Abstract
Induced pluripotent stem cell-derived endothelial progenitors (iPSC-EPs) have emerged as a promising candidate cell source for patient-specific ischemic therapies. Before these cells can be appropriately deployed in a clinical setting, it is imperative to study their assembly into functional vascular networks in extracellular matrix (ECM)-mimicking, three-dimensional (3D) microenvironments. To elucidate the interactions of iPSC-EPs with the ECM, we examined how in vitro modulation of structural protein density, the presence of angiogenic growth factors, and relative proteolytic activity affected the vasculogenic potential of these progenitors, that is, their ability to self-assemble into vessel-like networks. We found that the addition of a ROCK pathway inhibitor and exogenous vascular endothelial growth factor (VEGF) are imperative for inducing robust iPSC-EP vasculogenesis in collagen hydrogels. Under these conditions, 3D vascular-like networks containing VE-cadherin-expressing lumens formed within a week of culture. To quantify this 3D vessel-like network, we developed a computational pipeline to analyze network length, connectivity, and average lumen diameter. Increasing the concentration of collagen in the hydrogels abrogated network formation and encouraged the formation of disconnected, large-diameter lumens. This phenomenon was in part related to the cells' proteolytic capacity and the hydrogels' properties, specifically hydrogel deformability and pore size. In conclusion, we demonstrate that the vasculogenic potential of iPSC-EPs is regulated by cell–matrix interactions and the matrix properties of collagen hydrogels.
Impact Statement
Our work reinforces the role of extracellular matrix (ECM) density and matrix metalloprotease activity on the formation of microvasculature from induced pluripotent stem cell (iPSC)-derived vascular cells. The cell–matrix interactions discussed in this study underscore the importance of understanding the role of mechanoregulation and matrix degradation on vasculogenesis and can potentially drive the development of ECM-mimicking angiogenic biomaterials. Furthermore, our work has broader implications concerning the response of iPSC-derived cells to the mechanics of engineered microenvironments. An understanding of these interactions will be critical to creating physiologically relevant transplantable tissue replacements.
Keywords: IPS cells, angiogenesis and vasculogenesis, blood vessel, hydrogels, 3D cell culture
Introduction
The revascularization of lab-grown and native ischemic tissue remains one of the most critical challenges facing the fields of tissue engineering and regenerative medicine. A practical model of vascular development promises to inspire new vascular therapies by enhancing our fundamental understanding of vasculogenic/angiogenic mechanisms and constructing faithful disease models for therapeutic drug testing.1–3 Endothelial progenitors (EPs) have been considered a promising cell source for both vascular modeling and therapy since Asahara et al. first demonstrated that EPs could differentiate into functional endothelial cells (ECs) and participate in murine angiogenesis in 1997.4 To date, adult EPs have been isolated from the spleen, bone marrow, peripheral blood, and cord blood.5–9 However, the critically low number of functional EPs present in these tissues is therapeutically limiting. In addition, patients with cardiovascular diseases and diabetes often have EPs with reduced functionality.10–13 EPs derived from induced pluripotent stem cells (iPSCs) have been proposed as an alternative cell source that can sidestep the current limitations of adult and cord-blood EPs. In addition to their high proliferation capability and pluripotency, iPSCs are a patient-specific, largely untapped cell source that can generate functional EPs from patients with risk factors, as was shown for iPSCs derived from patients with diabetes.14
Different protocols to differentiate iPSCs into EPs have emerged in recent years, and EP differentiation efficacy has steadily risen.15–18 Furthermore, iPSC-EPs embedded in hyaluronic acid hydrogels have been shown to undergo vasculogenesis and form self-assembling vascular networks in vivo.19,20 Recently, Bezenah et al. demonstrated that iPSC-EC-coated Cytodex beads undergo sprouting angiogenesis when cocultured in the presence of primary fibroblasts.21 However, the response of iPSC-EPs to variations of physical and chemical characteristics in the local extracellular matrix (ECM) remains relatively unknown and is a significant barrier to translation.
The goal of this research was to elucidate the interactions of iPSC-EPs with the ECM by examining how in vitro modulation of collagen hydrogels affected their vasculogenic potential, that is, the ability of these progenitors to develop into vessel-like structures. Specifically, we modulated collagen hydrogel density, the concentration of angiogenic growth factors, and proteolytic activity. To compare the resulting vascular network topologies generated under these varying conditions, we developed a computational pipeline that tracked three-dimensional (3D) network connectivity, length, and average lumen diameter with minimal user input.
Materials and Methods
Maintenance of iPSCs
iPSCs, derived from human dermal foreskin fibroblasts (DF19-19-9-11T) using an episomal delivery of seven factors (SOKMNLT: SOX2, OCT4 (POU5F1), KLF4, MYC, NANOG, LIN28, and SV40L T antigen), were purchased from WiCell. iPSCs were cultured under feeder-free conditions on vitronectin-coated six-well plates in complete Essential 8 (E8) medium (ThermoFisher). iPSCs were passaged upon reaching 70–80% confluency. In brief, iPSCs were rinsed with Dulbecco's phosphate-buffered saline (DPBS) and then treated with 0.5 mM ethylenediaminetetraacetic acid (EDTA) for 5 min at 37°C. The EDTA solution was then removed, and the iPSC colonies were gently resuspended in complete E8 medium and seeded on freshly coated vitronectin plates.
Differentiation of iPSCs to CD34+ iPSC-EPs
iPSCs were differentiated into iPSC-EPs following an established protocol with small modifications22 (Fig. 1A). In brief, iPSCs were manually dissociated into a single-cell suspension in E8 medium supplemented with 10 μM ROCK inhibitor (Y-27632; Selleckchem) and plated at 20,000 cells/cm2 on Matrigel-coated 24-well tissue culture plates. Y-27632 was removed after 24 h, and fresh E8 medium was added to the cells. Forty-eight hours after seeding, iPSCs were treated with 6 μM of CHIR99021 (LC Technologies) in Advanced Dulbecco's Modified Eagle Medium (DMEM)/F12 (ThermoFisher Scientific) supplemented with 60 μg/mL ascorbic acid (hereafter LaSR Basal). Forty-eight hours after CHIR99021 induction, media were replaced with fresh LaSR Basal without CHIR99021. Cells were cultured for an additional 3 days with daily media changes.
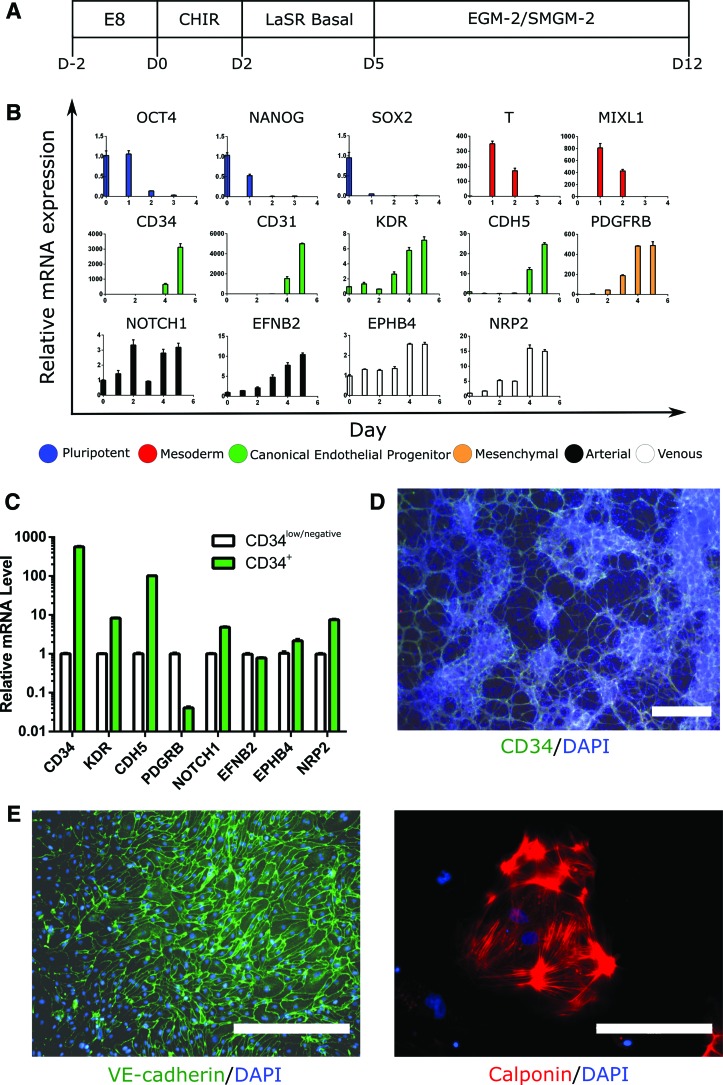
FIG. 1.
iPSC differentiation and CD34+ iPSC-EP biopotency. (A) iPSC colonies were differentiated into mesoderm with CHIR99021 (D2) and then further differentiated into CD34+ iPSC-EPs by exposing the cells to advanced DMEM/F12 and ascorbic acid (i.e., LaSR Basal) (D5). Further culture in proendothelial or promesenchymal media for one additional week was used to evaluate the bipotency of these progenitors (D12). (B) mRNA was isolated from the unsorted cells every 24 h after CHIR99021 induction. Quantitative polymerase chain reaction was performed to evaluate temporal gene expression profiles during the 5 days of differentiation into iPSC-EPs. (C) Expression levels of EP and EC markers of sorted CD34+ iPSC-EPs were compared with the counterpart subpopulation that is low or negative for CD34 expression (CD34low/negative). (D) Five days after being induced with CHIR, iPSC-EPs were fixed and stained with an anti-CD34 antibody and the nuclei were counterstained with DAPI. (E) Sorted CD34+ iPSC-EPs were further cultured in EGM-2 (left) or SMGM-2 (right). Scale bars are 200 μm (D, E right) and 400 μm (E, left). iPSC-EP, induced pluripotent stem cell-derived endothelial progenitors; EC, endothelial cell; EGM-2, endothelial growth media 2; SMGM-2, smooth muscle growth media 2. Color images are available online.
Quantitative reverse transcription–polymerase chain reaction
Messenger RNA (mRNA) combined from at least three wells of confluent differentiating cells was isolated with an RNeasy Mini Kit (Qiagen) and then reverse transcribed into cDNA with a High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific) according to manufacturer instructions. Quantitative reverse transcription–polymerase chain reaction was performed with PowerUp SYBR green (ThermoFisher Scientific) using the StepOne Plus system (Applied Biosystems). Twenty-five nanograms of cDNA and 500 nM of primers were used for each reaction. cDNA solutions were activated at 50°C for 2 min and then at 95°C for 2 min. Forty amplification cycles (95°C for 15 s, followed by annealing at 60°C for 60 s) were performed.
Relative expression of mRNA was quantified using the ΔΔCT method with GAPDH as the endogenous control: ΔCt = Cttarget − CtGAPDH; ΔΔCt = ΔCtsample − ΔCtreference; and relative expression = 2−ΔΔCt. Results are presented as mean and standard deviation of three technical replicates unless indicated otherwise. A list of primers used in these experiments is given in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/tea).
Fluorescence-activated cell sorting of CD34-expressing iPSC-EPs (CD34+ iPSC-EPs)
iPSCs were differentiated into iPSC-EPs as described previously. Five days after CHIR99021 induction, the differentiated cells were incubated with Accutase (ThermoFisher Scientific) for 10 min at 37°C and manually dissociated into single cells with a P1000 pipette tip. Cells were then spun down at 300 g for 5 min and resuspended in 200 μL of ice-cold sorting buffer containing DPBS, 2 mM EDTA, and 0.5% bovine serum albumin (BSA). Cells were filtered with a 35 μm cell strainer (Corning), incubated with 5 μL of CD34-PE antibody (Miltenyi Biotec) for 10 min at 4°C, and isolated with a fluorescence-activated cell sorting instrument (FACS) (S3e; Bio-Rad). Gating and population analysis were performed with Bio-Rad software native to the S3e cell sorter (Supplementary Fig. S1A).
Preparation of ECM protein-coated tissue culture plates
To evaluate the impact of ECM coatings on the biopotency of CD34+ iPSC-EPs, the following ECM coatings were prepared: laminin, fibronectin, vitronectin, and gelatin as described herein. Laminin (isolated from Engelbreth–Holm–Swarm murine sarcoma basement membrane; Sigma-Aldrich) was diluted in DPBS to a final concentration of 6 μg/mL and was used to coat tissue culture plates for 2 h at room temperature. Fibronectin (isolated from bovine plasma; Sigma-Aldrich) and vitronectin (recombinant; ThermoFisher) were diluted in DPBS to a final concentration of 6 and 0.6 μg/mL, respectively. These solutions were used to coat tissue culture plates for 1 h at room temperature. Gelatin solution (0.1% in water; STEMCELL Technologies) was diluted to a concentration of 125 μg/mL and was used to coat tissue culture plates for 30 min at room temperature. All coating solutions were aspirated before seeding to avoid desiccation and replaced with appropriate media.
Immunocytochemistry
To check for CD34 protein expression, differentiating iPSCs were fixed in 4% paraformaldehyde (PFA), rinsed three times with 300 mM glycine (Sigma-Aldrich), and then incubated in blocking buffer containing 1% BSA and 0.1% Tween-20 (Fisher Scientific) for 30 min. Cells were then incubated for 90 min at room temperature with mouse anti-human CD34 antibody (BD Biosciences; 1:200). A secondary antibody, anti-mouse AF488 (1:200; ThermoFisher Scientific), was added for 60 min at room temperature and the nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
To evaluate CD34+ iPSC-EP bipotency in two-dimensional (2D), FACS-isolated CD34+ iPSC-EPs were centrifuged twice to remove residual sorting buffer and seeded at 10,000 cells/cm2 on ECM protein-coated 24-well tissue culture plates. The cells were subsequently cultured in complete endothelial growth media 2 (EGM-2; Lonza), smooth muscle growth media 2 (SMGM-2; Lonza), a medium consisting of 50:50 EGM-2:SMGM-2, or a medium consisting of 50:50 EGM-2:DMEM supplemented with 10% fetal bovine serum (FBS) to generate iPSC-ECs and smooth muscle cells (iPSC-SMCs).
After 7 days of culture with daily media changes, cells were fixed, washed, and blocked as described previously; the cells were then permeabilized with 0.5% Triton-X for 10 min. Permeabilized cells were incubated for 90 min at room temperature with mouse anti-human VE-cadherin antibody (CD144; Santa Cruz Biotechnology; 1:200), rabbit anti-human alpha-smooth muscle actin (α-SMA) (Abcam; 1:100), and rabbit anti-human calponin (Abcam; 1:50) primary antibodies. The cells were then incubated with anti-mouse Alexa Fluor 488, anti-sheep Alexa Fluor 555, and anti-rabbit Alexa Fluor 647 secondary antibodies, respectively (ThermoFisher Scientific) for 60 min at room temperature. Cells were counterstained with DAPI, rinsed in PBS, and imaged on a Zeiss Apotome.2 fluorescent microscope.
Encapsulation of CD34+ iPSC-EPs in collagen hydrogels
A total of 500,000 FACS-isolated CD34+ iPSC-EPs were centrifuged twice and resuspended in EGM-2 supplemented with penicillin–streptomycin (100 U/mL; ThermoFisher Scientific), 10 μM ROCK inhibitor, and 10 × Medium 199 (M199). Ice-cold 10 mg/mL collagen (Type 1, rat tail; Corning) was added to the cell suspension and neutralized with 1 M sodium hydroxide (NaOH; Sigma-Aldrich), turning the solution bright pink. The final concentrations of each hydrogel component are summarized in Supplementary Table S2. Fifty-six microliters of the neutralized collagen-cell suspension was then pipetted into individual wells of an ultra-low attachment round-bottomed 96-well plate and allowed to solidify for 30 min at 37°C and 5% CO2. CD34+ iPSC-EP-laden hydrogels were immersed in 100 μL of EGM-2, SMGM-2 or 50:50 EGM-2:SMGM-2. These media were supplemented with penicillin–streptomycin, 10 μM ROCK inhibitor, and 50 ng/mL vascular endothelial growth factor (VEGF) or 50 ng/mL platelet-derived growth factor (PDGF). Twenty-four hours after cell encapsulation, ROCK inhibitor and antibiotics were removed from the culture medium, which was changed daily thereafter. To check cell viability postencapsulation, cell-laden hydrogels were cultured in fresh EGM-2 supplemented with 2 μM calcein-AM (ThermoFisher Scientific; to identify live cells) and 2 μM ethidium homodimer-1 (EthD-1; ThermoFisher Scientific; to detect dead cells) for 30 min. To track the changes in encapsulated cell morphology, brightfield images were acquired every 24 h on an EVOS FL fluorescent microscope.
Vascular network visualization through confocal fluorescence
The vessel-like networks generated within collagen hydrogels were visualized by following an established immunocytochemistry protocol for cells embedded in thick collagen hydrogels.23 In brief, CD34+ iPSC-EP-laden hydrogels were fixed in a 4% PFA and 5% sucrose solution for 20 min, washed three times with PBS and then permeabilized with 0.5% Triton-X for 10 min. After three washes with PBS supplemented with 0.5% Tween-20, CD34+ iPSC-EP-laden hydrogels were incubated in blocking buffer for 30 min and then with 1:40 rhodamine–phalloidin (ThermoFisher Scientific) and 1:200 VE-cadherin antibodies in blocking buffer. Nuclei were counterstained with DAPI and then rinsed with PBS.
CD34+ iPSC-EP-laden hydrogels were transferred to an μ-Plate Angiogenesis 15-well imaging slide (Ibidi). Z-stacks were acquired at 15–16 μm intervals on a spinning disk confocal microscope (Zeiss Axio Observer Z1 with Yokogawa CSU-X1M). At least three regions of interest (∼1.3 mm2 cross-sections) across the full depth of the hydrogel were acquired per technical replicate; at least three technical replicates were analyzed per experimental condition.
Length and connectivity analysis of vessel-like networks
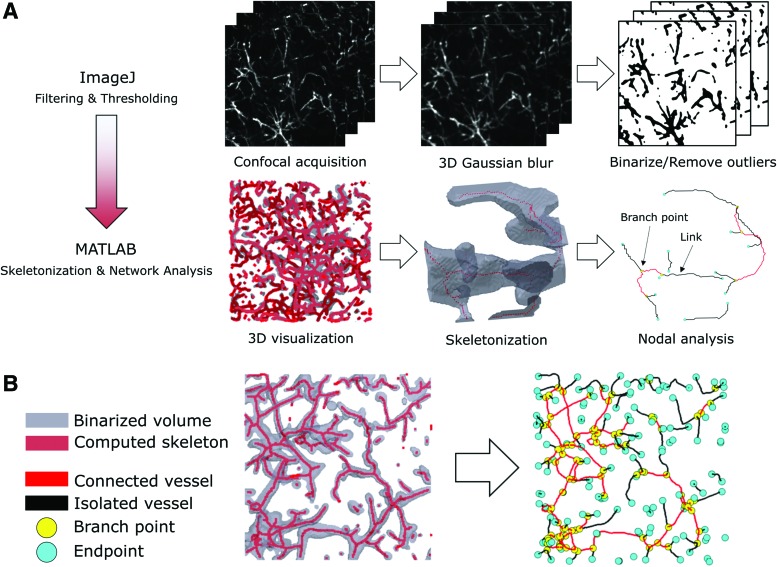
To analyze the total length, connectivity, and lumen diameter of the networks, we developed a computational pipeline using both ImageJ and MATLAB (Fig. 2). In brief, confocal z-stacks were edited to remove slices that contained ECs displaying a cobblestone morphology on the surface of the gel (as previously observed by Stamati et al.24). The edited stacks were then blurred with a 2-pixel 3D Gaussian filter and binarized by applying an ImageJ implementation of the minimum cross-entropy thresholding algorithm.25 The “Remove Outliers” ImageJ plugin was used to remove holes with a radius of 4 pixels or less and speckles with a radius of 5 pixels or less.
FIG. 2.
A schematic outlining the computational pipeline used to quantitatively compare network topologies. (A) Z-stacks were acquired on a spinning disk confocal microscope, blurred, filtered, and then skeletonized. The resulting skeleton was analyzed to determine the number of endpoints, branch points, links (i.e., branches), total network length, and link diameter. (B) To validate the pipeline, small sections (100 × 100 pixels) of the hydrogel were processed with this pipeline and the results were nearly identical compared with a manual count. Color images are available online.
The filtered, binarized images were skeletonized and analyzed with a modified MATLAB algorithm that has been previously applied to analyze the quality of osteocyte networks in bone tissue.26 In brief, the filtered skeleton was converted into a network topology that describes the connected capillary-like structures in terms of nodes (i.e., branch/endpoints) and links (vessels or linear clumps of cells). From this topology, the number of branch points, lumen, and the total length of the network were extracted by using a MATLAB script (available upon request). Total network length included the entirety of the vessel network skeleton (i.e., including lumens that were unconnected to the longest connected network). Each z-stack (∼15 MB) required ∼45 s to process on a Lenovo X250 ThinkPad equipped with 8 GB RAM and an Intel Core i5-5200 CPU.
Collagen fibril visualization via confocal reflectance
Collagen (Ibidi) hydrogels were prepared as described previously. The fiber structure of the collagen hydrogels was imaged using an upright confocal laser scanning microscope (Zeiss LSM 510 Meta) equipped with 63 × , NA 1.0 water dipping objective (Zeiss). The 488 nm laser line was used to illuminate the hydrogel and reflected light was acquired by setting the Meta channel to detect wavelength between 475 and 514 nm to work in reflectance mode. The pinhole was set such that each axial slice was 1.5 μm thick, and all images were 512 × 512 pixels. The pixel sizes in the images were 0.280 × 0.280, 0.140 × 0.140, or 0.070 × 0.070 μm2, depending on the zoom factor (1, 2, or 4, respectively). The scan speed was set to 1.6 μs/pixel, and each line was averaged four times to produce the final images. As the reflectance signal is quite strong, care was taken to ensure that detector saturation did not occur.
To determine the total fiber volume of each z-stack, the final images were binarized, and the total number of pixels occupied by fibers was extracted from the resulting histogram. To determine the average fiber orientation, we used the OrientationJ plugin in ImageJ27,28; this plugin generated a distribution of fiber angles by evaluating a structural tensor in the local neighborhood of collagen fibers (we used a Gaussian gradient). To compare these distributions, we condensed this distribution into a single orientation index that scales from 0 (isotropic) to 1 (anisotropic). The orientation index (S) is given by S = 2 < cos2(α) > − 1, where α is the difference between an individual fiber angle and the mean angle of all fibers.29
Bulk rheological measurements on collagen hydrogels
The bulk elastic modulus and viscoelasticity were measured on an Anton Paar MCR101 rheometer with an 8 mm parallel plate geometry. A frequency sweep was performed from 0.5 to 50 Hz to determine the linear range; all subsequent elastic modulus measurements were acquired at 1.99 Hz. To measure the relative viscoelasticity of the collagen hydrogels, a 10% strain was applied in shear and the force needed to maintain this strain was recorded for the duration of the experiment.
Statistical analysis
Statistical analysis was performed using a Student's t-test (two experimental conditions) or a one-way analysis of variance (three or more experimental conditions) followed by a Tukey's multiple comparison test in Prism 6 (GraphPad). Significance is denoted as follows: *p < 0.05, **p < 0.01, ***p < 0.005, and ****p < 0.001. Data are presented as mean ± standard deviation of at least two biological replicates unless indicated otherwise.
Results
Gene expression profiles of differentiating CD34+ iPSC-EPs
To analyze gene expression patterns during the differentiation of iPSCs to iPSC-EPs, we monitored daily changes in the expression of pluripotency, mesoderm, and endothelial markers over 6 days. The expression of the three tested pluripotency genes OCT4 (octamer-binding transcription factor 4), NANOG, and SOX2 (sex determining region Y-box 2) was downregulated by >100-fold 3 days after CHIR99021 induction. Twenty-four hours after treatment with CHIR99021, genes typically associated with mesoderm formation (T (brachyury) and MIXL1) were upregulated by about three orders of magnitude. Four days after CHIR99021 induction, genes usually associated with EPs (CD34, CDH5 [VE-cadherin], KDR [VEGF receptor 2], and CD31) were significantly upregulated. CD34 and CD31 expression levels increased by approximately three orders of magnitude, whereas CDH5 and KDR levels increased by roughly an order of magnitude.
Interestingly, the expression of PDGFRB, a mesenchymal marker, was upregulated by two orders of magnitude at 72 h after CHIR99021 induction. Arterial genes (NOTCH1 [neurogenic locus notch homolog protein 1] and EFNB2 [ephrin-B2]) and venous genes (EPHB4 [ephrin type-B receptor 4] and NRP2 [neuropilin-2]) all increased in expression by approximately a factor of 3, 10, 3, and 15, respectively, on day 5 of differentiation (summarized in Fig. 1B).
To further characterize iPSC-EPs and to control for possible subpopulation heterogeneity, we sorted a subpopulation of CD34-expressing cells (CD34+ iPSC-EPs) and quantified their EP and EC gene expression profiles in comparison with the counterpart subpopulation of cells, expressing low levels of CD34 (CD34low/negative) (Fig. 1C and Supplementary Fig. S1A). Nearly all EP and EC tested markers in the CD34+ EPs were elevated compared with the CD34low/negative subpopulation. Of note, PDGFRB expression was significantly downregulated in the CD34+ subpopulation compared with the CD34low/negative subpopulation.
CD34+ iPSC-EPs are bipotent and can differentiate into iPSC-ECs and iPSC-SMCs
To establish that the observed gene expression levels led to detectable changes in protein expression, we immunostained iPSC-EPs, iPSC-ECs, and iPSC-SMCs for CD34, VE-cadherin, and calponin, respectively. Differentiated iPSC-EPs exhibited a branched, elongated morphology and stained positively for CD34 (Fig. 1D). In addition, iPSC-EPs highly coexpressed CD34 and VE-cadherin, as has been previously reported22 (Supplementary Fig. S1B). After sorting, CD34+ iPSC-EPs were seeded on tissue culture plates coated with laminin, fibronectin, vitronectin, or gelatin and subjected to either proendothelial or pro-SMC medium for 7 days to verify their bipotency. We tested two types of pro-SMC medium: DMEM supplemented with 10% FBS (that has been shown to be conducive to the generation of iPSC-derived pericytes19,30) and commercially available SMGM-2 (PromoCell). As anticipated, CD34+ iPSC-EPs cultured in endothelial medium formed colonies with VE-cadherin-expressing intercellular junctions. In contrast, CD34+ iPSC-EPs cultured in smooth muscle medium stained positively for calponin and did not display tight junctions, as seen with iPSC-ECs (Fig. 1E and Supplementary Fig. S2A). Of interest, we noted that α-SMA could not be used to distinguish iPSC-SMCs from iPSC-ECs, as generated iPSC-ECs expressed detectable levels of α-SMA (Supplementary Fig. S2A). When cultured in DMEM supplemented with 10% FBS alone, CD34+ iPSC-EPs failed to attach and underwent rapid apoptosis within 24 h.
However, when combined with EGM-2, DMEM supplemented with FBS promoted the survival and proliferation of calponin-positive iPSC-SMCs (Supplementary Fig. S2B). In contrast, SMGM-2 alone stimulated the growth of iPSC-SMCs and limited iPSC-EC proliferation (Supplementary Fig. S2C). In all media, the ECM coating did not increase CD34+ iPSC-EP proliferation; however, laminin tended to promote differentiation of an endothelial subtype, whereas fibronectin, vitronectin, and gelatin all markedly increased the iPSC-SMC to total differentiated cell ratio (Supplementary Fig. S2C). In summary, the differentiation of bipotent CD34+ iPSC-EPs into iPSC-ECs and iPSC-SMCs was dependent on cell culture media and ECM substrate.
VEGF and Y-27632 mediate CD34+ iPSC-EP vasculogenesis in collagen hydrogels
To evaluate the vasculogenic potential of CD34+ iPSC-EPs in microenvironments that simulate the native ECM, CD34+ iPSC-EPs were encapsulated in collagen hydrogels, and changes in cellular morphology were monitored daily. We found that ∼81% of the CD34+ iPSC-EPs were viable 1 h postencapsulation, indicating that CD34+ iPSC-EPs can survive the relatively harsh processes of cell dissociation and sorting (Supplementary Fig. S3).
CD34+ iPSC-EPs maintained a rounded morphology for the first 24–48 h after encapsulation. In the days following, the cells began to take on extended morphologies; initial lumen formation was visible 4 days after encapsulation. The lumen started to coalesce and form a highly branched network 6 days after encapsulation (Fig. 3A, Supplementary Video S1). Staining these vessel-like networks with VE-cadherin and F-actin revealed the presence of lumens and highly branched structures (Fig. 3B); interestingly, we observed that calponin-positive cells often remained in the interstitial space between VE-cadherin-positive lumens (Supplementary Fig. S4).
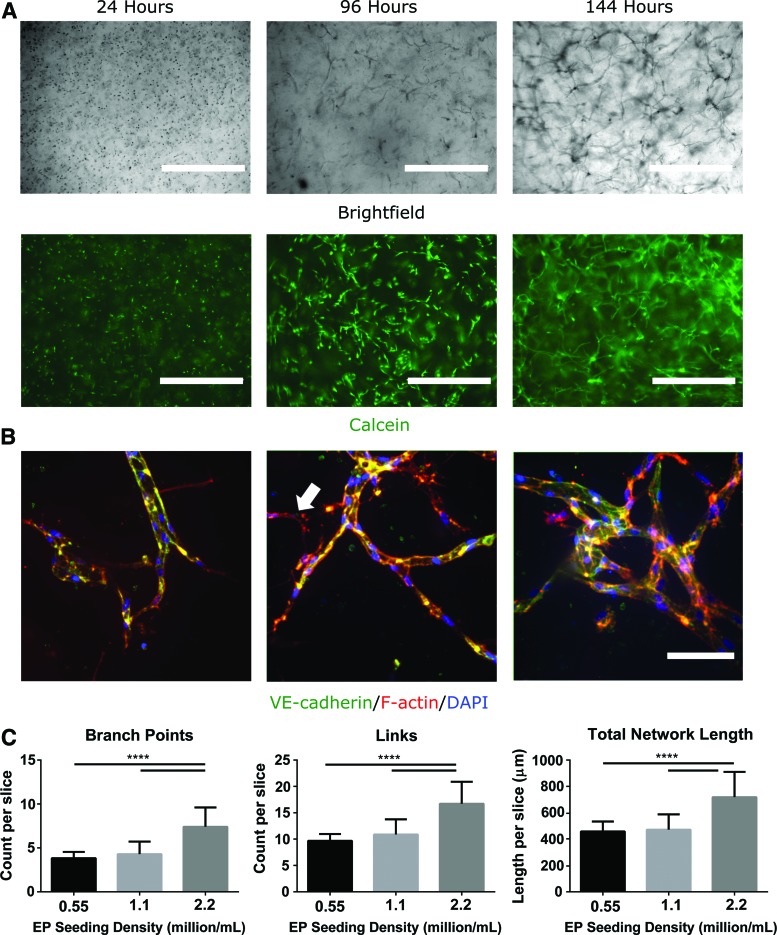
FIG. 3.
CD34+ iPSC-EPs undergo vasculogenesis and self-assemble into a capillary plexus in collagen hydrogels. (A) The assembly of the capillary plexus was tracked with brightfield and epifluorescent microscopy throughout the experiment. Calcein was used to confirm viability. (B) After 1 week in culture, the vascular networks were fixed and stained for VE-cadherin, F-actin, and DAPI. White arrow indicates the presence of SMCs. (C) CD34+ iPSC-EP seeding density was varied from 0.55 to 2 million cells/mL; the resulting networks were evaluated and compared using our computational pipeline. Scale bars are 400 μm (A) and 100 μm (B). ****p < 0.0001. SMCs, smooth muscle cells. Color images are available online.
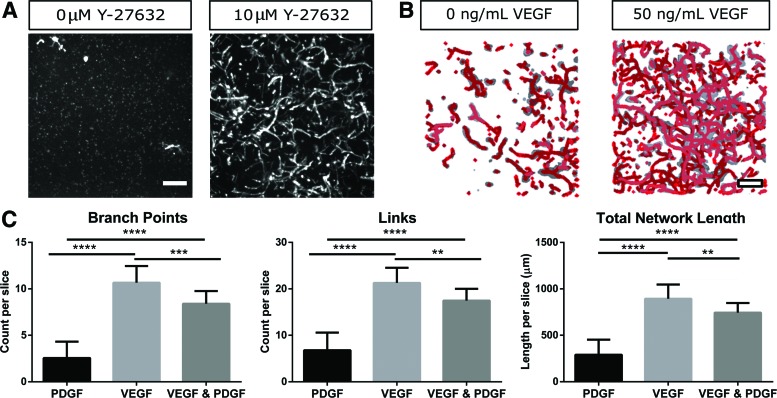
To self-assemble into robust vessel-like networks, CD34+ iPSC-EP-laden hydrogels required culturing in EGM-2 supplemented with exogenous VEGF for the duration of the experiment and ROCK pathway inhibitor (Y-27632) for 24 h. When the sorted CD34+ iPSC-EPs were seeded in hydrogels without being exposed to Y-27632, the progenitors did not undergo an additional morphological transition after 1 week in culture (Fig. 4A). Whereas Y-27632 was necessary for cell survival upon encapsulation within collagen hydrogels, soluble VEGF (50 ng/mL) was observed to be essential for network formation. Cell-laden hydrogels cultured in EGM-2, which contains VEGF at 5 ng/mL, contained only isolated, unconnected lumen (Fig. 4B). Furthermore, the addition of PDGF did not aid vessel growth and led to a slight decrease in vessel network length and connectivity (Fig. 4C).
FIG. 4.
iPSC-EPs require Y-27632 and exogenous VEGF to undergo robust vasculogenesis in collagen hydrogels. CD34+ iPSC-EPs were isolated and embedded in collagen hydrogels at 2.2 million cells/mL (A) with/without Y-27632 for 24 h (B) with/without exogenous VEGF for 1 week (C) with/without VEGF and PDGF. Scale bars are 200 μm. **p < 0.01, ***p < 0.001, ****p < 0.0001. PDGF, platelet-derived growth factor; VEGF, vascular endothelial growth factor. Color images are available online.
To determine the number of cells needed to vascularize a collagen hydrogel, we varied the CD34+ iPSC-EP seeding density from 550,000 cells/mL to 2.2 million cells/mL. We found that increasing this seeding density to 2.2 million cells/mL doubled the length and connectivity of the vascular-like networks compared with the 550,000 cells/mL experimental condition (Fig. 3C); however, further increasing the CD34+ iPSC-EP seeding density resulted in premature degradation of the collagen hydrogel.
Collagen density regulates vascular network length, connectivity, and lumen diameter
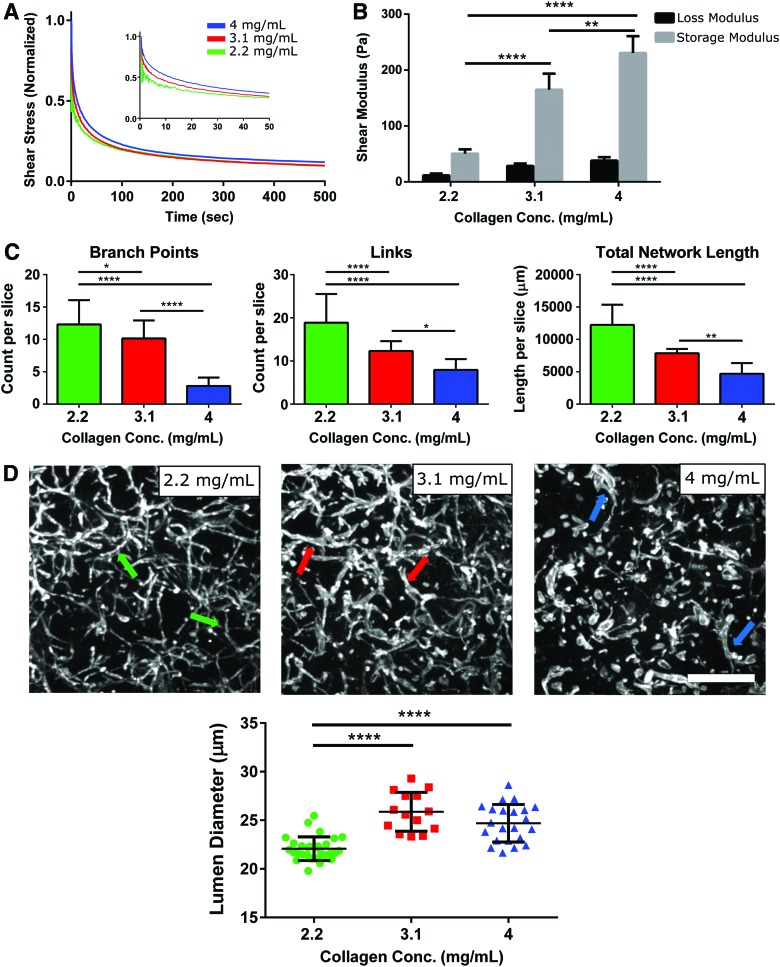
The density of collagen hydrogels has been demonstrated to have a significant impact on the topology of vascular networks in 2D tissue culture plates and murine models.31,32 We hypothesized that varying the density of collagen hydrogels would similarly impact the vasculogenic potential of embedded CD34+ iPSC-EPs. Three collagen concentrations were tested: 2.2, 3.1, and 4 mg/mL. When the density of the collagen hydrogels was lowered below 2 mg/mL, the hydrogels degraded significantly within 72 h and were therefore not usable for extended CD34+ iPSC-EP culture. However, CD34+ iPSC-EPs encapsulated in hydrogels with a higher concentration of collagen remained stable for at least 1 week.
All three hydrogels concentrations exhibited similar viscoelastic properties, as evaluated by their ability to undergo stress relaxation in response to an applied strain; within 10 s, all three hydrogels had dissipated at least half of the initial shear stress (Fig. 5A). As expected, increasing the collagen concentration increased the elastic modulus of the hydrogel; the 2.2, 3.1, and 4 mg/mL conditions displayed a storage modulus of 53 ± 7.35, 165 ± 28.62, and 230 ± 30.07 Pa, respectively (Fig. 5B).
FIG. 5.
Increasing the density of collagen hydrogels limits CD34+ iPSC-EP vasculogenesis and increases average lumen diameter. (A) The stress relaxation profiles of collage hydrogels of different densities were compared by applying 10% strain on an 8 mm parallel plate rheometer (n = 3). (B) The loss and storage moduli of the collagen hydrogels were measured at 1.99 Hz after performing a frequency sweep to ensure modulus linearity (n = 3). (C) CD34+ iPSC-EPs were seeded in collagen hydrogels containing different concentrations of collagen. The resulting vascular networks were evaluated and compared with the computational pipeline. (D) The same pipeline was then used to determine the average lumen diameter in these hydrogels. Scale bar is 400 μm. Arrows indicate distinguishable lumen. *p < 0.05, **p < 0.01, ****p < 0.0001. Color images are available online.
We next examined the impact of collagen density on the collagen fibril microstructure in these hydrogels, as collagen microstructure is correlated with the bulk mechanical properties of the hydrogel33 (Supplementary Fig. S5A). We determined that the collagen fibrils in these hydrogels were not well-aligned, irrespective of density (the orientation index ranged from 0.032 ± 0.013 to 0.084 ± 0.012) (Fig. S5B). In contrast, the total collagen fibril volume increased with increasing collagen density, ranging from ∼16% to ∼31.5% (Supplementary Fig. S5C). Qualitatively, we observed that differentiating CD34+ iPSC-EPs began extending and migrating rapidly in the lower density hydrogels. A nodal analysis of the vascular networks revealed that the 2.2 mg/mL collagen hydrogels contained the highest number of branches (∼12 branch points per planar section), vessels (i.e., links as defined by the algorithm), and total network length (∼12 mm) (Fig. 5C). In addition, the well-connected, longer networks also contained lumens with significantly smaller diameters. It was determined that vessels in 2.2 mg/mL collagen hydrogels were ∼20% smaller than vessels found in 3.1 and 4 mg/mL hydrogels (Fig. 5D).
GM6001 abrogates network formation through dose-dependent matrix metalloprotease inhibition
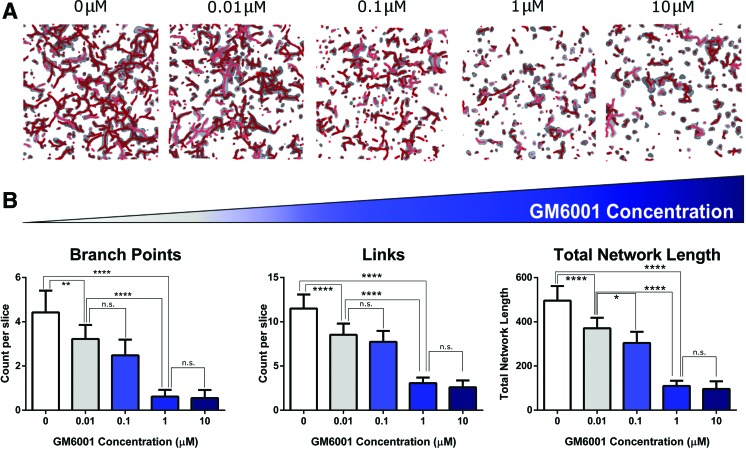
It has been demonstrated that the sprouting of human umbilical vein endothelial cells and iPSC-ECs is highly dependent on the production and proteolytic activity of secreted and membrane-bound matrix metalloproteases (MMPs).21,34 Given that the CD34+ iPSC-EP-laden collagen hydrogels degraded within a week, it was clear that derived CD34+ iPSC-EPs secreted MMPs; to test if derived CD34+ iPSC-EPs secreted the specific class of MMPs necessary for vasculogenesis, we exposed CD34+ iPSC-EPs encapsulated in collagen hydrogels to varying concentrations of GM6001, a broad-spectrum MMP inhibitor.
Micromolar concentrations of GM6001 completely abrogated network formation; cells formed small clumps and did not form enclosed, connected lumen. The total branch points, links, and network length all decreased as the GM6001 concentration was raised from 10 to 10 μM (Fig. 6). Lower concentrations of GM6001 revealed a dose dependency, as viable lumens were visible in both the 10 and 100 nM GM6001-treated samples.
FIG. 6.
The effect of a broad-spectrum MMP inhibitor (GM6001) on CD34+ iPSC-EP vasculogenesis. (A) The concentration of GM6001 was increased from 0.01 to 10 μM; (B) the length and connectivity of the resulting networks were analyzed with the computational pipeline. MMP, matrix metalloprotease. *p < 0.05,**p < 0.01, ****p < 0.0001. NS, not significant. Color images are available online.
Discussion
To date, most studies related to iPSCs-EPs have focused on generating functional iPSC-ECs and interrogating iPSC-EC functionality in 2D assays.35,36 Here, we studied the vasculogenic potential of CD34+ iPSC-EPs in collagen hydrogels to elucidate their self-assembly into 3D vascular networks, which remains poorly understood. We chose to use type I collagen to simulate an ECM-like microenvironment because it displays microscale fibrillarity, readily dissipates stress, contains integrin-binding sequences, and is the most abundant fibrous protein in the ECM.37 In addition, collagen was recently determined to be one of the two bioinks most suitable for the bioprinting of ECs.38 We observed that the ability of CD34+ iPSC-EPs to form lumen-containing vascular networks was sensitive to variations in collagen density, proteolytic activity, and the presence of angiogenic growth factors. To quantify these networks, we developed a computational pipeline that includes algorithms based in ImageJ and MATLAB. The pipeline was able to measure network length, connectivity, and diameter, thereby allowing us to quantify the relative 3D vasculogenic potential of CD34+ iPSC-EPs in response to physical and chemical modulation of their microenvironment. The advantage of our pipeline is that it preserved the z-dimension and thus avoids errors commonly found in methods that compress 3D images to a single plane. Moreover, this pipeline reduces user bias as it is automatically programmed to identify branching points or vessels.
Both primary and pluripotent-derived EPs represent heterogeneous populations; Medina et al. recently reviewed the differences between myeloid angiogenic cells, which indirectly participate in angiogenesis via paracrine action, and endothelial colony forming cells, which can directly incorporate into neovessels.39 We decided to utilize CD34+ iPSC-EPs because these have been shown to give rise to both endothelial and SMCs,22 the building blocks of vasculature.
To further characterize these EPs, we conducted a comprehensive gene expression study to distinguish between unsorted day 5 differentiated iPSC-EPs, sorted CD34+ iPSC-EPs and their counterpart subpopulation displaying very low levels of CD34 expression (CD34low/negative). We found that nearly all tested markers that were elevated in the unsorted EP population were also highly upregulated in the population of CD34+ iPSC-EPs. Of interest, both unsorted iPSC-EPs and the sorted CD34low/negative population had high expression levels of PDGFRB compared with the CD34+ iPSC-EPs (approximately three orders of magnitude difference). Still, when CD34+ iPSC-EPs were exposed to SMGM-2 or a 50:50 mixture of EGM2 and DMEM supplemented with 10% FBS, we observed that these cells differentiated into calponin-expressing SMCs. CD34+ iPSC-EPs also differentiated into VE-cadherin-expressing ECs when exposed to EGM-2, demonstrating their bipotent nature (as previously shown).22,40
We noted that these iPSC-ECs expressed α-SMA, which is a marker that is typically associated with smooth muscle and mesenchymal cells; however, α-SMA expression has previously observed for ECs derived from CD34+ human umbilical cord blood cells.41 Although Patsch et al.42 showed that iPSC-EPs could differentiate into proliferating vascular SMCs in response to 10 ng/mL of PDGF-BB, we noted that differentiated iPSC-ECs were significantly more proliferative compared with differentiated iPSC-SMCs. These results suggest that iPSC-EPs generated from this differentiation protocol (using CHIR99021 and ascorbic acid) may be phenotypically distinct than iPSC-EPs generated by Patsch et al.42 (using CHIR99021 and bone morphogenetic protein 4 [BMP4]). Further work is needed to establish phenotypic differences between subpopulations of EPs generated from different differentiation protocols.
Finally, we noted that both CD34+ iPSC-EP proliferation and the final ratio of iPSC-SMCs to the total number of differentiated CD34+ iPSC-EPs were closely correlated with the type of ECM protein coating used. As expected, laminin most significantly promoted the differentiation of iPSC-ECs; surprisingly, polystyrene (i.e., no coating) was almost as effective, implying that scaling iPSC-ECs production may not require coating with costly ECM proteins. Although fibronectin, vitronectin, and gelatin limited the total number of cells present in each well, all three protein coatings increased the ratio of iPSC-SMCs to total differentiated cells. In summary, we confirmed the bipotent nature of CD34+ iPSC-EPs and demonstrated that their differentiation into iPSC-ECs and iPSC-SMCs is dependent on chemical signaling (e.g., cell culture medium) and integrin signaling (e.g., ECM protein coating).
Upon encapsulation in collagen hydrogels containing Y-27632 and VEGF (50 ng/mL), we noted robust vasculogenesis of CD34+ iPSC-EPs and observed their development into connected, lumen-containing 3D networks. Y-27632 was critical for the initial survival of iPSC-EPs in the collagen hydrogels. Y-27632 is commonly used to inhibit the apoptosis of human pluripotent stem cells that often occurs upon dissociating colonies into single cells.43 Therefore, it is possible that the dissociation step, necessary for cell sorting of the CD34+ population, disrupts cadherins and results in actin hyperactivation in CD34+ iPSC-EPs, leading to downstream apoptosis.44
VEGF has long been recognized as a critical mediator of angiogenic processes and has been identified as one of the most significant mitogens secreted by mural cells.45 Of interest, we noted that complete endothelial media supplemented with VEGF alone was sufficient to form a vascular network in collagen hydrogels; other studies have often incorporated fibroblasts or other supporting cell types to add a biological source of angiogenic growth factor secretion.
Surprisingly, when CD34+ iPSC-EP-laden collagen hydrogels were exposed to PDGF alone or in combination with VEGF, network formation was significantly reduced compared with CD34+ iPSC-EP-laden collagen hydrogels exposed to VEGF alone. In addition, in all cell culture media, relatively few cells differentiated toward a mesenchymal or perivascular lineage compared with ECs. A small number of cells (<1%) displayed an extended phenotype, stained positively for calponin, and were largely present in the interstitial space; we speculate that these cells may contribute to vascular development through paracrine signaling, which may be the focus of a future study.
We hypothesize that the supplemented VEGF needed to drive vascular network formation also biased most CD34+ iPSC-EPs to differentiate along an endothelial lineage, which limited the proliferation of iPSC-SMCs. It is also possible that the simultaneous administration of VEGF and PDGF had an inhibitory effect on vessel formation, as was previously shown by Greenberg et al.46
Most critically, we found that the formation of a complex 3D capillary plexus from CD34+ iPSC-EPs was inhibited by increasing matrix density that also encouraged the formation of wider lumen. Our observations closely match other studies that have reported that increasing the density of collagen31,32,47 or fibrin48,49 reduced primary EC network length and connectivity in in vitro studies and murine vascular models.
A range of mechanisms has been proposed to explain how ECM density impacts vascular morphogenesis, including local diffusivity/pore size,48 matrix anisotropy,50 and relative proteolytic activity.51 In brief, decreasing pore size is thought to slow the diffusion of angiogenic growth factors and thereby reduce the effective concentration presented to embedded ECs. By increasing collagen density, the collagen fibrils become more condensed and aligned, reducing the local isotropy needed for branching. MMPs have been well-studied and shown to be vital for capillary tube formation; an insufficient concentration of soluble MMPs will slow EC migration and block network development.
We focused on elucidating which one of these mechanisms explains the observed sensitivity of CD34+ iPSC-EPs to collagen density. By interrogating hydrated hydrogels with confocal reflectance imaging, we observed the native collagen fibril microstructure and determined that pore size (i.e., fiber density), not fiber alignment, varied significantly among the different density hydrogels. The phenomenon that correlates increasing collagen density with increased collagen fiber orientation seems to be most relevant at fiber densities of 60% or greater52; as our measured fiber densities ranged from ∼16% (2.2 mg/mL) to ∼31.5% (4 mg/mL), we did not observe a correlation between the calculated orientation index and collagen density.
The increase in storage modulus that we observed among the varying density collagen hydrogels is consistent with the scaling expected for densely cross-linked semiflexible collagen networks ([concentrationprotein]2.5).53 Two notable studies have used ribose-mediated glycation and an interpenetrating network of GelMA and alginate to decouple the impact of stiffness from the effect of varying matrix density on sprouting angiogenesis in 3D microenvironments54,55; however, the two studies reached opposing conclusions. Therefore, the effect of stiffness on the vasculogenesis of CD34+ iPSC-EPs in 3D microenvironments remains in need of further elucidation.
To determine the relative proteolytic activity of CD34+ iPSC-EPs, we discovered that adding a broad-spectrum MMP inhibitor to CD34+ iPSC-EP-laden collagen hydrogels nearly abrogated lumen development. We speculate that this inhibition is closely tied to the effect of varying density; as density increases, soluble MMPs may experience limited diffusion through smaller pores, thereby limiting proteolytic activity and matrix degradation. We conclude that CD34+ iPSC-EPs can differentiate into ECs that secret MMPs essential for vascular development.
In conclusion, we have found that a low concentration of collagen that maintained the structural integrity of the hydrogel, coupled with the addition of exogenous VEGF and Y-27632, encouraged robust, 3D capillary plexus formation from CD34+ iPSC-EPs. Pore size and proteolytic activity partially guide these morphogenic processes in collagen hydrogels. In light of these findings, CD34+ iPSC-EPs must continue to be evaluated as a candidate cell source for vascular modeling and therapy; it is of particular interest to track their downstream differentiation into ECs of different subtypes (e.g., arterial vs. venous) and the role this differentiation plays in vascular development.
The cell–matrix interactions discussed in this study underscore the importance of understanding the role of mechanoregulation on vasculogenesis so that ECM-mimicking angiogenic biomaterials can be effectively deployed in the clinic.
Supplementary Material
Acknowledgments
The authors acknowledge Prof. Jeanne Stachowiak (The University of Texas at Austin) for her technical advice on the acquisition of high-resolution confocal images. The authors thank Christine Wei (The University of Texas at Austin) and Adam Jerome Poole (Stanford University) for their help in maintaining lab iPSC cultures. The authors are also grateful for discussions with Samuel Mihelic (The University of Texas at Austin), Dr. Alicia Allen (The University of Texas at Austin), Dr. Julie Rytlewski (Adaptive Biotech), and Dr. Leon Bellan (Vanderbilt University) on the computational analysis of 3D networks. Finally, we thank Dr. Xiaoping Bao (University of California, Berkeley) and Prof. Sean Palecek (University of Wisconsin-Madison) for their advice on differentiating iPSCs into iPSC-EPs.
The authors gratefully acknowledge the financial support of the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health (EB007507, awarded to C.C.), the Alexander von Humboldt Foundation (awarded to S.K.), the Deutsche Forschungsgemeinschaft (#PA:252611-1, awarded to S.H.P.), and the American Heart Association (15SDG25740035, awarded to J.Z.).
Disclosure Statement
No competing financial interests exist.
Supplementary Material
References
- 1. Bogorad M.I., DeStefano J., Karlsson J., Wong A.D., Gerecht S., and Searson P.C. Review: in vitro microvessel models. Lab Chip 15, 4242, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rouwkema J., and Khademhosseini A. Vascularization and angiogenesis in tissue engineering: beyond creating static networks. Trends Biotechnol 34, 733, 2016 [DOI] [PubMed] [Google Scholar]
- 3. Kant R.J., and Coulombe K.L.K. Integrated approaches to spatiotemporally directing angiogenesis in host and engineered tissues. Acta Biomater 69, 42, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Asahara T., Murohara T., Sullivan A., et al. Isolation of putative progenitor endothelial cells for angiogenesis. Science 275, 964, 1997 [DOI] [PubMed] [Google Scholar]
- 5. Urbich C., and Dimmeler S. Endothelial progenitor cells: characterization and role in vascular biology. Circ Res 95, 343, 2004 [DOI] [PubMed] [Google Scholar]
- 6. Schmidt D., Breymann C., Weber A., et al. Umbilical cord blood derived endothelial progenitor cells for tissue engineering of vascular grafts. Ann Thorac Surg 78, 2094, 2004 [DOI] [PubMed] [Google Scholar]
- 7. Takahashi T., Kalka C., Masuda H., et al. Ischemia- and cytokine-induced mobilization of bone marrow-derived endothelial progenitor cells for neovascularization. Nat Med 5, 434, 1999 [DOI] [PubMed] [Google Scholar]
- 8. Sharpe E.E., Teleron A.A., Li B., et al. The origin and in vivo significance of murine and human culture-expanded endothelial progenitor cells. Am J Pathol 168, 1710, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rafii S., and Lyden D. Therapeutic stem and progenitor cell transplantation for organ vascularization and regeneration. Nat Med 9, 702, 2003 [DOI] [PubMed] [Google Scholar]
- 10. Chavakis E., Urbich C., and Dimmeler S. Homing and engraftment of progenitor cells: a prerequisite for cell therapy. J Mol Cell Cardiol 45, 514, 2008 [DOI] [PubMed] [Google Scholar]
- 11. Hristov M., Fach C., Becker C., et al. Reduced numbers of circulating endothelial progenitor cells in patients with coronary artery disease associated with long-term statin treatment. Atherosclerosis 192, 413, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Choi J.H., Kim K.L., Huh W., et al. Decreased number and impaired angiogenic function of endothelial progenitor cells in patients with chronic renal failure. Arterioscler Thromb Vasc Biol 24, 1246, 2004 [DOI] [PubMed] [Google Scholar]
- 13. Fadini G.P., Miorin M., Facco M., et al. Circulating endothelial progenitor cells are reduced in peripheral vascular complications of type 2 diabetes mellitus. J Am Coll Cardiol 45, 1449, 2005 [DOI] [PubMed] [Google Scholar]
- 14. Millman J.R., Xie C., Van Dervort A., Gürtler M., Pagliuca F.W., and Melton D.A. Generation of stem cell-derived β-cells from patients with type 1 diabetes. Nat Commun 7, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bao X., Lian X., and Palecek S.P. Directed Endothelial Progenitor Differentiation from Human Pluripotent Stem Cells Via Wnt Activation Under Defined Conditions. In: Barrett Q., and Lum L., eds. Wnt Signaling: Methods and Protocols. New York, NY: Humana Press, 2016, pp. 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoo C.H., Na H., Lee D., et al. Endothelial progenitor cells from human dental pulp-derived iPS cells as a therapeutic target for ischemic vascular diseases. Biomaterials 34, 8149, 2013 [DOI] [PubMed] [Google Scholar]
- 17. Kusuma S., Macklin B., and Gerecht S. Derivation and network formation of vascular cells from human pluripotent stem cells. Methods Mol Biol 1204, 1, 2014 [DOI] [PubMed] [Google Scholar]
- 18. Prasain N., Lee M.R., Vemula S., et al. Differentiation of human pluripotent stem cells to cells similar to cord-blood endothelial colony-forming cells. Nat Biotechnol 32, 1151, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kusuma S., Shen Y.-I., Hanjaya-Putra D., Mali P., Cheng L., and Gerecht S. Self-organized vascular networks from human pluripotent stem cells in a synthetic matrix. Proc Natl Acad Sci U S A 110, 12601, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan X.Y., Black R., Dickerman K., et al. Three-Dimensional Vascular Network Assembly from Diabetic Patient-Derived Induced Pluripotent Stem Cells. Arterioscler Thromb Vasc Biol 35, 2677, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bezenah J.R., Kong Y.P., and Putnam A.J. Evaluating the potential of endothelial cells derived from human induced pluripotent stem cells to form microvascular networks in 3D cultures. Sci Rep 8, 2671, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lian X., Bao X., Al-Ahmad A., et al. Efficient differentiation of human pluripotent stem cells to endothelial progenitors via small-molecule activation of WNT signaling. Stem Cell Rep 3, 804, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Artym V., and Matsumoto K. Imaging cells in three-dimensional collagen matrix. Curr Protoc Cell Biol 2010 [Epub ahead of print]; DOI: 10.1002/0471143030.cb1018s48.Imaging [DOI] [PMC free article] [PubMed]
- 24. Stamati K., Priestley J.V., Mudera V., and Cheema U. Laminin promotes vascular network formation in 3D in vitro collagen scaffolds by regulating VEGF uptake. Exp Cell Res 327, 68, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li C.H., and Tam P.K.S. An iterative algorithm for minimum cross entropy thresholding. Pattern Recognit Lett 19, 771, 1998 [Google Scholar]
- 26. Kerschnitzki M., Kollmannsberger P., Burghammer M., et al. Architecture of the osteocyte network correlates with bone material quality. J Bone Miner Res 28, 1837, 2013 [DOI] [PubMed] [Google Scholar]
- 27. Rezakhaniha R., Agianniotis A., Schrauwen J.T.C., et al. Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomech Model Mechanobiol 11, 461, 2012 [DOI] [PubMed] [Google Scholar]
- 28. Püspöki Z., Storath M., Sage D., and Unser M. Transforms and operators for directional bioimage analysis : a survey. Adv Anat Embryol Cell Biol 219, 69, 2016 [DOI] [PubMed] [Google Scholar]
- 29. Ferdman A.G., and Yannas I.V. Scattering of light from histologic sections: a new method for the analysis of connective tissue. J Invest Dermatol 100, 710, 1993 [DOI] [PubMed] [Google Scholar]
- 30. Orlidge A., and D'Amore P.A. Inhibition of capillary endothelial cell growth by pericytes and smooth muscle cells. J Cell Biol 105, 1455, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edgar L.T., Underwood C.J., Guilkey J.E., Hoying J.B., and Weiss J.A. Extracellular matrix density regulates the rate of neovessel growth and branching in sprouting angiogenesis. PLoS One 9, e85178, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Critser P.J., Kreger S.T., Voytik-Harbin S.L., and Yoder M.C. Collagen matrix physical properties modulate endothelial colony forming cell-derived vessels in vivo. Microvasc Res 80, 23, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jansen K.A., Licup A.J., Sharma A., Rens R., MacKintosh F.C., and Koenderink G.H. The role of network architecture in collagen mechanics. Biophys J 114, 2665, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ghajar C.M., Kachgal S., Kniazeva E., et al. Mesenchymal cells stimulate capillary morphogenesis via distinct proteolytic mechanisms. Exp Cell Res 316, 813, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Halaidych O.V., Freund C., van den Hil F., et al. Inflammatory responses and barrier function of endothelial cells derived from human induced pluripotent stem cells. Stem Cell Rep 10, 1642, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Orlova V.V., Drabsch Y., Freund C., et al. Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts. Arterioscler Thromb Vasc Biol 34, 177, 2014 [DOI] [PubMed] [Google Scholar]
- 37. Frantz C., Stewart K.M., and Weaver V.M. The extracellular matrix at a glance. J Cell Sci 123, 4195, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benning L., Gutzweiler L., Tröndle K., et al. Assessment of hydrogels for bioprinting of endothelial cells. J Biomed Mater Res 106, 935, 2018 [DOI] [PubMed] [Google Scholar]
- 39. Medina R.J., Barber C.L., Sabatier F., et al. Endothelial progenitors : a consensus statement on nomenclature. Stem Cells Transl Med 6, 1316, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bao X., Lian X., Dunn K.K., et al. Chemically-defined albumin-free differentiation of human pluripotent stem cells to endothelial progenitor cells. Stem Cell Res 15, 122, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu X., Dunn J., Dickinson A.M., Gillespie J.I., and Baudouin S.V. Smooth muscle alpha-actin expression in endothelial cells derived from CD34+ human cord blood cells. Stem Cells Dev 13, 521, 2004 [DOI] [PubMed] [Google Scholar]
- 42. Patsch C., Challet-Meylan L., Thoma E.C., et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol 17, 994, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watanabe K., Ueno M., Kamiya D., et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol 25, 681, 2007 [DOI] [PubMed] [Google Scholar]
- 44. Ohgushi M., Matsumura M., Eiraku M., et al. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 7, 225, 2010 [DOI] [PubMed] [Google Scholar]
- 45. Evensen L., Micklem D.R., Blois A., et al. Mural cell associated VEGF is required for organotypic vessel formation. PLoS One 4, e5798, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Greenberg J.I., et al. A role for VEGF as a negative regulator of pericyte function and vessel maturation. Nature 457, 809, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shamloo A., and Heilshorn S.C. Matrix density mediates polarization and lumen formation of endothelial sprouts in VEGF gradients. Lab Chip 10, 3061, 2010 [DOI] [PubMed] [Google Scholar]
- 48. Ghajar C.M., Chen X., Harris J.W., et al. The effect of matrix density on the regulation of 3-D capillary morphogenesis. Biophys J 94, 1930, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kniazeva E., and Putnam A.J. Endothelial cell traction and ECM density influence both capillary morphogenesis and maintenance in 3-D. AJP Cell Physiol 297, C179, 2009 [DOI] [PubMed] [Google Scholar]
- 50. Bauer A., Gu L., Kwee B., et al. Hydrogel substrate stress-relaxation regulates the spreading and proliferation of mouse myoblasts. Acta Biomater 62, 82, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ghajar C.M., Blevins K.S., Hughes C.C., George S.C., and Putnam A.J. Mesenchymal stem cells enhance angiogenesis early matrix metalloproteinase upregulation. Tissue Eng 12, 2875, 2006 [DOI] [PubMed] [Google Scholar]
- 52. Bauer A.L., Jackson T.L., and Jiang Y. Topography of extracellular matrix mediates vascular morphogenesis and migration speeds in angiogenesis. PLoS Comput Biol 5, e1000445, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. MacKintosh F.C., Käs J., and Janmey P.A. Elasticity of semiflexible biopolymer networks. Phys Rev Lett 75, 4425, 1995 [DOI] [PubMed] [Google Scholar]
- 54. Mason B.N., Starchenko A., Williams R.M., Bonassar L.J., and Reinhart-King C.A. Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater 9, 4635, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Berger A.J., Linsmeier K.M., Kreeger P.K., and Masters K.S. Decoupling the effects of stiffness and fiber density on cellular behaviors via an interpenetrating network of gelatin-methacrylate and collagen. Biomaterials 141, 125, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.