SUMMARY
The intestinal microbiota produces tens of thousands of metabolites. Here, we used host sensing of small molecules by G-protein coupled receptors (GPCRs) as a lens to illuminate bioactive microbial metabolites that impact host physiology. We screened 144 human gut bacteria against the non-olfactory GPCRome and identified dozens of bacteria that activated both well-characterized and orphan GPCRs, including strains that converted dietary histidine into histamine and shaped colonic motility; a prolific producer of the essential amino acid L-Phe, which we identified as an agonist for GPR56 and GPR97; and a species that converted L-Phe into the potent psychoactive trace amine phenethylamine, which crosses the blood-brain barrier and triggers lethal phenethylamine poisoning after monoamine oxidase inhibitor administration. These studies establish an orthogonal approach for parsing the microbiota metabolome and uncover multiple biologically relevant host-microbiota metabolome interactions.
In Brief
Metabolites produced by the human microbiota are able to function as agonists for a range of G protein coupled receptors, making metabolome screening a useful tool to both de-orphan human GPCRs and identify metabolic exchanges between commensal microbes in the gut with effects on host physiology.
INTRODUCTION
The human gut microbiota produces thousands of unique small molecules that can potentially affect nearly all aspects of human physiology, from regulating immunity in the gut to shaping mood and behavior (Donia and Fischbach, 2015; Husted et al., 2017; Smith, 2013; Tan, 2017; Yano et al., 2015). These metabolites can act locally in the intestine or can accumulate up to millimolar concentrations in the serum (Donia and Fischbach, 2015; Fischbach, 2018; Pedersen et al., 2016; Perry et al., 2016). Recent studies employing state-of-the-art genomic and metabolomic approaches have begun to reveal the enormously complex intra- and inter-species microbial chemistries that potentially impinge on host physiology, as well as the impact of gut microbes on the processing of dietary small molecules and medical drugs (Dodd et al., 2017; Guo et al., 2017; Haiser et al., 2013; Larsbrink et al., 2014; Milshteyn et al., 2018). In addition, they underscore the importance of continuing to develop new approaches to explore the bioactive microbiota metabolome (Fischbach, 2018).
G-protein coupled receptors (GPCRs) are the largest family of membrane proteins encoded in the human genome (including over 350 conventional non-olfactory GPCRs), are critical sensors of diverse small molecules, and regulate various aspects of host physiology, including vision, mood, pain, and immunity (Wacker et al., 2017). Specific GPCRs are also known to sense microbial metabolites, such as microbiota-derived short-chain fatty acids (Husted et al., 2017; Tan, 2017), and recent studies have continued to reveal novel microbiota-derived GPCR ligands that can shape host physiology (Cohen et al., 2017; Cohen et al., 2015). Thus, the microbiota metabolome is a rich source of potential GPCR ligands.
Here, by building on recent developments in high-throughput screening of the complete GPCRome and activity-guided microbial metabolite identification approaches (Donia and Fischbach, 2015; Fischbach, 2018; Kroeze et al., 2015; Milshteyn et al., 2018), we developed a pipeline to screen human gut microbes for the ability to produce ligands that activate human GPCRs. In so doing, we established an orthogonal approach for elucidating biologically-relevant microbiota metabolite-host interactions and, in the process, uncovered multiple diet-microbe-host and microbe-microbe-host metabolic axes that shape both local and systemic host physiology.
RESULTS
A forward chemical genetic screen to identify bioactive microbiota metabolites
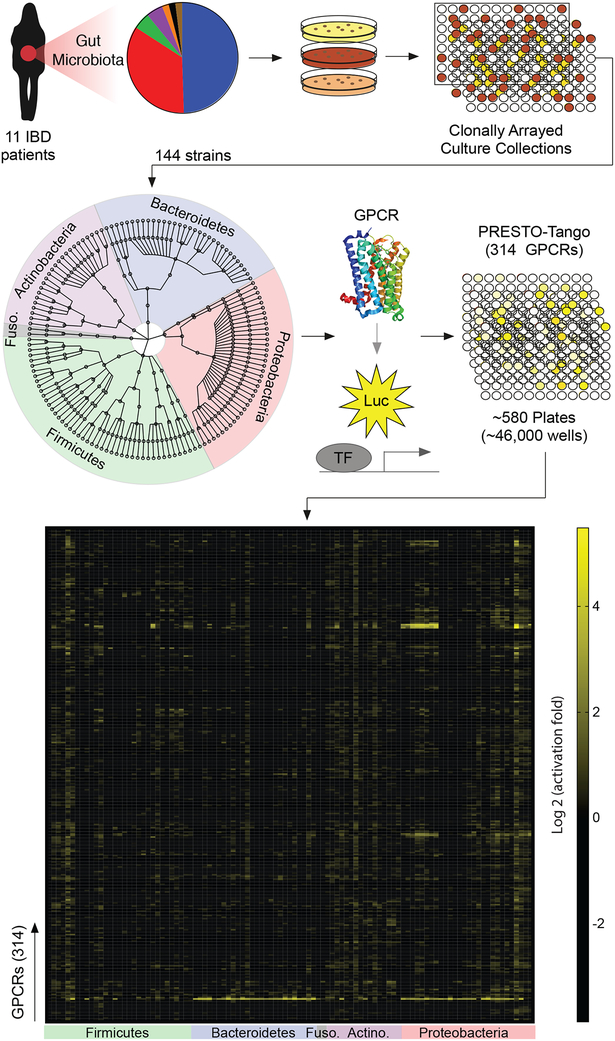
We set out to establish a high-throughput screening system to identify specific human gut microbes that produce agonists or antagonists of conventional GPCRs. We developed a pipeline for parsing the microbiota metabolome based on the GPCR screening technology Parallel Receptor-ome Expression and Screening via Transcriptional Output-Tango (PRESTO-Tango) (Kroeze et al., 2015). This technology leverages the Tango β-arrestin recruitment assay to simultaneously measure the activation of nearly all non-olfactory GPCRs (Figure 1, S1A, B) (Barnea et al., 2008; Kroeze et al., 2015). We thus proceeded to exploit this assay to perform a broad-ranging screen of bioactive metabolites produced by diverse members of the human gut microbiota.
Figure 1. A forward chemical genetic screen identifies human gut microbes that activate GPCRs.
We isolated 144 unique human gut bacteria spanning five phyla, nine classes, eleven orders, and twenty families from 11 inflammatory bowel disease patients via high-throughput anaerobic culturomics and massively barcoded 16S rRNA gene sequencing. Bacterial isolates were grown in monoculture in a medium specialized for the cultivation of human gut microbes (gut microbiota medium) and supernatants from individual bacterial monocultures were screened against the near-complete non-olfactory GPCRome (314 conventional GPCRs) using Parallel Receptor-ome Expression and Screening via Transcriptional Output-Tango (PRESTO-Tango).
We previously assembled personalized gut microbiota culture collections from eleven inflammatory bowel disease patients through high-throughput anaerobic culture methods and next-generation sequencing (Palm et al., 2014). This collection yielded 144 unique bacterial isolates from five phyla, nine classes, eleven orders, and twenty families, as well as multiple strains that were assigned to the same species (Table S1). We cultured all members of our collection individually in a medium specialized for the cultivation of gut commensals (gut microbiota medium; GMM) (Goodman et al., 2011) and screened their supernatants for activation or inhibition of nearly all conventional GPCRs (as compared to media alone) using PRESTO-Tango (see methods for details) (Figure 1, 2).
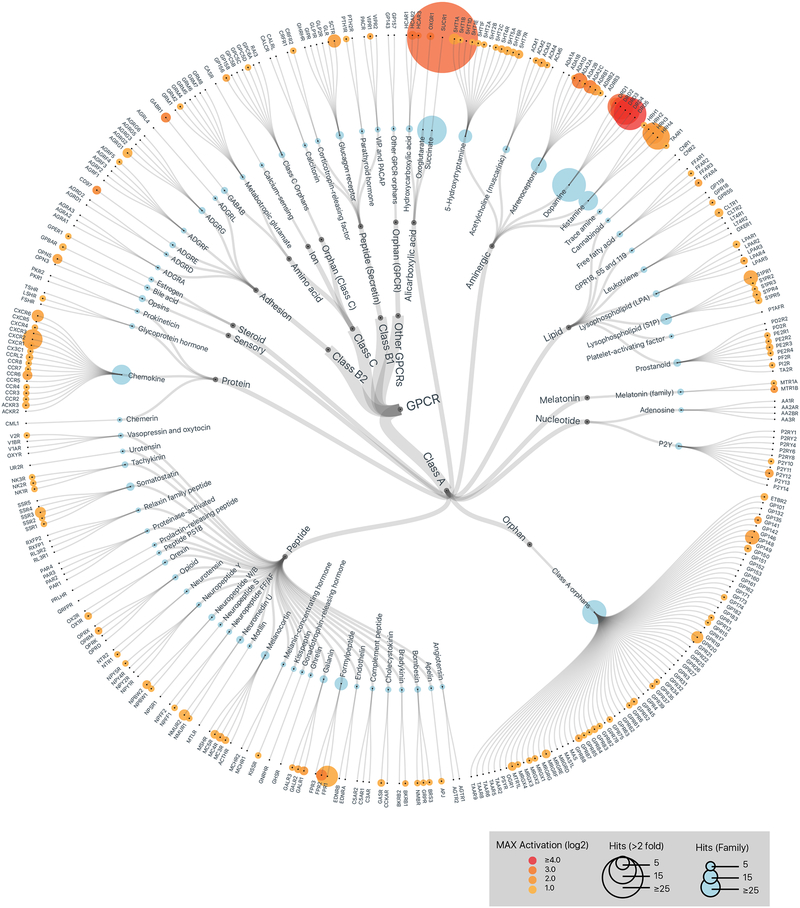
Figure 2. Members of the human gut microbiota produce metabolites that activate diverse human GPCRs.
GPCR activation by metabolomes from a human gut microbiota culture collection consisting of 144 strains isolated from 11 IBD patients. Data is displayed on a hierarchical tree of GPCRs organized by class, ligand type, and receptor family. Color intensity represents the maximum magnitude of activation (log 2) over background (gut microbiota medium alone) across the complete data set. Radii of the circles at each tip represent the number of strains that activated a given receptor or receptor family by more than two-fold over background. Graphics were generated in collaboration with visavisllc using d3.js.
Human gut microbes produce compounds that activate both well-characterized and orphan GPCRs
PRESTO-Tango screening revealed that bacterial-derived metabolite mixtures activated both well-characterized GPCRs as well as orphan receptors, including GPCRs from nearly every class (Figure 2). One specific pattern of activation tracked closely with gross phylogeny—Bacteroidetes and Proteobacteria potently activated the succinate receptor (Sucr1), while Firmicutes, Fusobacteria and Actinobacteria largely failed to activate this receptor (Figure 1 and Table S2). However, most activation patterns did not correlate with phylogeny, and multiple bacterial strains assigned to the same species exhibited unique GPCR agonist activities (Table S2). GMM alone also activated select GPCRs when compared to PBS, and supernatants from specific microbes sometimes reversed these effects either due to bacterial consumption of GPCR ligands in the media or bacterial production of GPCR antagonists (Table S3). Given the exquisite sensitivity of PRESTO-Tango, the veracity of all individual hits will need to be confirmed using alternative methods. Nonetheless, these data demonstrate that human gut microbes produce a remarkable array of GPCR ligands.
Human gut microbes produce compounds that activate aminergic receptors
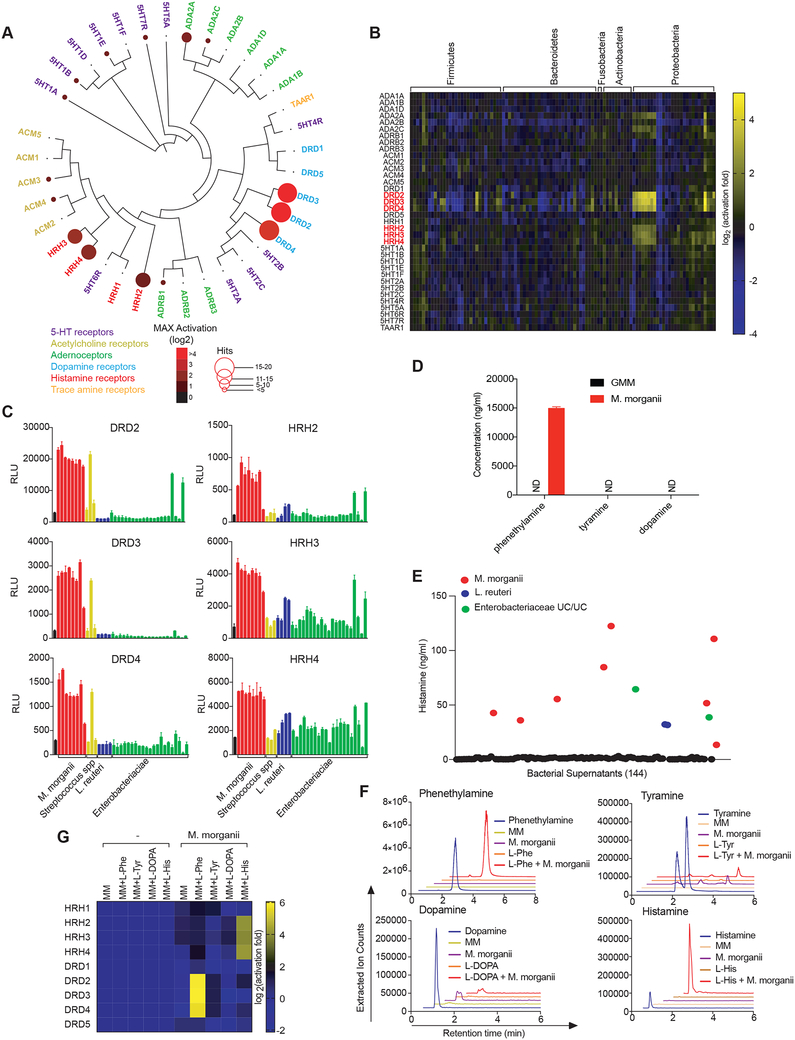
Besides the succinate receptor, the next most prevalent class of GPCRs activated by gut commensals was the aminergic receptors, which are expressed in diverse tissues and cell types and regulate a variety of core physiological processes ranging from neurotransmission to immunity (Figure 2) (Albuquerque et al., 2009; Beaulieu and Gainetdinov, 2011; Thurmond et al., 2008). More than a dozen commensal supernatants activated the dopamine (DRDs) or histamine (HRHs) receptors (Figure 3A, S2). For example, ten Proteobacteria strains activated both DRDs and HRHs, including all eight Morganella morganii strains in our collection (Figure 3B, C, S2). In contrast, two Lactobacillus reuteri strains activated HRHs, while two distinct L.reuteri strains failed to activate HRHs despite displaying similar growth kinetics (Figure 3C, S2). Finally, one Streptococcus strain, but not two related isolates of Streptococcus, activated DRD2–4, and two unclassified Enterobacteriaceae strains activated HRH1–4 and DRD2 but failed to activate other DRDs (Figure 3C, S2).
Figure 3. Diverse human gut bacteria activate aminergic GPCRs.
(A) Activation of aminergic GPCRs by metabolomes from a human gut microbiota culture (see Figure 1). GPCR activation was measured by PRESTO-Tango. Screening results are displayed on a phylogenetic tree of aminergic GPCRs. Color intensity represents magnitude of activation over media alone and radii of the circles represents the number of bacteria that activated a given GPCR by more than two-fold over media alone.
(B) Heatmap depicting the activation of aminergic GPCRs by metabolites from a human gut microbiota culture collection as measured by PRESTO-Tango. Fold induction over stimulation with bacterial media alone is depicted on a log2 scale.
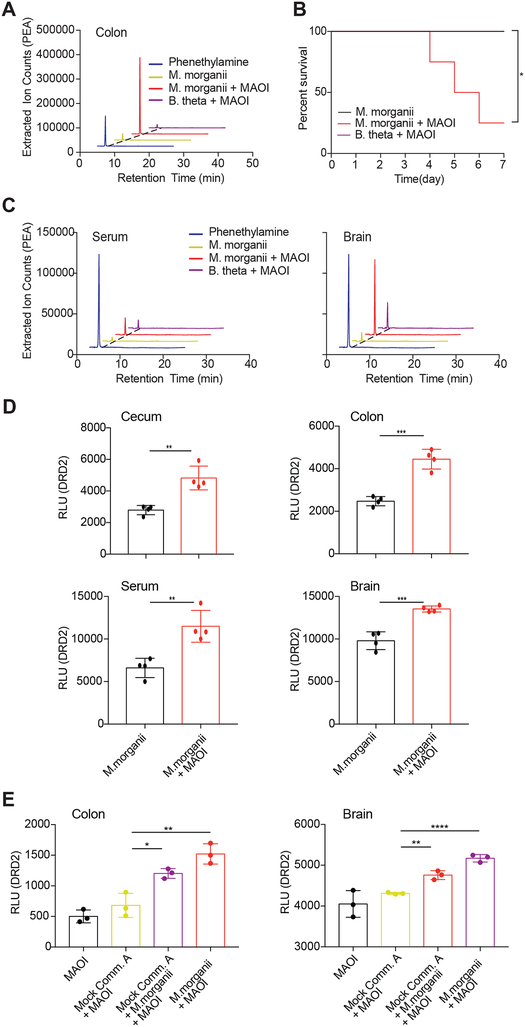
(C) Activation of DRD2–4 and HRH2–4 by select species and strains as measured by Tango assays.
(D) Quantification of dopamine, phenethylamine and tyramine production by M. morganii. Supernatants from 24-hour cultures of M. morganii C135 in gut microbiota medium were analyzed by Triple Quadrupole-Mass Spectrometry (QQQ-MS/MS).
(E) Quantification of histamine production by 144 isolates of human gut bacteria by ELISA (48 hr. cultures).
(F) Mass spectrometric traces of metabolite production by M. morganii C135. M. morganii was cultured in minimal medium (MM) with or without additional L-Phe, L-His, L-Tyr or L-DOPA for 48 hours. Metabolite production was analyzed by Liquid Chromatography-Mass Spectrometry (LC-MS).
(G) M. morganii-derived phenethylamine and histamine activate DRD2–4 and HRH2–4, respectively. M. morganii C135 were cultured as described in F and supernatants were screened for activity against DRDs and HRHs by PRESTO-Tango.
Data for all panels other than A and B are representative of at least three independent experiments. Data are presented as mean ± SEM. n=3 replicates per group (C-G).
M. morganii was previously reported to produce various biogenic amines, including dopamine, tyramine, and phenethylamine (PEA) (Kim et al., 2000; Özoğul, 2004). We noted that all M. morganii supernatants activated DRD2–4, but not DRD1 and 5 (Figure 3A, B, S2). In contrast, dopamine itself activated all five dopamine receptors (Figure S1A). Therefore, we suspected that M. morganii might produce a metabolite that is structurally related to dopamine and can act as a selective ligand for DRD2–4 but not DRD1 or 5 (Figure S3A). We examined the ability of all possible upstream and downstream metabolites in the mammalian dopamine pathway to activate DRDs and found that PEA and tyramine showed identical activation patterns to M. morganii supernatant (Figure S3A–C). Accordingly, metabolomic analyses revealed that M. morganii produced only trace amounts of dopamine and no detectable tyramine, but instead secreted significant quantities of the potent trace amine PEA which, unlike dopamine and tyramine, can readily cross the blood-brain barrier (Figure 3D, S3D, E) (Oldendorf, 1971).
We next used the cyclic AMP response element-secreted human placental alkaline phosphatase (CRE-SEAP) assay to examine whether PEA and related chemicals also activate G proteins (in addition to β-arrestin) downstream of DRDs (Durocher et al., 2000). To facilitate use of the CRE-SEAP assay for GPCRs that couple to G proteins other than Gαs, such as DRD2–4, we used a Gαs-Gαo fusion protein (a kind gift of Stephen Liberles) to redirect DRD2–4 signaling to Gαs (Liberles and Buck, 2006). PEA activated G protein signaling downstream of all five dopamine receptors, which suggests that PEA is a full agonist for DRD2–4 and may be a biased agonist for DRD1 and 5 (Figure S4A, B).
Previous reports have also suggested that M. morganii produces histamine (Özoğul, 2004). We confirmed that M. morganii secreted significant amounts of histamine by ELISA and that our M. morganii strains encode a previously described histidine decarboxylase; furthermore, 48 of 49 previously deposited M. morganii strains also encoded this histidine decarboxylase (Figure 3E and Table S4, S5). Two L. reuteri strains and two Enterobacteriaceae strains from our collections also secreted histamine (Figure 3E). Based on whole genome sequencing, both the histamine-producing and non-histamine-producing strains of L. reuteri encoded an identical histidine decarboxylase proenzyme (Table S4, S5). Together, these data reveal that M. morganii secretes high levels of PEA, which acts as a potent dopamine receptor agonist, and that M. morganii and select strains of L. reuteri secrete histamine.
In mammals, PEA, dopamine, and tyramine are produced via decarboxylation of L-Phe, LDOPA, and L-Tyr, respectively, by the aromatic L-amino acid decarboxylase (AADC; Figure S3A) (Lovenberg, 1962). Thus, we tested whether M. morganii would similarly process these amino acids into their respective biogenic amines using a minimal medium (MM) lacking L-Phe, L-DOPA, L-Tyr, and L-His. Despite normal M. morganii growth in this medium, we could not detect any production of PEA, tyramine, dopamine, or histamine (Figure 3F). However, supplementation with L-Phe or L-His led to the production of high levels of PEA or histamine, and activation of DRD2–4 or HRH2–4 (Figure 3F, G). In contrast, supplementation with LDOPA or L-Tyr failed to lead to the production of dopamine or tyramine, or activation of DRDs (Figure S4C). Thus, unlike mammalian AADC, M. morganii selectively converts L-Phe into PEA and cannot efficiently convert L-DOPA or L-Tyr into dopamine or tyramine. While it is currently unclear which genes are involved in production of PEA by M. morganii, whole genome sequencing revealed the presence of at least 17 decarboxylases that are shared between the two strains of M. morganii that we sequenced (Table S4, S5).
Microbiota-derived histamine promotes increased colonic motility through activation of the histamine receptors
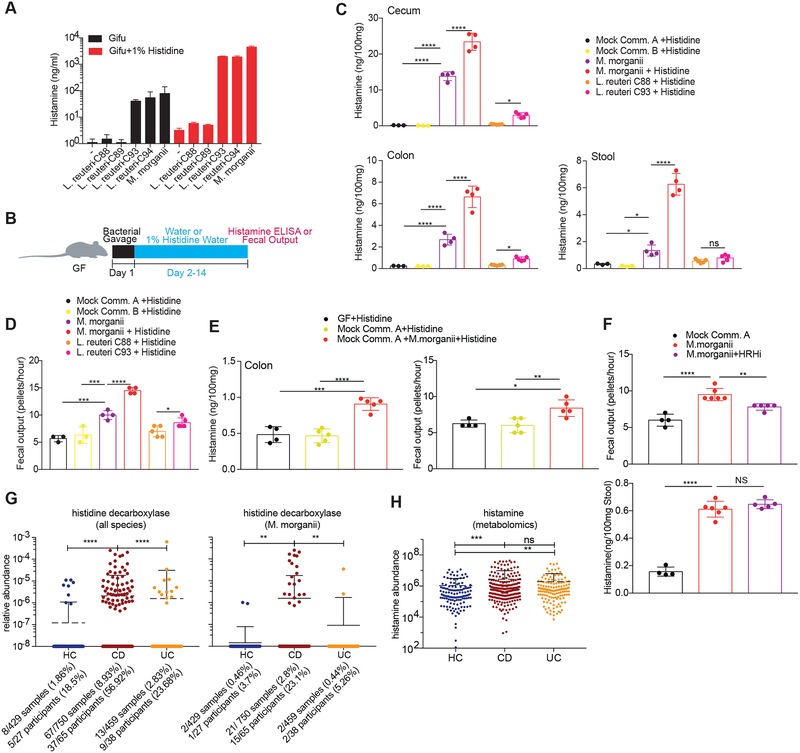
All M. morganii strains and two L. reuteri strains in our collection generated histamine in vitro and supplementation with additional L-His significantly increased histamine production by these strains; in contrast, two distinct strains of L. reuteri failed to produce histamine regardless of supplementation with L-His (Figure 4A). To test whether M. morganii can also produce histamine in vivo, we colonized germ-free mice with two distinct mock communities containing 9 or 10 diverse human gut microbes or with M. morganii C135 with or without supplementation of 1% L-His in the drinking water to approximate an L-His-rich diet (e.g., a meat-heavy diet) (Figure 4B). In addition, we monocolonized mice with two L. reuteri strains with divergent histamine production capabilities: L. reuteri C93, which produced significant histamine in vitro, and L. reuteri C88, which failed to produce histamine in vitro. Mice colonized with M. morganii C135 or L. reuteri C93 exhibited high levels of intestinal histamine production, while mice colonized with the two mock communities or L. reuteri C88 showed nearly undetectable intestinal histamine (Figure 4C). In addition, supplementation with dietary L-His increased histamine production in M. morganii monocolonized mice (Figure 4C). Finally, we also detected increased histamine in the serum of mice colonized with M. morganii (Figure S5A).
Figure 4. Commensal-derived histamine promotes colon motility.
(A) Production of histamine by M. morganii and L. reuteri. L. reuteri and M. morganii strains were cultured in Gifu medium with or without supplemental L-His and histamine concentrations in the supernatants were measured by ELISA after 48 hours (background levels in controls containing supplemental histidine are due to slight cross-reactivity).
(B) Experimental design to test in vivo histamine production and the effects of histamine-producing bacteria on colon motility.
(C) M. morganii- and L. reuteri-derived histamine accumulates in vivo in monocolonized mice. Female germ-free C57Bl/6 mice were colonized with mock communities of 9 or 10 phylogenetically diverse human gut bacteria (Mock Community A or B) or monocolonized with M. morganii C135, L. reuteri C88 or C93. Mice were fed a conventional diet with or without administration of 1% L-His ad libitum in the drinking water. Histamine concentrations in cecal and colonic extracts and feces were measured via ELISA. n=3–5 mice per group.
(D) M. morganii C135- and L. reuteri C93-derived histamine enhances colon motility. Fecal output for mice treated as described in B was measured by counting the number of fecal pellets produced by a single mouse in one hour. n=3–5 mice per group.
(E) M. morganii increases colon motility in the context of a mock gut microbial community. Female germ-free C57Bl/6 mice were colonized Mock Community A with or without M. morganii C135 and administered 1% L-His ad libitum in the drinking water. Histamine concentrations in colonic extracts were measured via ELISA and fecal output was measured as in (D). n=4–5 mice per group.
(F) Histamine receptor inhibition partially reverses the impact of M. morganii on colon motility. Female germ-free C57Bl/6 mice were colonized with Mock Community A or monocolonized with M. morganii C135 for two weeks. Mice were then treated with or without a cocktail of four histamine receptor inhibitors (targeting HRH1–4) in the drinking water for one week. Histamine concentrations in feces were measured via ELISA and fecal output was measured as in (D). n=4–6 mice per group.
(G and H) Relative abundances of genes encoding histidine decarboxylases (from all bacteria or M. morganii) are increased in the microbiomes of patients with Crohn’s disease as compared to healthy controls (G). Relative abundance of histamine is increased in IBD patients as compared to healthy controls as measured by metabolomics (H). Data are from longitudinal stool samples from IBD patients publically available from the Human Microbiome Project 2 (iHMP). Total numbers of samples or subjects with detectable M. morganii are denoted below each plot; a subject was considered positive if M. morganii was detectable in one or more samples from that patient across the complete dataset.
Data in all panels are representative of at least two independent experiments. Data are presented as mean ± SEM. One-way ANOVA with Tukey’s post-hoc test (C-F) or Kruskall-Wallis with Dunn’s multiple comparisons (G-H), *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, NS not significant (p > 0.05).
To determine the location of M. morganii in vivo, we used modified Niven’s agar to enumerate M. morganii CFUs in gnotobiotic mice colonized with two mock communities of diverse human gut microbes plus M. morganii C135 (Mavromatis, 2002). We found that M. morganii constitutes approximately 5% of the microbiota in the context of a mock community, and primarily inhabits the cecum and colon (Figure S5C and Table S3). Notably, M. morganii also preferentially localizes in tissue- or mucus-associated niches in the colon in humans (Eun et al., 2016).
Oral gavage with histamine increases colon motility in rodents (Kim et al., 2011; Tyagi et al., 2009). We thus hypothesized that gut microbe-derived histamine might also increase intestinal motility. We monitored intestinal motility in gnotobiotic mice colonized with two mock communities (which do not produce histamine) or M. morganii C135 with or without administration of 1% L-His in the water and found that M. morganii induced a significant increase in fecal output, which was further increased upon supplementation with L-His (Figure 4D). Similarly, mice colonized with L. reuteri C93 exhibited increased fecal output as compared to mice colonized with L. reuteri C88 (Figure 4D).
To test whether M. morganii can impact host physiology in the context of a more diverse microbial community, we colonized germ-free mice with a mock community of nine human gut microbes with or without M. morganii C135 and examined histamine accumulation in the gut and serum, as well as fecal output. Although the effects were less profound than those observed with monocolonizations, the addition of M. morganii to a mock community also led to an accumulation of histamine in the colon and serum (Figure 4E and S5D) as well as increased fecal output (Figure 4E). Finally, we found that treatment with histamine receptor antagonists could largely block the effects of M. morganii on fecal output despite accumulation of similar levels of histamine in the gut (Figure 4F).
To examine the potential importance of histamine production by M. morganii (or other microbes) in human physiology, we mined publicly available metagenomic and metabolomic data from the integrative Human Microbiome Project (see methods for details) to determine the relative abundance of histamine producing enzymes or histamine itself in microbiomes from patients with IBD versus healthy controls (Integrative, 2014). We found that CD patients exhibited an increased prevalence and abundance of histidine decarboxylase genes, including M. morganii-encoded histidine decarboxylase, as compared to healthy controls or UC patients (Figure 4G and Figure S5E), and that histamine itself was increased in fecal samples from CD and UC patients as compared to controls (Figure 4H). This observation is in line with previous studies demonstrating increased intestinal histamine in patients with IBD, but this was largely attributed to host-derived histamine production (Smolinska et al., 2014).
Together, these data demonstrate that M. morganii impacts intestinal motility through histamine secretion and activation of histamine receptors, that dietary histidine can enhance these effects, and that bacterial histidine decarboxylases (both generally and from M. morganii) are enriched in patients with CD.
M. morganii can trigger ‘phenethylamine poisoning’ when combined with monoamine oxidase inhibition
Unlike histamine, we observed only low levels of colonic PEA in M. morganii monocolonized mice (Figure 5A). One potential explanation for this observation is that many biogenic amines, including PEA, are rapidly degraded in the intestine by host monoamine oxidases (MAOs) (Glover, 1977). MAO inhibitors (MAOIs) were the first FDA-approved antidepressants (Fiedorowicz, 2004). Thus, to reveal the potential production of PEA in vivo, we treated germ-free mice or mice monocolonized with M. morganii C135 or a Bacteroides thetaiotaomicron strain (B. theta C34) that does not produce DRD agonists with the irreversible MAOI phenelzine. While colonic PEA remained undetectable in germ-free mice or mice colonized with B. theta even after phenelzine treatment, M. morganii-colonized mice treated with phenelzine exhibited high levels of colonic PEA (Figure 5A). M. morganii-colonized mice also became lethargic within days after MAOI treatment, and more than half of all M. morganii-colonized mice died by day seven post MAOI administration, while germ-free and B. theta C34-colonized mice remained healthy (Figure 5B). Morbidity and mortality after MAOI treatment correlated with elevated levels of PEA in the colon, serum, and brains of M. morganii-colonized mice (Figure 5C and S5F, G), and cecal and colonic contents, as well as serum and brain extracts from these mice also activated DRD2 (Figure 5D). Finally, mice colonized with a mock community plus M. morganii also exhibited increased PEA in the colon and brain, as measured by DRD2 activation (Figure 5E). Together, these data show that M. morganii-derived phenethylamine can accumulate systemically and cross the blood-brain barrier in mice treated with MAOIs.
Figure 5. M. morganii-derived phenethylamine combined with MAOI triggers lethal phenethylamine poisoning.
(A) M. morganii produces phenethylamine in vivo. Female germ-free C57Bl/6 mice were colonized with M. morganii C135 and treated with or without the MAOI phenelzine. Phenethylamine concentration in colonic extracts was examined using QQQ-MS/MS.
(B) Mice colonized with M. morganii exhibit lethal phenethylamine poisoning after treatment with the MAOI phenelzine. Female germ-free C57Bl/6 mice were monocolonized with M. morganii C135 for one week before treatment with phenelzine in the drinking water. Survival is depicted on a Kaplan-Meier curve. n=4 mice per group.
(C and D) M. morganii-colonized mice treated with phenelzine accumulated phenethylamine in the cecum, colon, serum and brain. M. morganii C135 and B. theta C34 monocolonized female C57Bl/6 mice were treated with or without the MAOI phenelzine in the drinking water. Phenethylamine was measured via QQQ-MS/MS (C) or DRD2 PRESTO-Tango (D). n=4 mice per group.
(E) M. morganii-derived phenethylamine accumulates in the sera and brains of mice colonized with Mock Community A plus M. morganii. Germ-free female C57Bl/6 mice were colonized with Mock Community A with or without M. morganii C135, or monocolonized with M. morganii C135. All mice were treated with the MAOI phenelzine in the drinking water for one week and phenethylamine accumulation was detected using DRD2-Tango as a proxy.
Data in all panels are representative of at least two independent experiments. Data are presented as mean ± SEM. One-way ANOVA with Tukey’s post-hoc test (D-E), Kaplan meier and Log rank analysis (B), *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
A unique Bacteroides isolate activates GPR56/AGRG1
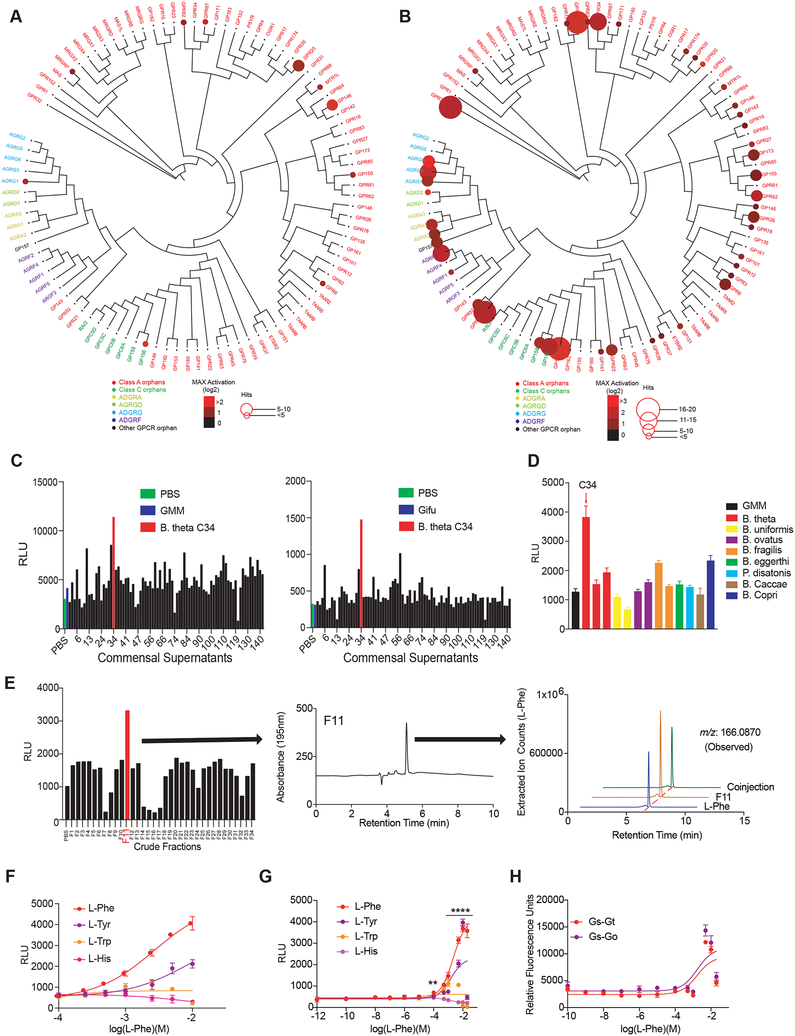
We observed that specific bacterial supernatants activated select orphan GPCRs (Figure 6A). To confirm these hits, we repeated our PRESTO-Tango screening procedure using a richer culture medium (Gifu) that supports more robust growth of many of the human gut microbes in our collection. This modified procedure significantly expanded the number of positive hits against orphan GPCRs: 17 orphan GPCRs showed greater than four-fold activation in response to at least one bacterial supernatant (Figure 6B). Metabolites from a strain assigned to the species B. theta (B. theta C34) activated GPR56/AGRG1 under both culture conditions (Figure 6C). In contrast, other strains of B. theta as well as multiple related Bacteroides strains failed to activate GPR56/AGRG1 (Figure 6D) despite similar bacterial growth (Figure S6A).
Figure 6. A unique strain of B. thetaiotaomicron C34 is a prolific producer of LPhe and activates GPR56/AGRG1.
(A and B) Activation of orphan GPCRs by metabolomes from a human gut microbiota culture (see Figure 1) grown in gut microbiota medium (A) or Gifu (B) as measured by PRESTO-Tango. Screening results are displayed on a phylogenetic tree of orphan GPCRs that was constructed and visualized with equal branch lengths using gpcrdb.org, PHYLIP and jsPhyloSVG. Color intensities represent the magnitude of activation over media and radii of circles represent the number of bacteria that activated a given GPCR by more than two-fold.
(C) A single isolate C34 assigned to the species Bacteroides thetaiotaomicron activates GPR56/AGRG1 when cultured in gut microbiota medium (GMM: top panel) or Gifu medium (bottom panel). Activation of GPR56/AGRG1 by supernatants from 144 human gut isolates was measured via GPR56-Tango.
(D) B. theta strain C34 uniquely activates GPR56/AGRG1. Activation of GPR56/AGRG1 by supernatants from diverse species and strains from the genera Bacteroides and Parabacteroides cultured in GMM was measured via GPR56 PRESTO-Tango.
(E) B. theta C34-produced L-Phe activates GPR56/AGRG1. B. theta C34 supernatants were fractionated via reversed-phase HPLC and fractions were evaluated for activation of GPR56/AGRG1 via GPR56-Tango. The active fraction (F11) contained a primary constituent that was identified via LC-MS, HRMS-ESI-QTOF, NMR, and advanced Marfey’s analyses as LPhe.
(F and G) L-Phe activates the orphan receptor GPR56/AGRG1. Activation of GPR56/AGRG1 by titrating doses of pure L-Phe, L-Tyr, L-Trp, and L-His was measured via GPR56-Tango using RPMI 1640 medium (F) or a custom medium lacking L-Phe and L-Tyr (G).
(H) L-Phe activates G protein-dependent signaling downstream of GPR56/AGRG1 as measured by the CRE-SEAP assay. Gαs-Gαt and Gαs-Gαo chimeras were used to redirect GPR56/AGRG1 signaling to Gαs.
Data in all panels except for A, B, and E are representative of at least three independent experiments. Data are presented as mean ± SEM. One-way ANOVA with Tukey’s post-hoc test **p < 0.01, ****p < 0.0001.
The essential amino acid L-Phe activates GPR56/AGRG1 and GPR97/AGRG3
Since there was no known endogenous small molecule ligand for GPR56/AGRG1 (Purcell, 2018), we next attempted to identify the specific metabolite produced by B. theta C34 that activated GPR56/AGRG1. B. theta C34 supernatants were extracted and subjected to fractionation by reversed-phase HPLC and all fractions were analyzed for activity via GPR56-Tango (Figure 6E). High resolution mass spectrometry, NMR and coinjection analyses of the active fraction (F11) revealed the essential amino acid phenylalanine (Phe) as the primary constituent of F11 (Figure 6E and S6B) and structural characterization using advanced Marfey’s analysis confirmed that L-Phe is the likely bioactive ligand (Figure S6C) (Bhushan and Bruckner, 2011). Accordingly, pure L-Phe and, to a lesser extent, L-Tyr stereoselectively activated GPR56/AGRG1, while L-Trp and L-His, D-Phe, D-Trp, D-His and D-Tyr showed no activity (Figure 6F and S6D). We hypothesized that L-Phe and L-Tyr in the medium used for the Tango assay might obscure the full extent of GPR56/AGRG1 activation by L-Phe. Indeed, removal of endogenous L-Phe and L-Tyr from the culture medium greatly increased the sensitivity and magnitude of GPR56 activation by L-Phe and L-Tyr as measured by GPR56-Tango (Figure 6G and S6E); furthermore, GPR56 expression was essential for this response (Figure S6E). Finally, despite their differential secretion of L-Phe, the genomes of B. theta C34 and two strains of Bacteroides that failed to activate GPR56/AGRG1 all encoded the full suite of enzymes in the shikimate pathway that synthesize L-Phe (Table S4, S5).
Using the promiscuous Gαs-Gαt and Gαs-Gαo chimeras and the CRE-SEAP assay described above (Liberles and Buck, 2006), we found that L-Phe activated G protein-dependent signaling downstream of GPR56/AGRG1 (Figure 6H and Figure S6F); however, since high concentrations of L-Phe (>1mM) were required to activate GPR56/AGRG1, it remains unclear whether physiological concentrations of L-Phe will engage GPR56/AGRG1-mediated G protein signaling in vivo. GPR56/AGRG1 belongs to the adhesion GPCR family, whose members possess large extracellular domains that mediate interactions with a variety of protein ligands (Purcell, 2018). However, we found that the extracellular domain of GPR56/AGRG1 is also required for L-Phe-induced activation of GPR56/AGRG1 (Kishore et al., 2016) (Figure S6E). Together, our data demonstrate that a unique strain of B. theta secretes high levels of L-Phe and that L-Phe is a novel agonist of the adhesion GPCR GPR56/AGRG1.
We next examined whether other orphan GPCRs might also respond to L-Phe by stimulating all adhesion and orphan GPCRs with pure L-Phe (Figure S7A). We found that GPR97/AGRG3 also responded to L-Phe and showed greater selectivity toward L-Phe than GPR56/AGRG1—L-Phe, but not L-Tyr, L-Trp, or L-His, activated GPR97/AGRG3 (Figure S7B). Like GPR56/AGRG1, the extracellular domain of GPR97/AGRG3 was required for its ability to respond to L-Phe (Figure S7C), and removal of L-Phe and L-Tyr from the medium increased the magnitude of activation of GPR97/AGRG3 by exogenous L-Phe (Figure S7B and S7D). Furthermore, L-Phe also activated G protein-dependent signaling downstream of GPR97/AGRG3 (Figure S7E). Notably, GPR56/AGRG1 and GPR97/AGRG3 are evolutionarily related (Figure S7F), which may explain their shared ability to detect L-Phe.
Bacterial metabolic exchange can contribute to in vivo production of phenethylamine
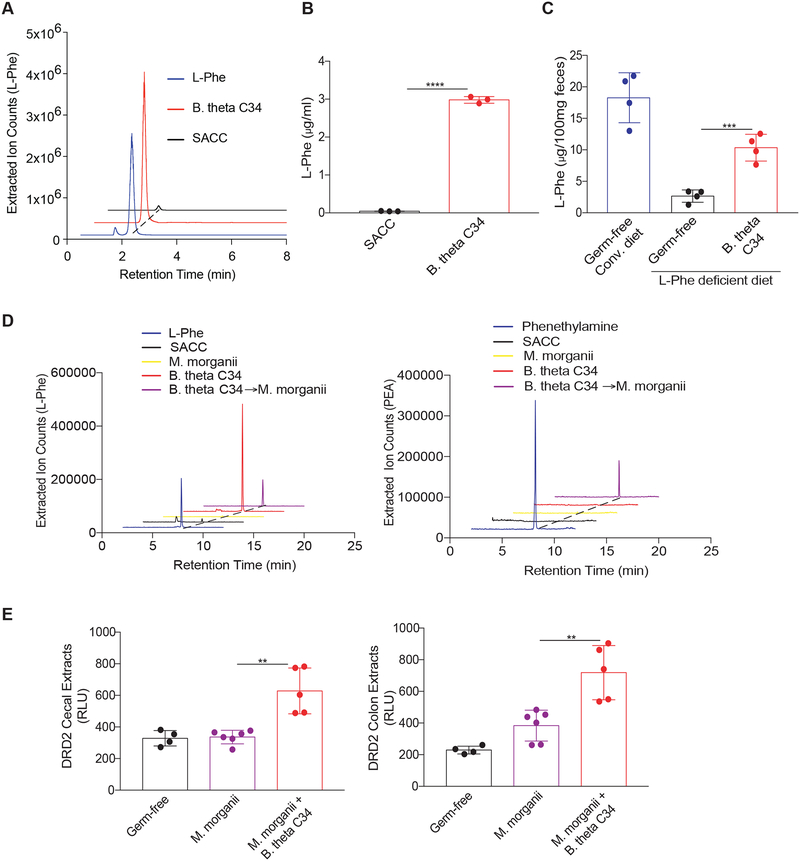
Our reductionist studies revealed that B. theta C34 produces large amounts of L-Phe while M. morganii C135 can process L-Phe into PEA. Thus, we wished to address whether these two bacteria might participate in an active metabolic exchange in vivo. Using a defined minimal bacterial medium that lacks L-Phe (standard amino acid complete medium or SACC; see methods) (Dodd et al., 2017; Lovitt et al., 1987), we observed that B. theta C34 can directly synthesize large amounts of L-Phe in vitro (Figure 7A, B). Furthermore, mice monocolonized with B. theta C34 and fed an L-Phe-deficient diet displayed significant intestinal accumulation of L-Phe as compared to GF mice (Figure 7C).
Figure 7. Active metabolic exchange between two commensals supports production of phenethylamine.
(A and B) B. theta C34 can directly synthesize L-Phe. L-Phe concentrations in supernatants from C34 grown in a minimal medium (SACC) lacking L-Phe were evaluated by LC-MS (A) and quantitated by QQQ-MS/MS (B).
(C) B. theta C34 produces L-Phe in vivo. Germ-free female C57Bl/6 mice fed a conventional diet or a defined diet lacking L-Phe were colonized with or without B. theta C34. Fecal L-Phe concentrations were measured by QQQ-MS/MS one week after colonization. n=4 mice per group.
(D) M. morganii C135 consumes B. theta C34-derived L-Phe to produce phenethylamine in vitro. B. theta C34 cultures grown in SACC medium lacking L-Phe and then incubated with M. morganii C135. L-Phe and phenethylamine (PEA) as measured by QQQ-MS/MS.
(E) B. theta C34 and M. morganii C135 can participate in active metabolic exchange to produce phenethylamine in vivo. Germ-free C57Bl/6 mice were monocolonized with M. morganii C135 or co-colonized with B. theta C34 and M. morganii C135, fed a diet lacking L-Phe, and treated with the MAOI phenelzine. Activation of DRD2 by phenethylamine in cecal and colonic extracts was measured by DRD2-Tango. n=4–6 mice per group.
Data in all panels are representative of at least two independent experiments. Data are presented as mean ± SEM. One-way ANOVA with Tukey’s post-hoc test (B-C and E-F), **p < 0.01, ***p < 0.001, ****p < 0.0001.
We next examined whether M. morganii would process B. theta C34-derived L-Phe into PEA in vitro and observed that B. theta C34-derived L-Phe was efficiently converted into PEA by M. morganii C135 (Figure 7D). To test whether metabolic exchange occurs in vivo, we colonized GF mice with either M. morganii C135 alone or with both B. theta C34 and M. morganii C135, fed these mice a simplified diet lacking L-Phe, and then treated them with phenelzine. Mice colonized with M. morganii C135 alone remained healthy and produced minimal PEA in the absence of dietary L-Phe (Figure 7E). In contrast, mice co-colonized with B. theta C34 and M. morganii C135 became lethargic by day 4 after MAOI treatment and exhibited significant accumulation of PEA (Figure 7E). This demonstrates that metabolic exchange between B. theta C34 and M. morganii C135 can contribute to the production of a bioactive trace amine that can have potent effects on systemic host physiology.
DISCUSSION
The overwhelming complexity of the gut microbiota metabolome often obscures facile recognition of chemical communication between microbes and their hosts (Donia and Fischbach, 2015; Fischbach, 2018). Here, we used host GPCR activation as a lens to detect bioactive metabolites produced by individual gut microbes. We found that dozens of phylogenetically diverse human gut bacteria produced small molecules that activated various GPCRs, including both well-characterized GPCRs and orphan GPCRs. We observed patterns of metabolite production that were largely predictable based on phylogeny, as well as strain-specific differences within a given species. Our approach thus revealed a plethora of novel microbiota metabolite-GPCR interactions of potential physiological importance. For example, we uncovered a diet-microbe-host axis that influences intestinal motility through microbial production of histamine and a tri-partite microbe-microbe-host relationship that results in the production of the potent trace amine phenethylamine. These chemicals exerted profound effects on both local and systemic host physiology. Overall, our results further support the notion that human-associated microbes represent a remarkably rich source of small molecules that impact human biology.
Prior studies have employed functional metagenomic screens as well as bioinformatics- and bioassay-guided natural product discovery approaches to uncover novel microbial-derived ligands for host GPCRs, including orphans (e.g., SCFA and GPR41 and 43, and commendamide and G2A and GPR119; (Brown et al., 2003; Cohen et al., 2017; Cohen et al., 2015; Le Poul et al., 2003); however, these approaches also have notable limitations (Donia and Fischbach, 2015; Milshteyn et al., 2018). For example, while functional metagenomic screens enable identification of novel biosynthetic gene clusters and their products from unculturable microorganisms, they are restricted in scope to contiguous biosynthetic gene clusters that are active in heterologous hosts, require large-scale library construction, and necessitate extensive follow up to identify specific host receptors that recognize novel bioactive compounds. Similarly, while bioassay-guided natural product discovery efforts enable identification of compounds produced by native sources that engage a specific receptor or pathway, their utility is largely restricted to cultivatable microorganisms and they typically examine only a single receptor or activity at a time. In contrast, the high-throughput functional profiling approach that we employ here enables simultaneous interrogation of hundreds of receptors and thousands of chemicals and is unconstrained by prior annotations of biosynthetic gene clusters or metabolites (although still dependent on microbial cultivation). We thus anticipate that future expansions of our overall approach will continue to uncover microbial metabolites that impact host physiology and reveal novel natural ligands for orphan receptors.
We were particularly interested in examining the possibility that the microbiota-derived GPCR agonists we identified in vitro would also shape host physiology in vivo. We found that histamine production by M. morganii or L. reuteri increased colonic motility, that feeding with exogenous histidine enhanced this phenotype, and that histamine receptor inhibition reversed these effects. Since fecal output can be impacted by multiple factors (e.g., fluid secretion and modulation of the enteric nervous system), future studies will be necessary to determine the mechanistic basis of this phenotype. M. morganii monocolonized mice also exhibited elevated levels of serum histamine, indicating a potential systemic role for microbiota-derived histamine. Notably, a recent study found that M. morganii relative abundance was increased in asthmatics as compared to healthy controls (Barcik et al., 2016). Finally, we found that histamine decarboxylases (specifically from M. morganii) are enriched in patients with Crohn’s disease, which suggests that histamine production by the microbiota may directly impact IBD (Smolinska et al., 2014).
We found that all of our isolates of M. morganii produced PEA in vitro and that M. morganii monocolonized mice treated with an MAOI exhibited systemic accumulation of PEA and mortality. PEA is a potent neuroactive chemical that, unlike dopamine and tyramine, can readily cross the blood-brain barrier (Oldendorf, 1971). The effects of PEA are thought to be mediated primarily through activation of trace amine-associated receptors and subsequent release of norepinephrine and dopamine (Borowsky et al., 2001; Bunzow, 2001; Sotnikova et al., 2004). However, our studies suggest that PEA can also act as a full agonist for DRD2–4 and potentially a biased agonist for DRD1 and 5. While MAOIs were the first FDA-approved antidepressants (Ramachandraih, 2011), their current usage is limited due to dangerous diet- and drug-drug interactions (Fiedorowicz, 2004). Nonetheless, MAOIs remain an important treatment option for patients with refractory depression and other psychiatric disorders (Fiedorowicz, 2004). Since PEA enhances mood and can readily cross the blood-brain barrier (Irsfeld et al., 2013), our findings imply that inter-individual variability in microbial production of PEA could potentially explain the variable efficacy of MAOIs on depression.
Our studies also uncovered a specific Bacteroides strain that produces high levels of the essential amino acid L-Phe and revealed that L-Phe activates the orphans GPR56/AGRG1 and GPR97/AGRG3. These findings raise multiple intriguing possibilities. GPR56/AGRG1 is highly expressed in the small intestine and human pancreatic islets (Amisten et al., 2013; Duner et al., 2016), and L-Phe concentrations in the jejunum can reach concentrations up to 2 mM after a meal (Adibi, 1973). Thus, GPR56/AGRG1 may act as a nutrient sensor to regulate digestion and satiety. In addition, although L-Phe concentrations in the serum are typically well below the levels necessary to activate GPR56/97, patients with phenylketonuria (PKU) can exhibit serum L-Phe concentrations greater than 1 mM (Williams, 2008). Thus, it is also theoretically possible that GPR56/AGRG1 and/or GPR97/AGRG3 may be activated in extraintestinal tissues in PKU (e.g., GPR56/AGRG1 is highly expressed in the central nervous system).
The natural microbiota metabolome results from a complex web of interactions between diverse microbial species and strains, environmental inputs (e.g., diet), and host factors. Using a reductionist approach, we discovered two bacterial isolates that traffic in the same small molecule: a unique strain of B. theta that is a prolific producer of L-Phe and M. morganii, which converts L-Phe into PEA. These studies thus demonstrate that reductionist approaches can reveal metabolic exchanges that would be missed when examining endpoint microbiota metabolomes produced by complex mixtures of microorganisms. Understanding metabolic exchange networks will be essential to understand the effects of the microbiota metabolome on host biology under more physiological settings (i.e., in the context of complete gut microbial communities) and to eventually leverage microbial chemistries for therapeutic benefit. Towards these ends, we examined the effects of M. morganii on host physiology in the context of a mock gut microbial community consisting of nine phylogenetically diverse human gut microbes. We found that M. morganii continued to exhibit measurable (albeit more modest) metabolite-dependent impacts on the host in the context of this simplified community. However, there are almost certainly other gut microbial community contexts where competition for ecological space or metabolic precursors (e.g., L-His or L-Phe), or active degradation of M. morganii-derived metabolites may reduce or eliminate the impact of M. morganii on the host (or, conversely, may enhance the effects of M. morganii).
Our studies underscore the role of dietary amino-acids (e.g., L-His) in microbial production of biogenic amines. However, they also highlight the role of other members of the microbiota as sources of compounds that are often thought of as primarily derived from diet (e.g., essential amino acids). This leads to the question of when microbial-produced amino acids may potentially supplement or even replace dietary amino acids in microbial biotransformations. We modeled the possibility that microbe-derived L-Phe can be biotransfored by M. morganii using a simplified diet that lacks L-Phe. However, bacterial L-Phe may also be important under more physiological conditions. For example, dietary amino acids are largely absorbed in the small intestine and thus free amino acid concentrations in the colon are often limiting (Adibi, 1973); also, low-protein diets and fasting can dramatically reduce intestinal amino acid availability (Pezeshki et al., 2016). Thus, microbial production of amino acids may play a critical role in the production of bioactive microbiota metabolites.
While our reductionist approach revealed multiple potentially physiologically important host-microbiota metabolome interactions, it also suffers from notable limitations. For example, microbial metabolite production varies substantially depending on the media used for cultivation, and in vitro monoculture conditions fail to capture metabolites that result from interactions with the host organism, biotransformations of compounds absent from the cultivation medium, or interactions with other microbes. Furthermore, the metabolite concentrations produced during in vitro cultivation may not reflect in vivo metabolite production. Finally, in vitro screens fail to reveal the natural tissue distributions of gut microbiota metabolites. Understanding these distributions will be particularly important for metabolites that activate host receptors that are expressed in diverse cell types and tissues. For example, histamine and dopamine receptors are expressed on cells as diverse as immune cells, central and peripheral neurons, smooth muscle, epithelial and endothelial cells, and in essentially all tissues including the gut, lung, and brain (Beaulieu and Gainetdinov, 2011; Jutel et al., 2009; Smolinska et al., 2014).
In conclusion, while the human gut microbiota metabolome is dauntingly complex and diverse, emerging approaches have begun to reveal key chemical interactions at the host-microbiota interface. We show here that high-throughput activity-based screening using potential host receptors as a lens can highlight physiologically relevant microbiota metabolites from complex metabolite mixtures. Such host-centric, functional profiling approaches can thus facilitate a mechanistic understanding of how we interact with and are affected by our microbial inhabitants, and have the potential to yield targeted therapeutic interventions aimed at the interface between indigenous microbes and their hosts.
STAR METHODS
See KRT Table Appended Separately
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| B.fragilis | ATCC | Cat# 25285 |
| B.ovatus | ATCC | Cat# 8483 |
| B. thetaiotaomicron | ATCC | Cat# 29741 |
| B.uniformis | ATCC | Cat# 8492 |
| M.morganii | ATCC | Cat# 25830 |
| M.morganii | ATCC | Cat# 49948 |
| Human Gut Microbiota Culture Collections from IBD Patients | Isolated from patients (Palm et al., 2014) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Gifu Anaerobic Broth | VWR | Cat# 11007–214 |
| Desloratadine | Tocris | Cat# 5958 |
| Tiotidine | Tocris | Cat# 0826 |
| Iodophenpropit | Tocris | Cat# 0779 |
| A987306 | Tocris | Cat# 3640 |
| Phenethylamine | Sigma-Aldrich | Cat# 241008 |
| Tyramine | Sigma-Aldrich | Cat# T90344 |
| Dopamine | Sigma-Aldrich | Cat# H8502 |
| Histamine | Sigma-Aldrich | Cat# H7129 |
| Acetylcholine | Sigma-Aldrich | Cat# A2661 |
| L-DOPA | Sigma-Aldrich | Cat# D9628 |
| Succinate | Sigma-Aldrich | Cat# 398055 |
| Serotonin | Sigma-Aldrich | Cat# 14927 |
| Gastrin | Sigma-Aldrich | Cat# G9145 |
| Peptide YY | Anaspec | Cat# AS024401 |
| Pancreatic polypeptide | Anaspec | Cat# AS-22866 |
| Cholecystokinin | Sigma-Aldrich | Cat# C2175 |
| Trace mineral supplement | ATCC | Cat# MD-TMS |
| Vitamin supplement | ATCC | Cat# MD-VS |
| L-glycine | Sigma-Aldrich | Cat# G8898 |
| L-valine | Sigma-Aldrich | Cat# 94619 |
| L-leucine | Sigma-Aldrich | Cat# L8000 |
| L-isoleucine | Sigma-Aldrich | Cat# 12752 |
| L-methionine | Sigma-Aldrich | Cat# 64319 |
| L-histidine | Sigma-Aldrich | Cat# H8000 |
| L-arginine | Sigma-Aldrich | Cat# A5131 |
| L-phenylalanine | Sigma-Aldrich | Cat# P2126 |
| L-tyrosine | Sigma-Aldrich | Cat# T3754 |
| L-tryptophan | Sigma-Aldrich | Cat# T0254 |
| N-methylphenethylamine | Sigma-Aldrich | Cat# M68423 |
| Octopamine | Sigma-Aldrich | Cat# O0250 |
| Synephrine | Sigma-Aldrich | Cat# S0752 |
| Epinephrine | Sigma-Aldrich | Cat# E4250 |
| Norepinephrine | Sigma-Aldrich | Cat# A7257 |
| 3-Methoxytyramine | Sigma-Aldrich | Cat# M4251 |
| DMEM | Sigma-Aldrich | Cat# D6429 |
| RPMI 1640 | Thermo Fisher | Cat# 21870092 |
| D-phenylalanine | Sigma-Aldrich | Cat# 673-06-3 |
| FDAA (Marfey’s Reagent) | Thermo Fisher | Cat# 48895 |
| Critical Commercial Assays | ||
| Bright-Glo™Luciferase Assay System | Promega | Cat# E2620 |
| Histamine Elisa Kit | Enzo Life Sciences | Cat# ENZ-KIT140-0001 |
| Deposited Data | ||
| Shotgun whole-genome sequencing data | This paper | PRJNA512876 |
| iHMP metabolome data | NIH Integrative Human Microbiome Project (Integrative, 2014) | https://ibdmdb.org/tunnel/public/HMP2/Metabolites/1723/products |
| iHMP metagenome data | NIH Integrative Human Microbiome Project (Integrative, 2014) | https://ibdmdb.org/tunnel/public/HMP2/Metabolites/1723/products |
| M. morganii whole genomes (previously deposited) | NCBI | https://www.ncbi.nlm.nih.gov/genome |
| Experimental Models: Cell Lines | ||
| HTLA cells | (Barnea et al., 2008) | N/A |
| HEK293T | ATCC | Cat# CRL-3216 |
| Experimental Models: Organisms/Strains | ||
| Germ-free C57Bl/6 mice | University of Michigan Gnotobiotics Center | https://microbe.med.umich.edu/services/germ-free-gnotobiotic-mouse-facilities |
| Recombinant DNA | ||
| PRESTO-Tango Plasmids | Addgene; (Kroeze et al., 2015) | Kit #1000000068 |
| Gαs-Gαt and Gαs-Gαo chimeras | (Liberles and Buck, 2006) | N/A |
| CRE-SEAP reporter plasmid | (Liberles and Buck, 2006) | N/A |
| Software and Algorithms | ||
| QIIME2 2018.8 | QIIME 2 development team, 2018 | https://qiime2.org/ |
| DADA2 (QIIME2 plugin) | (Callahan et al., 2016) | http://benjjneb.github.io/dada2/ |
| Trimmomatic v.0.38 | (Bolger et al., 2014) | http://www.usadellab.org/cms/?page=trimmomatic |
| SPAdes 3.13.0 | (Bankevich et al., 2012; Nurk et al., 2013) | http://cab.spbu.ru/files/release3.13.0/manual.html |
| RAST server | (Aziz et al., 2008) | http://rast.nmpdr.org/rast.cgi?page=Jobs |
| BBMap | DOE Joint Genome Institute | https://sourceforge.net/projects/bbmap/ |
| Other | ||
| 600 MHz NMR system with a cryoprobe | Agilent | N/A |
| iFunnel 6550 ESI-HRMS-QTOF | Agilent | N/A |
| 1260 Infinity system | Agilent | N/A |
| Prepstar HPLC | Agilent | N/A |
| Anaerobic Chambers | Coy | Custom built |
| Spectramax i3x plate reader | Molecular Devices | I3X |
| Gnotobiotic isolators | Class Biologically Clean | N/A |
| Isocage P Microisolator Caging System | Techniplast | ISO72P |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to the Lead Contact, Noah W. Palm (noah.palm@yale.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice.
6–12 week old germ-free wild-type C57Bl/6 mice were used in all experiments. Both male and female mice were used for these studies and mice were age and sex matched within each experiment (only one sex was used for each independent experiment). We did not observe any obvious sex-specific differences in in vivo phenotypes in any of these studies.
Bacteria.
All strains were cultured in gut microbiota medium (Goodman et al., 2011) or Gifu broth at 37 °C under anaerobic conditions and the identities of all strains were confirmed by 16S rRNA gene sequencing.
Media Formulations.
Custom (L-Phe- and L-Tyr-free) Dulbecco’s Modified Eagle’s Medium (DMEM) formulation
| Ingredients | Concentration in Medium (g/L) |
|---|---|
| Inorganic Salts | |
| Calcium Chloride | 0.2 |
| Ferric Nitrate•9H2O | 0.0001 |
| Magnesium Sulfate(anhydrous) | 0.09767 |
| Potassium Chloride | 0.4 |
| Sodium Bicarbonate | 3.7 |
| Sodium Chloride | 6.4 |
| Sodium Phosphate Monobasic(anhydrous) | 0.109 |
| Amino Acids | |
| L-Arginine•HCl | 0.084 |
| L-Cystine•2HCl | 0.0626 |
| L-Glutamine | 0.584 |
| Glycine | 0.03 |
| L-Histidine•HCl•H2O | 0.042 |
| L-Isoleucine | 0.105 |
| L-Leucine | 0.105 |
| L-Lysine•HCl | 0.146 |
| L-Methionine | 0.03 |
| L-Phenylalanine | 0 |
| L-Serine | 0.042 |
| L-Threonine | 0.095 |
| L-Tryptophan | 0.016 |
| L-Tyrosine•2Na•2H2O | 0 |
| L-Valine | 0.094 |
| Vitamins | |
| Choline Chloride | 0.004 |
| Folic Acid | 0.004 |
| myo-Inositol | 0.0072 |
| Niacinamide | 0.004 |
| D-Pantothenic Acid (hemicalcium) | 0.004 |
| Pyridoxine•HCl | 0.004 |
| Riboflavin | 0.0004 |
| Thiamine•HCl | 0.004 |
| Other | |
| Glucose | 4.5 |
| Phenol Red•Na | 0.0159 |
| Pyruvic Acid•Na | 0.11 |
Minimal Medium
| Ingredients | Concentration in Medium (g/L) |
|---|---|
| Resazurin | 0.0001 |
| KH2PO4 | 2 |
| K2HPO4 | 2 |
| MgCl2•6H2O | 0.2 |
| (NH4)2SO4 | 5 |
| L-Glycine | 0.075 |
| L-Valine | 0.117 |
| L-Leucine | 0.131 |
| L-Isoleucine | 0.131 |
| L-Methionine | 0.149 |
| L-Histidine | 0 |
| L-Arginine | 0.174 |
| L-Phenylalanine | 0 |
| L-Tyrosine | 0 |
| L-Tryptophan | 0 |
| NaHCO3 | 2.5 |
| Cysteine HCl | 0.5 |
| Glucose | 3.603 |
| Trace Mineral Supplement(g/L) | 10ml |
| EDTA | 0.5 |
| MgSO4•7H2O | 3.0 |
| MnSO4•H2O | 0.5 |
| NaCl | 1.0 |
| FeSO4•7H2O | 0.1 |
| Co(NO3)2•6H2O | 0.1 |
| CaCl2 (anhydrous) | 0.1 |
| ZnSO4•7H2O | 0.1 |
| CuSO4•5H2O | 0.010 |
| AlK(SO4)2 (anhydrous) | 0.010 |
| H3BO3 | 0.010 |
| Na2MoO4•2H2O | 0.010 |
| Na2SeO3 (anhydrous) | 0.001 |
| Na2WO4•2H2O | 0.010 |
| NiCl2•6H2O | 0.020 |
| Vitamin Supplement (mg/L) | 10ml |
| Folic acid | 2.0 |
| Pyridoxine hydrochloride | 10.0 |
| Riboflavin | 5.0 |
| Biotin | 2.0 |
| Thiamine | 5.0 |
| Nicotinic acid | 5.0 |
| Calcium Pantothenate | 5.0 |
| Vitamin B12 | 0.1 |
| p-Aminobenzoic acid | 5.0 |
| Thioctic acid | 5.0 |
| Monopotassium phosphate | 900.0 |
Standard Amino Acid Complete (SACC) medium
| Ingredients | Concentration in Medium (g/L) |
|---|---|
| Resazurin | 0.0001 |
| KH2PO4 | 2 |
| K2HPO4 | 2 |
| MgCl2•6H2O | 0.2 |
| (NH4)2•SO4 | 5 |
| L-Glycine | 0.075 |
| L-Valine | 0.117 |
| L-Leucine | 0.131 |
| L-Isoleucine | 0.131 |
| L-Methionine | 0.149 |
| L-Histidine | 0.155 |
| L-Arginine | 0.174 |
| L-Phenylalanine | 0 |
| L-Tyrosine | 0.181 |
| L-Tryptophan | 0.204 |
| NaHCO3 | 2.5 |
| Cysteine HCl | 0.5 |
| Glucose | 3.603 |
| Trace Mineral Supplement(g/L) | 10ml |
| EDTA | 0.5 |
| MgSO4•7H2O | 3.0 |
| MnSO4•H2O | 0.5 |
| NaCl | 1.0 |
| FeSO4•7H2O | 0.1 |
| Co(NO3)2•6H2O | 0.1 |
| CaCl2 (anhydrous) | 0.1 |
| ZnSO4•7H2O | 0.1 |
| CuSO4•5H2O | 0.010 |
| AlK(SO4)2 (anhydrous) | 0.010 |
| H3BO3 | 0.010 |
| Na2MoO4•2H2O | 0.010 |
| Na2SeO3 (anhydrous) | 0.001 |
| Na2WO4•2H2O | 0.010 |
| NiCl2•6H2O | 0.020 |
| Vitamin Supplement (mg/L) | 10ml |
| Folic acid | 2.0 |
| Pyridoxine hydrochloride | 10.0 |
| Riboflavin | 5.0 |
| Biotin | 2.0 |
| Thiamine | 5.0 |
| Nicotinic acid | 5.0 |
| Calcium Pantothenate | 5.0 |
| Vitamin B12 | 0.1 |
| p-Aminobenzoic acid | 5.0 |
| Thioctic acid | 5.0 |
| Monopotassium phosphate | 900.0 |
DATA AND SOFTWARE AVAILABILITY
Data deposition.
The raw sequence reads for all bacterial genomes have been deposited in the NCBI Sequence Read Archive (SRA) database in FASTQ format and genome assemblies have been deposited in NCBI genomes database in FASTA format. The accession number of the connected BioProject is PRJNA512876. The accession numbers for each raw sequence file are indicated in Table S4.
METHOD DETAILS
Bacterial growth conditions
For PRESTO-Tango screening, all commensals were cultured in gut microbiota medium or Gifu broth for 2 days in an anaerobic chamber (Coy) and commensal supernatants were sterilized by high-speed centrifugation and sterile filtration (0.22 μm). For in vitro studies, M. morganii was cultured in minimal medium (MM), or MM with 10 mM L-Phe, 2.5mM L-Tyr, 10 mM L-DOPA, or 10mM L-His for 24 hours. Bacterial supernatants were analyzed by LC-MS.
PRESTO-Tango Assay
HTLA cells, a HEK293 cell line that stably expresses β-arrestin-TEV and tTA-Luciferase (a kind gift from Gilad Barnea, Brown University), were plated in 96-well tissue culture plates (Eppendorf) in DMEM containing 10% FBS and 1% Penicillin/Streptomycin. One day after plating (after reaching approximately 90% confluence), 200 ng per well GPCR-Tango plasmids (Kroeze et al., 2015) in 20 μl DMEM were mixed with 400 ng polyethylenimine (Polysciences) in an equal volume of DMEM and incubated for 20 minutes at room temperature before adding the transfection mixture to the HTLA cells. 16–24 hrs after transfection, medium was replaced with 180μl fresh DMEM containing 1% Penicillin/Streptomycin and 10mM HEPES and 20μl bacterial medium alone or bacterial supernatant. All bacteria were cultured in gut microbiota medium or Gifu under anaerobic conditions for 2 days and bacterial supernatant was isolated via high-speed centrifugation followed by filtration with a 0.22 μm filter. Bacteria that failed to reach an OD of 0.5 after 2 days or caused obvious cell toxicity (e.g., Clostridum perfringens isolates) were excluded from further analysis. Supernatants were aspirated 16–24 hr after stimulation and 50 μl per well of Bright-Glo solution (Promega) diluted 20-fold with PBS containing 20mM HEPES was added into each well. After 20 min incubation at room temperature, luminescence was quantified using a Spectramax i3x (Molecular Devices). Activation fold for each sample was calculated by dividing relative luminescence units (RLU) for each condition by RLUs from media alone controls. No stimulation and bacterial media alone controls were included as separate conditions in each experiment to correct for day to day and experiment to experiment variability across the screen.
Histamine ELISA
All strains were cultured in gut microbiota medium with or without 1% L-His for 24 hours and supernatants were collected via high-speed centrifugation. Cecal and colonic contents and fecal samples were collected and weighed; all samples were suspended in PBS at a ratio of 1:2 (w/v) and were homogenized by vortexing. Serum and brains were collected, weighed and suspended in PBS at a ratio of 1:2 (w/v). Brains were homogenized by passing through a 21G needle fifty times. All samples were centrifuged and supernatants were collected for histamine ELISA according to the manufacturer’s protocol.
Colonization of germ-free mice
Germ-free C57Bl/6 mice were colonized via oral gavage with 200μl of individual bacterial cultures or mixed bacterial consortia. Mock communities A and B consisted of the following taxa: Community A: Bacteroides spp; P. distasonis; Peptoniphilus spp; B. ovatus; Clostridiales UC/UC; Lachnospiraceae UC/UC; C. stercoris; B. uniformis and Parabacteroides spp.; and Community B: Streptococcus spp; C. perfringens; B. fragilis; Erisipelotrichaceae spp; C. aerofaciens; Bacteroides UC; B. producta; Allobaculum spp and Oscillospira spp. All strains were grown to roughly mid-log phase in GMM, mixed in equal ratios based on optical density, and then frozen at −80 °C in GMM containing 20% glycerol in rubber capped 2ml Wheaton vials until use. All gnotobiotic mice were maintained in Techniplast P Isocages and manipulated aseptically for the duration of the experiment. 16S rRNA gene sequencing of the V4 region to confirm colonization and microbial composition was performed essentially as described previously (Palm et al., 2014) except that data processing and analysis was done using QIIME2-DADA2 (Caporaso et al., 2010).
Fecal output assay.
Individual mice were housed in an empty container (1/4 gallon) for 1 hour after which time the fecal pellets were counted and weighed. For mice fed with L-His, mice were given water containing 1% L-His ad libitum for 2 weeks before fecal output measurements. Based on an average water intake of 4 ml/ms/day combined with the existing dietary histidine present in conventional mouse chow (5 g/kg; Envigo), feeding of 1% L-His ad libitum in the water is equivalent to an overall histidine concentration of ~15g/kg in the diet. For reference, histidine rich foods such as soy protein, egg white, parmesan cheese, cured pork, and beef contain roughly 23, 20.5, 16, 16, and 14 g/kg of histidine, respectively (https://ndb.nal.usda.gov/ndb/nutrients/report/).
CRE-SEAP Assay.
96-well plates were pretreated with 30 μl poly-D-lysine (10ug/ml in water) and incubated at room temperature for 30 minutes. Plates were washed twice with 100 μl PBS and HEK293T were seeded in 100 μl DMEM+10% FBS+1% Penicillin/Streptomycin in each well. When cells were 90% confluent, plasmids were transfected using PEI reagent at a ratio of 1:2. For DRD1, DRD5 and TA1 (which couple to Gαs protein), HEK293T cells were transfected with 50 ng GPCR and 50 ng CRE-SEAP reporter plasmid per well; For DRD2, DRD3, DRD4, GPR56 and GPR97, cells were transfected with 50 ng GPCR, 50 ng CRE-SEAP reporter plasmid (BD Biosciences) and 2.5 ng Gαs- Gαo or Gαs- Gαt chimeras (a kind gift from Stephen Liberles, Harvard; (Liberles and Buck, 2006) per well. 6 hours after transfection, medium from the wells was replaced with 180 μl serum-free DMEM and 20 μl DMEM containing putative GPCR ligands was added to each well. After incubating for 48 hours at 37°C, followed by 2 hours of incubation at 70°C, supernatants from each well were mixed with an equal volume of 0.12 mM 4-methylumbelliferyl phosphate in 2 M diethanolamine bicarbonate, pH 10, and incubated at room temperature for 10 minutes. Fluorescence was measured using a SpectraMax plate reader (Molecular Devices).
Histamine Receptor Antagonist Treatment.
For histamine receptor antagonist treatment, 20 uM Desloratadine (HRH1 antagonist), Tiotidine (HRH2 antagonist), Iodophenpropit (HRH3 antagonist) and A987306 (HRH4 antagonist) were added to the drinking water. Colon motility was measured after one week of ad libitum treatment.
General Metabolomic Procedures.
NMR spectra were taken using an Agilent 600 MHz NMR system with a cryoprobe. High-resolution MS and tandem MS (MS/MS) data were obtained using an Agilent iFunnel 6550 ESIHRMS-QTOF (Electron Spray Ionization-High Resolution Mass Spectrometry-Quadrupole Time-of-Flight) instrument on Phenomenex Kinetex 5 μm C18 100Å (4.6 × 250 mm) columns. The Agilent 1260 Infinity system with a Phenomenex Kinetex 5 μm C18 100Å column (4.6 × 250 mm) or an Agilent Poroshell 120 EC-C18 2.7 μm (3.0 × 50 mm) column and a photodiode array (PDA) detector was used for routine sample analysis. An Agilent Prepstar HPLC system with an Agilent Polaris C18-A 5 μm (21.2 × 250 mm) column were used for sample fractionation and purification.
Metabolite Isolation and Purification.
B. thetaiotaomicron strain C11 was grown in 10 mL of gut microbiota medium under anaerobic conditions at 37°C for 24hr. Supernatant was harvested, lyophilized and extracted with 2mL methanol. The crude extract was then dried and fractionated using an Agilent Prepstar HPLC system with an Agilent Polaris C18-A 5 μm (21.2 × 250 mm) column. The gradient used was 10–50% acetonitrile in water (with 0.01% trifluoroacetic acid) for 30min, then 100% for 5min. The fractions, which were collected every minute, were dried, resuspended in PBS buffer, and tested for bioactivity using PRESTO-Tango. The active fraction was characterized using ESI-HRMSQTOF and NMR analyses (instruments listed as above). Stereochemistry was confirmed by advanced Marfey’s analysis (Figure S5D) (Bhushan and Bruckner, 2011).
Metabolite Quantitation.
Electro Spray Ionization-Triple Quadrupole-Tandem Mass Spectrometry ESI-QQQ-MS/MS was run using Multiple Reaction Monitoring (MRM) mode. An Agilent 6490 ESI-QQQ-MS/MS instrument with a Phenomenex Kinetex 1.7 μm C18 100Å (100 × 2.10mm) column was used for quantitation and calibration. Each standard (L-Phenylalanine, Phenethylamine, Histamine, Tyramine, Dopamine) was optimized using an Agilent Mass Hunter Optimizer. A calibration curve for each standard was established using various concentrations (0 – 25μM range) in triplicate. The gradient constituted 10–100% acetonitrile in ddH2O (with 0.1% Formic Acid), then a wash step with 100% acetonitrile. The triplicate data was then subjected to linear regression analysis to produce a linear calibration curve. Processing of the experimental samples involved lyophilization and extraction with 100% MeOH (20% volume of original culture volume) before injecting samples. Sample absorbance was subjected to linear calibration to calculate concentrations.
Whole-Genome Sequencing and Annotation, HMP2 Data Mining and Data Deposition.
Whole-genome sequencing.
Bacterial DNA was extracted using the DNeasy Ultraclean Microbial Kit (Qiagen) according to the manufacturer’s instructions. Sequencing libraries were prepared using the Nextera XT library preparation kit (Illumina) according to the manufacturer’s instructions and sequenced on a NovaSeq (2×150; Illumina).
De novo genome assembly.
Genome assembly and annotation were performed essentially as described in Dodd et al. (Dodd et al., 2017). Briefly, all Illumina paired-end reads were filtered and trimmed using Trimmomatic v.0.38 (Bolger et al., 2014) with the following parameters: ILLUMINACLIP: NexteraPE-PE.fa:2:30:12:1:true LEADING:3 TRAILING:3 MAXINFO:40:0.994 MINLEN:36. The four output files after trimming included two (forward and reverse) FASTQ files with paired reads and two FASTQ files with unpaired reads. All four files from each strain were assembled into contigs using SPAdes 3.13.0 (Nurk et al., 2013) with the default parameters for paired-end libraries. Genome coverage was calculated by BBMap (http://sourceforge.net/projects/bbmap/). Summary statistics for each genome assembly and alignment are shown in Table S3.
Genome annotation.
For each genome assembly, scaffolds longer than 2000 bp were uploaded to the Rapid Annotation using Subsystem Technology (RAST) server (Aziz et al., 2008) for annotation (using the default RASTtk pipeline). The annotated genomes were downloaded as spreadsheets (Excel XLS format), and the summary of genome annotations as well as the detailed annotations for each strain are shown in Table S3.
HMP2 data acquisition and analysis.
Publicly available metagenome and metabolome data from the HMP2 were downloaded from The Inflammatory Bowel Disease Multi’omics Database (IBDMDB), which was funded by the NIH Human Microbiome Project NIDDK U54DE023798 (https://ibdmdb.org). The IBDMDB provides longitudinal meta’ome data on the microbiome of subjects with three clinical diagnoses: nonIBD, CD and UC. Raw metagenomic sequencing data was pre-processed and used to generate taxonomic and functional profiles by the HMP team. The pipeline employed two steps: (1) MetaPhlAn2 (Truong DT, 2015)-based taxonomic profiling, which uses clade-specific marker genes to identify species-level microbial taxa and their relative abundances using metagenomic data; and (2) HUMAnN2 (Franzosa EA, 2018)-based functional profiling. Briefly, HUMAnN2 implements a tiered search strategy to profile the functional content (gene family, functional pathway, etc.) of a meta’ome sample at species-level resolution. In the first tier, based on the known microbial species in a sample (as identified by MetaPhlAn2), HUMAnN2 constructs a custom gene sequence database for the samples by concatenating precomputed, functionally annotated pangenomes of detected species. In the second tier, nucleotide-level mapping of all reads against the sample’s pangenome database is performed. In the final search tier, reads that do not align to an identified species’ pangenome are then subjected to translated search against a comprehensive, non-redundant protein sequence database (UniRef90 or UniRef50). Per-gene alignment statistics are weighted based on alignment quality, coverage and sequence length to yield gene abundance values. Both taxonomic determinations and functional gene abundances are normalized as relative abundances to facilitate comparisons between samples with different sequencing depths.
The merged tables of taxonomic profiles (https://ibdmdb.org/tunnel/products/HMP2/WGS/1818/taxonomic_profiles.tsv.gz) and functional profiles (https://ibdmdb.org/tunnel/products/HMP2/WGS/1818/ecs.tsv.gz) from metagenomic analyses, the merged table from metabolomics analysis (https://ibdmdb.org/tunnel/products/HMP2/Metabolites/1723/HMP2_metabolomics.csv.gz), and the HMP2 metadata (https://ibdmdb.org/tunnel/products/HMP2/Metadata/hmp2_metadata.csv), were downloaded and analyzed to evaluate the distribution of abundance of histidine decarboxylase and histamine in this cohort. For participants whose clinical diagnosis changed during the course of the study, data points were assigned to the clinical diagnosis at the time of sample collection.
Quantification and Statistical Analysis.
Statistical analysis was performed using Graphpad Prism version 7.0. Data were assessed for normal distribution and plotted in the figures as mean ± SEM. No samples or animals were excluded from the analyses. One-way ANOVA and post hoc analysis with Tukey’s test was used to compare the difference between treatment groups. Kaplan-Meier and Log rank analysis was used for survival experiments. Kruskall-Wallis with Dunn’s multiple comparisons was used to analyze metagenomic and metabolomic data from the HMP; p-values for Kruskall-Wallis are approximated based on the chi-squared distribution and account for rank ties. Samples sizes are indicated in each figure legend and significant differences are indicated in the figures by *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
Table S1. Taxonomic assignments of the gut microbiota culture collection, related to Figure 1.
Activation of DRD1–5 (A) and HRH1–4 (B) by supernatants from 144 human gut bacteria cultured in gut microbiota medium (GMM) as measured by PRESTO-Tango.
(A) Mammalian dopamine metabolism.
(B) Phenethylamine and tyramine serve as selective DRD2/DRD3/DRD4 agonists. Activation of DRD1–5 by metabolites in the mammalian dopamine metabolism pathway was measured via PRESTO-Tango.
(C) DRD1–5 activation by titrating doses of tyramine, dopamine and phenethylamine was measured by PRESTO-Tango (D). n=3 replicates per sample.
(D) Calibration curve for phenethylamine and tyramine on QQQ-MS/MS instrument.
(E) Quantification of phenethylamine production by M. morganii strains via QQQ-MS/MS.
Data in all panels are representative of at least two independent experiments.
(A) Activation of Gαs-dependent signaling downstream of DRD1, 5 and TAAR1 by titrating doses of phenethylamine and related chemicals was measured by the CRE-SEAP assay. n=3 replicates per sample.
(B) Activation of G protein-dependent signaling downstream of DRD2–4 by titrating doses phenethylamine and related chemicals was measured by the CRE-SEAP assay. A Gαs-Gαo fusion was used to redirect DRD2–4 to Gαs and enable use of the CRE-SEAP assay. n=3 replicates per sample.
(C) OD values for 24 hour cultures of M. morganii grown in minimal medium (MM) with or without L-Phe, L-Tyr, L-DOPA or L-His. n=3 replicates per sample.
Data in all panels are representative of at least two independent experiments.
(A) Groups of female germ-free C57Bl/6 mice were colonized with mock communities of 9 or 10 phylogenetically diverse human gut bacteria (Mock Community A or B) or monocolonized with M. morganii C135. Mice were fed a conventional diet with or without administration of 1% L-His ad libitum in the drinking water. Histamine concentrations in serum were measured via ELISA. n=3–5 mice per group.
(B-C) M. morganii primarily inhabits the cecum and colon. Groups of female germ-free C57Bl/6 mice were colonized with mock communities of 9 or 10 phylogenetically diverse gut microbes (Mock community A and B, respectively) with or without M. morganii C135. M. morganii CFUs can be distinguished from other bacteria based on their purple halos when plated on modified Niven’s agar. Gastric, small intestinal, cecal and colonic contents from mice colonized with Mock communities A or B and M. morganii were plated on Modified Niven’s agar to determine M. morganii colonization levels at various intestinal loci. Stacked barplot represents relative abundance of bacterial taxa in mice colonized with Mock community A plus M. morganii based on 16S rRNA gene sequencing (see also Table S3). n=4 mice per group.
(D) Groups of female germ-free C57Bl/6 mice were colonized with a mock community of 9 phylogenetically diverse human gut bacteria (Mock Community A) with or without M. morganii C135. Mice were fed a conventional diet and administered 1% L-His ad libitum in the drinking water. Histamine concentrations in serum were measured via ELISA. n=3–5 mice per group.
(E) Contribution of individual species to the relative abundance of histidine decarboxylase genes in the microbiomes of patients with IBD (CD and UC) as compared to controls (non-IBD). Metagenomic data from longitudinal stool samples from IBD patients (publicly available from the Human Microbiome Project 2; iHMP) were analyzed for the presence and relative abundance of histidine decarboxylase genes (see methods for details). Data shown are a compilation of all data across multiple collection timepoints.
(F) Quantification of phenethylamine (PEA) in cecum, colon, serum, and brain from mice monocolonized with M. morganii C135 and treated with or without phenelzine (MAOI) via QQQ-MS/MS. n=4 mice per group.
(G) Accumulation of phenethylamine (PEA) in serum and brains of mice monocolonized with M. morganii C135 and treated with or without phenelzine (MAOI) as measured via QQQ-MS/MS. n=4 mice per group.
Data in all panels are representative of at least two independent experiments.
Data are presented as mean ± SEM. One-way ANOVA with Tukey’s post-hoc test (A and E), *p < 0.05, ***p < 0.001.
(A) OD600 values of indicated Bacteroides and Parabacteroides strains cultured in gut microbiota medium (GMM) for 24 hours. n=3 replicates per isolate.
(B) 1H NMR spectrum of active fraction 11 in MeOD revealed Phe as the major component.
(C) Advanced Marfey’s analysis verified the stereochemistry of Phe in fraction 11 to be L-Phe. D-Phe in the active fraction was not detected. FDAA is 1-fluoro-2,4-dinitrophenyl-5-L-alanine amide (Marfey’s Reagent).
(D) L-Phe and L-Tyr stereoselectively activate the orphan receptor GPR56/AGRG1. Activation of GPR56/AGRG1 by titrating doses of pure L-Phe, L-Tyr, D-Phe, and D-Tyr (in L-Phe and LTyr-free medium) was measured via GPR56-Tango. n=3 replicates per sample.
(E) L-Phe-induced Tango activation is GPR56/AGRG1-dependent. Luciferase expression (RLU) was measured after stimulation of cells transfected with GPR56-Tango or empty vector with titrating doses of L-Phe. n=3 replicates per sample.
(F) L-Phe-induced activation of G protein-dependent signaling in HEK cells is GPR56-dependent. Activation of G proteins downstream of GPR56/AGRG1 by L-Phe as measured by the CRE-SEAP assay. Gαs-Gαt and Gαs-Gαo chimeras were used to redirect GPR56/AGRG1 signaling to Gαs and enable use of the CRE-SEAP assay. Cells transfected with DRD2-Tango and Gαs-Gαt and Gαs-Gαo chimeras failed to respond to L-Phe. n=3 replicates per sample.
(G) The extracellular domain of GPR56/AGRG1 is indispensable for GPR56/AGRG1 activation by L-Phe. Activation of GPR56 or GPR56-NT (a mutant lacking the extracellular domain) by titrating doses of L-Phe was measured via PRESTO-Tango. n=3 replicates per sample.
Data in all panels except B and C are representative of at least three independent experiments. Data are presented as mean ± SEM. One-way ANOVA with Tukey’s post-hoc test *p < 0.05, ****p < 0.0001.
(A) L-Phe activates GPR56/AGRG1 and GPR97/AGRG3. Activation of all orphan, adhesion and other potential amino acid-sensing GPCRs by L-Phe was evaluated via PRESTO-Tango. n=3 replicates per sample.
(B) L-Phe specifically activates GPR97/AGRG3. Activation of GPR97/AGRG3 by titrating doses of L-Phe, L-Tyr, L-Trp, and L-His was measured via GPR97 PRESTO-Tango. n=3 replicates per sample.
(C) The extracellular domain of GPR97/AGRG3 is indispensable for GPR97/AGRG3 activation by L-Phe. Activation of GPR97 or GPR97-NT (a mutant lacking the extracellular domain) by titrating doses of L-Phe was measured via PRESTO-Tango. n=3 replicates per sample.
(D) L-Phe specifically activates GPR97/AGRG3. Activation of GPR97/AGRG3 by titrating doses of L-Phe, L-Tyr, L-Trp, and L-His was measured via GPR97 PRESTO-Tango in media lacking L-Phe and L-Tyr. n=3 replicates per sample.
(E) L-Phe activates G protein-dependent signaling downstream of GPR97/AGRG3. Activation of G proteins downstream of GPR97/AGRG3 by L-Phe as measured by the CRE-SEAP assay. Gαs-Gαt and Gαs-Gαo chimeras were used to redirect GPR97/AGRG3 signaling to Gαs and enable use of the CRE-SEAP assay. n=3 replicates per sample.
(F) GPR56/AGRG1 and GPR97/AGRG3 are evolutionarily related. A phylogenetic tree for a subset of GPCRs, including all adhesion GPCRs, was constructed and visualized with equal branch lengths using gpcrdb.org, PHYLIP and jsPhyloSVG.
Data in all panels are representative of at least three independent experiments. Data are presented as mean ± SEM.
Table S2. PRESTO-Tango screening data, related to Figure 2 and Figure 6. GPCR activation or inhibition was determined by comparing stimulation with a given bacterial supernatant to stimulation of the same GPCR with bacterial media alone (GMM, Tab 1; or Gifu, Tab 2) and is reported as magnitude of activation (log 2).
Tab 1. Potential GPCR activation by components of GMM was determined by comparing stimulation with GMM to stimulation of the same GPCR with PBS. Specific GPCRs that were activated by GMM by over two-fold are displayed here along with corresponding activation by supernatants from commensal microbes (as compared to PBS).
Tab 2. Gut microbiota composition in mice colonized with Mock Consortium A plus M. morganii (displayed graphically in Figure S5C).
Table S4. Whole genome assembly metrics, accession numbers and RAST annotations for M. morganii, L. reuteri and B. theta strains, related to Figures 3 and 6.
Tab 1. Predicted histidine decarboxylases in M. morganii and L. reuteri strains.
Tab 2. Histidine decarboxylases in all published M. morganii genomes (homology relative to the identified histidine decarboxylase from M. morganii C135).
Tab 3. Predicted decarboxylases present in both M. morganii C135 and C28.
Tab 4. Enzymes involved in L-Phe synthesis (shikimate pathway) in B. theta strains C34, C80, and ATCC 29741.
(A) Activation of CHRMs and DRDs by titrating doses of acetylcholine and dopamine as measured by PRESTO-Tango. n=3 replicates per sample.
(B) Activation of GPCRs by defined GPCR ligands as measured by PRESTO-Tango. Activation is depicted on a log 2 scale as a heatmap of 314 GPCRs versus ligands.
Highlights.
Screening of gut microbiota metabolomes against hundreds of G protein-coupled receptors
Phylogenetically diverse gut microbes produce agonists for many GPCRs including orphans
Bioactivity-based screening reveals diet-microbe-host and microbe-microbe-host axes
Microbiota-derived GPCR ligands impact local and systemic host physiology
ACKNOWLEDGEMENTS
The authors thank members of the Palm laboratory for useful discussions and J. Henao-Mejia, A. Wang, and R. Medzhitov for critical reading of the manuscript. The NWP lab acknowledges support from Yale University School of Medicine and the Kingsley Award in Biomedical Research, the Helmsley Foundation, the Richard and Susan Smith Family Foundation, the Rainin Foundation, the Global Probiotics Council, the National Institutes of Health (K22 AI123477; R21 AI137935), TATA Sons Limited and Artizan Biosciences. The JMC lab acknowledges support from the Camille and Henry Dreyfus Foundation (TC-17–011), the Damon Runyon Cancer Research Foundation (DRR-39–16), the National Institutes of Health (1DP2-CA186575), the Burroughs Wellcome Foundation (1016720) and Yale University. The AMR lab acknowledges support for this work from an NIH Director’s Early Independence Award (DP5-OD023088) and the Robert T. McCluskey Foundation.
DECLARATION OF INTERESTS
NWP is a co-founder of, consultant for, and receives research funding from Artizan Biosciences. HC, AMR, and NWP are inventors on a patent application based on these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adibi S M. DW. (1973). Protein Digestion in Human Intestine as Reflected in Luminal, Mucosal, and Plasma Amino Acid Concentrations after Meals. The Journal of Clinical Investigation 52, 1586–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque EX, Pereira EF, Alkondon M, and Rogers SW (2009). Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev 89, 73–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amisten S, Salehi A, Rorsman P, Jones PM, and Persaud SJ (2013). An atlas and functional analysis of G-protein coupled receptors in human islets of Langerhans. Pharmacol Ther 139, 359–391. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19, 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcik W, Pugin B, Westermann P, Perez NR, Ferstl R, Wawrzyniak M, Smolinska S, Jutel M, Hessel EM, Michalovich D, et al. (2016). Histamine-secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol 138, 1491–1494 e1497. [DOI] [PubMed] [Google Scholar]
- Barnea G, Strapps W, Herrada G, Berman Y, Ong J, Kloss B, Axel R, and Lee KJ (2008). The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci U S A 105, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, and Gainetdinov RR (2011). The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev 63, 182–217. [DOI] [PubMed] [Google Scholar]
- Bhushan R, and Bruckner H (2011). Use of Marfey’s reagent and analogs for chiral amino acid analysis: assessment and applications to natural products and biological systems. J Chromatogr B Analyt Technol Biomed Life Sci 879, 3148–3161. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowsky B, Adham N, Jones KA, Raddatz R, Artymyshyn R, Ogozalek KL, Durkin MM, Lakhlani PP, Bonini JA, Pathirana S, et al. (2001). Trace amines: identification of a family of mammalian G protein-coupled receptors. Proc Natl Acad Sci U S A 98, 8966–8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, et al. (2003). The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem 278, 11312–11319. [DOI] [PubMed] [Google Scholar]
- Bunzow J S. MS; Arttamangkul S; Harrison LM; Zhang G; Quigley DI; Darland T; Suchland KL; Pasumanmula S; Kennedy JL; Olson SB; Magenis RE; Amara SG; Grandy DK. (2001). Amphetamine, 3,4-Methylenedioxymethamphetamine, Lysergic Acid Diethylamide, and Metabolites of the Catecholamine Neurotransmitters Are Agonists of a Rat Trace Amine Receptor. Molecular Pharmacology 60, 1181–1188. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, and Holmes SP (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen LJ, Esterhazy D, Kim SH, Lemetre C, Aguilar RR, Gordon EA, Pickard AJ, Cross JR, Emiliano AB, Han SM, et al. (2017). Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 549, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen LJ, Kang HS, Chu J, Huang YH, Gordon EA, Reddy BV, Ternei MA, Craig JW, and Brady SF (2015). Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc Natl Acad Sci U S A 112, E4825–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, et al. (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, and Fischbach MA (2015). HUMAN MICROBIOTA. Small molecules from the human microbiota. Science 349, 1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duner P, Al-Amily IM, Soni A, Asplund O, Safi F, Storm P, Groop L, Amisten S, and Salehi A (2016). Adhesion G Protein-Coupled Receptor G1 (ADGRG1/GPR56) and Pancreatic beta-Cell Function. J Clin Endocrinol Metab 101, 4637–4645. [DOI] [PubMed] [Google Scholar]
- Durocher Y, Perret S, Thibaudeau E, Gaumond MH, Kamen A, Stocco R, and Abramovitz M (2000). A reporter gene assay for high-throughput screening of G-protein-coupled receptors stably or transiently expressed in HEK293 EBNA cells grown in suspension culture. Anal Biochem 284, 316–326. [DOI] [PubMed] [Google Scholar]
- Eun CS, Kwak MJ, Han DS, Lee AR, Park DI, Yang SK, Kim YS, and Kim JF (2016). Does the intestinal microbial community of Korean Crohn’s disease patients differ from that of western patients? BMC Gastroenterol 16, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedorowicz J S. KL. (2004). The Role of Monoamine Oxidase Inhibitors in Current Psychiatric Practice. J Psychiatr Pract 10, 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA (2018). Microbiome: Focus on Causation and Mechanism. Cell 174, 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover V S. M; Owen F; Riley GJ. (1977). Dopamine is a monoamine oxidase B substrate in man. Nature 265, 80–81. [DOI] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, and Gordon JI (2011). Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A 108, 6252–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo CJ, Chang FY, Wyche TP, Backus KM, Acker TM, Funabashi M, Taketani M, Donia MS, Nayfach S, Pollard KS, et al. (2017). Discovery of Reactive Microbiota-Derived Metabolites that Inhibit Host Proteases. Cell 168, 517–526 e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, and Turnbaugh PJ (2013). Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husted AS, Trauelsen M, Rudenko O, Hjorth SA, and Schwartz TW (2017). GPCR-Mediated Signaling of Metabolites. Cell Metab 25, 777–796. [DOI] [PubMed] [Google Scholar]
- Integrative, H.M.P.R.N.C. (2014). The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe 16, 276–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irsfeld M, Spadafore M, and Pruss BM (2013). beta-phenylethylamine, a small molecule with a large impact. Webmedcentral 4. [PMC free article] [PubMed] [Google Scholar]
- Jutel M, Akdis M, and Akdis CA (2009). Histamine, histamine receptors and their role in immune pathology. Clin Exp Allergy 39, 1786–1800. [DOI] [PubMed] [Google Scholar]
- Kim H, Dwyer L, Song JH, Martin-Cano FE, Bahney J, Peri L, Britton FC, Sanders KM, and Koh SD (2011). Identification of histamine receptors and effects of histamine on murine and simian colonic excitability. Neurogastroenterol Motil 23, 949–e409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Ben-Gigirey B, Barros-Velazquez J, Price RJ, and An H (2000). Histamine and biogenic amine production by Morganella morganii isolated from temperature-abused albacore. J Food Prot 63, 244–251. [DOI] [PubMed] [Google Scholar]
- Kishore A, Purcell RH, Nassiri-Toosi Z, and Hall RA (2016). Stalk-dependent and Stalk-independent Signaling by the Adhesion G Protein-coupled Receptors GPR56 (ADGRG1) and BAI1 (ADGRB1). J Biol Chem 291, 3385–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeze WK, Sassano MF, Huang XP, Lansu K, McCorvy JD, Giguere PM, Sciaky N, and Roth BL (2015). PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol 22, 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, et al. (2014). A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, Brezillon S, Dupriez V, Vassart G, Van Damme J, et al. (2003). Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem 278, 25481–25489. [DOI] [PubMed] [Google Scholar]
- Liberles SD, and Buck LB (2006). A second class of chemosensory receptors in the olfactory epithelium. Nature 442, 645–650. [DOI] [PubMed] [Google Scholar]
- Lovenberg W W. H & Udenfriend S (1962). Aromatic amino acid decarboxylase. The Journal of Biological Chemistry 237, 89–93. [PubMed] [Google Scholar]
- Lovitt RW, Morris JG, and Kell DB (1987). The growth and nutrition of Clostridium sporogenes NCIB 8053 in defined media. J Appl Bacteriol 62, 71–80. [DOI] [PubMed] [Google Scholar]
- Mavromatis P Q. PC. (2002). ModifiIcation of Niven’s Medium for the Enumeration of Histamine-Forming Bacteria and Discussion of the Parameters Associated with Its Use. Journal of Food Protection 65, 546–551. [DOI] [PubMed] [Google Scholar]
- Milshteyn A, Colosimo DA, and Brady SF (2018). Accessing Bioactive Natural Products from the Human Microbiome. Cell Host Microbe 23, 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20, 714–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldendorf W (1971). Brain uptake of radiolabeled amino acids, amines, and hexoses after arterial injection. American Journal of Physiology 221, 1629–1639. [DOI] [PubMed] [Google Scholar]
- Özoğul F (2004). Production of biogenic amines by Morganella morganii, Klebsiella pneumoniae and Hafnia alvei using a rapid HPLC method. European Food Research and Technology 219, 465–469. [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, Forslund K, Hildebrand F, Prifti E, Falony G, et al. (2016). Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381. [DOI] [PubMed] [Google Scholar]
- Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, and Shulman GI (2016). Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature 534, 213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezeshki A, Zapata RC, Singh A, Yee NJ, and Chelikani PK (2016). Low protein diets produce divergent effects on energy balance. Scientific Reports 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell R H. RA. (2018). Adhesion G Protein–Coupled Receptors as Drug Targets. Annu Rev Pharmacol Toxicol 58, 429–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandraih C S. N; Bar KJ; Baker G; Yeragani VK. (2011). Antidepressants: From MAOIs to SSRIs and more. Indian J Psychiatry 53, 180–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PH, MR; Panikov N; Michaud M; Gallini CA; Bohlooly-Y M; Glickman JN; Garret WS. (2013). The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 341, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolinska S, Jutel M, Crameri R, and O’Mahony L (2014). Histamine and gut mucosal immune regulation. Allergy 69, 273–281. [DOI] [PubMed] [Google Scholar]
- Sotnikova TD, Budygin EA, Jones SR, Dykstra LA, Caron MG, and Gainetdinov RR (2004). Dopamine transporter-dependent and -independent actions of trace amine beta-phenylethylamine. J Neurochem 91, 362–373. [DOI] [PubMed] [Google Scholar]
- Tan JK M. C; Mariño E; Macia L and Mackay CR. (2017). Metabolite-Sensing G Protein-Coupled Receptors-Facilitators of Diet-Related Immune Regulation. AnnuRevImmunol 35, 371–402. [DOI] [PubMed] [Google Scholar]
- Thurmond RL, Gelfand EW, and Dunford PJ (2008). The role of histamine H1 and H4 receptors in allergic inflammation: the search for new antihistamines. Nat Rev Drug Discov 7, 41–53. [DOI] [PubMed] [Google Scholar]
- Tyagi P, Mandal MB, Mandal S, Patne SC, and Gangopadhyay AN (2009). Pouch colon associated with anorectal malformations fails to show spontaneous contractions but responds to acetylcholine and histamine in vitro. J Pediatr Surg 44, 2156–2162. [DOI] [PubMed] [Google Scholar]
- Wacker D, Stevens RC, and Roth BL (2017). How Ligands Illuminate GPCR Molecular Pharmacology. Cell 170, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RM, CDS; Burnett JR. (2008). Phenylketonuria- An Inborn Error of Phenylalanine Metabolism. Clin Biochem Rew 29, 31–41. [PMC free article] [PubMed] [Google Scholar]
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, and Hsiao EY (2015). Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Taxonomic assignments of the gut microbiota culture collection, related to Figure 1.
Activation of DRD1–5 (A) and HRH1–4 (B) by supernatants from 144 human gut bacteria cultured in gut microbiota medium (GMM) as measured by PRESTO-Tango.
(A) Mammalian dopamine metabolism.
(B) Phenethylamine and tyramine serve as selective DRD2/DRD3/DRD4 agonists. Activation of DRD1–5 by metabolites in the mammalian dopamine metabolism pathway was measured via PRESTO-Tango.
(C) DRD1–5 activation by titrating doses of tyramine, dopamine and phenethylamine was measured by PRESTO-Tango (D). n=3 replicates per sample.
(D) Calibration curve for phenethylamine and tyramine on QQQ-MS/MS instrument.
(E) Quantification of phenethylamine production by M. morganii strains via QQQ-MS/MS.
Data in all panels are representative of at least two independent experiments.
(A) Activation of Gαs-dependent signaling downstream of DRD1, 5 and TAAR1 by titrating doses of phenethylamine and related chemicals was measured by the CRE-SEAP assay. n=3 replicates per sample.
(B) Activation of G protein-dependent signaling downstream of DRD2–4 by titrating doses phenethylamine and related chemicals was measured by the CRE-SEAP assay. A Gαs-Gαo fusion was used to redirect DRD2–4 to Gαs and enable use of the CRE-SEAP assay. n=3 replicates per sample.
(C) OD values for 24 hour cultures of M. morganii grown in minimal medium (MM) with or without L-Phe, L-Tyr, L-DOPA or L-His. n=3 replicates per sample.
Data in all panels are representative of at least two independent experiments.
(A) Groups of female germ-free C57Bl/6 mice were colonized with mock communities of 9 or 10 phylogenetically diverse human gut bacteria (Mock Community A or B) or monocolonized with M. morganii C135. Mice were fed a conventional diet with or without administration of 1% L-His ad libitum in the drinking water. Histamine concentrations in serum were measured via ELISA. n=3–5 mice per group.
(B-C) M. morganii primarily inhabits the cecum and colon. Groups of female germ-free C57Bl/6 mice were colonized with mock communities of 9 or 10 phylogenetically diverse gut microbes (Mock community A and B, respectively) with or without M. morganii C135. M. morganii CFUs can be distinguished from other bacteria based on their purple halos when plated on modified Niven’s agar. Gastric, small intestinal, cecal and colonic contents from mice colonized with Mock communities A or B and M. morganii were plated on Modified Niven’s agar to determine M. morganii colonization levels at various intestinal loci. Stacked barplot represents relative abundance of bacterial taxa in mice colonized with Mock community A plus M. morganii based on 16S rRNA gene sequencing (see also Table S3). n=4 mice per group.
(D) Groups of female germ-free C57Bl/6 mice were colonized with a mock community of 9 phylogenetically diverse human gut bacteria (Mock Community A) with or without M. morganii C135. Mice were fed a conventional diet and administered 1% L-His ad libitum in the drinking water. Histamine concentrations in serum were measured via ELISA. n=3–5 mice per group.
(E) Contribution of individual species to the relative abundance of histidine decarboxylase genes in the microbiomes of patients with IBD (CD and UC) as compared to controls (non-IBD). Metagenomic data from longitudinal stool samples from IBD patients (publicly available from the Human Microbiome Project 2; iHMP) were analyzed for the presence and relative abundance of histidine decarboxylase genes (see methods for details). Data shown are a compilation of all data across multiple collection timepoints.
(F) Quantification of phenethylamine (PEA) in cecum, colon, serum, and brain from mice monocolonized with M. morganii C135 and treated with or without phenelzine (MAOI) via QQQ-MS/MS. n=4 mice per group.
(G) Accumulation of phenethylamine (PEA) in serum and brains of mice monocolonized with M. morganii C135 and treated with or without phenelzine (MAOI) as measured via QQQ-MS/MS. n=4 mice per group.
Data in all panels are representative of at least two independent experiments.
Data are presented as mean ± SEM. One-way ANOVA with Tukey’s post-hoc test (A and E), *p < 0.05, ***p < 0.001.
(A) OD600 values of indicated Bacteroides and Parabacteroides strains cultured in gut microbiota medium (GMM) for 24 hours. n=3 replicates per isolate.
(B) 1H NMR spectrum of active fraction 11 in MeOD revealed Phe as the major component.
(C) Advanced Marfey’s analysis verified the stereochemistry of Phe in fraction 11 to be L-Phe. D-Phe in the active fraction was not detected. FDAA is 1-fluoro-2,4-dinitrophenyl-5-L-alanine amide (Marfey’s Reagent).
(D) L-Phe and L-Tyr stereoselectively activate the orphan receptor GPR56/AGRG1. Activation of GPR56/AGRG1 by titrating doses of pure L-Phe, L-Tyr, D-Phe, and D-Tyr (in L-Phe and LTyr-free medium) was measured via GPR56-Tango. n=3 replicates per sample.
(E) L-Phe-induced Tango activation is GPR56/AGRG1-dependent. Luciferase expression (RLU) was measured after stimulation of cells transfected with GPR56-Tango or empty vector with titrating doses of L-Phe. n=3 replicates per sample.
(F) L-Phe-induced activation of G protein-dependent signaling in HEK cells is GPR56-dependent. Activation of G proteins downstream of GPR56/AGRG1 by L-Phe as measured by the CRE-SEAP assay. Gαs-Gαt and Gαs-Gαo chimeras were used to redirect GPR56/AGRG1 signaling to Gαs and enable use of the CRE-SEAP assay. Cells transfected with DRD2-Tango and Gαs-Gαt and Gαs-Gαo chimeras failed to respond to L-Phe. n=3 replicates per sample.
(G) The extracellular domain of GPR56/AGRG1 is indispensable for GPR56/AGRG1 activation by L-Phe. Activation of GPR56 or GPR56-NT (a mutant lacking the extracellular domain) by titrating doses of L-Phe was measured via PRESTO-Tango. n=3 replicates per sample.
Data in all panels except B and C are representative of at least three independent experiments. Data are presented as mean ± SEM. One-way ANOVA with Tukey’s post-hoc test *p < 0.05, ****p < 0.0001.
(A) L-Phe activates GPR56/AGRG1 and GPR97/AGRG3. Activation of all orphan, adhesion and other potential amino acid-sensing GPCRs by L-Phe was evaluated via PRESTO-Tango. n=3 replicates per sample.
(B) L-Phe specifically activates GPR97/AGRG3. Activation of GPR97/AGRG3 by titrating doses of L-Phe, L-Tyr, L-Trp, and L-His was measured via GPR97 PRESTO-Tango. n=3 replicates per sample.
(C) The extracellular domain of GPR97/AGRG3 is indispensable for GPR97/AGRG3 activation by L-Phe. Activation of GPR97 or GPR97-NT (a mutant lacking the extracellular domain) by titrating doses of L-Phe was measured via PRESTO-Tango. n=3 replicates per sample.
(D) L-Phe specifically activates GPR97/AGRG3. Activation of GPR97/AGRG3 by titrating doses of L-Phe, L-Tyr, L-Trp, and L-His was measured via GPR97 PRESTO-Tango in media lacking L-Phe and L-Tyr. n=3 replicates per sample.
(E) L-Phe activates G protein-dependent signaling downstream of GPR97/AGRG3. Activation of G proteins downstream of GPR97/AGRG3 by L-Phe as measured by the CRE-SEAP assay. Gαs-Gαt and Gαs-Gαo chimeras were used to redirect GPR97/AGRG3 signaling to Gαs and enable use of the CRE-SEAP assay. n=3 replicates per sample.
(F) GPR56/AGRG1 and GPR97/AGRG3 are evolutionarily related. A phylogenetic tree for a subset of GPCRs, including all adhesion GPCRs, was constructed and visualized with equal branch lengths using gpcrdb.org, PHYLIP and jsPhyloSVG.
Data in all panels are representative of at least three independent experiments. Data are presented as mean ± SEM.
Table S2. PRESTO-Tango screening data, related to Figure 2 and Figure 6. GPCR activation or inhibition was determined by comparing stimulation with a given bacterial supernatant to stimulation of the same GPCR with bacterial media alone (GMM, Tab 1; or Gifu, Tab 2) and is reported as magnitude of activation (log 2).
Tab 1. Potential GPCR activation by components of GMM was determined by comparing stimulation with GMM to stimulation of the same GPCR with PBS. Specific GPCRs that were activated by GMM by over two-fold are displayed here along with corresponding activation by supernatants from commensal microbes (as compared to PBS).
Tab 2. Gut microbiota composition in mice colonized with Mock Consortium A plus M. morganii (displayed graphically in Figure S5C).
Table S4. Whole genome assembly metrics, accession numbers and RAST annotations for M. morganii, L. reuteri and B. theta strains, related to Figures 3 and 6.
Tab 1. Predicted histidine decarboxylases in M. morganii and L. reuteri strains.
Tab 2. Histidine decarboxylases in all published M. morganii genomes (homology relative to the identified histidine decarboxylase from M. morganii C135).
Tab 3. Predicted decarboxylases present in both M. morganii C135 and C28.
Tab 4. Enzymes involved in L-Phe synthesis (shikimate pathway) in B. theta strains C34, C80, and ATCC 29741.
(A) Activation of CHRMs and DRDs by titrating doses of acetylcholine and dopamine as measured by PRESTO-Tango. n=3 replicates per sample.
(B) Activation of GPCRs by defined GPCR ligands as measured by PRESTO-Tango. Activation is depicted on a log 2 scale as a heatmap of 314 GPCRs versus ligands.
Data Availability Statement
Data deposition.
The raw sequence reads for all bacterial genomes have been deposited in the NCBI Sequence Read Archive (SRA) database in FASTQ format and genome assemblies have been deposited in NCBI genomes database in FASTA format. The accession number of the connected BioProject is PRJNA512876. The accession numbers for each raw sequence file are indicated in Table S4.