Abstract
Throughout heart development, cardiomyocytes differentiate and mature in direct contact with nonparenchymal cell types, such as cardiac fibroblasts. Thus, when modeling myocardial tissue in vitro, tissue engineers include a supporting stromal cell population that is necessary for tissue formation, although the source of stromal cells has varied widely. This study systematically characterized the phenotype of commonly used stromal cell populations and analyzed the differential impacts of stromal phenotype on cardiac microtissue phenotype and function. Quantitative morphometric analysis, flow cytometry, unbiased morphological feature clustering, and RNA sequencing of the different stromal populations revealed variable cell morphologies, surface marker expression, and gene signatures, with primary adult stromal populations exhibiting more similar phenotypes to each other than to stem cell-derived and progenitor populations. The ability of self-assembled cardiac microtissues to consistently form tissues was highly dependent on the stromal population mixed with stem cell-derived cardiomyocytes, with cardiac fibroblasts and dermal fibroblasts (DFs) forming the most robust tissues compared with mesenchymal stromal cells and induced pluripotent stem cell-derived fibroblasts. Cardiac fibroblasts and DFs also resulted in cardiac microtissues displaying a more mature calcium handling profile, with increased amplitude and upstroke velocity. These results demonstrate the breadth of phenotypic variation across stromal populations owing to cell and tissue source, with certain primary populations, such as cardiac fibroblasts and DFs, supporting cardiac microtissue phenotype and improved calcium handling function.
Impact Statement
Understanding the relationship between parenchymal and supporting cell populations is paramount to recapitulate the multicellular complexity of native tissues. Incorporation of stromal cells is widely recognized to be necessary for the stable formation of stem cell-derived cardiac tissues; yet, the types of stromal cells used have varied widely. This study systematically characterized several stromal populations and found that stromal phenotype and morphology was highly variable depending on cell source and exerted differential impacts on cardiac tissue function and induced pluripotent stem cell–cardiomyocyte phenotype. Therefore, the choice of supporting stromal population can differentially impact the phenotypic or functional performance of engineered cardiac tissues.
Keywords: cardiomyocytes, fibroblasts, cardiac microtissues, heterotypic interactions, stromal cells, stem cell-derived tissues
Introduction
The heart is composed of multiple cell types that interact through numerous heterotypic interactions and collectively impact cardiac tissue structure and function. During normal heart development, cardiomyocytes (CMs) differentiate and mature in intimate contact with nonmyocytes, such as cardiac fibroblasts, which concurrently increase in number and ultimately constitute >50% of the total number of cells in the heart.1 Cardiac fibroblasts interact with cardiomyocytes through direct cell–cell contacts,2–6 are the primary producers of extracellular matrix,7,8 and secrete paracrine mediators9–12 that affect cardiac homeostasis, cardiomyocyte phenotype, hypertrophy, and tissue remodeling. The wide variety of mechanisms by which cardiac fibroblasts can influence the microenvironment of cardiomyocytes and the close spatial relationship throughout pre- and postnatal development reflect the importance of heterotypic interactions in influencing cardiomyocyte phenotype and cardiac tissue function. Despite the well-known physical juxtaposition between the two cell types, the specific mechanisms by which cardiac fibroblasts influence cardiomyocytes have not been rigorously interrogated.
Recent advances in pluripotent stem cell-directed differentiation protocols13,14 have provided a robust source of human cardiomyocytes (hPSC-CMs) that can be used to study heterotypic interactions in vitro. Tissue engineering approaches utilizing hPSC-CMs to mimic the myocardium enable precise control over the cellular composition. Cardiac tissue biologists have long recognized the need for using heterogeneous, nonpurified differentiations of hPSC-CMs or including nonmyocyte populations along with cardiomyocytes to enable stable tissue formation.15–17 A wide variety of different stromal cell types has been mixed with hPSC-CMs in engineered constructs including cardiac fibroblasts,18 dermal fibroblasts (DFs),19,20 mesenchymal stromal cells (MSCs),21 induced pluripotent stem (iPS) cell-derived fibroblasts,15,17 endothelial cells (EC),16,18 and/or combinations of these cell types.22–25 Generally speaking, the inclusion of stromal populations increases the expression of cardiac-specific genes16,18 and improves functional properties, such as calcium handling, action potential kinetics,16 and contractile force.18 However, the variability of stromal cell type has led to inconsistencies in cardiac tissue function and cardiomyocyte pharmacological responses reported by independent studies, thus limiting our understanding of the impact of different stromal cells on engineered cardiac tissues.
Because of the variability in stromal cell use throughout the field, this study systematically evaluated the effects of commonly used stromal populations on cardiac microtissue formation, phenotype, and physiological function through a series of quantitative phenotypic and transcriptomic approaches. This study (i) characterizes the phenotypic differences between stromal cells obtained from different cell and tissue sources and (ii) examines their respective consequences on hPSC-CM phenotype and cardiac microtissue function. These results emphasize the importance of heterotypic mixing with nonmyocytes in complex three-dimensional (3D) cardiac models and highlight the need for detailed characterization of starting cell populations to better define their functional impacts on engineered cardiac tissue calcium handling.
Materials and Methods
Stromal cell culture
See Supplementary Table S1 for cell source and donor information. Human fetal cardiac fibroblasts (fCFs) were purchased from Cell Applications (San Diego, CA) and human adult cardiac fibroblasts (aCFs) were purchased from Cell Applications and PromoCell (aCFPC) (Heidelberg, DE). Cardiac fibroblasts were seeded at a concentration of 1 × 104 cells/cm2 and expanded in cardiac fibroblast growth medium (Cell Applications) or fibroblast growth medium 3 (PromoCell) for up to 10 passages. Human bone marrow-derived MSCs were purchased from RoosterBio (Frederick, MD), maintained in Rooster-Nourish MSC medium, replated at 1 × 104 cells/cm2, and cultured for up to 10 passages. Human DFs were purchased from Cell Applications, maintained in fibroblast medium (KnockOut Dulbecco's modified Eagle's medium [KO DMEM; Thermo Fisher, Waltham, MA], 10% fetal bovine serum [FBS; Atlanta Biologicals, Flowery Branch, GA], 1 × nonessential amino acid [NEAA], 1 × l-glut, 0.1 mM β-mercaptoethanol [Bio-Rad, Hercules, CA]), replated at 1 × 104 cells/cm2, and cultured for up to 10 passages. iPS cell-derived teratoma outgrowth stromal cells (iPS-Ftera)26 were maintained in KO DMEM containing 10% FBS, 1 mM NEAA, 1 mM l-glut, and 1 mM antibiotic–antimycotic (Thermo Fisher), replated at 1 × 104 cells/cm2, and cultured for up to 15 passages. iPS cell-derived embryoid body outgrowth stromal cells (iPS-FEB)17 were maintained in fibroblast medium, replated at 1 × 104 cells/cm2, and cultured for up to 10 passages.
Flow cytometry
Stromal cells were fixed with 4% paraformaldehyde for 15–20 min at room temperature (RT). Based on the standards issued by the International Society for Cellular Therapy for mesenchymal stromal markers,27 stromal cells were stained for CD166, CD90, CD73, CD45, and CD34 (Supplementary Table S3) using Stain Buffer (FBS; BD Biosciences). Samples were analyzed on a BD FACSCalibur DxP8 and the resulting data were analyzed using FlowJo software (v.10).
Immunocytochemistry
Stromal cells were fixed with 4% paraformaldehyde for 15–20 min at RT and stained with antibodies against vimentin overnight at 4°C. Alexa Fluor 647, phalloidin 488, HCS Cell Mask Red, and Hoechst were added for 45 min at RT (Supplementary Table S3 for antibody information and concentrations).
Image acquisition and morphometric assessment
A minimum of 150 stained cells was imaged for each stromal cell population on the Cellomics ArrayScan XTI (Thermo Fisher) and then analyzed with HCS Studio Cell Analysis Software using the Morphology Assay (v.6.0.3.4024) to obtain measurements of the following morphological features: cell length, cell width, cell area, fiber area, cell perimeter, nuclear area, fiber alignment 1, and fiber alignment 2. Fiber alignment 1 corresponds to the standard deviation of each fiber measurement with the axis of the image. Fiber alignment 2 is related to the anisotropy of a cell that corresponds to the ratio of second moments of the principle axes. The Kolmogorov–Smirnov statistical test with Holm–Bonferroni multiple comparison correction was performed to compare empirical distributions of morphometric measurements between stromal cell populations. Statistical significance was determined at p < 0.05 for Holm–Bonferroni corrected p-values.
Principle component analysis
Principal component analysis (PCA) was performed with a Python (v.3.6) script implementing the scikit-learn package28 to analyze primary morphological feature measurements (cell length, cell width, cell perimeter, cell area, nuclear area, fiber area, fiber alignment 1, and fiber alignment 2) of ∼100–150 cells per stromal cell population obtained from the HCS Studio Cell Analysis Morphology Assay.
PhenoRipper
PhenoRipper software (v.1.20)29 was used to compare stromal cell images in an unbiased fashion with minimal user input. Eight-bit tiff images acquired by Cellomics ArrayScan XTI (n > 100 for each stromal cell population) were analyzed with the following parameters specified within the software: threshold intensity: 16; block size: 10; number of colors used: 10; number of block types: 10; number of superblock types: 30; training images: 50–100; use background images to obtain a multidimensional scaling plot and clustergram of top ranking superblocks found within the dataset.
RNA sequencing
Stromal cells were lysed with Trizol for RNA extraction with the Direct-zol RNA Miniprep kit (ZymoResearch, Irvine, CA). All groups were collected in duplicate or triplicate. RNA was quantified using the NanoDrop 2000c (Thermo Fisher). RNAseq libraries were created using the SMARTer Stranded Total RNA Sample Prep Kit (Takara Bio, Kusatsu, JP) and sequenced on the NextSeq 500 (Illumina, San Diego, CA) to a minimum depth of 25 million reads per sample. The sequences were aligned to hg19 using TopHat2,30 reads were quantified using featureCounts,31 and differential expression analysis was determined using edgeR.32,33 Hierarchical clustering was carried out using the Hierarchical Ordered Partitioning and Collapsing Hybrid (HOPACH) package in R.34 Raw data will be available at GEO (Accession #: GSE129058).
PSC culture
Human PSCs (WTC11 cells modified with GCaMP6f reporter; generously donated by Dr. Bruce Conklin)35,36 were cultured on Matrigel-coated (80 μg/mL; Corning, Corning, NY) plates in mTeSR medium (Stem Cell Technologies, Vancouver, CA). Cells were grown to 70% confluence, passaged using Accutase (Innovative Cell Technologies, San Diego, CA), and seeded in mTeSR medium with 10 μM ROCK inhibitor (Y27632; SelleckChem, Houston, TX) at a density of 1 × 104 cells/cm2.
Cardiomyocyte differentiation
Differentiation of CM from human induced pluripotent stem cells (hiPSCs) was achieved with a serum-free, chemically defined protocol on monolayer cultures.13,14 In brief, GCaMP6f WTC11 hiPSCs were seeded onto Matrigel-coated dishes at 3 × 104 cells/cm2 in mTeSR medium and grown to confluence for 3 days. On day 0 of differentiation, the cultures were fed with RPMI 1640 medium (Thermo Fisher) and B27 supplement minus insulin (referred to as RPMI/B27− medium; Life Technologies, Grand Island, NY) supplemented with 12 μM CHIR99021 (SelleckChem). The initial induction medium was completely removed after 24 h and exchanged with RPMI/B27− medium. On day 3 of differentiation, the medium was changed to RPMI/B27− medium supplemented with 5 μM IWP2 (Tocris, Bristol, United Kingdom) for 48 h and then replaced with fresh RPMI/B27− on day 5. On day 7 of differentiation, medium was changed to RPMI 1640 medium with B27 supplement plus insulin (referred to as RPMI/B27+ medium; Life Technologies) and refreshed every 3 days thereafter. Cells were replated at a concentration of 80,000 cells/cm2 on day 15 onto Matrigel-coated dishes in RPMI/B27+ medium. Purification of cardiomyocytes occurred on days 20–24 by feeding cells with lactate purification medium37 (no-glucose DMEM [Thermo Fisher] with 1 × NEAA [Corning], 1 × GlutaMAX [l-glut; Life Technologies], and 4 mM lactate), refreshing the medium once on day 22. On day 24, the medium was refreshed with RPMI/B27+ and hPSC-CMs were subsequently maintained with RPMI/B27+ medium completely exchanged every 3 days thereafter until harvest (D35 ± 7).
Cardiac microtissue formation
Lactate purified cardiomyocytes (>85% cTnT+; day 35 ± 7) were mixed at a 3:1 ratio with human aCFs, DFs, MSCs, or iPS cell-derived fibroblasts (teratoma outgrowth stromal cells) before seeding ∼2000 cells per inverted 400 μm pyramidal agarose microwell.38 Cells self-assembled over the course of 18–24 h and the resultant microtissues were then removed from the microwells and maintained in rotary orbital suspension culture for 7 days in RPMI/B27+ medium at a density of ∼2000 microtissues per 100 mm Petri dish.38
Gene expression analysis
Stromal cells and cardiac microtissues were lysed with Trizol (Invitrogen, Carlsbad, CA) for RNA extraction with the RNeasy Mini Kit (Qiagen, Hilden, DE). All groups were collected in duplicate or triplicate. RNA was converted to cDNA following the iScript cDNA Synthesis Kit (Bio-Rad). Forward and reverse primers (Supplementary Table S2) were designed with the NCBI Primer-BLAST. Preamplified cDNA samples and primers were mixed with Sso Fast EvaGreen Supermix (Bio-Rad), loaded onto a 96.96 Dynamic Array integrated fluidic circuit (Fluidigm, San Francisco, CA), and run on a BioMark HD system. Relative gene expression levels were normalized to reference gene 18s rRNA, and fold changes were calculated using the ΔΔCt method39 compared with expression in 3D microtissues composed of CMs alone. Results were plotted using Genesis software (Graz University of Technology40).
Calcium imaging analysis
WTC11 hiPSCs with genetically encoded GCaMP6f calcium indicator were used to enable visualization of calcium transients in hPSC-CMs. Day 7 aggregates were equilibrated in Tyrode's solution (137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 0.2 mM Na2HPO4, 12 mM NaHCO3, 5.5 mM d-glucose, and 1.8 mM CaCl2; Sigma-Aldrich) in 35 mm Petri dishes for 30 min at 37°C before measuring calcium transients. Samples were mounted on a Zeiss Axio Observer Z1 inverted microscope equipped with an Orca Flash Hammamatsu camera. Electrodes were placed in the Petri dish and electrical field stimulation was applied at 1 Hz (MyoPacer, IonOptix). Video acquisition of calcium flux was obtained using Zen Professional software (v.2.0.0.0) with 10 ms exposure and 100 fps. One circular 120-pixel region of interest was selected at the center of each aggregate and the mean fluorescent intensity values were plotted. Calcium fluorescence intensity profiles were further analyzed through a custom R-script to quantify the kinetics of each transient (i.e., amplitude, time, velocity, and beat rate).
Histology and immunofluorescent staining
Microtissues were fixed for 1 h in 10% neutral-buffered formalin at RT. Microtissues were then embedded in HistoGel Specimen Processing Gel (Thermo Fisher), paraffin processed, sectioned at 5 μm-thickness, and adhered to SuperFrost Plus microscope slides (Thermo Fisher). Samples were deparaffinized with xylene, rehydrated by a series of graded ethanol steps, and washed in phosphate-buffered saline. Slides were immersed in citrate buffer (Vector Laboratories, Burlingame, CA) at pH 6.0 in a 95°C water bath for 35 min for heat-induced epitope retrieval. Samples were then permeabilized in 0.2% Triton X-100 (Sigma-Aldrich), blocked (1.5% normal donkey serum) for 1 h at RT, probed with primary and secondary antibodies against cardiac troponin T, slow+fast troponin I, and cardiac troponin I, and counterstained with Hoechst and wheat germ agglutinin (WGA; antibody information in Supplementary Table S3). Coverslips were mounted with antifade mounting medium (ProlongGold; Life Technologies). All samples were imaged on a Zeiss Axio Observer Z1 inverted microscope equipped with an Orca Flash Hamamatsu camera.
Statistics
The mean ± standard deviation was calculated from at least 10 biological replicates for all data unless otherwise noted. When comparing three or more groups, one-way analysis of variance (ANOVA) followed by Tukey's post hoc analysis was performed. For all comparisons, statistical significance was determined at p < 0.05. All statistical analyses were performed using GraphPad Prism 7.0 software.
Results
Stromal populations have distinct expression and morphological profiles
Phenotypic profiling of the stromal populations was performed to gain insight into the differences between fibroblast populations. To more completely reflect the breadth of stromal populations used in heterotypic co-culture studies across the field, a wide variety of stromal cell types (aCFs, fCFs, DFs, MSCs, iPS-Ftera, and iPS-FEB) were examined by the following phenotypic analyses. A panel of common stromal surface markers was measured in individual cell types by flow cytometry to reveal expression levels across stromal populations. As expected, no cell types expressed CD45 and most cells types did not express CD34 at appreciable amounts except for iPS-Ftera (>50% of cells), MSCs (∼20%), and DFs (∼25%) (Supplementary Fig. S1 and Supplementary Table S4). All stromal populations contained many CD166+ cells; however, fCFs and aCFs had moderate expression (∼23% and ∼45%, respectively) compared with DFs, MSCs, iPS-Ftera, and iPS-FEB, which contained >98% CD166+ cells. MSCs were the only cell population that expressed CD73 (∼97%). CD90 expression was highly variable among the different cell populations with very little expression observed in aCFs and fCFs (Supplementary Table S4).
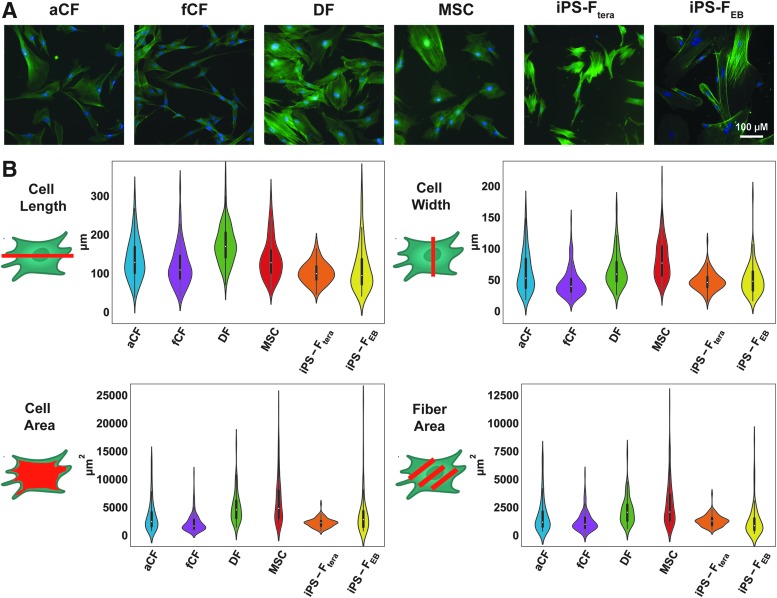
Dramatic differences among stromal cell morphometry were revealed by fluorescent labeling of F-actin with phalloidin (Fig. 1A). MSCs and DFs were the largest cells that spread out the most (average area >5000 μm2) in contrast to the iPS-Ftera and fCFs that exhibited a small, compact morphology (average area <2500 μm2; Fig. 1B). MSCs were the most variable with regard to cell size and shape, as indicated by the wide distribution with each measured parameter (Fig. 1B). Cultures of both aCFs and fCFs included many long, thin spindle-shaped cells, whereas DFs contained long, wide cells, giving rise to apparent differences in cell area across stromal type (Fig. 1B). Automated imaging of each stromal cell type enabled high throughput analysis of population morphometry. Specific features of individual cells (length, width, perimeter, area, fiber area, nuclear area, and fiber alignment) extracted from the images were compared between the different stromal populations (Fig. 1B and Supplementary Fig. S2). iPS-FEB displayed the widest distribution in length and area, indicating more morphological variability within this population. This variability reflects the iPS-FEB differentiation itself, which was inconsistent and did not yield large quantities of stromal cells as easily as the iPS-Ftera differentiation did. DFs and MSCs exhibited the largest nuclear area (>295 μm2), but the smallest nuclear area-to-cell area ratio given that they also had the largest average cell areas. MSCs displayed the greatest cell area (>6200 μm2) and corresponding fiber area, although actin fibers were more unaligned compared with the other stromal populations (Fig. 1B and Supplementary Fig. S2). The iPS-FEB exhibited the highest actin fiber alignment despite having the lowest fiber area (Fig. 1B and Supplementary Fig. S2).
FIG. 1.
Morphometric analysis of stromal cell populations. (A) F-actin labeling (phalloidin) of all stromal cell populations. (B) Violin plots of morphological features: cell length, cell width, cell area, and fiber (F-actin) area. Kolmogorov–Smirnov statistical test with Holm–Bonferroni multiple comparison correction was used to compare empirical distributions of morphological feature measurements for each stromal cell type with significance determined at p < 0.05 (Supplementary Fig. S2 for table of p-values). Color images are available online.
Although the individual morphometric parameters illustrate some differences between cell types, the correlated image-extracted measurements were subjected to PCA to reveal uncorrelated variables contributing to morphological differences (Supplementary Fig. S3). The overall variance in cell morphology was separated along two principal components on the PCA biplot based on their morphologic characteristics. PC1 captured 91% of the variance and was primarily correlated to differences in cell area (Supplementary Fig. S3), demonstrated by individual images of smaller cells on the left of the plot compared with the larger cells on the right of the plot. PC2 captured only 4% of the variance and was largely driven by fiber area. Cells did not completely separate by stromal type, but MSCs, DFs, and aCFs appeared to cluster more closely together, perhaps reflecting a more similar morphology.
The PCA was run on a small number of known (i.e., biased) parameters that were predetermined and measured from the images to quantify cell morphometrics. To contrast this approach, PhenoRipper was used to evaluate cell morphology in an unbiased manner (Fig. 2 and Supplementary Fig. S4).29 The individual cells were analyzed based on similarity of like features within the images (Fig. 2, examples of features are shown in images along the top of the heatmap). Clustering stromal cells based on like features indicated that primary isolated adult cells (DFs, aCFs, MSCs) shared greater morphological similarity than the fetal or iPS cell-derived fibroblasts (fCFs, iPS-Ftera, iPS-FEB) (Fig. 2B). Taken together, these phenotypic data suggest that cultured stromal cells adopt characteristic physical features that enable distinction based on morphometric analysis.
FIG. 2.
PhenoRipper analysis of images of F-actin-labeled stromal cell populations. (A) MDS plots of individual cell images colored by stromal cell identity. (B) Clustergram of differentially expressed superblocks. Expanded views of superblocks representing (i) cell perimeter, (ii) actin fiber-rich cell interior, (iii) nucleus, and (iv) cell interior with less defined actin fibers. MDS, multidimensional scaling. Color images are available online.
Stromal populations have distinct transcriptomic profiles
Transcriptional analysis was performed to characterize phenotypic differences between stromal cell populations. Not surprisingly, all the stromal populations were more transcriptionally similar to each other than to hPSC-CMs (Supplementary Fig. S5A, B). As illustrated in the PCA, the variance in cell phenotype was separated along two principal components based on their subtype. PC1 captured 62% of the variance and was based largely on the differences between cardiomyocytes and all stromal populations combined (Supplementary Fig. S5B). PC2 captured 12% of the variance and separated the stromal cells into three clusters based on their similarity with one another. DF clustered individually, fCFs and aCFs clustered together, and MSCs, iPS-Ftera, and iPS-FEB clustered together in the third group. To enable better separation of the individual stromal populations, the transcriptomic data were reclustered without the cardiomyocytes. This reclustering further highlighted the similarities among MSCs, iPS-Ftera, and iPS-FEB compared with the other stromal cell populations (Fig. 3A, B), indicating a transcriptional difference in stromal cells isolated from progenitor or stem cell-based differentiations. Adult CF populations from different vendors (aCFs that were used throughout these studies vs. aCFPC) did not cluster together with the PCA, indicating variability in fibroblasts even from the same tissue source, perhaps as a result of different isolation and culture methods.
FIG. 3.
RNA sequencing analysis of stromal cell populations through (A) differential gene expression heatmap and (B) PCA plot. PCA, principal component analysis. Color images are available online.
Nine individual clusters of genes that shared similar expression patterns across the cell populations were identified using the HOPACH clustering algorithm on the RNAseq results.34 The separation of five clusters (2, 3, 5, 6, and 7) was driven entirely by the comparison of stromal cells with cardiomyocyte transcriptomes (Supplementary Fig. S5C). Several of the top GO terms associated with these clusters included cardiac developmental and regulatory pathways such as muscle contraction, conduction, and sarcomeric organization. However, the differences between the remaining HOPACH clusters (1, 4, 8, and 9) could be attributed to other specific stromal populations. For example, the differential genes in cluster 4 were linked with MSCs, iPS-Ftera, and iPS-FEB, which contained several homeotic genes (i.e., HOXA9, HOXA13, HOXC10, HOXC11, and HOXD10) with associated GO terms involved in embryonic morphogenesis and pattern formation. In contrast, the differential genes in cluster 8 were associated with DFs and aCFPC, which contained several extracellular matrix-related genes (i.e., HAS1, ELN, COL22A1, VIT, and ACAN) and associated with GO terms involved in tissue regeneration and collagen organization (Supplementary Fig. S5C). The genes driving clusters 1 and 9 were largely expressed in the cardiac fibroblast populations (aCFs, fCFs, and aCFPC) and associated with GO terms related to chemokine signaling, inflammatory response, and peptide signaling (Supplementary Fig. S5C). Taken together, these results demonstrate that stromal cells obtained from various sources are transcriptionally distinct from one another, but that some gene expression patterns may be conserved across primary isolated stromal cells (aCFs, fCFs, and DFs) compared with progenitor or stem cell-derived stromal cells (MSCs, iPS-Ftera, and iPS-FEB).
Stromal cells enable variable cardiac microtissue formation
Given the observed variability in stromal phenotypes, we hypothesized each population would differentially impact cardiac microtissue formation, phenotype, and function. To determine the required stromal fraction for consistent cardiac microtissue formation, lactate-purified hPSC-CMs were mixed with varying ratios of aCFs (0%, 5%, 10%, 20%, 25% fibroblasts). Aggregation with ratios <10% aCFs did not yield microtissues, only small clusters containing a few cells. Larger, more uniform aggregates were observed using >10% stromal fraction. However, the most consistent, robust formation was observed with 25% stromal incorporation (Fig. 4A). Thus, to ensure consistency across studies, lactate-purified hPSC-CMs were combined with 25% stromal fraction for all subsequent experiments.
FIG. 4.
Stromal cells differentially impact cardiac microtissue formation. (A) Ability of tissues to form was based on the incorporation fraction of stromal cells (0–25% adult cardiac fibroblasts). (B) Graphical representation of heterotypic cardiac microtissue groups. (C) Tissue formation at day 0 and 3 of cardiomyocytes alone and cardiomyocytes with adult cardiac fibroblasts, dermal fibroblasts, mesenchymal stromal cells, and iPS cell-derived fibroblasts. (D) Average diameter of cardiac microtissues comprised cardiomyocytes with different stromal cell populations (n > 125; *p < 0.05; ***p < 0.001; ****p < 0.0001). iPS, induced pluripotent stem. Color images are available online.
As different stromal populations have been used in engineered cardiac tissues, microtissue aggregation potential was compared using cardiomyocytes mixed with the most commonly used stromal types: aCFs, DFs, MSCs, and iPS-Ftera (Fig. 4B). The resulting microtissues were highly variable in size across groups. Microtissues composed of only cardiomyocytes did not form well with many small clusters of only a few cells, similar to that observed in the cell ratio study. Microtissues generated with MSCs or iPS-derived fibroblasts also contained small clusters of cells in addition to larger, nonuniform aggregates leading to highly heterogeneous size distributions based on measured aggregate diameter (+MSC: ∼55–250 μm; +iPS-Ftera: ∼50–180 μm) (Fig. 4C, D). The variability in size of aggregates led to an overall decrease in the total number of cardiac microtissues observed in groups with CM alone, MSCs, or iPS-Ftera compared with groups with aCFs or DFs. The inclusion of aCFs and DFs led to the formation of large, uniform aggregates with consistent size (∼180 μm in diameter) (Fig. 4C, D). These data suggest that some stromal populations (MSCs and iPS-Ftera) do not support uniform formation of microtissues, whereas other stromal populations (aCFs and DFs) readily support consistent generation of cardiac tissues.
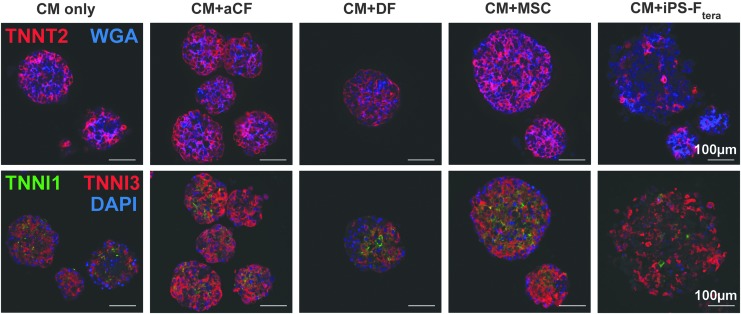
To determine structural uniformity between different heterotypic mixes, cardiac troponin T (TNNT2) distribution was analyzed across fixed, 7-day-old microtissues (Fig. 5). Even distribution of cardiomyocytes (TNNT2+) was observed in all aggregates. Stromal cells (WGA+) appeared to be evenly distributed within individual aggregates, outnumbered by TNNT2+ cells in all groups except microtissues with iPS-Ftera. Differences in expression of cardiac troponin I isoforms were observed between groups. Microtissues formed with aCFs and MSCs contained the most robust TNNI3 staining throughout, whereas TNNI1 and TNNI3 were more diffuse in microtissues without stromal cells, with DFs, or with iPS-Ftera (Fig. 5). These data reflect the similarity in microtissue composition between groups and suggest that different stromal interactions may lead to CM phenotypic changes (TNNI1 vs. TNNI3 expression).
FIG. 5.
Immunofluorescent staining of day 7 heterotypic cardiac microtissues. Top row: cardiac-specific marker cardiac troponin T (TNNT2, red) and membrane marker WGA (blue). Bottom row: slow+fast troponin I (TNNI1, green) and cardiac troponin I (TNNI3, red) and Hoechst nuclear stain (blue). WGA, wheat germ agglutinin. Color images are available online.
Stromal cell inclusion yields phenotypic variation among cardiac microtissues
In addition to differences in aggregation potential resulting from heterotypic interactions, CM phenotype was compared across 7-day-old microtissues to analyze changes in cardiac gene expression as a result of culture with various stromal populations (Fig. 6). A panel of primarily cardiac-specific gene markers was assessed for the cardiac microtissues and normalized to microtissues comprised entirely from cardiomyocytes. The same panel of markers was characterized on individual stromal cell populations and, as expected, cardiac-related genes were not expressed in the stromal cells (Supplementary Fig. S6). Commonly expressed stroma-related genes (i.e., THY1, VIM, GJA1, and GJC1) were abundant, although differentially expressed, by the different stromal cell populations. Gene expression profiles of cardiac microtissues comprising only CMs or with iPS-Ftera were clearly distinguished from the other stromal cell populations through hierarchical clustering of the gene expression profiles (Fig. 6). The stark contrast in gene expression profiles between the heterotypic cardiac microtissues and their respective individual stromal populations allowed for direct comparison of the specific influence of stromal cell type on cardiomyocyte phenotype within the microtissues.
FIG. 6.

Gene expression analysis of cardiac- and stromal-specific markers on day 7 heterotypic microtissues compared with day 7 homotypic microtissues comprised only cardiomyocytes. Color images are available online.
Increased expression of genes mediating ion exchange (SLN and KCNH2) was observed in all microtissues containing stromal cells compared with microtissues consisting of only CMs (Fig. 6). Several genes involved in cardiomyocyte development (TBX5, MESP1, MEF2C, GATA4, ISL1, and LEF1), contractility (MYH6, MYH7, MYL7, TNNT2, TNNI1, and TNNI3), and metabolism (PGC1B and PPARA) were expressed at higher levels in the microtissues cultured with MSCs, aCFs, and DFs than those containing iPS-Ftera. Microtissues containing MSCs expressed higher levels of most genes, including regulators of metabolism (FABP2 and PGC1A) and sodium ion channels (SCN5A and SLC8A1), than microtissues with aCFs or DFs. Of interest, a subset of genes related to early cardiac development (TBX5 and MESP1) were expressed at higher levels in microtissues containing DFs than aCFs, whereas some genes related to myocyte contractility (ACTN2 and MYBPC3) were expressed at higher levels in microtissues containing aCFs than DFs. Overall, microtissues containing iPS-Ftera expressed lower levels of most cardiac-related genes, suggesting that iPS-Ftera cells may not support cardiac phenotype as well as the stromal cell populations sourced from primary tissues. Together, these data demonstrate that cardiomyocyte phenotype is altered by heterotypic interactions and that culture with primary-derived stromal cells (aCFs, DFs, and MSCs) promotes a more robust cardiac phenotype compared with culture without exogenous fibroblasts or with iPS-derived fibroblasts.
Stromal cell modulation of cardiac microtissue calcium handling
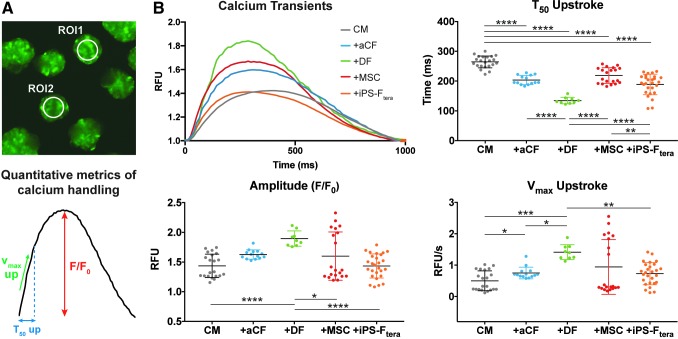
Calcium transients of individual aggregates were optically monitored for 7 days after tissue formation to evaluate the influence of various stromal populations on cardiac tissue calcium handling properties (Fig. 7A). All the heterotypic microtissues exhibited a decreased T50 upstroke compared with homotypic microtissues formed only with CMs, demonstrating improved kinetics of calcium influx with the inclusion of stromal cells. The amplitude of calcium transients was greatest for microtissues containing DFs and the Vmax upstroke was significantly higher in microtissues containing aCFs or DFs compared with CM-alone microtissues (Fig. 7B). The calcium handling properties of microtissues containing CM alone, +MSC, or +iPS-Ftera exhibited greater variability with increased standard deviations for each measurement, reflecting inconsistent responses between microtissues within these groups. These results demonstrate that calcium handling properties of engineered microtissues are modulated by heterotypic interactions with different stromal cell populations and that microtissues comprising CM with aCFs or DFs yield the most consistent microtissues with accelerated functional maturation with respect to their calcium handling properties.
FIG. 7.
Stromal cells impacted day 7 cardiac microtissue function. (A) ROIs (120-pixel diameter) were selected at the center of each tissue (n > 10). (B) Representative calcium transients were determined by averaging normalized (F/F0) fluorescent intensity values from all tissues within each group. Calcium handling properties were assessed by quantifying amplitude (F/F0), maximum upstroke velocity (Vmax up), and time to 50% of upstroke amplitude (T50 up). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ROIs, regions of interest. Color images are available online.
Discussion
The observed phenotypic differences between fibroblastic populations instigated assessment of the structural and functional benefits of heterotypic microtissues. The International Society for Cellular Therapy along with researchers working with MSCs have come to consensus on a typical panel of marker expression expected within the cell population27: CD166+, CD90+, CD73+, CD34−, and CD45− (Supplementary Table S4 and Supplementary Fig. S1). However, stromal cells in general are broadly defined as adherent cells that secrete extracellular matrix and are usually characterized by a spindle-shaped morphology. Morphometric profiling of the different stromal populations examined in this study revealed significant variability in cell shape, size, and actin fiber organization (Fig. 1 and Supplementary Fig. S2), indicating that qualitative assessment of cell appearance is insufficient to describe fibroblast phenotype. Furthermore, flow cytometry data demonstrated variable surface marker expression across stromal populations (Supplementary Fig. S1). CD90 is generally thought to be an identifying marker of fibroblasts, yet reports of CD90 expression in cardiac fibroblasts is variable, from high expression levels41,42 to mixed expression,43 and in this study, a distinct lack of CD90 expression in both the fetal and adult cardiac fibroblast populations. The reported variability could be because of in vivo versus in vitro phenotype of cardiac fibroblasts and changes that arise during in vitro culture.44 For example, basic fibroblast growth factor (FGF2) specifically decreases CD90 expression in MSC populations,45 indicating that specific culture conditions can lead to changes in surface marker expression. Altogether, these data demonstrate that singular metrics are insufficient to characterize stromal cells and predict their suitability for use in engineered tissues.
In recent years, there has been an increase in studies using PSC-derived cardiomyocytes as in vitro models of cardiac behavior given the advancement of differentiation protocols, which yield more robust cardiomyocyte populations. Several studies have shown that engineered 3D cardiac tissues generated from hPSC-CMs serve as better predictive models of cardiac physiology and drug response than two-dimensional (2D) monolayers.17,46–49 Within the context of 3D-engineered tissues, strong evidence has been presented for the inclusion of a nonmyocyte fraction to form stable cardiac tissues.15–17 However, the requisite ratio of stromal cells and the effects of different stromal cell populations on multicellular cardiac tissue function have not been fully explored. Consistent with previous reports,15,19,20,50,51 the data presented here demonstrate the need for a minimum 10–25% stromal fraction to ensure robust tissue formation and stability (Fig. 4A). The variability in stromal fraction observed across studies can be explained in part by the variety of stromal cell types used in different engineered constructs, and also inconsistencies with CM differentiation efficiencies from batch-to-batch and derivation protocols between laboratories.52 In this study, microtissues generated from CM alone resulted in inconsistent tissue formation despite following the same differentiation and purification protocols, highlighting the variability in pure hPSC-CMs (Fig. 4A, C). This study aimed to systematically compare the impacts of multiple commonly used stromal populations on 3D-engineered cardiac tissue to determine if specific sources of stromal cells influenced cardiac tissue formation and function differentially.
To date, cardiac tissues have been generated using cardiomyocytes alone,48,53–55 or by mixing with cardiac fibroblasts,18 DFs,18,19,47,56 MSCs,21,57 and, more recently, PSC-derived fibroblasts.17,25,50,58 In a subset of these studies, EC have also been combined with cardiomyocytes and stromal cells to generate complex tricellular cardiac tissues.16,24,59,60 However, as a result of cell type and source variability, the functional consequences of stromal inclusion have remained elusive. Cardiac tissues comprising CM+CF+EC exhibited appropriate contractile responses to inotropic chemical agents compared with microtissues comprising CM+DF+EC that did not respond to these drugs, suggesting that inclusion of DFs may not support proper pharmacological response of cardiac tissues.18 However, these results are confounded by the fact that the specific influences of the fibroblasts on the cardiomyocytes cannot be dissected apart from the cardiomyocyte–endothelial cell interactions. Therefore, in the study described here, only one nonmyocyte population was paired with cardiomyocytes to study heterotypic impacts in a more controlled manner. In contrast to the reports mentioned previously, DFs in this two-population system supported cardiac microtissue formation (Fig. 4C), cardiac gene expression (Fig. 6), and calcium handling similar to or better than CFs (Fig. 7B). Moreover, a recent report of engineered cardiac tissues comprising CM+DF under electrical stimulation demonstrated the greatest extent of CM phenotypic and functional maturation seen to date,19 suggesting that DFs are able to support cardiac function. It is important to note that the use of primary human cells is accompanied by inherent limitations in availability and donor matching. The CFs and DFs in this study came from donors of different ages and sexes, making it challenging to dissect the exact mechanism as to why DFs showed greater improvements in calcium handling compared with CFs. One other limitation to this study is the in vitro passage artifact associated with primary cells. Although restrictions on passage use (<10 passages) were implemented in this study, primary stromal cells expanded in culture may exhibit varying degrees of phenotypic drift leading to further observed differences in performance.
It is also highly possible to imagine that individual stromal cells contribute to cardiac microenvironments in different ways. MSCs in co-culture models have been reported to deliver paracrine signals to cardiomyocytes.61 In this study, a global increase in cardiac gene expression was observed in co-culture of CM+MSC (Fig. 6); however, functional benefits of MSCs were variable (Fig. 7B), mirroring the inconsistent ability of MSCs to form robust cardiac tissues (Fig. 4C, D). This variability in MSCs microtissue function may be a direct result of the variability in formation given that previous reports have demonstrated a link between microtissue size and resulting functional consequences,62 highlighting the importance of robust microtissue formation for consistent functional outcomes. The variable impact on cardiac microtissue calcium handling properties imparted by MSCs (Fig. 7) was consistent with reports of MSCs exerting little effect on cardiac contractile function in other engineered systems.60 Also consistent with previous reports that suggest MSCs exert paracrine effects on tissue regulation and organization,63,64 we observed improved cardiac gene expression with the presence of MSCs, suggesting that MSCs may contribute to phenotypic changes in CMs but have limited functional consequences on engineered cardiac tissues. This highlights the idea that stromal populations may serve different functions within the context of microtissues and that there is not a single stromal cell type that will outperform others in all aspects.
As the field moves toward engineering heterotypic tissues derived entirely from iPS cell sources to enable isogenic modeling, how to best define iPS cell-derived stromal cells remains a major question. There are several reports of hPSC-derived MSC-like cells65,66 and fibroblastic cells using 2D-directed approaches,67 3D embryoid body-based approaches,17,68 and in vivo-based derivation methods.69 However, differences between these hPSC-derived stromal populations' morphology, characteristic phenotypic marker expression, and ability to support formation and function of engineered tissues remain unclear. In this study we compared iPS-F using two different derivation methods from the same isogenic iPS cell line as the cardiomyocytes: an in vivo-based derivation capturing teratoma outgrowth cells (iPS-Ftera)26 and an in vitro-based derivation using an embryoid body outgrowth method (iPS-FEB).17 The resulting cells exhibited different morphologies with the iPS-Ftera adopting a small and spindle-like shape, whereas the iPS-FEB were large and flat (Fig. 1 and Supplementary Fig. S2). The iPS cell-derived fibroblasts also had distinct surface marker expression with ∼60% of iPS-Ftera expressing CD90 compared with no expression in the iPS-FEB (Supplementary Fig. S1) and iPS-Ftera did not support cardiac tissue formation, gene expression, or functional properties (Figs. 4B, 6, and 7B). Despite their morphological differences, transcriptomic analysis of the iPS-Ftera and iPS-FEB suggested that both stem cell-derived stromal populations were more closely related to each other than to either the CF or DF populations, reflecting that the iPS-F cells may represent a more immature stromal population. Image analysis software, PhenoRipper, classified the two iPS cell-derived fibroblast populations as more similar to each other and to fetal CFs than any of the primary-derived adult populations, suggesting that the iPS cell-derived cells may be morphologically more similar to an early “immature” developmental stromal cell. The concept of cell maturation has been a focus of iPS cell-derived parenchymal populations, but its relevance to supporting stromal cells has largely been overlooked. A recent study comparing the transcriptome of fCFs versus aCFs revealed distinct signatures between them with genes related to muscle development and function as well as immune cell trafficking upregulated in aCFs, further highlighting the phenotypic changes that occur throughout stromal maturation.41 The distinctions observed in our study between early developmental stromal populations and adult tissue-derived fibroblasts indicate the need to further develop fibroblast differentiation protocols to yield homogeneous, mature populations of stromal cells that consistently support all aspects of cardiac tissue development and function.
In conclusion, this study provides a direct comparison of individual stromal populations through quantitative phenotypic and transcriptomic approaches, revealing distinct differences between primary adult fibroblasts and iPS cell-derived stromal cells. These phenotypic differences were also demonstrated through the inability of iPS cell-derived stromal cells to support consistent cardiac microtissue formation and function. With the current shift toward engineering complex heterotypic tissues from isogenic sources, ongoing development of robust directed fibroblast differentiation protocols from iPS cell sources should remain a focus of research. Recent reports of iPS cell-derived cardiac fibroblasts have been described,70,71 but the ability of these cells to contribute to cardiac function remains unknown. Given the observed benefits of adult tissue-derived fibroblasts in contributing to improved cardiac microtissue function seen in this study, identifying methods to “mature” iPS cell-derived fibroblasts toward an adult-like phenotype may provide the key to improved function of isogenic tissues.
Supplementary Material
Acknowledgments
The authors acknowledge funding support from the California Institute of Regenerative Medicine (LA1-08015) and the Gladstone BioFulcrum Heart Failure Research Program. T.A.H. was supported by an American Heart Association Postdoctoral Fellowship (15POST22750003). O.B.M. is a National Science Foundation Graduate Research Fellow (1650113). The authors thank the Gladstone Flow Cytometry Core (Dr. Marielle Cavrois; NIH P30 AI027763), Gladstone Assay Development and Drug Discovery Core (Dr. Anke Meyer-Franke), Gladstone Bioinformatics Core (Dr. Reuben Thomas), Gladstone Genomics Core (Jim McGuire), and the Gladstone Stem Cell Core (Dr. Po-Lin So; Roddenberry Stem Cell Foundation) for assisting with experimental analysis. The authors also thank Dr. Nate Huebsch and Dr. Bruce Conklin for providing the iPS-FEB and the WTC11 GCaMP iPS cell lines, and Dr. Tim Rand and Dr. Shinya Yamanaka for providing the iPS-Ftera cells for these studies. The authors also acknowledge Michael Olvera for providing the R-script for calcium handling analysis and the Gladstone Illustration and Design department (Giovanni Maki, Sarah Gardner) for assistance with graphics.
Disclosure Statement
T.C.M. is a consultant for Tenaya Therapeutics. The other authors declare that no competing financial interests exist.
Supplementary Material
References
- 1. Zhou P., and Pu W.T. Recounting cardiac cellular composition. Circ Res 118, 368, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Souders C.A., Bowers S.L., and Baudino T.A. Cardiac fibroblast: the renaissance cell. Circ Res 105, 1164, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang P., Su J., and Mende U. Cross talk between cardiac myocytes and fibroblasts: from multiscale investigative approaches to mechanisms and functional consequences. Am J Physiol Heart Circ Physiol 303, H1385, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rother J., Richter C., Turco L., et al. Crosstalk of cardiomyocytes and fibroblasts in co-cultures. Open Biol 5, 150038, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Camelliti P., Green C.R., and Kohl P. Structural and functional coupling of cardiac myocytes and fibroblasts. Adv Cardiol 42, 132, 2006 [DOI] [PubMed] [Google Scholar]
- 6. Gaudesius G., Miragoli M., Thomas S.P., and Rohr S. Coupling of cardiac electrical activity over extended distances by fibroblasts of cardiac origin. Circ Res 93, 421, 2003 [DOI] [PubMed] [Google Scholar]
- 7. Camelliti P., Borg T.K., and Kohl P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res 65, 40, 2005 [DOI] [PubMed] [Google Scholar]
- 8. Zeng Q.C., Guo Y., Liu L., et al. Cardiac fibroblast-derived extracellular matrix produced in vitro stimulates growth and metabolism of cultured ventricular cells. Int Heart J 54, 40, 2013 [DOI] [PubMed] [Google Scholar]
- 9. Cartledge J.E., Kane C., Dias P., et al. Functional crosstalk between cardiac fibroblasts and adult cardiomyocytes by soluble mediators. Cardiovasc Res 105, 260, 2015 [DOI] [PubMed] [Google Scholar]
- 10. Tomoda Y., Kikumoto K., Isumi Y., et al. Cardiac fibroblasts are major production and target cells of adrenomedullin in the heart in vitro. Cardiovasc Res 49, 721, 2001 [DOI] [PubMed] [Google Scholar]
- 11. Bang C., Batkai S., Dangwal S., et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest 124, 2136, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lyu L., Wang H., Li B., et al. A critical role of cardiac fibroblast-derived exosomes in activating renin angiotensin system in cardiomyocytes. J Mol Cell Cardiol 89, 268, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lian X., Zhang J., Azarin S.M., et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc 8, 162, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lian X., Hsiao C., Wilson G., et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A 109, E1848, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thavandiran N., Dubois N., Mikryukov A., et al. Design and formulation of functional pluripotent stem cell-derived cardiac microtissues. Proc Natl Acad Sci U S A 110, E4698, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giacomelli E., Bellin M., Sala L., et al. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 144, 1008, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huebsch N., Loskill P., Deveshwar N., et al. Miniaturized iPS-cell-derived cardiac muscles for physiologically relevant drug response analyses. Sci Rep 6, 24726, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ravenscroft S.M., Pointon A., Williams A.W., Cross M.J., and Sidaway J.E. Cardiac non-myocyte cells show enhanced pharmacological function suggestive of contractile maturity in stem cell derived cardiomyocyte microtissues. Toxicol Sci 152, 99, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ronaldson-Bouchard K., Ma S.P., Yeager K., et al. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kensah G., Roa Lara A., Dahlmann J., et al. Murine and human pluripotent stem cell-derived cardiac bodies form contractile myocardial tissue in vitro. Eur Heart J 34, 1134, 2013 [DOI] [PubMed] [Google Scholar]
- 21. Stevens K.R., Kreutziger K.L., Dupras S.K., et al. Physiological function and transplantation of scaffold-free and vascularized human cardiac muscle tissue. Proc Natl Acad Sci U S A 106, 16568, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nunes S.S., Miklas J.W., Aschar-Sobbi R., et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods 10, 781, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tulloch N.L., Muskheli V., Razumova M.V., et al. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res 109, 47, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caspi O., Lesman A., Basevitch Y., et al. Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ Res 100, 263, 2007 [DOI] [PubMed] [Google Scholar]
- 25. Masumoto H., Nakane T., Tinney J.P., et al. The myocardial regenerative potential of three-dimensional engineered cardiac tissues composed of multiple human iPS cell-derived cardiovascular cell lineages. Sci Rep 6, 29933, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rand T.A., Sutou K., Tanabe K., et al. MYC releases early reprogrammed human cells from proliferation pause via retinoblastoma protein inhibition. Cell Rep 23, 361, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dominici M., Le Blanc K., Mueller I., et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8, 315, 2006 [DOI] [PubMed] [Google Scholar]
- 28. Pedregosa F.V.G., Gramfort A., Michel V., et al. Scikit-learn: machine learning in python. J Mach Learn Res 12, 2825, 2011 [Google Scholar]
- 29. Rajaram S., Pavie B., Wu L.F., and Altschuler S.J. PhenoRipper: software for rapidly profiling microscopy images. Nat Methods 9, 635, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim D., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liao Y., Smyth G.K., and Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923, 2014 [DOI] [PubMed] [Google Scholar]
- 32. Robinson M.D., McCarthy D.J., and Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCarthy D.J., Chen Y., and Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van der Laan M.P.K. Hybrid clustering of gene expression data with visualization and the bootstrap. J Stat Plan Inference 117, 275, 2003 [Google Scholar]
- 35. Huebsch N., Loskill P., Mandegar M.A., et al. Automated video-based analysis of contractility and calcium flux in human-induced pluripotent stem cell-derived cardiomyocytes cultured over different spatial scales. Tissue Eng Part C Methods 21, 467, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma Z.H.N., Koo S., Mandegar M.A., et al. Contractile deficits in engineered cardiac microtissues as a result of MYBPC3 deficiency and mechanical overload. Nat Biomed Eng 2, 955, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tohyama S., Hattori F., Sano M., et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 12, 127, 2013 [DOI] [PubMed] [Google Scholar]
- 38. Hookway T.A., Butts J.C., Lee E., Tang H., and McDevitt T.C. Aggregate formation and suspension culture of human pluripotent stem cells and differentiated progeny. Methods 101, 11, 2016 [DOI] [PubMed] [Google Scholar]
- 39. Livak K.J., and Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402, 2001 [DOI] [PubMed] [Google Scholar]
- 40. Sturn A., Quackenbush J., and Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics 18, 207, 2002 [DOI] [PubMed] [Google Scholar]
- 41. Jonsson M.K.B., Hartman R.J.G., Ackers-Johnson M., et al. A transcriptomic and epigenomic comparison of fetal and adult human cardiac fibroblasts reveals novel key transcription factors in adult cardiac fibroblasts. JACC Basic Transl Sci 1, 590, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hudon-David F., Bouzeghrane F., Couture P., and Thibault G. Thy-1 expression by cardiac fibroblasts: lack of association with myofibroblast contractile markers. J Mol Cell Cardiol 42, 991, 2007 [DOI] [PubMed] [Google Scholar]
- 43. Furtado M.B., Costa M.W., Pranoto E.A., et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res 114, 1422, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cho N., Razipour S.E., and McCain M.L. Featured article: TGF-beta1 dominates extracellular matrix rigidity for inducing differentiation of human cardiac fibroblasts to myofibroblasts. Exp Biol Med (Maywood) 243, 601, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hagmann S., Moradi B., Frank S., et al. FGF-2 addition during expansion of human bone marrow-derived stromal cells alters MSC surface marker distribution and chondrogenic differentiation potential. Cell Prolif 46, 396, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nguyen D.C., Hookway T.A., Wu Q., et al. Microscale generation of cardiospheres promotes robust enrichment of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Reports 3, 260, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vuorenpaa H., Penttinen K., Heinonen T., et al. Maturation of human pluripotent stem cell derived cardiomyocytes is improved in cardiovascular construct. Cytotechnology 69, 785, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Correia C., Koshkin A., Duarte P., et al. 3D aggregate culture improves metabolic maturation of human pluripotent stem cell derived cardiomyocytes. Biotechnol Bioeng 115, 630, 2018 [DOI] [PubMed] [Google Scholar]
- 49. Kerscher P., Tumbull I.C., Hodge A.J., et al. Direct hydrogel encapsulation of pluripotent stem cells enables ontomimetic differentiation and growth of engineered human heart tissues. Biomaterials 83, 383, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mills R.J., Titmarsh D.M., Koenig X., et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci U S A 114, E8372, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ariyasinghe N.R., Reck C.H., Viscio A.A., et al. Engineering micromyocardium to delineate cellular and extracellular regulation of myocardial tissue contractility. Integr Biol (Camb) 9, 730, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Laco F., Woo T.L., Zhong Q., et al. Unraveling the inconsistencies of cardiac differentiation efficiency induced by the GSK3beta inhibitor CHIR99021 in human pluripotent stem cells. Stem Cell Reports 10, 1851, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Balistreri M., Davis J.A., Campbell K.F., Da Rocha A.M., Treadwell M.C., and Herron T.J. Effect of glucose on 3D cardiac microtissues derived from human induced pluripotent stem cells. Pediatr Cardiol 38, 1575, 2017 [DOI] [PubMed] [Google Scholar]
- 54. Kofron C.M., Kim T.Y., King M.E., et al. Gq-activated fibroblasts induce cardiomyocyte action potential prolongation and automaticity in a three-dimensional microtissue environment. Am J Physiol Heart Circ Physiol 313, H810, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim T.Y., Kofron C.M., King M.E., et al. Directed fusion of cardiac spheroids into larger heterocellular microtissues enables investigation of cardiac action potential propagation via cardiac fibroblasts. PLoS One 13, e0196714, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Noguchi R., Nakayama K., Itoh M., et al. Development of a three-dimensional pre-vascularized scaffold-free contractile cardiac patch for treating heart disease. J Heart Lung Transplant 35, 137, 2016 [DOI] [PubMed] [Google Scholar]
- 57. Emmert M.Y., Wolint P., Wickboldt N., et al. Human stem cell-based three-dimensional microtissues for advanced cardiac cell therapies. Biomaterials 34, 6339, 2013 [DOI] [PubMed] [Google Scholar]
- 58. Nakane T., Masumoto H., Tinney J.P., et al. Impact of cell composition and geometry on human induced pluripotent stem cells-derived engineered cardiac tissue. Sci Rep 7, 45641, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Archer C.R., Sargeant R., Basak J., Pilling J., Barnes J.R., and Pointon A. Characterization and validation of a human 3D cardiac microtissue for the assessment of changes in cardiac pathology. Sci Rep 8, 10160, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Burridge P.W., Metzler S.A., Nakayama K.H., et al. Multi-cellular interactions sustain long-term contractility of human pluripotent stem cell-derived cardiomyocytes. Am J Transl Res 6, 724, 2014 [PMC free article] [PubMed] [Google Scholar]
- 61. Mureli S., Gans C.P., Bare D.J., Geenen D.L., Kumar N.M., and Banach K. Mesenchymal stem cells improve cardiac conduction by upregulation of connexin 43 through paracrine signaling. Am J Physiol Heart Circ Physiol 304, H600, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tan Y., Richards D., Coyle R.C., et al. Cell number per spheroid and electrical conductivity of nanowires influence the function of silicon nanowired human cardiac spheroids. Acta Biomater 51, 495, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Takebe T., Enomura M., Yoshizawa E., et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell 16, 556, 2015 [DOI] [PubMed] [Google Scholar]
- 64. Richards D.J., Coyle R.C., Tan Y., et al. Inspiration from heart development: biomimetic development of functional human cardiac organoids. Biomaterials 142, 112, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Boyd N.L., Robbins K.R., Dhara S.K., West F.D., and Stice S.L. Human embryonic stem cell-derived mesoderm-like epithelium transitions to mesenchymal progenitor cells. Tissue Eng Part A 15, 1897, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Barberi T., Willis L.M., Socci N.D., and Studer L. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med 2, e161, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shamis Y., Hewitt K.J., Bear S.E., et al. iPSC-derived fibroblasts demonstrate augmented production and assembly of extracellular matrix proteins. In Vitro Cell Dev Biol Anim 48, 112, 2012 [DOI] [PubMed] [Google Scholar]
- 68. Itoh M., Umegaki-Arao N., Guo Z., Liu L., Higgins C.A., and Christiano A.M. Generation of 3D skin equivalents fully reconstituted from human induced pluripotent stem cells (iPSCs). PLoS One 8, e77673, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chan S.S., Arpke R.W., Filareto A., et al. Skeletal muscle stem cells from PSC-derived teratomas have functional regenerative capacity. Cell Stem Cell 23, 74, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bao X., Lian X., Qian T., Bhute V.J., Han T., and Palecek S.P. Directed differentiation and long-term maintenance of epicardial cells derived from human pluripotent stem cells under fully defined conditions. Nat Protoc 12, 1890, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bao X., Lian X., Hacker T.A., et al. Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nat Biomed Eng 1, 2016. doi: 10.1038/s41551-016-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.