Abstract
Objective:
The topographic distribution of atherosclerosis in vasculature underscores the importance of shear stress in regulating endothelium. With a systems approach integrating sequencing data, the current study aims to explore the link between shear stress-regulated master transcription factor and its regulation of endothelial cell (EC) function via epigenetic modifications.
Approach and Results:
H3K27ac-ChIP-seq, ATAC-seq, and RNA-seq were performed to investigate the genome-wide epigenetic regulations in ECs in response to atheroprotective pulsatile shear stress (PS). In silico prediction revealed that KLF4 binding motifs were enriched in the PS-enhanced H3K27ac regions. By integrating PS- and KLF4-modulated H3K27ac, we identified 18 novel PS-upregulated genes. The promoter regions of these genes showed an overlap between the KLF4-enhanced ATAC signals and the PS-induced H3K27ac peaks. Experiments using ECs isolated from mouse aorta, lung ECs from EC-KLF4-TG vs. EC-KLF4-KO mice, and atorvastatin-treated ECs showed that ITPR3 was robustly activated by KLF4 and statins. KLF4 ATAC-qPCR and ChIP-qPCR further demonstrated that a specific locus in the promoter region of the ITPR3 gene was essential for KLF4 binding, H3K27ac enrichment, chromatin accessibility, RNA polymerase II recruitment, and ITPR3 transcriptional activation. Deletion of this KLF4 binding locus in ECs by using CRISPR-Cas9 resulted in blunted calcium influx, reduced expression of endothelial nitric oxide synthase, and diminished NO bioavailability.
Conclusions:
These results from a novel multi-omics study suggest that KLF4 is crucial for PS-modulated H3K27ac that allow the transcriptional activation of ITPR3. This novel mechanism contributes to the Ca2+-dependent eNOS activation and EC homeostasis.
Keywords: shear stress, histone modifications, KLF4, ITPR3, eNOS
Subject codes: Vascular Biology, Hemodynamics, Gene Expression and Regulation, Epigenetics, Endothelium, Vascular Type, Nitric Oxide
Introduction
Distinct flow patterns cause different gene expression profiles in vascular endothelial cells (ECs), which lead to variations in atheroprotective vs. atheroprone phenotypes1–3. Previous studies with the use of microarray and RNA-seq have generated a plethora of data to decipher gene expression profiles in ECs in response to atheroprotective vs. atheroprone flows4–6. In principle, the atheroprotective flow pattern upregulates genes that are anti-inflammatory and anti-oxidative and enhance nitric oxide (NO) bioavailability. In contrast, atheroprone flow impairs EC function by inducing genes that increase the inflammatory and oxidative burden7.
Recent advances in studies on mechanotransduction have revealed that epigenetics, including DNA methylation, histone modifications, and long non-coding RNAs, are integral to shear-stress–regulated gene expression8–12. Covalent modifications of histones at adjacent regions of encoded genes govern the status of transcription13. For example, mono-methylation of histone 3 lysine 4 (H3K4me1) marks the loci as poised, whereas acetylation of H3K27 (H3K27ac) results in a loosely packed euchromatic state, which allows for access by transcription factors (TFs) and recruitment of RNA polymerase II (Pol II) to initiate transcription14. Pioneer TFs bind to the condensed chromatin to modify chromatin accessibility, thereby facilitating the recruitment and binding of other TFs and the transcriptional machinery15.
The emerging techniques of next-generation sequencing (NGS), including chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq) and assay for transposase-accessible chromatin-seq (ATAC-seq), allow for studying epigenetic regulation at the genome-wide level16,17. Transcriptionally activated loci, defined by H3K4me1+/H3K27ac+18, at the basal state in human umbilical vein ECs (HUVECs) have been profiled in the ENCODE project (Histone Modifications by ChIP-seq from ENCODE/Broad Institute, GSE29611)19. By using ChIP-seq and ATAC-seq, the current study aimed to explore the link between epigenetic regulation and gene expression in ECs responding to distinct flow patterns. Such analyses revealed that Krüppel-like factor 4 (KLF4) functions as a pioneer TF in ECs to de-condense chromatin, thus allowing for transcriptional activation of atheroprotective flow-responsive genes, exemplified by inositol 1,4,5-trisphosphate (IP3) receptor 3 (ITPR3) encoding ITPR3.
In ECs, intracellular calcium concentration ([Ca2+]i) can be regulated in part by the release of calcium from ITPR stores20. There is substantial evidence that the activation of the IP3-ITPR pathway, with an ensuing increase in [Ca2+]i, leads to increased production of NO via activated endothelial nitric oxide synthase (eNOS), thereby inducing the relaxation of vascular smooth muscle cells21,22. [Ca2+]i has been reported to be modulated by ITPR1 in ECs and ITPR2 in cardiomyocytes to maintain normal blood pressure and regulate mitochondrial function, respectively22,23. The Human Protein Atlas project (www.proteinatlas.org) demonstrated that among ITPR family members, ITPR3 is predominantly expressed in ECs24. However, the epigenetic and transcriptional regulation of ITPR3 is largely unknown. Our data from high-throughput unbiased profiling (i.e., ChIP-seq, ATAC-seq, and RNA-seq) followed by functional validation demonstrate that atheroprotective flow is a principal mechanosensitive cue for activating the ITPR3-eNOS axis in ECs via KLF4-regulated ITPR3 transcription.
Material and Methods
The authors declare that all supporting data are available within the article and its online supplementary files. The associated ChIP-seq and ATAC-seq data have been made publicly available at the NCBI’s GEO and can be accessed at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128391
Cell culture, shear stress, and antibodies
HUVECs were obtained from Cell Applications (San Diego, CA) and were maintained and exposed to shear stress as previously described32. Briefly, a circulating flow system was used to impose atheroprotective pulsatile shear stress (PS, 12 ± 4 dyn/cm2) or atheroprone oscillatory shear stress (OS, 1 ± 4 dyn/cm2). The flow system was held at 37°C and ventilated with 95% humidified air plus 5% CO2. Antibodies against H3K4me1 and H3K27ac were from Active Motif (Carlsbad, CA); anti-eNOS, anti-phospho-eNOS (Ser1177), and anti-calmodulin antibodies were from Abcam (Cambridge, MA); anti-α-tubulin, anti-phospho-eNOS (Thr495), and horseradish peroxide-conjugated anti-rabbit and anti-mouse antibodies were from Cell Signaling Technology (Beverly, MA); anti-caveolin-1 antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). The atorvastatin hemicalcium salt was from Tocris Biosciences (Bristol, UK).
ChIP-seq, RNA-seq, and ATAC-seq
Cells were fixed at 37°C with 1% paraformaldehyde for 15 min, and then washed twice in cold phosphate buffered saline (PBS) and scraped and pelleted at 3000 rpm for 5 min; cell pellets were resuspended in 50 μl lysis buffer, then diluted with 500 μl TE buffer before sonication. For antibody and chromatin binding, 20 μg chromatin, 3 μg antibody, and 11 μl dynabeads (Thermo Fisher Scientific, Waltham, MA) were incubated in a master mix buffer at 4°C overnight. The chromatin were eluted by 150 μl elution buffer followed by crosslinking at 65℃ overnight. After incubation with RNase A and Proteinase K, DNA was precipitated by phenol:chloroform:isoamyl (25:24:1, v/v), dissolved in TE buffer, and stored at −80°C. For RNA-seq, total RNA from HUVECs was extracted by using the mirVana miRNA Isolation Kit (Thermo Fisher) according to the manufacturer’s instructions. For ATAC-seq, ECs were washed twice with cold PBS, and nuclei were isolated by resuspending ~5000 cells in 50 μl cold lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, and 0.1% NP-40). The nuclear pellets were resuspended in 50 μL transposition buffer (Nextera DNA Library Preparation Kit (Illumina Inc. San Diego, CA) and 22.5 μL H2O, and then incubated at 37°C for 30 min. After incubation, the DNA samples were purified with Qiagen MinElute PCR purification kit. The preparation of the various sequencing libraries and sequencing by use of an Illumina HiSeq 2000 sequencer were performed in the IGM Genomics Core, University of California, San Diego. The resulting sequencing raw data have been made publicly available at the NCBI’s GEO (accession No. GSE128391).
Analysis of sequencing libraries
Trimmomatic v0.35 was used to process the fastq files to remove poor-quality and adapter reads26. RNA reads were aligned by using HISAT2 with a pre-built reference human UCSC hg19 transcriptome index27. To evaluate gene expression, gene information was obtained from GENCODE (https://www.gencodegenes.org/, version GRCh37) and count per gene was calculated by featureCount28. Statistics analysis of differentially expressed genes was performed with the use of DESeq2 to process the raw counts of genes29. We used Benjamini-Hochberg method at level of q<0.01 to limit false positive rate in multiple testing. DNA reads were aligned to the pre-built UCSC hg19 reference human genome index by bowtie230. For DNA alignment, PCR duplicate reads were further removed by Picard-tools (https://broadinstitute.github.io/picard/). To identify ChIP- and ATAC-enriched loci, the de-duplicated alignments were subjected to peak calling and annotated by the homer suite31. Homer-idr (https://github.com/karmel/homer-idr) was used to process ChIP-seq data for high reproducibility among biological duplicates. Loci with overlapped ChIP-seq and ATAC-seq data were identified by bedtools (http://bedtools.readthedocs.io/en/latest/). The resulting profiles were visualized by use of the UCSC genome browser (https://genome.ucsc.edu). To identify ChIP/ATAC read enrichment, the identified peaks were split into 500-bp windows and processed by featureCount and DESeq2. Heatmaps and binding profiles of ChIP- and ATAC-seq data were generated by using deeptools32.
Motif analysis
To define enriched TF binding sites, RSAT Metazoa was used to process H3K27 hyperacetylated loci identified in ChIP-seq analysis33. Genomic sequences of PS and OS loci were fetched separately and used as mutual background models in the peak-motif program of RSAT. The motifs enriched by PS or OS vs. background model were corrected for positional bias and then output for follow-up analyses.
RT-qPCR, western blot analysis, and RNA silencing
Total RNA was isolated from cultured ECs, mouse lung ECs, and the intima of the mouse aorta, aortic arch, and thoracic aorta by using Trizol reagent (Invitrogen) and protein was isolated with RIPA lysis buffer. Animal experiments were approved by the institutional animal care and use committee of the University of California, San Diego. C57BL/6j mice were obtained from The Jackson Laboratory (Sacramento, CA). At 6–8 weeks of age, both sexes (3 male and 3 female) of mice were intraperitoneally injected with atorvastatin (50 mg/kg) or an equal volume of saline (500 μL) randomly. After 24 hr, mice were sacrificed by CO2 inhalation, lung ECs were harvested for RNA isolation. For intimal mRNA isolation, the aortic arch (AA) and thoracic aorta (TA) were isolated and peri-adventitial fat was removed, the aortic lumen was quickly flushed with 250 μl TRIzol reagent with use of a 28 1/2-gauge syringe. Due to the limited amount of mRNA that can be extracted from TA and AA, the intimal eluates from five C57BL/6j mice (3 male and 2 female) were collected and pooled for RNA isolation. The small number of animals used for isolating intimal mRNA limited the separation of male and female mice for tissue collection. Total RNA from lung ECs of EC-KLF4−/− and EC-KLF4-Tg mice was a gift from Dr. Mukesh K. Jain34. The extracted RNA was reverse-transcribed and analyzed by qPCR with β-actin as an internal control. The primer sequences used for qPCR are in Supplemental Table 2. For western blot analysis, protein extracts were separated by SDS-PAGE and transferred to PVDF membranes, which were incubated with primary antibodies. Protein bands were detected by using Immobilon chemiluminescent substrate (Millipore). Human THBD, DAG1, CAMKKα, ITGB4, ALS2CL, and ITPR3 siRNA and control siRNA (Ctrl siRNA) were obtained from Qiagen. All RNAs were transfected into ECs by using Lipofectamine RNAi max (Invitrogen).
Immunostaining and eNOS activity assay
ECs were fixed in 4% paraformaldehyde at 4°C for 10 min. Triton X-100 (0.1% in PBS) was then added. Specimens were then incubated with primary antibody for 1 hr, then for 2 hr with Alex-488 (anti-mouse)- or Alex-568 (anti-rabbit)-conjugated secondary antibody. Fluorescent images were acquired under an Olympus FV100 confocal microscope. The colocalization of eNOS and caveolin-1 was quantified by Image J. NO production was assessed as nitrite (NO2−) accumulated in culture media by nitrite/nitrate fluorometric assay (Cayman Chemicals).
CRISPR/Cas9-mediated KLF4 binding site disruption
Single-guide RNAs (sgRNAs) were designed by using the online tool provided by the Church laboratory35. Oligonucleotide pairs with BsmBI-compatible overhangs were annealed and cloned into the lentiviral vector lentiCRISPR v2 (Addgene plasmid #52961)36. For virus production, HEK293T cells were transfected with lentiCRISPR v2, psPAX2 (Addgene #12260) and pMD2.G (Addgene plasmid #12259) at a 8:4:1 ratio with 100 μl Plus Reagent (Life Technologies) and 50 μl Lipofectamine 2000 (Life Technologies) and cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum, 1% sodium pyruvate, 1% non-essential amino acids, 1% kanamycin, 50 units/ml penicillin/streptomycin and 50 μM β-mercaptoethanol for 48 hr before cell lysis. Cell debris was removed by centrifugation followed by ultra-centrifugation (2.5 hr at 24,000 rpm) with a sucrose cushion. Early passage HUVECs were lentivirally transduced and cultured for 2 days in medium supplemented with 1 μg/ml puromycin to select cells expressing the lentiCRISPR V2 vector. A mixed population of lentiCRISPR V2–positive ECs was selected.
Calcium imaging
For Fura2-AM experiments, cells were pre-incubated with 0.5 μM Fura2-AM (Molecular Probes) for 15 min at 37°C before imaging. Cells were washed twice with modified HBSS and imaged in the dark at room temperature. Images were acquired under a Zeiss Observer Z1 microscope with a cooled charge-coupled device camera (Roper). Fura2 dual excitation ratio imaging involved two excitation filters, ET340x and ET380x for 340-nm and 380-nm excitation, and a HQ535/45m emission filter. Exposure times were 200 ms, and images were taken every 30 s. Imaging data were analyzed with Metafluor 6.2 software (Universal Imaging). Fluorescence images were background-corrected by deducting the background (regions with no cells) from the emission intensities of cells loaded with Fura2-AM. The 340/380-nm emission ratio was calculated at different times. Traces were normalized by setting the emission ratio to 1 before the addition of drugs.
Statistical analysis
All statistical analyses were performed using SPSS version 14.0 or R project version 2.10. Statistical analyses of sequencing data were performed using standard codes for the packages indicated above. Wet bench experiments results were expressed as means ± SEM. Initially, data were tested for normality and equal variance to confirm the appropriateness of parametric tests. Parametric data was analyzed by 2-tailed Student’s t test. If normality or equal variance test failed, non-parametric data were analyzed using Kruskal-Wallis with Dunn’s post hoc test or 2-tailed Mann-Whitney U test. P < 0.05 was considered statistically significant.
Results
Shear stress regulation of histone modifications
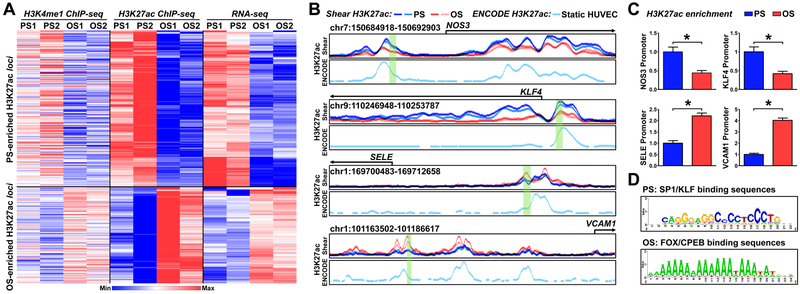
ECs subjected to atheroprotective PS or atheroprone OS were used for H3K4me1 ChIP-seq and H3K27ac ChIP-seq to explore the histone modifications in ECs responding to distinct flow patterns. We first identified the cis-elements involved in the active chromatin state at the genome-wide scale, represented by H3K4me1+/H3K27ac+ signals. Because H3K4me1+ is a prerequisite for the active cis-element and H3K27ac+ is more prominent for defining the activated chromatin state18, we quantified the chromatin activation by assessing the acetylation level of H3K27 (i.e., H3K4me1+/H3K27achigh vs H3K4me1+/H3K27aclow), which depicts a chromatin relaxation state with the ensuing transcriptional activation37. Changes in H3K27ac levels were then cross-referenced with those from RNA-seq expression profiles under PS or OS for exploring the expression of genes regulated by the active chromatin state. Fig. 1A shows the heatmap generated from such analysis and includes genes that showed both enriched H3K27ac+ and higher RNA levels. As positive controls, PS-responsive genes such as NOS3 (encoding eNOS) and KLF4 showed enhanced H3K27ac signals in their promoter regions under the PS condition. Conversely, OS-induced genes such as selectin E (SELE; encoding E-selectin) and vascular cell adhesion protein 1 (VCAM1) showed enhanced H3K27ac signals in their promoter regions under the OS condition (Fig. 1B). These flow-induced H3K27ac loci overlapped with those defined in ENCODE data for static HUVECs (GSM733691)38. The H3K27ac enrichment in the promoter regions of NOS3, KLF4, SELE, and VCAM1 (highlighted in green in Fig. 1B) in ECs was indeed regulated by shear stress, as verified by H3K27ac ChIP-qPCR (Fig. 1C).
Figure 1. Shear stress regulation of histone modifications, assessed by H3K4me1 ChIP-seq, H3K27ac ChIP-seq, and RNA-seq.
(A, B) Human umbilical vein endothelial cells (HUVECs) were exposed to pulsatile flow (PS; 12 ± 4 dyn/cm2) or oscillatory flow (OS; 1 ± 4 dyn/cm2) for 16 hr. Cells from two biological repeats (PS1, OS1, PS2, OS2) were subjected to H3K4me1 ChIP-seq, and H3K27ac ChIP-seq, and RNA-seq analyses. (A) Heat map of the normalized H3K4me1 and H3K27ac enrichment and RNA-seq signal of the nearest expressed genes. The genes represented were those with H3K27ac and mRNA enrichment by PS. (B) Normalized PS (blue)- or OS (red)-associated H3K27ac ChIP signals at the promoter regions of NOS3, KLF4, SELE, and VCAM1. The H3K27ac enrichment in static HUVECs (light turquoise, below the PS and OS H3K27ac) were adopted from ENCODE (GSM733691)38. (C) HUVECs were exposed to PS or OS. H3K27ac enrichment in the region highlighted in green (in B) was evaluated by H3K27ac ChIP-qPCR. Data are mean ± SEM from 3 independent experiments. Statistical significance was determined by 2-tailed Mann-Whitney U test between PS and OS. *P < 0.05. (D) PS- or OS-enhanced H3K27ac hyper-condensed regions were analyzed by RSAT to decipher TF binding sites.
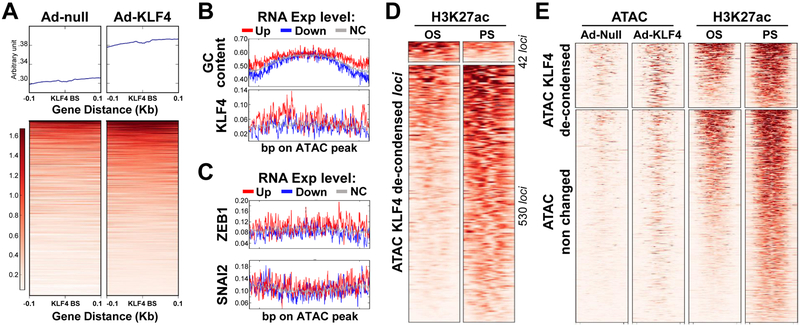
To deduce the TFs involved in the transcription of these differentially regulated genes under PS vs. OS, we analyzed TF binding sequences located at these differentially enriched H3K27ac loci. Sp1-like/KLF family and FOX/CPEB binding sequences were among the top-ranked cis-elements involved in PS- and OS-enriched H3K27ac loci, respectively (Fig. 1D). KLF4 is a master regulator of EC homeostasis that is PS-inducible. As a pioneer TF, KLF4 recruits histone acetyltransferases and deacetylases to modify histones, thereby changing the chromatin accessibility39. Thus, we used data from ATAC-seq to compare the chromatin accessibility in control ECs and ECs overexpressing KLF4. The loci close to the KLF4 binding sites were largely less condensed in ECs overexpressing KLF4 (Fig. 2A), suggesting that KLF4 increased the chromatin accessibility of its targeted genes. In line with the GC-enriched KLF4 binding sequence shown in Fig. 1D, the GC content and percentage of KLF4 binding sites were higher in these decondensed loci of PS-inducible genes, than that of PS-suppressed or non-effected genes (Fig. 2B). Such enrichments were not found in binding sequence for other GC-bound TFs such as ZEB1 and SNAI2 (Fig. 2C). The KLF4-decondensed loci had a higher degree of superimposition with the PS-enriched H3K27ac loci than those of OS-enriched H3K27ac (Fig. 2D). Furthermore, ectopically expressed KLF4 decondensed ~25% of the PS-enhanced H3K27ac loci, as revealed by superimposing the Ad-KLF4-ATAC and H3K27ac ChIP-seq data (Fig. 2E). These results confirmed that KLF4 is a pioneer TF in ECs that regulates multiple PS-responsive genes via chromatin remodeling.
Figure 2. KLF4 regulation of PS-induced genes via chromatin remodeling, assessed by ATAC-seq.
HUVECs were transfected with Ad-null or Ad-KLF4 for 24 hr. Cells were subjected to ATAC-seq. (A) ATAC peaks from Ad-null or Ad-KLF4-infected ECs were initially merged into a union set of ATAC peaks. The KLF4 putative binding sites within the ATAC peaks were then predicted. The upper and lower panels show the overall and per-loci ATAC signals at ± 0.1 kb flanking the putative KLF4 binding sites. (B, C) ATAC loci were first divided into 3 groups by the expression levels of their closest genes induced by PS. The assessed GC ratio or frequency of putative TF binding in these loci are summarized in the representative heat maps. The x-axis represents the position of the de-condensed loci and the y-axis the frequency of changes. (D) Heat maps showing 572 loci de-condensed by KLF4, as assessed by ATAC-seq. These loci coincide with the PS- or OS-associated H3K27ac enrichment. (E) Heat maps of the ATAC-seq and H3K27ac ChIP-seq signals in loci de-condensed or not by KLF4. Distance at the abscissa denotes the relative location within the loci, and each row represents a locus.
PS-induced transcription via KLF4
Next, we set three criteria to screen the PS-inducible genes via KLF4 regulation: (1) inducible by PS, as revealed by RNA-seq; (2) containing putative KLF4 binding sites in the promoter region; and (3) containing enhanced H3K27ac signal near the KLF4 binding sites under PS (Fig. 3A). Consequently, we selected 18 genes that were putatively induced by PS via KLF4 (Table 1). We first examined the dynamic regulation of these 18 genes by PS vs. OS in time-series RNA-seq6. From these 18 genes, all showed higher expression under PS than OS except for ST6GAL1 and TBC1D7 (Fig. 3B and Sup. Fig. 1). As positive controls, KLF4 and NOS3 (encoding eNOS), known to be PS-responsive, were substantially induced by PS for up to 24 hr. Next, these 18 genes were found to have at least one putative KLF4 binding site in the promoter regions, with H3K27ac enrichment in the flanking region (Fig. 3C and Sup. Fig. 2A). Further validation by Ad-KLF4-ATAC-seq revealed that the promoter regions of these 18 genes had a KLF4-enhanced ATAC signal in at least one set of ATAC-seq data (Fig. 3D and Sup. Fig. 2B). Importantly, these enhanced ATAC signals had considerable overlap with the PS-induced H3K27ac peaks.
Figure 3. PS-induced genes via KLF4-dependent chromatin remodeling.
(A) Three selective criteria for the PS-induced genes (see text). (B) Heat map indicating differentially expressed mRNAs for the 18 selected genes. The presented data were selected from the 10-time point RNA-seq reported by Ajami et. al6. (C) Normalized PS (blue)- or OS (red)-induced H3K27ac enrichment in the promoter regions of ALS2CL, ITPR3, B4GALT3, CAMKK1, CBX2, FAM167B, ITGB4, and THBD. The putative KLF4 binding sites in the respective promoter regions are illustrated in green. (D) HUVECs were infected with Ad-null or Ad-KLF4 for 24 hr in two biological repeats. ATAC-seq was performed to evaluate the chromatin accessibility. Lines in gray (from Ad-null–infected ECs) and blue (from Ad-KLF4–infected ECs) represent ATAC signals in the same loci as those in (C).
Table 1,
Characteristics of 18 PS-induced genes
| Symbol | Aliases | Full name | Location | Gene ID |
|---|---|---|---|---|
| ITGB4 | CD104; GP150 | Integrin subunit beta 4 | 17q25.1 | 3691 |
| ZNF467 | EZI; Zfp467 | Zinc finger protein 467 | 7q36.1 | 168544 |
| ST6GALNAC1 | STYI; SIAT7A; ST6GalNAcI | ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 | 17q25.1 | 55808 |
| CCM2L | C20orf160; dJ310O13.5 | CCM2 like scaffolding protein | 20q11.21 | 140706 |
| FAM167B | C1orf90 | Family with sequence similarity 167 member B | 1p35.2 | 84734 |
| MYOM3 | - | Myomesin 3 | 1p36.11 | 127294 |
| ALS2CL | RN49018 | ALS2 C-terminal like | 3p21.31 | 259173 |
| CBX2 | M33; CDCA6; SRXY5 | Chromobox 2 | 17q25.3 | 84733 |
| THBD | TM; THRM; AHUS6 | Thrombomodulin | 20p11.21 | 7056 |
| DAG1 | A3a; DAG; AGRNR; | Dystroglycan 1 | 3p21.31 | 1605 |
| TBC1D7 | TBC7; MGCPH; PIG51 | TBC1 domain family member 7 | 6p24.1 | 51256 |
| CAMKK1 | CAMKKA | Calcium/calmodulin dependent protein kinase kinase 1 | 17p13.2 | 84254 |
| B4GALT3 | β4Gal-T3 | β−1,4-galactosyltransferase 3 | 1q23.3 | 8703 |
| ARAP3 | DRAG1; CENTD3 | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 | 5q31.3 | 64411 |
| RNASE1 | RAC1;RIB1;RNS1 | Ribonuclease A family member 1 | 14q11.2 | 6035 |
| SYNE3 | NET53; Nesp3 | Spectrin repeat containing nuclear envelope family member 3 | 14q32.13 | 161176 |
| TRIB3 | NIPK; SINK; TRB3; SKIP3 | Tribbles pseudokinase 3 | 20p13 | 57761 |
| ITPR3 | IP3R3 | Inositol 1,4,5-trisphosphate receptor type 3 | 6p21.31 | 3710 |
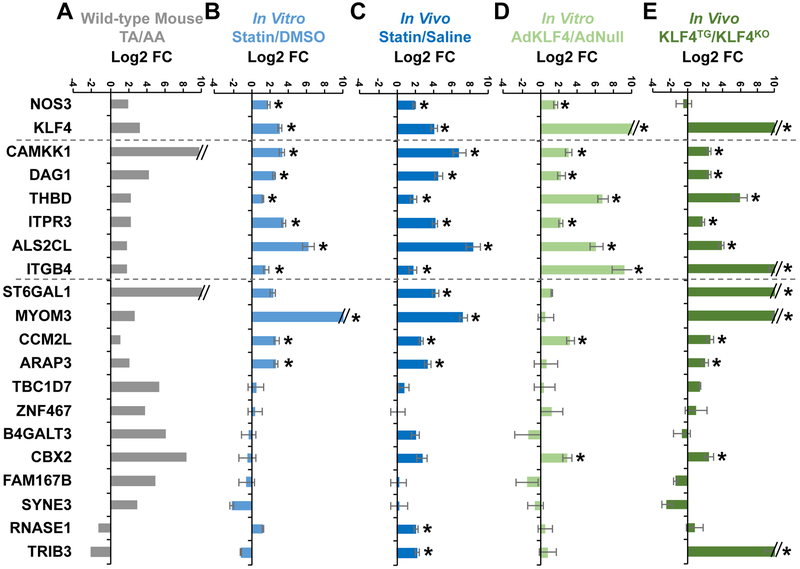
The tunica intima tissue isolated from the thoracic aorta (TA, atheroprotective region) and aortic arch (AA, atheroprone region) of C57BL/6j mice was used first for biological validation. Overall, the mRNA level for 16 of the 18 genes was higher in TA than AA intima (Fig. 4A). The cholesterol-lowing drugs statins have a profound effect on maintaining a functional endothelium and have been shown to induce KLF4 in ECs in vitro and in vivo40. Atorvastatin treatment of cultured ECs for 48 hr induced 9 of the 18 genes (Fig. 4B). Furthermore, 12 genes were significantly induced in lung ECs isolated from mice receiving atorvastatin for 3 days (Fig. 4C). To explore the KLF4 inducibility, we overexpressed KLF4 in cultured ECs and examined the mRNA level of the 18 genes. Eight genes were induced by KLF4 overexpression (Fig. 4D). For in vivo validation of KLF4 induction of these genes, lung ECs were isolated from EC-KLF4-Tg and EC-KLF4−/− mice34; 12 of the 18 genes were induced in a KLF4-dependent manner in vivo (Fig. 4E). Overall, 6 genes in ECs, viz. CAMKK1, DAG1, THBD, ITPR3, ALS2CL, and ITGB4, were commonly induced by PS, atorvastatin, and KLF4 in vitro and in vivo.
Figure 4. KLF4-dependent gene expression in ECs.
(A) Intima isolated from thoracic aorta (TA) and aortic arch (AA) of five C57BL/6j mice (3 male and 2 female) were pooled. (B) HUVECs were treated with atorvastatin (5 μM) or DMSO for 24 hr. (C) C57BL/6j mice were intraperitoneally injected with atorvastatin (50 mg/kg) or an equal volume of saline (500 μL) randomly. Lung ECs were harvested 24 hr post-injection, n=6 mice (3 male and 3 female) in each group. (D) HUVECs were infected with Ad-null or Ad-KLF4 for 24 hr. (E) Lung ECs were isolated from EC-KLF4−/− or EC-KLF4-Tg mice, n=3 mice in each group. The expression of the identified 18 genes (shown in Fig. 3 and Table 1) was detected by RT-qPCR and presented by Log 2 fold change (Log 2 FC). Data are mean ± SEM from 3 independent experiments for (B) and (D). Statistical significance was determined by 2-tailed Mann-Whitney U test. *P < 0.05 compared with control.
ITPR3 regulation of eNOS in ECs
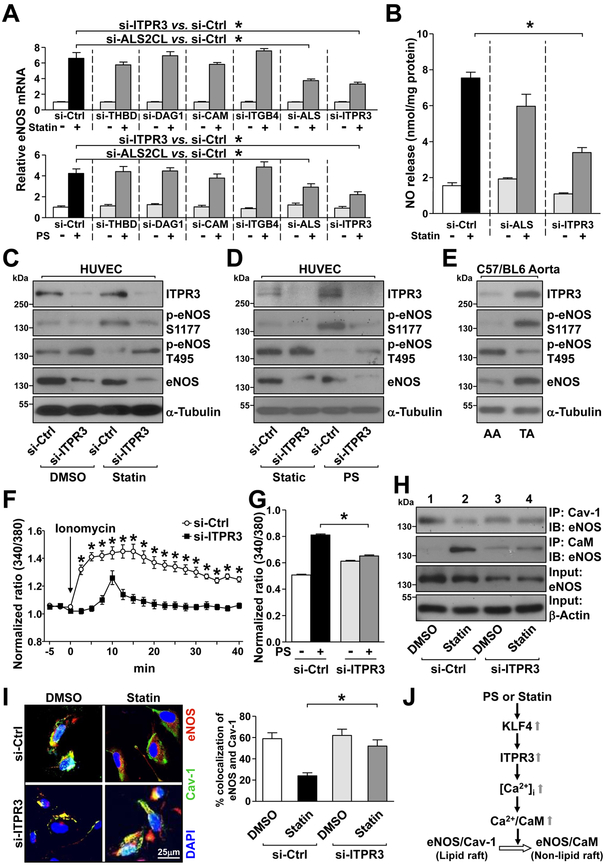
We next explored whether changes in the expression of these 6 genes affected EC functions by using eNOS-derived NO bioavailability as a functional readout. ECs in which the 6 commonly induced genes had been individually knocked down were treated with atorvastatin or PS for 16 hr. eNOS expression and NO production was assessed accordingly. siRNA knockdown of ITPR3 or ALS2CL attenuated the atorvastatin- or PS-induced eNOS mRNA level in ECs (Fig. 5A). However, ITPR3 knockdown impaired the atorvastatin-increased NO production more drastically and hence we focused on ITPR3 for further functional characterization (Fig. 5B).
Figure 5. Atorvastatin and PS upregulate the ITPR3-eNOS axis in ECs.
(A) HUVECs were transfected with siRNA against THBD, DAG1, CAMKK1, ITGB4, ALS2CL, or ITPR3 for 48 hr. In control experiments, HUVECs were transfected with control siRNA. After atorvastatin (5 μM) or PS stimulation for the last 16 hr, cells were lysed and eNOS mRNA level was detected by RT-qPCR. (B) HUVECs were transfected with ALS2CL siRNA, ITPR3 siRNA, or control siRNA. After atorvastatin stimulation for 1 hr, NO level was detected by Griess reagent. (C, D, F, G, H, I) HUVECs were transfected with ITPR3 siRNA or control siRNA, and then stimulated with atorvastatin (C, H, I) for 1 hr, PS for 1 hr (D), or PS for 10 mins (G). (E) Tissues isolated from TA and AA of C57BL/6j mice (pooled from 3 male and 2 female mice). In (C-E), the cellular or tissue extracts were examined by western blot analysis for protein levels of ITPR3, p-eNOS T495, p-eNOS S1177, eNOS, and α-tubulin. In (F, G), ECs were treated with Fura2-AM for 1 hr before stimulation with ionomycin (2 μM, F) or PS (G). The calcium influx rate was detected by comparing the 340/380-nm ratio under a fluorescence microscope. Overall, 12–15 cells (F) or 130–160 cells (G) were randomly chosen to detect the fluorescence in each group. In (H), cell lysates were immunoprecipitated (IP) with anti-Cav-1 or anti-calmodulin (CaM) antibody. The co-immunoprecipitated eNOS was evaluated by immunoblotting (IB), with β-actin as a loading control. In (I), immunostaining was performed with anti-eNOS (red) or anti-Cav-1 (green) antibody. Nuclei were counterstained with DAPI (blue). Fluorescence was detected under an Olympus confocal microscope. The colocalization of eNOS and caveolin-1 was quantified by ImageJ. (J) A working model of atorvastatin and PS regulation of the ITPR3-eNOS axis via KLF4. Data are mean ± SEM from at least 3 independent experiments. For parametric data (F, G, I), statistical significance was determined by 2-tailed Student’s t test between two indicated groups. Non parametric data (A, B) were analyzed using Kruskal-Wallis with Dunn’s post-hoc test. *P < 0.05.
A hallmark of eNOS activation is its phosphorylation at Ser-117741,42. Concurrently, eNOS Thr-495 dephosphorylation is positively associated with stimuli that elevate [Ca2+]i43. siRNA knockdown of ITPR3 impaired the eNOS Ser-1177 phosphorylation increased by atorvastatin and PS (Fig. 5C, 5D). As well, ITPR3 knockdown abolished the eNOS Thr-495 dephosphorylation in response to atorvastatin or PS (Fig. 5C, 5D). In vivo, the differential regulation of ITPR3 by atheroprotective vs. atheroprone flows was evident by a higher level of ITPR3 in the TA than AA region (Fig. 5E). The increased ITPR3 expression was also accompanied by decreased eNOS Thr-495 phosphorylation and increased Ser-1177 phosphorylation and eNOS level (Fig. 5E).
Functioning as a calcium channel on the ER membrane, ITPR regulates [Ca2+]i44. ITPR3 knockdown indeed decreased the ionomycin- and PS-induced calcium influx in ECs (Fig. 5F, 5G). Changes in [Ca2+]i modulate eNOS phosphorylation status and also eNOS subcellular compartmentation in ECs, which plays an essential role in regulating eNOS activity43,45. The underlying mechanism involves the disassembly of the eNOS–caveolin-1 (Cav-1) complex in the lipid raft and assembly of the eNOS–calmodulin complex in the non-lipid raft (illustrated in Fig. 5J). With atorvastatin treatment of wild-type ECs, eNOS was translocated from lipid rafts to non-lipid rafts. This was revealed by the dissociation of eNOS from Cav-146, a structural protein in caveolae, and the association with calmodulin (lane 2 vs 1 in Fig. 5H). Notably, such eNOS translocation and eNOS-calmodulin association were not seen in ECs with ITPR3 knocked down (lane 4 vs 3 in Fig. 5H). We performed immunofluorescence staining to confirm that ITPR3 is necessary for statin-activated eNOS. The co-localization of eNOS with Cav-1 at the basal level, presumably in lipid rafts, is disrupted by atorvastatin treatment (Fig. 5I).
KLF4 regulates the ITPR3 chromatin accessibility
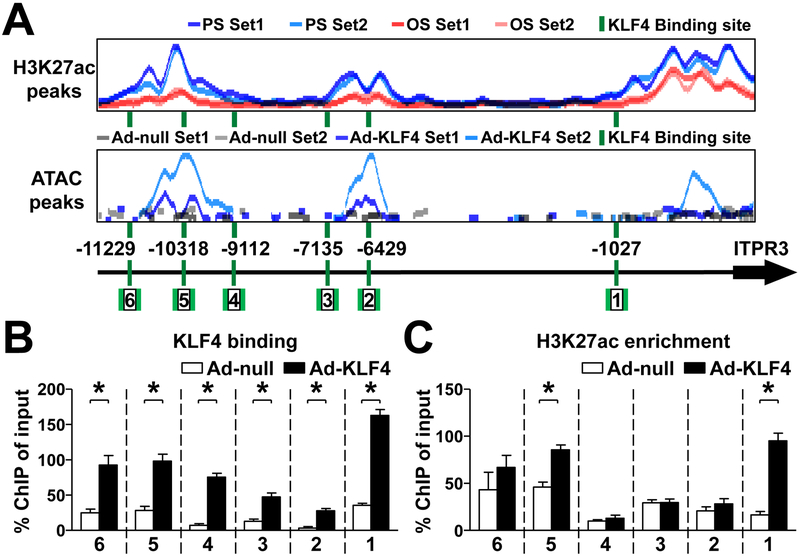
In the promoter region of ITPR3, we identified 6 putative KLF4 binding sites (shown in Sup. Table 1). DNA segments flanking these KLF4 cognate binding sequence showed enriched H3K27ac signals and increased chromatin accessibility, as revealed by ChIP-seq and ATAC-seq, respectively (Fig. 6A). By using KLF4 ChIP-qPCR, we verified that KLF4 overexpression increased KLF4 binding to all these 6 KLF4 binding sites (Fig. 6B). However, H3K27ac ChIP-PCR revealed the greatest enrichment of H3K27ac near the 1st and 5th sites (Fig. 6C).
Figure 6. KLF4 regulates ITPR3 by modulating chromatin accessibility.
(A) Putative KLF4 binding sites, PS- or OS-induced H3K27ac enrichment, and Ad-null- or Ad-KLF4-induced ATAC signals in the ITPR3 promoter. (B, C) HUVECs were infected with Ad-null or Ad-KLF4 for 24 hr. KLF4 binding to the ITPR3 promoter and H3K27ac enrichment of the ITPR3 promoter were detected by KLF4 ChIP-qPCR and H3K27ac ChIP-qPCR, respectively. The used PCR primers were depicted by arrows in Panel A. Data are mean ± SEM from at least 3 independent experiments. Statistical significance was determined by 2-tailed Mann-Whitney U test between two indicated groups. *P < 0.05.
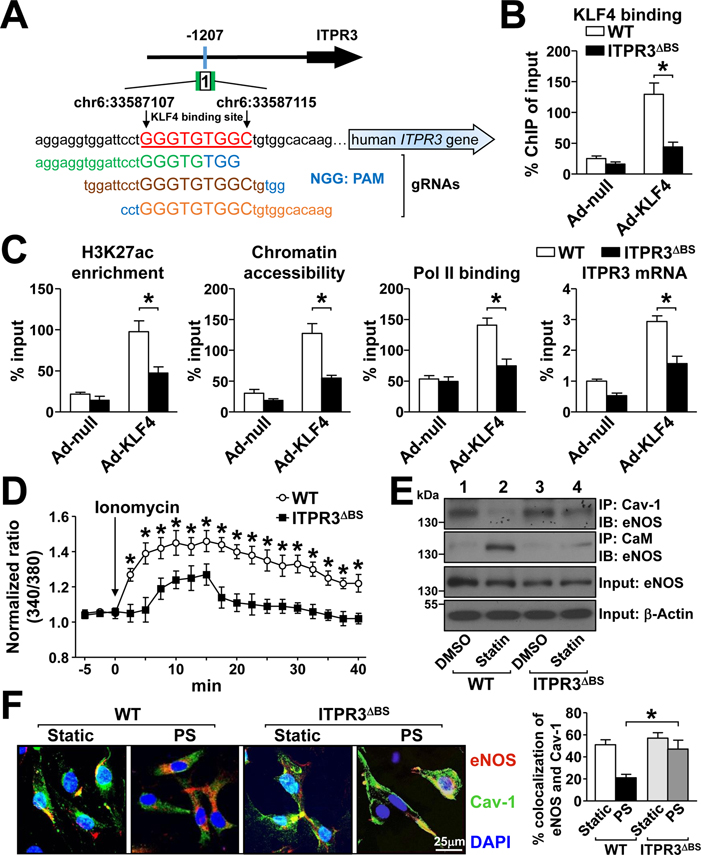
To further test whether KLF4 binding to its cognate binding sequence affects histone H3K27ac modification, we deleted the 1st KLF4 binding site in the ITPR3 promoter in ECs (ITPR3ΔBS) by using CRISPR-Cas9 genomic editing (Fig. 7A). Deletion of this binding site indeed decreased KLF4 binding, with concurrent attenuations of H3K27ac modification, chromatin accessibility, Pol II recruitment, and ITPR3 mRNA expression (Fig. 7B and 7C). These data suggest that KLF4, acting as a PS-inducible pioneer TF, regulates ITPR3 expression by remodeling histone and the consequent chromatin accessibility of the ITPR3 promoter. Functionally, the ionomycin-induced calcium influx was significantly lower in ITPR3ΔBS than wild-type ECs (Fig. 7D). Furthermore, the dissociation of eNOS-Cav-1 and association of eNOS-calmodulin seen in wild-type ECs stimulated with atorvastatin or PS were largely abolished in ITPR3ΔBS ECs (Fig. 7E, 7F). Thus, the physiological (i.e., PS) and pharmacological (i.e., statins) stimulations of eNOS-derived NO bioavailability are contributed, at least in part, by KLF4 regulation of ITPR3 via chromatin remodeling.
Figure 7. KLF4-dependent activation of the ITPR3-eNOS axis.
(A) CRISPR-Cas9 strategy for targeting the 1st KLF4 binding site (chr6:33587106-chr6:33587115) in the ITPR3 promoter. (B, C) Wild-type HUVECs or CRISPR-Cas9-edited HUVECs (ITPR3ΔBS) were infected with Ad-null or Ad-KLF4. KLF4 binding, H3K27ac enrichment, and chromatin accessibility near the KLF4 binding site 1 were detected by KLF4-ChIP, H3K27ac-ChIP, and ATAC-qPCR, respectively. Pol II recruitment near the TSS site was detected by Pol II-ChIP-qPCR, and the ITPR3 mRNA level was assayed by qPCR. (D, E, F) Wild-type and ITPR3ΔBS HUVECs were stimulated with ionomycin, atorvastatin, or PS. The calcium influx rate (D), Cav-1-eNOS or CaM-eNOS co-immunoprecipitation (E), and immunostaining (F) were as described in Figure 5. Data are mean ± SEM from at least 3 independent experiments. For parametric data (D, F), statistical significance was determined by 2-tailed Student’s t test between two indicated groups. Non parametric data (B, C) were analyzed using 2-tailed Mann-Whitney U test between 2 indicated groups. *P < 0.05.
Discussion
Vascular tone is regulated by EC responses to mechanical and biochemical stimuli, and [Ca2+]i-modulated eNOS activity plays a significant role in this regulation. By using NGS, including RNA-seq, ChIP-seq, and ATAC-seq, we have found that KLF4, functioning as a PS-induced pioneer TF, activates ITPR3 via epigenetic regulation. This finding indicates that PS and statins augment the eNOS-derived NO bioavailability, at least in part, via KLF4-induced ITPR3.
[Ca2+]i in ECs can be regulated by voltage-dependent calcium channels, Ca2+-induced calcium release from ryanodine receptors, and Ca2+ release from ITPR stores. There is substantial evidence that the increase in [Ca2+]i with the ensuing activation of the IP3-ITPR pathway leads to an increase in the production of NO and other vasodilatory factors21,22. In a recent study by Yuan et al., mice with endothelial deletion of Itpr1 showed reduced eNOS activity, blunted vasodilation, and elevated blood pressure22. Our current study suggested a mechanism by which the PS-induced KLF4 increased its binding to the promoter region of ITPR3 in a trans manner, altered chromatin accessibility in a cis manner (as revealed by enrichment of H3K27ac and ATAC-seq), and then recruited Pol II to transactivate ITPR3.
Three ITPR subtypes have been reported: ITPR1, ITPR2, and ITPR347. Our RNA-seq data indicate that ITPR3 is the major type in cultured ECs (Sup. Fig. 3). Furthermore, the upstream promoter region of ITPR3 seems to have more putative KLF4 binding sites and PS-enriched H3K27ac signals than ITPR1and ITPR2 (Sup. Fig. 4). Functionally, the KLF4-transactivated ITPR3 regulates the IP3-ITPR pathway both at the basal level and under PS. These two modes of regulation would constitute lineage- and signal-dependent ITPR3 transcription, respectively, to provide the basal and stimulatory regulation of [Ca2+]i signaling essential for maintaining vascular tone.
The selection of the 18 PS-induced genes for further validation was based on the results of KLF4 binding, histone remodeling by ChIP-seq, and expression levels by RNA-seq. As one of the four Yamanaka factors, KLF4 binds to the target DNA sequence via its zinc-finger domain, which enhances transcription by recruiting transcription machinery and Pol II48. Acting as a master TF in ECs49, KLF4 not only is important for the endothelial lineage but also activates genes essential for EC homeostasis [e.g., eNOS and cholesterol-25-hydroxylase]34,50. Functionally, KLF4 is upregulated by atheroprotective flow and cholesterol-lowering drugs (e.g., statins)4,40. Mechanistically, KLF4 binds to Sp1-like binding sites on the promoter regions of KLF4-targeted genes51. Furthermore, KLF4 interacts with histone acetyltransferases or deacetylases to increase or attenuate chromatin acetylation, respectively, thereby affecting transcription. For example, KLF4 can regulate chromatin remodeling of interleukin 6 promoter in dendritic cells by changing the adjunct histone modification52. In normal breast epithelial cells, KLF4 recruitment of HDAC2/3 to the vascular endothelial growth factor (VEGF) promoter represses the expression of VEGF53. KLF4-driven histone modification in transactivating ITPR3 was further supported by the decondensed chromatin in loci adjacent to the KLF4 binding sites. In the context of PS induction of ITPR3 in ECs, Pol II is recruited to the KLF4-occupied ITPR3 promoter for transcriptional initiation (Fig. 7C).
Although we focused on KLF4 as the key TF, KLF4 and KLF2 bind to similar cis-elements (e.g., Sp1-like binding site) to transactivate commonly targeted genes such as eNOS and thrombomodulin (THBD)49. Atheroprotective flow and pharmacological agents such as statins induce optimal levels of KLF2 and/or KLF4 to maintain EC homeostasis4,40,54,55. While KLF2 and KLF4 seem to be compensatory for each other in maintaining EC function via the commonly transactivated genes, ablations of both lead to EC dysfunction more than either alone16. Thus, KLF2 and KLF4 likely synergize each other to orchestrate the transcriptional machinery in regulating genes such as NOS3, THBD, and ITPR3. The detailed mechanism underlying this synergism deserves further investigation by including KLF2 ChIP-seq and ATAC-seq analyses in addition to KLF4.
Approximately 85% of the coding regions in the human and mouse genome are similar, but conservation between the non-coding regions of human and mouse genomes is moderate56. Nevertheless, mouse models with EC-specific overexpression of klf4 decreases atherosclerosis in atheroprone areas, whereas EC-specific ablation of klf4 increases atherosclerosis in atheroprotective areas34. Indeed, opposite trends of KLF4 and ITPR3 expression are evident in mouse AA vs. TA areas (Fig. 4A). These opposing transcriptional and phenotypic outcomes demonstrate the importance of flow regulation of KLF4 in the mouse vasculature. We found five putative KLF4 binding sites predicted in the promoter region of the mouse Itpr3 gene (Sup. Fig. 5), which suggests that mouse Itpr3 may also undergo chromatin remodeling initiated by KLF4 binding. Data in Fig. 5B show that KLF4 binding to site 1 at −1207 and site 5 at −10318 of the ITPR3 promoter caused chromatin remodeling to a reduced compact state, as shown by H3K27ac enrichment and ATAC-seq peaks. Interestingly, deletion of site 1 affected KLF4 binding and chromatin remodeling at site 5 (Sup. Fig. 6). Hence, these two KLF4 binding sites, separated by ~9 kb, may crosstalk to co-regulate the expression of ITPR3.
In summary, using a multi-layer and genome-wide approach, we have made the novel discovery that the PS-induced KLF4 regulates the transcriptome in ECs epigenetically and transcriptionally. Furthermore, we have demonstrated that ITPR3 plays a critical role as a KLF4-regulated gene in mediating H3K27ac enrichment and chromatin accessibility, and hence Ca2+-dependent eNOS activation and EC homeostasis.
Supplementary Material
Highlights.
By using RNA-seq, ChIP-seq, and ATAC-seq, we found that KLF4, functioning as a PS-induced transcription factor in ECs, activates ITPR3 transcriptionally and epigenetically.
KLF4 regulates ITPR3 expression by mediating H3K27ac enrichment and chromatin accessibility in the promoter region.
PS induction of ITPR3 augments Ca2+-dependent eNOS activation and EC homeostasis.
PS and statins increase the eNOS-derived NO bioavailability, at least in part, by KLF4-induction of ITPR3.
This innovative systemic approach identifies many other PS-inducible molecules and pathways through the similar mechanism.
Acknowledgments
The authors thank Drs. Brendan Gongol, Traci Marin, Nathan Spann, Phu Nguyen, Julie Yi-Shuan Li, Mano R. Maurya, and Shankar Subramaniam at University of California, San Diego, and Chun-San Tai at National Chiao Tung University for technical assistance, useful discussions, and their valuable suggestions, and Drs. Mukesh Jian, Panjamaporn Sangwung, and Guangjin Zhou, School of Medicine, Case Western University, for providing lung ECs from EC-KLF4-Tg and EC-KLF4−/− mice.
Sources of Funding
This work was supported in part by NIH research grants R01HL108735 (to J.Y-J.S. and S.C.), R01HL125643 (to J.Y-J.S.), R01GM111665 (to Jin Zhang), K99/R00HL122368 (to Z.C.), 5T32HL007444 (to M.M.), American Heart Association Postdoctoral Fellowship 17POST33410101 (to T.-S.H.), Taiwan Ministry of Science and Technology grants MOST 106–2917-I-009–016 (to H.-D.H.), MOST 107–2633-B-009–003 (to H.-D.H., W.-T.W.).
Abbreviations
- AA
aortic arch
- ATAC-seq
assay for transposase-accessible chromatin-sequencing
- Cav-1
Caveolin-1
- ChIP-Seq
chromatin immunoprecipitation followed by high throughput sequencing
- EC
endothelial cell
- eNOS
Endothelial nitric oxide synthase
- HUVECs
human umbilical vein endothelial cells
- H3K4me1
mono-methylation of histone 3 lysine 4
- H3K27ac
acetylation of histone 3 lysine 27
- ITPR3
Inositol 1,4,5-trisphosphate receptor 3
- KLF4
Krüppel-like factor 4
- NGS
next-generation sequencing
- NO
nitric oxide
- OS
oscillatory shear stress
- Pol II
RNA polymerase II
- PS
pulsatile shear stress
- TA
thoracic aorta
- TF
transcription factors
- THBD
Thrombomodulin
- VEGF
Vascular endothelial growth factor
Footnotes
Disclosures
None.
References
- 1.Chiu JJ, Chien S. Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives. Physiol. Rev 2011;91:327–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gimbrone MA Jr., Garcia-Cardena G Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ. Res 2016;118:620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baeyens N, Bandyopadhyay C, Coon BG, Yun S, Schwartz MA. Endothelial fluid shear stress sensing in vascular health and disease. J. Clin. Invest 2016;126:821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCormick SM, Eskin SG, McIntire LV, Teng CL, Lu CM, Russell CG, Chittur KK. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc. Natl. Acad. Sci. U. S. A 2001;98:8955–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen BP, Li YS, Zhao Y, Chen KD, Li S, Lao J, Yuan S, Shyy JY, Chien S. DNA microarray analysis of gene expression in endothelial cells in response to 24-h shear stress. Physiol. Genomics 2001;7:55–63. [DOI] [PubMed] [Google Scholar]
- 6.Ajami NE, Gupta S, Maurya MR, Nguyen P, Li JY, Shyy JY, Chen Z, Chien S, Subramaniam S. Systems biology analysis of longitudinal functional response of endothelial cells to shear stress. Proc. Natl. Acad. Sci. U. S. A 2017;114:10990–10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J, Li YS, Chien S. Shear stress-initiated signaling and its regulation of endothelial function. Arterioscler. Thromb. Vasc. Biol 2014;34:2191–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee DY, Lee CI, Lin TE, Lim SH, Zhou J, Tseng YC, Chien S, Chiu JJ. Role of histone deacetylases in transcription factor regulation and cell cycle modulation in endothelial cells in response to disturbed flow. Proc. Natl. Acad. Sci. U. S. A 2012;109:1967–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang YZ, Jimenez JM, Ou K, McCormick ME, Zhang LD, Davies PF. Hemodynamic disturbed flow induces differential DNA methylation of endothelial kruppel-like factor 4 promoter in vitro and in vivo. Circ. Res 2014;115:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn J, Qiu H, Kim S, Jjingo D, Hoffman R, Kim CW, Jang I, Son DJ, Kim D, Pan C, Fan Y, Jordan IK, Jo H. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J. Clin. Invest 2014;124:3187–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang TS, Wang KC, Quon S, Nguyen P, Chang TY, Chen Z, Li YS, Subramaniam S, Shyy J, Chien S. Linc00341 exerts an anti-inflammatory effect on endothelial cells by repressing vcam1. Physiol. Genomics 2017;49:339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miao Y, Ajami NE, Huang TS, Lin FM, Lou CH, Wang YT, Li S, Kang J, Munkacsi H, Maurya MR, Gupta S, Chien S, Subramaniam S, Chen Z. Enhancer-associated long non-coding rna leene regulates endothelial nitric oxide synthase and endothelial function. Nat Commun. 2018;9:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donaghey J, Thakurela S, Charlton J, et al. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet 2018;50:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA. Genome-wide location and function of DNA binding proteins. Sci. 2000;290:2306–2309. [DOI] [PubMed] [Google Scholar]
- 17.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013;10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone h3k27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A 2010;107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santulli G, Nakashima R, Yuan Q, Marks AR. Intracellular calcium release channels: An update. J. Physiol 2017;595:3041–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singleton PA, Bourguignon LY. Cd44 interaction with ankyrin and ip3 receptor in lipid rafts promotes hyaluronan-mediated ca2+ signaling leading to nitric oxide production and endothelial cell adhesion and proliferation. Exp. Cell Res 2004;295:102–118. [DOI] [PubMed] [Google Scholar]
- 22.Yuan Q, Yang J, Santulli G, Reiken SR, Wronska A, Kim MM, Osborne BW, Lacampagne A, Yin Y, Marks AR. Maintenance of normal blood pressure is dependent on ip3r1-mediated regulation of enos. Proc. Natl. Acad. Sci. U. S. A 2016;113:8532–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S, Lu Q, Wang Q, Ding Y, Ma Z, Mao X, Huang K, Xie Z, Zou MH. Binding of fun14 domain containing 1 with inositol 1,4,5-trisphosphate receptor in mitochondria-associated endoplasmic reticulum membranes maintains mitochondrial dynamics and function in hearts in vivo. Circulation. 2017;136:2248–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thul PJ, Akesson L, Wiking M, et al. A subcellular map of the human proteome. Sci. 2017;356. [DOI] [PubMed] [Google Scholar]
- 25.Xiao H, Lu M, Lin TY, et al. Sterol regulatory element binding protein 2 activation of nlrp3 inflammasome in endothelium mediates hemodynamic-induced atherosclerosis susceptibility. Circulation. 2013;128:632–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, Langmead B, Salzberg SL. Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 2015;12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao Y, Smyth GK, Shi W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. [DOI] [PubMed] [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012;9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and b cell identities. Mol. Cell 2010;38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. Deeptools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen NTT, Contreras-Moreira B, Castro-Mondragon JA, Santana-Garcia W, Ossio R, Robles-Espinoza CD, Bahin M, Collombet S, Vincens P, Thieffry D, van Helden J, Medina-Rivera A, Thomas-Chollier M. Rsat 2018: Regulatory sequence analysis tools 20th anniversary. Nucleic Acids Res. 2018;46:W209–W214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou G, Hamik A, Nayak L, et al. Endothelial kruppel-like factor 4 protects against atherothrombosis in mice. J. Clin. Invest 2012;122:4727–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chari R, Yeo NC, Chavez A, Church GM. Sgrna scorer 2.0: A species-independent model to predict crispr/cas9 activity. ACS Synth Biol. 2017;6:902–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for crispr screening. Nat. Methods 2014;11:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kilpinen H, Waszak SM, Gschwind AR, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Sci. 2013;342:744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu C, Liu M, Zhang W, Xu Q, Ma K, Chen L, Wang Z, He S, Zhu H, Xu N. Upregulation of klf4 by methylseleninic acid in human esophageal squamous cell carcinoma cells: Modification of histone h3 acetylation through hat/hdac interplay. Mol. Carcinog 2015;54:1051–1059. [DOI] [PubMed] [Google Scholar]
- 40.Maejima T, Inoue T, Kanki Y, et al. Direct evidence for pitavastatin induced chromatin structure change in the klf4 gene in endothelial cells. PLoS One. 2014;9:e96005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fulton D, Gratton JP, McCabe TJ, Fontana J, Fujio Y, Walsh K, Franke TF, Papapetropoulos A, Sessa WC. Regulation of endothelium-derived nitric oxide production by the protein kinase akt. Nature. 1999;399:597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dimmeler S, Fleming I, Fisslthaler B, Hermann C, Busse R, Zeiher AM. Activation of nitric oxide synthase in endothelial cells by akt-dependent phosphorylation. Nature. 1999;399:601–605. [DOI] [PubMed] [Google Scholar]
- 43.Fleming I, Fisslthaler B, Dimmeler S, Kemp BE, Busse R. Phosphorylation of thr(495) regulates ca(2+)/calmodulin-dependent endothelial nitric oxide synthase activity. Circ. Res 2001;88:E68–75. [DOI] [PubMed] [Google Scholar]
- 44.Foskett JK, White C, Cheung KH, Mak DO. Inositol trisphosphate receptor ca2+ release channels. Physiol. Rev 2007;87:593–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boo YC, Kim HJ, Song H, Fulton D, Sessa W, Jo H. Coordinated regulation of endothelial nitric oxide synthase activity by phosphorylation and subcellular localization. Free Radic. Biol. Med 2006;41:144–153. [DOI] [PubMed] [Google Scholar]
- 46.Michel JB, Feron O, Sacks D, Michel T. Reciprocal regulation of endothelial nitric-oxide synthase by ca2+-calmodulin and caveolin. J. Biol. Chem 1997;272:15583–15586. [DOI] [PubMed] [Google Scholar]
- 47.Taylor CW, Genazzani AA, Morris SA. Expression of inositol trisphosphate receptors. Cell Calcium. 1999;26:237–251. [DOI] [PubMed] [Google Scholar]
- 48.Liu L, Xu Y, He M, et al. Transcriptional pause release is a rate-limiting step for somatic cell reprogramming. Cell Stem Cell. 2014;15:574–588. [DOI] [PubMed] [Google Scholar]
- 49.Sangwung P, Zhou G, Nayak L, et al. Klf2 and klf4 control endothelial identity and vascular integrity. JCI Insight. 2017;2:e91700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z, Martin M, Zhang J, et al. Kruppel-like factor 4 regulation of cholesterol-25-hydroxylase and liver x receptor mitigates atherosclerosis susceptibility. Circulation. 2017;136:1315–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaczynski J, Cook T, Urrutia R. Sp1- and kruppel-like transcription factors. Genome Biol. 2003;4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenzweig JM, Glenn JD, Calabresi PA, Whartenby KA. Klf4 modulates expression of il-6 in dendritic cells via both promoter activation and epigenetic modification. J. Biol. Chem 2013;288:23868–23874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ray A, Alalem M, Ray BK. Loss of epigenetic kruppel-like factor 4 histone deacetylase (klf-4-hdac)-mediated transcriptional suppression is crucial in increasing vascular endothelial growth factor (vegf) expression in breast cancer. J. Biol. Chem 2013;288:27232–27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, Pannekoek H, Horrevoets AJ. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung kruppel-like factor (klf2). Blood. 2002;100:1689–1698. [DOI] [PubMed] [Google Scholar]
- 55.Parmar KM, Nambudiri V, Dai G, Larman HB, Gimbrone MA Jr., Garcia-Cardena G Statins exert endothelial atheroprotective effects via the klf2 transcription factor. J. Biol. Chem 2005;280:26714–26719. [DOI] [PubMed] [Google Scholar]
- 56.Dietrich WF, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain Z, Dredge R, Daly MJ, Ingalls KA, O’Connor TJ. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.