r-project.org/), and the results are presented as the mean ± standard error of the mean (SEM). The statistical significance of differences between two groups was compared with an unpaired two-tailed Student t-test. The differences were considered statistically significant when ∗ P < 0.05, ∗∗ P < 0.01, and ∗∗∗ P < 0.001.
3. Results
3.1. Growth Performance
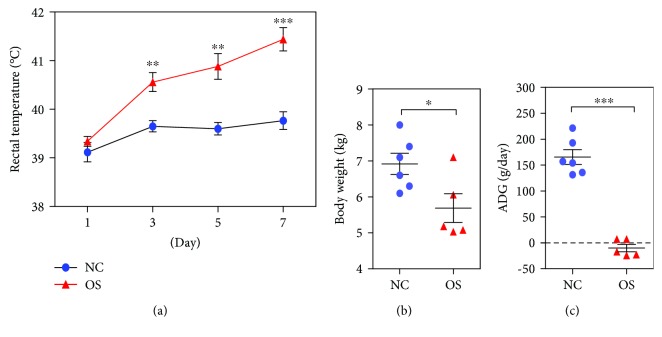
The average rectal temperature of the oxidative stress group was significantly higher than that of the negative control group on days 3, 5, and 7 (Figure 1(a)). However, the injection of DQ significantly decreased the average body weight (Figure 1(b)), and the average daily gain (ADG) of the oxidative stress group was significantly lower than that of the negative control group (Figure 1(c)).
Figure 1.
The effects of DQ on piglet performance between the oxidative stress group (n = 5) and the negative control group (n = 6). (a) The rectal temperature of piglets changes at days 1, 3, 5, and 7. (b) The last body weight at the end of the experiment. (c) The effect of DQ on average daily gain (ADG). OS: oxidative stress group; NC: negative control group. ∗ P < 0.05, ∗∗ P < 0.01, and ∗∗∗ P < 0.001.
3.2. Activities of Antioxidant Enzymes and MDA Concentrations in the Liver
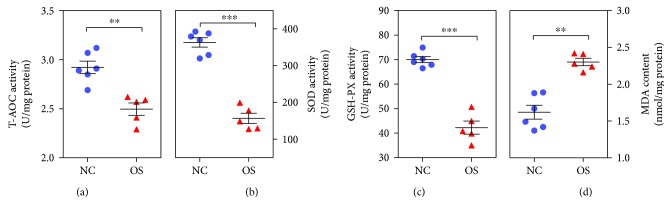
It was observed that animals injected with DQ at 10 mg/kg of body weight presented significantly lower activity of T-AOC (Figure 2(a)), SOD (Figure 2(b)), and GSH-PX (Figure 2(c)) than the negative control group pigs. However, the MDA contents in oxidative stress group piglets were significantly higher than those in the negative control piglets (Figure 2(d)).
Figure 2.
The effects of DQ on total antioxidant capacity (T-AOC) activity (a), superoxide dismutase (SOD) activity (b), glutathione peroxidase (GSH-Px) activity (c), and malondialdehyde (MDA) content (d) between oxidative stress (n = 5) and negative control (n = 6) groups. OS: oxidative stress group; NC: negative control group. ∗ P < 0.05, ∗∗ P < 0.01, ∗∗∗ P < 0.001.
3.3. Effects of DQ on Liver Morphological Structure
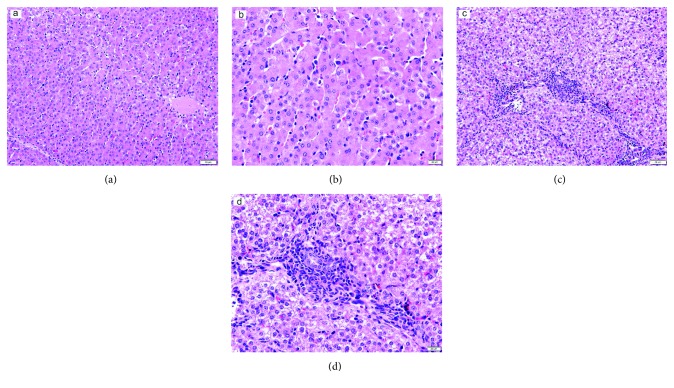
The liver is the major target organ of DQ [31]; therefore, the injection of DQ significantly influenced the liver morphological structure. Compared to the negative control group (Figures 3(a) and 3(b)), the liver of oxidative stress group piglets was in an oxidative stress state, and H&E staining results showed extensive empty package degeneration, hepatocyte disintegration, seriously disordered hepatic cords, hepatic sinusoidal congestion, and the interstitial infiltration of red blood cells and inflammatory cells. The liver was in an inflammatory state, accompanied by hepatocyte vacuolar degeneration in the DQ group. The hepatocytes were swollen, granular, and partially necrotic (Figures 3(c) and 3(d)).
Figure 3.
Histological evaluation of liver tissues after exposure to diquat: (a) negative control group (HE, ×200), (b) negative control group (HE, ×400), (c) oxidative stress group (HE, ×200), and (d) oxidative stress group (HE, ×400).
3.4. Overview of the Sequencing Data
To understand which DQ-sensitive RNAs were involved in the liver, RNA-Seq analysis was performed in the oxidative stress group and negative control group (three samples per group). After filtering the 3′ adaptor sequence and discarding low-quality reads, a high percentage of the reads was mapped to the pig reference genome, ranging from 82.52 to 84.48%. Among the mapped reads, 69.09 to 74.65% were mapped uniquely to the pig reference genome. All six samples had nearly 95% of reads equal to or exceeding Q30 (Table 1).
Table 1.
A summary of the sequencing read alignment to the Sus scrofa genome.
| Sample | NC1 | NC2 | NC3 | OS1 | OS2 | OS3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Count | % | Count | % | Count | % | Count | % | Count | % | Count | % | |
| Total reads | 56879106 | 36876312 | 31523852 | 23752128 | 33136858 | 34721210 | ||||||
| Total mapped | 47350312 | 83.25 | 30742830 | 83.37 | 26632830 | 84.48 | 20008310 | 84.24 | 27346751 | 82.53 | 28653139 | 82.52 |
| Multiple mapped | 7509799 | 13.20 | 4866020 | 13.20 | 3100574 | 9.84 | 3598810 | 15.15 | 2678440 | 8.08 | 2916956 | 8.40 |
| Uniquely mapped | 39840513 | 70.04 | 25876810 | 70.17 | 23532256 | 74.65 | 16409500 | 69.09 | 24668311 | 74.44 | 25736183 | 74.12 |
| %≥Q30 | 95.33 | 95.02 | 95.00 | 94.84 | 95.23 | 95.28 | ||||||
| GC content (%) | 61.98 | 62.92 | 62.06 | 67.01 | 57.83 | 57.31 | ||||||
3.5. Identification and Analysis of DE lncRNAs and mRNAs
Compared to the negative control group, eight lncRNAs were differentially expressed in the DQ-induced liver samples, in which six lncRNAs were upregulated and the other two lncRNAs were downregulated (Table S2). Additionally, a total of 30 mRNAs were differentially expressed in the liver of the DQ-induced oxidative stress group: eight genes (PCK1, HMGCS2, VCL, HAMP, APOA4, GNMT, ENSSSCG00000015054, and ILF3) were upregulated, and 22 genes were downregulated in the oxidative stress group (Table S2).
3.6. GO and KEGG Pathway Enrichment Analysis of DE lncRNAs and mRNAs
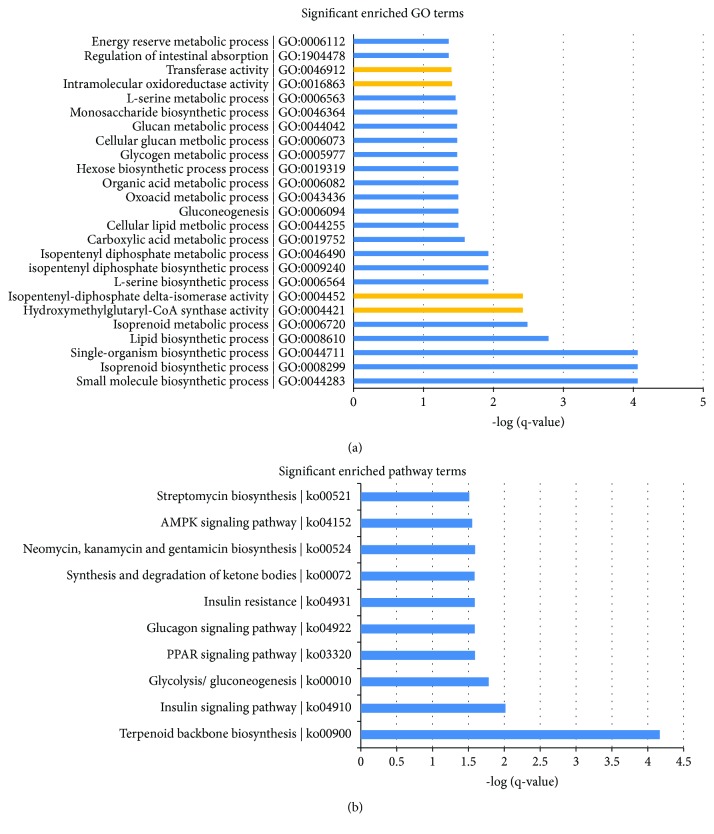
To further understand the functions of these DE genes, we performed GO enrichment analysis, which produced terms arranged and displayed according to biological processes, molecular functions, and cellular components (Figure 4(a)) to identify the gene expression changes associated with DQ treatment. The significantly enriched categories of our gene set were small-molecule biosynthetic process, isoprenoid biosynthetic process, extracellular matrix structural constituent, hydroxymethylglutaryl-CoA synthase activity, and isopentenyl-diphosphate delta-isomerase activity. In addition to these terms, we were also interested to find that the oxidation-reduction process played an important role in the biological processes involved in these altered genes. The biological process of the oxidation-reduction process serves to protect the body from oxidative damage, maintaining a dynamic balance.
Figure 4.
GO (a) and KEGG pathway (b) enrichment analysis of DE lncRNAs and mRNAs.
We determined the KEGG pathways related to the differentially expressed genes in the liver. The results, as shown in Figure 4(b), indicated that DQ caused a change in 10 signaling pathways involved in the sensitive-expressed RNAs in piglets. Among these, the insulin signaling pathway; glycolysis/gluconeogenesis; the PPAR signaling pathway; the glucagon signaling pathway; insulin resistance; the synthesis and degradation of ketone bodies; neomycin, kanamycin, and gentamicin biosynthesis; and the AMPK signaling pathway are oxidative stress-relevant pathways.
3.7. DE Genes Associated with Oxidative Stress
Among the DE mRNAs, we found that GNMT, protein phosphatase 1 regulatory subunit 3C (PPP1R3C), SCD, APOA4, GCK, and ENSSSCG00000027643 were enriched under the oxidation-reduction process term. GNMT and APOA4 were upregulated genes in the DQ-induced oxidative stress group, and the other genes were downregulated. Other oxidative stress-related genes, including CHAC1, HMGCS1, PSAT1, and PSPH, were all differentially expressed between the two groups. ILF3, as an immune-related gene, was upregulated in the oxidative stress group and was also associated with oxidative stress.
3.8. qPCR Validation of the Gene Expression Data from RNA-Seq
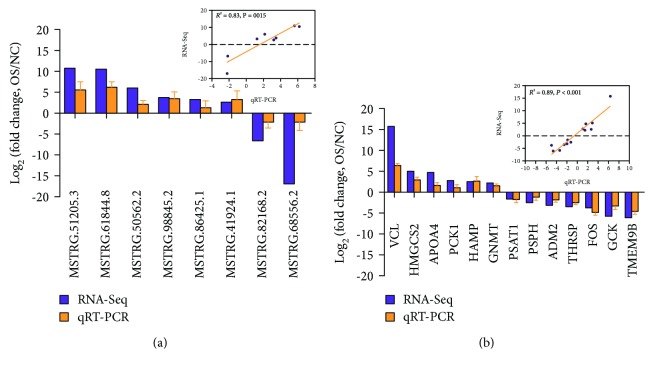
To verify the gene and lncRNA expression data by RNA-Seq, qPCR was performed for eight DE lncRNAs and 13 DE mRNAs for oxidative stress, including six upregulated genes and seven downregulated genes in oxidative stress group piglets (n = 5) compared to negative control group piglets (n = 6). The results showed that all tested lncRNAs and mRNAs were consistent between RT-qPCR and sequencing analysis, and there was a significant correlation (R 2 = 0.83 for DE lncRNAs and 0.89 for DE genes; Figures 5(a) and 5(b), respectively). Besides, the expression levels of the DE lncRNAs and mRNAs were also performed in primary porcine hepatocytes after DQ stimulation 12 h, and the results showed that there was the same expression tend (Figure S1). These results suggest that our estimation of abundance was accurate.
Figure 5.
Validation of eight DE lncRNAs and 13 DE mRNAs identified from RNA-Seq results using qPCR. There was a significant correlation between transcript fold-change values determined by RNA-Seq and qPCR, and R 2 = 0.83 for DE lncRNAs (a) and 0.89 for DE mRNAs (b).
3.9. Oxidation-Reduction Process-Related Gene Expression
Because the GO term oxidation-reduction process is very important, we selected GNMT (upregulated) and GCK (downregulated) from the GO term and further studied their expression in primary porcine hepatocytes after DQ stimulation. The expression level of the GNMT gene after 6 (P = 0.037) and 12 h (P = 0.021) stimulation with DQ was significantly higher than that in the control group (Figure 6(a)). After stimulation with DQ, the GCK mRNA level was significantly decreased in the treatment group at 12 h (P < 0.05); however, there was no difference between the control group and treatment group at 6 h (Figure 6(b)). These results suggest that the DQ treatment had a significant effect on oxidation-reduction process signaling.
Figure 6.
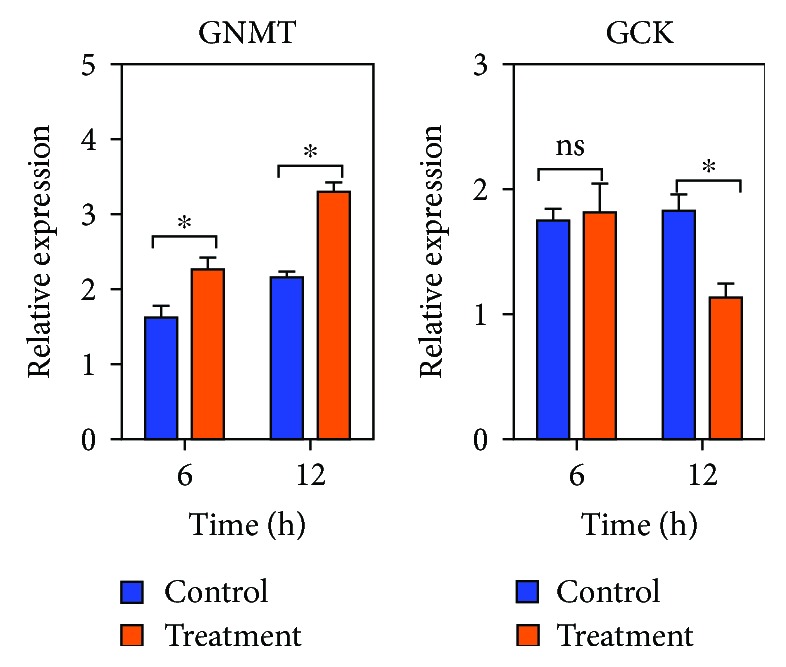
The expression of GNMT and GCK in primary porcine hepatocytes (n = 5). Treatment: the isolated hepatocytes were treated with 50 μM DQ for 6 and 12 h. Control: the control cells were cultured under the same conditions except without DQ treatment. ∗ P < 0.05; ns: not significant.
4. Discussion
DQ, as a hepatotoxic bipyridyl herbicide [32, 33], is widely applied in the field and is used to construct animal oxidative stress models in vivo [17]. Previous research has reported that DQ-induced oxidative stress decreases porcine growth performance and nutritional metabolism ability [4, 34, 35]. In the present study, we also found that the ADG was decreased in oxidative stress group pigs, which disrupted the redox state. The activity of T-AOC, SOD, and GSH-Px in the liver was decreased after DQ injection in piglets. Oxidative stress results from the failure of antioxidant enzymes to eliminate free radicals [36]. Thus, the ROS produced in mitochondria cannot decompose superoxide anions (O2 ·−) to hydrogen peroxide (H2O2) [37]; therefore, O2 ·− causes cellular injury and liver damage.
Although many studies have shown that DQ results in damage to the liver and the liver is the major target of DQ, the genetic and regulatory mechanisms resulting in oxidative stress remain unclear. As a kind of high-throughput detection technology, RNA-Seq has been widely used in many fields. To further analyze the molecular mechanism of damage induced by DQ, the livers of pigs were collected to screen the DE mRNAs, lncRNAs, and signaling pathways using RNA-Seq. Overall, there were significant differences in liver mRNA and lncRNA expression between the oxidative stress and NC groups. There were 30 DE mRNAs and 8 DE lncRNAs between the two groups.
lncRNAs have been identified in numerous porcine tissues, such as the skeletal muscles [38], lung [39], and spleen tissues [40], and play an important role in major mechanisms of gene expression, regulation, and cellular development. Despite the biological functions of lncRNAs, it is not yet clear whether lncRNAs are involved in the regulation of oxidative stress in the liver of piglets. In our present study, eight DE lncRNAs were identified, revealing the lncRNAs participating in the regulation of oxidative stress induced by DQ. In addition, the GO analysis also showed that the targets of lncRNAs were mainly enriched in lipoprotein oxidation. Our results provide a molecular basis for studying the function of lncRNAs in oxidative stress induced by DQ.
GNMT is the most abundant methyltransferase in the liver and plays an important role in S-adenosylmethionine homeostasis as it methylates glycine to form sarcosine [41]. Previous research has reported that GNMT regulates genes related to the antioxidation pathways [42, 43]. In the present study, GNMT was highly expressed in the oxidative stress group compared with the NC group. Furthermore, we detected GNMT expression levels in primary porcine hepatocytes in vitro, and the results showed that GNMT was also highly expressed in hepatocytes after stimulation with DQ. These findings agree with a previous study showing that GNMT may prevent oxidative stress-induced liver damage [44].
Because it can catalyze the conversion of glucose to glucose-6-phosphate, GCK plays an important role in glucose metabolism [45]. It has been reported that GCK is essential for the regulation of various glucose-responsive genes in hepatocytes [46]. In our present study, the expression of GCK was downregulated in the oxidative stress group and decreased significantly in hepatocytes after 12 h of treatment with DQ. These results suggest that the rate of both glucose uptake and glycogen synthesis in the liver was inhibited; therefore, the ADG of oxidative stress group piglets was decreased.
Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase (HMGCS2) delivers lipid-derived energy to liver cells, and suppression of HMGCS2 in the nonalcoholic steatohepatitis livers could indicate impairment of ketogenesis and cholesterol biosynthesis [47]. Additionally, the upregulated gene HMGCS2 is also correlated with fatty acid oxidation induced by a high-fat diet [48]. Moreover, ILF3 is an immune-related gene regulating p21 expression in leukemic cells and is associated with dichloroacetate-mediated cytotoxicity [49]. In our present study, these two genes displayed increased expression in oxidative stress group piglets, suggesting that the upregulation of HMGCS2 and ILF3 was resistant to oxidative stress in the liver.
5. Conclusion
In summary, the results of this study showed that DQ-induced oxidative stress decreased growth performance and led to the differential expression of lncRNAs and mRNAs in the livers of piglets. Eight DE lncRNAs and 30 DE mRNAs were identified, and the target genes of lncRNAs and mRNAs were enriched in the oxidation-reduction process. Moreover, our study also provides a reference for lncRNAs and mRNAs that can be used for biomedical research related to oxidative stress diseases in the future.
Acknowledgments
This study was supported by the funds of Shandong “Double Tops” Program (SYL2017YSTD12), the Shandong Provincial Modern Pig Technology and Industry System Project (SDAIT-08-02), the Shandong Provincial Natural Science Foundation (ZR2018BC046), and the Youth Foundation of Shandong Agricultural University.
Contributor Information
Yong-qing Zeng, Email: yqzeng@sdau.edu.cn.
Wei Chen, Email: wchen@sdau.edu.cn.
Data Availability
The data used to support the findings of this study are available from the corresponding authors upon request.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Supplementary Materials
Supplemental Table S1: primer information of differentially expressed lncRNAs and genes for qPCR. Supplemental Table S2: differentially expressed lncRNAs and mRNAs between the two groups. Supplemental Figure S1: the expression of DE lncRNAs and mRNAs in primary porcine hepatocytes.
References
- 1.Valko M., Leibfritz D., Moncol J., Cronin M. T. D., Mazur M., Telser J. Free radicals and antioxidants in normal physiological functions and human disease. The International Journal of Biochemistry & Cell Biology. 2007;39(1):44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Gutteridge J. M. Lipid peroxidation and antioxidants as biomarkers of tissue damage. Clinical Chemistry. 1995;41(12):1819–1828. [PubMed] [Google Scholar]
- 3.Duracková Z. Some current insights into oxidative stress. Physiological Research. 2010;59(4):459–469. doi: 10.33549/physiolres.931844. [DOI] [PubMed] [Google Scholar]
- 4.Zhu L. H., Zhao K. L., Chen X. L., Xu J. X. Impact of weaning and an antioxidant blend on intestinal barrier function and antioxidant status in pigs. Journal of Animal Science. 2012;90(8):2581–2589. doi: 10.2527/jas.2011-4444. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y. Q., Xing Y. Y., Wang Z. Q., Yan S. M., Shi B. L. Pre-protective effects of dietary chitosan supplementation against oxidative stress induced by diquat in weaned piglets. Cell Stress and Chaperones. 2018;23(4):703–710. doi: 10.1007/s12192-018-0882-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma Q. Transcriptional responses to oxidative stress: pathological and toxicological implications. Pharmacology & Therapeutics. 2010;125(3):376–393. doi: 10.1016/j.pharmthera.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Amaral P. P., Dinger M. E., Mattick J. S. Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Briefings in Functional Genomics. 2013;12(3):254–278. doi: 10.1093/bfgp/elt016. [DOI] [PubMed] [Google Scholar]
- 8.Hung T., Chang H. Y. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biology. 2014;7(5):582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer T. R., Dinger M. E., Mattick J. S. Long non-coding RNAs: insights into functions. Nature Reviews Genetics. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 10.Ponting C. P., Oliver P. L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 11.St. Laurent G., Wahlestedt C., Kapranov P. The landscape of long noncoding RNA classification. Trends in Genetics. 2015;31(5):239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature Reviews Genetics. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 13.Valadkhan S., Valencia-Hipólito A. lncRNAs in stress response. In: Morris K. V., editor. Long Non-coding RNAs in Human Disease. Cham: Springer International Publishing; 2016. pp. 203–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tehrani S. S., Karimian A., Parsian H., Majidinia M., Yousefi B. Multiple functions of long non-coding RNAs in oxidative stress, DNA damage response and cancer progression. Journal of Cellular Biochemistry. 2018;119(1):223–236. doi: 10.1002/jcb.26217. [DOI] [PubMed] [Google Scholar]
- 15.Giannakakis A., Zhang J., Jenjaroenpun P., et al. Contrasting expression patterns of coding and noncoding parts of the human genome upon oxidative stress. Scientific Reports. 2015;5, article 9737 doi: 10.1038/srep09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Askew E. W. Environmental and physical stress and nutrient requirements. The American Journal of Clinical Nutrition. 1995;61(3):631S–637S. doi: 10.1093/ajcn/61.3.631S. [DOI] [PubMed] [Google Scholar]
- 17.Yuan S.-b., Chen D.-w., Zhang K.-y., Yu B. Effects of oxidative stress on growth performance, nutrient digestibilities and activities of antioxidative enzymes of weanling pigs. Asian-Australasian Journal of Animal Sciences. 2007;20(10):1600–1605. doi: 10.5713/ajas.2007.1600. [DOI] [Google Scholar]
- 18.Zheng P., Yu B., Lv M., Chen D. Effects of oxidative stress induced by diquat on arginine metabolism of postweaning pigs. Asian-Australasian Journal of Animal Sciences. 2010;23(1):98–105. doi: 10.5713/ajas.2010.90270. [DOI] [Google Scholar]
- 19.Bolger A. M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D., Langmead B., Salzberg S. L. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T., Salzberg S. L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Risso D., Ngai J., Speed T. P., Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nature Biotechnology. 2014;32(9):896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang Y.-J., Yang D.-C., Kong L., et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Research. 2017;45(W1):W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun L., Luo H., Bu D., et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Research. 2013;41(17):p. e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn R. D., Coggill P., Eberhardt R. Y., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li A., Zhang J., Zhou Z. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics. 2014;15(1):p. 311. doi: 10.1186/1471-2105-15-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11(10, article R106) doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang X.-L., Shu G., Zhang Z.-Q., et al. Roles of α-linolenic acid on IGF-I secretion and GH/IGF system gene expression in porcine primary hepatocytes. Molecular Biology Reports. 2012;39(12):10987–10996. doi: 10.1007/s11033-012-2000-6. [DOI] [PubMed] [Google Scholar]
- 29.Untergasser A., Cutcutache I., Koressaar T., et al. Primer3—new capabilities and interfaces. Nucleic Acids Research. 2012;40(15, article e115) doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC T method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Burk R. F., Hill K. E., Awad J. A., et al. Pathogenesis of diquat-induced liver necrosis in selenium-deficient rats: assessment of the roles of lipid peroxidation and selenoprotein P. Hepatology. 1995;21(2):561–569. [PubMed] [Google Scholar]
- 32.Higuchi M., Kobayashi S., Kawasaki N., et al. Protective effects of wheat bran against diquat toxicity in male Fischer-344 rats. Bioscience, Biotechnology, and Biochemistry. 2007;71(7):1621–1625. doi: 10.1271/bbb.60698. [DOI] [PubMed] [Google Scholar]
- 33.Higuchi M., Oshida J., Orino K., Watanabe K. Wheat bran protects Fischer-344 rats from diquat-induced oxidative stress by activating antioxidant system: selenium as an antioxidant. Bioscience, Biotechnology, and Biochemistry. 2011;75(3):496–499. doi: 10.1271/bbb.100719. [DOI] [PubMed] [Google Scholar]
- 34.Lv M., Yu B., Mao X. B., Zheng P., He J., Chen D. W. Responses of growth performance and tryptophan metabolism to oxidative stress induced by diquat in weaned pigs. Animal. 2012;6(6):928–934. doi: 10.1017/S1751731111002382. [DOI] [PubMed] [Google Scholar]
- 35.Yin J., Liu M., Ren W., et al. Effects of dietary supplementation with glutamate and aspartate on diquat-induced oxidative stress in piglets. PLoS One. 2015;10(4, article e0122893) doi: 10.1371/journal.pone.0122893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan D., Hussain T., Tan B., Liu Y., Ji P., Yin Y. The evaluation of antioxidant and anti-inflammatory effects of Eucommia ulmoides flavones using diquat-challenged piglet models. Oxidative Medicine and Cellular Longevity. 2017;2017:9. doi: 10.1155/2017/8140962.8140962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadenbach B., Ramzan R., Vogt S. Degenerative diseases, oxidative stress and cytochrome c oxidase function. Trends in Molecular Medicine. 2009;15(4):139–147. doi: 10.1016/j.molmed.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Kern C., Wang Y., Chitwood J., et al. Genome-wide identification of tissue-specific long non-coding RNA in three farm animal species. BMC Genomics. 2018;19(1):p. 684. doi: 10.1186/s12864-018-5037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin L., Hu S., Tu T., et al. Global long noncoding RNA and mRNA expression changes between prenatal and neonatal lung tissue in pigs. Genes. 2018;9(9):p. 443. doi: 10.3390/genes9090443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Che T., Li D., Jin L., et al. Long non-coding RNAs and mRNAs profiling during spleen development in pig. PLoS One. 2018;13(3, article e0193552) doi: 10.1371/journal.pone.0193552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernández-Ramos D., Fernández-Tussy P., Lopitz-Otsoa F., et al. MiR-873-5p acts as an epigenetic regulator in early stages of liver fibrosis and cirrhosis. Cell Death & Disease. 2018;9(10):p. 958. doi: 10.1038/s41419-018-1014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deminice R., Cella P. S., Padilha C. S., et al. Creatine supplementation prevents hyperhomocysteinemia, oxidative stress and cancer-induced cachexia progression in Walker-256 tumor-bearing rats. Amino Acids. 2016;48(8):2015–2024. doi: 10.1007/s00726-016-2172-9. [DOI] [PubMed] [Google Scholar]
- 43.Yen C.-H., Lin Y.-T., Chen H.-L., Chen S.-Y., Chen Y.-M. A. The multi-functional roles of GNMT in toxicology and cancer. Toxicology and Applied Pharmacology. 2013;266(1):67–75. doi: 10.1016/j.taap.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 44.Liao Y.-J., Chen K.-H., Huang S.-F., et al. Deficiency of glycine N-methyltransferase results in deterioration of cellular defense to stress in mouse liver. Proteomics – Clinical Applications. 2010;4(4):394–406. doi: 10.1002/prca.200900074. [DOI] [PubMed] [Google Scholar]
- 45.Magnuson M. A. Glucokinase gene structure: functional implications of molecular genetic studies. Diabetes. 1990;39(5):523–527. doi: 10.2337/diab.39.5.523. [DOI] [PubMed] [Google Scholar]
- 46.Girard J., Ferré P., Foufelle F. Mechanisms by which carbohydrates regulate expression of genes for glycolytic and lipogenic enzymes. Annual Review of Nutrition. 1997;17(1):325–352. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- 47.Urasaki Y., Zhang C., Cheng J.-X., Le T. T. Quantitative assessment of liver steatosis and affected pathways with molecular imaging and proteomic profiling. Scientific Reports. 2018;8(1):p. 3606. doi: 10.1038/s41598-018-22082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikder K., Shukla S. K., Patel N., Singh H., Rafiq K. High fat diet upregulates fatty acid oxidation and ketogenesis via intervention of PPAR-γ . Cellular Physiology and Biochemistry. 2018;48(3):1317–1331. doi: 10.1159/000492091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castella S., Bernard R., Corno M., Fradin A., Larcher J.-C. Ilf3 and NF90 functions in RNA biology. Wiley Interdisciplinary Reviews: RNA. 2015;6(2):243–256. doi: 10.1002/wrna.1270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1: primer information of differentially expressed lncRNAs and genes for qPCR. Supplemental Table S2: differentially expressed lncRNAs and mRNAs between the two groups. Supplemental Figure S1: the expression of DE lncRNAs and mRNAs in primary porcine hepatocytes.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding authors upon request.