Abstract
The thalamus and its nuclei are largely indistinguishable on standard T1 or T2 weighted MRI. While diffusion tensor imaging based methods have been proposed to segment the thalamic nuclei based on the angular orientation of the principal diffusion tensor, these are based on echo planar imaging which is inherently limited in spatial resolution and suffers from distortion. We present a multi-atlas segmentation technique based on white-matter-nulled MP-RAGE imaging that segments the thalamus into 12 nuclei with computation times on the order of 10 minutes on a desktop PC; we call this method THOMAS (THalamus Optimized Multi Atlas Segmentation). THOMAS was rigorously evaluated on 7T MRI data acquired from healthy volunteers and patients with multiple sclerosis by comparing against manual segmentations delineated by a neuroradiologist, guided by the Morel atlas. Segmentation accuracy was very high, with uniformly high Dice indices: at least 0.85 for large nuclei like the pulvinar and mediodorsal nuclei and at least 0.7 even for small structures such as the habenular, centromedian, and lateral and medial geniculate nuclei. Volume similarity indices ranged from 0.82 for the smaller nuclei to 0.97 for the larger nuclei. Volumetry revealed that the volumes of the right anteroventral, right ventral posterior lateral, and both right and left pulvinar nuclei were significantly lower in MS patients compared to controls, after adjusting for age, sex and intracranial volume. Lastly, we evaluated the potential of this method for targeting the Vim nucleus for deep brain surgery and focused ultrasound thalamotomy by overlaying the Vim nucleus segmented from pre-operative data on post-operative data. The locations of the ablated region and active DBS contact corresponded well with the segmented Vim nucleus. Our fast, direct structural MRI based segmentation method opens the door for MRI guided intra-operative procedures like thalamotomy and asleep DBS electrode placement as well as for accurate quantification of thalamic nuclear volumes to follow progression of neurological disorders.
Keywords: thalamic nuclei, thalamic parcellation, multi-atlas segmentation, white matter nulled MP-RAGE
1. Introduction
The thalamus is a relay organ that has been implicated in a number of higher level neurological functions, including relaying of sensory and motor signals to the cerebral cortex, and regulation of consciousness, sleep, and alertness. It is a walnut-shaped bilateral structure, spanning an anterior-posterior length of 30mm and transverse length of 20mm on each side. Each thalamus can be subdivided broadly into anterior, lateral, medial, and posterior groups demarcated by bundles of white matter fibers called internal medullary laminae. Each of these groups can further be subdivided into smaller nuclei based on cytohistological differences, with the exact terminology and subdivision being dependent on the specific atlas used (Morel et al., 1997; Niemann et al., 1994).
The thalamus has been implicated in Alzheimer’s disease (Braak and Braak, 1991), schizophrenia (Andreasen, 1997; Parnaudeau et al., 2018) , essential tremor (Benabid et al., 1996), alcohol use disorder (Arts et al., 2017; Fama et al., 2014), multiple sclerosis (Minagar et al., 2013) and many other brain pathologies (Maggioni et al., 2017). These pathologies may differentially impact individual nuclei; hence, accurate volumetry of thalamic nuclei could be very useful in understanding the progression and effect of these neurological/neuropsychiatric disorders, as some preliminary studies seem to indicate (Braak and Braak, 1991; Coscia et al., 2009; Lee et al., 2016). Furthermore, accurate and patient-tailored localization of specific nuclei can minimize surgical targeting errors such as with the ventralis intermedius (Vim) nucleus for treatment of essential tremor using deep brain surgery (DBS). Fast and direct targeting can also improve the accuracy and efficacy of newer thalamus ablation methods like focused ultrasound thalamotomy (Elias et al., 2013). Accurate volumetry of thalamic nuclei would also be valuable for further understanding normal development and aging, given the critical role of the thalamus across a wide spectrum of brain functions.
Individual thalamic nuclei are not clearly visible on standard T1 or T2 weighted MRI sequences like Magnetization Prepared Rapid Gradient Echo (MP-RAGE) or Fast/Turbo Spin Echo (FSE/TSE), in part due to poor intra-thalamic nuclear contrast. As a result, clinical applications such as DBS and more recently, focused ultrasound thalamotomy, have relied on standard atlases as well as awake physiologic confirmation of nuclei localization. While there have been a few attempts at using T1 and T2 based discrimination of thalamic nuclei (Deoni et al., 2007; Traynor et al., 2011), most thalamic segmentation methods reported in literature to date (Battistella et al., 2017; Kumar et al., 2015; Mang et al., 2012; Wiegell et al., 2003; Ziyan et al., 2006) are based on diffusion tensor imaging (DTI), which provides voxel-level information on white matter anisotropy. Since DTI uses echo-planar imaging (EPI) for acquisition, its spatial resolution is restricted (typically 2×2×2 mm3) in order to reduce distortion, limit scan time, and improve signal-to-noise ratio (SNR). DTI-based methods are also suboptimal due to the predominance of grey matter in the thalamus, which results in low anisotropy and imprecise localization. Direct thalamic imaging methods based on susceptibility weighted imaging (SWI) (Abosch et al., 2010; Xiao et al., 2016) have been proposed for 7T MRI. However, these methods have only been useful in delineating a limited number of thalamic nuclei, mainly the Vim nucleus. Variants of MP-RAGE, where cortical grey matter (GM) or white matter (WM) are suppressed, have been proposed to visualize and target the Vim nucleus (Magnotta et al., 2000; Spiegelmann et al., 2006; Sudhyadhom et al., 2009; Vassal et al., 2012). These have mostly been used for DBS planning using manual segmentation and there has been no method developed for systematic and automated segmentation of all major thalamic nuclei based solely on structural MRI.
The thalamic nuclear segmentation methods that have been developed are specific to the acquisition methods described above. DTI-based methods involve clustering at a voxel level using properties like angular orientation of the principal diffusion tensor or information from the whole tensor. Small nuclei are difficult or impossible to resolve, probably because of the limited spatial resolution of EPI based methods. Many of these clustering methods also require good initialization parameters to perform optimally. Multiple contiguous nuclei such as those found in the ventrolateral complex could have similar diffusion tensor orientations while projecting to different cortical areas. As a result, DTI-based methods have been limited to resolving 4-7 clusters, loosely correlated to the 4 broad anatomical divisions of the thalamus (anterior, posterior, medial, and lateral), rather than specific nuclei within these groups, and almost none of these DTI-based methods report segmentation of important but small nuclei such as the centromedian (CM) and the habenular (Hb) nuclei. While some methods have reported parcellation of larger clusters of nuclei (e.g. 21 in (Kumar et al., 2015)), they do not appear to be consistent across multiple individuals. Stough et al. (Stough et al., 2013) used a multimodal approach which combined features derived from DTI with those from T1 and T2 MRI and a hierarchical random forest classifier to generate lateral geniculate nucleus (LGN) and medial geniculate nucleus (MGN) in addition to the 4 main nuclear groups, albeit with poor accuracy for the smaller nuclei compared to manual segmentation. Tractography based approaches (Schlaier et al., 2015; Sedrak et al., 2011; Yamada et al., 2010) have been clinically useful for DBS targeting as they directly map the fiber tracts running from the motor cortex via the Vim nucleus to the cerebellum. Generalization of this method to other nuclei requires prior knowledge of structural connectivity networks of the thalamic nuclei to different parts of the cortex as well as prior segmentation of those cortical regions, on a subject-specific basis. This has not been very successful in characterizing small nuclei (Behrens et al., 2003; Johansen-Berg et al., 2005). Liu et al. (Liu et al., 2015) used statistical shape models built on high contrast multi modal 7T MRI datasets to segment 3T MRI data. The 7T MRI included SWI and a series of MP-RAGE datasets acquired with different inversion times to generate a mosaic of intra-thalamic contrast. One common drawback of most current methods is lack of quantitative comparisons to a ground truth, such as segmentation based on an established anatomic atlas. Typically, validation has used qualitative comparisons to comparable slices from the Morel atlas, making rigorous evaluation of accuracy difficult.
Intra thalamic contrast is inherently low and any successful structural MRI segmentation method should be based on an acquisition technique that is able to generate sufficiently high contrast. We used a white-matter-nulled (WMn) variant of the commonly available MP-RAGE sequence (Saranathan et al., 2015; Tourdias et al., 2014a) for data acquisition, which maximizes the inter-thalamic nuclear contrast at 7T as well as provides excellent WM-thalamus contrast for delineation of the thalamic boundaries. We have previously demonstrated (Tourdias et al., 2014a) that such an acquisition could be used to uniquely identify boundaries between individual nuclei using manual segmentation, a laborious process, taking several hours and also requiring very specific domain expertise. In the present work, we leverage manual delineations that were derived from WMn MP-RAGE to fine tune an automatic pipeline. By combining information from a library of multiple WMn MP-RAGE datasets with prior manual segmentation by an expert guided by the Morel histological atlas, the thalamus is automatically segmented into 12 nuclei per side on the order of 10 minutes on a desktop PC. We call this method THalamus Optimized Multi Atlas Segmentation (THOMAS). THOMAS was quantitatively evaluated on WMn MP-RAGE data obtained from healthy volunteers and patients with multiple sclerosis (MS) using manual segmentation by an expert as ground truth. Volumetric analyses were performed to test for differences in thalamic nuclear volumes between MS patients and healthy controls after adjusting for age, sex, and intracranial volume (ICV) differences. In a small subset, the performance of THOMAS was compared to that of the very recently introduced thalamic segmentation module of FreeSurfer, which uses conventional MP-RAGE data to segment the thalamic nuclei. Finally, we illustrate, using example datasets, THOMAS applied retrospectively to targeting the Vim nucleus for treatment of essential tremor; one application being focused ultrasound ablation and the other being DBS electrode placement. These two examples demonstrate the potential prospective utility of our segmentation technique for precise targeting of thalamic nuclei in intraoperative MRI-based procedures.
2. Methods
2.1. Multi-atlas segmentation
While multi-atlas segmentation methods have been used for segmentation of hippocampal subfields (Yushkevich et al., 2010) and amygdala (Hanson et al., 2012), they have not been reported for segmentation of thalamic nuclei, presumably due to the lack of clear manually delineated thalamic nuclei. We have developed a multi-atlas segmentation method called THalamus Optimized Multi-Atlas Segmentation (THOMAS) which uses a multi-atlas prior dataset, comprising 20 WMn MP-RAGE datasets acquired with high contrast and spatial resolution, whose thalamic nuclei were manually segmented by an expert neuroradiologist, guided by the Morel atlas. THOMAS uses a template, created by registering and averaging the 20 prior datasets as an intermediate step, to reduce the number of computationally expensive nonlinear registration steps from 20 to 1, and thereby reducing the total processing time. Further reduction in processing times is achieved by automatically cropping the template and the input image, significantly speeding up the nonlinear registration step. These are described in detail in the following sections.
2.1.1. Building a library of prior atlases
To generate a library of 20 priors with labels, the whole thalamus and 12 thalamic nuclei were manually traced on 20 WMn MP-RAGE datasets acquired at 7T using the Morel atlas for guidance by an expert neuroradiologist (author XX). A reproducible manual segmentation delineation protocol was developed for this task with excellent intra-rater reliability as measured by intraclass correlation coefficient (ICC) and mean distance discrepancy between centers of mass ΔCoMs) at initial and repeat tracings 3 weeks later, yielding ICC = 0.997 (95% confidence interval 0.996-0.998) and ΔCoM = 0.69±0.38mm respectively (Tourdias et al., 2014b). Tracings were performed in 3D Slicer (http://www.slicer.org; Fedorov et al., 2012) using freehand drawing tools to build 3D models of each structure. More details of the manual segmentation procedure can be found here (Tourdias et al., 2014a).
2.1.2. THOMAS pipeline
a. Registration.
Image registration between the prior datasets and input dataset is the first major step in multi-atlas segmentation prior to warping of the labels from prior space to input space. We used Advanced Normalization Tools ANTs (Avants et al. 2008) and its Symmetric Normalization algorithm as the tool of choice for all nonlinear registration in our work. A mean brain template was first created from the library of 20 WMN MP-RAGE prior datasets. This was done using the buildtemplate feature available as part of ANTs, which iteratively registers each prior to an average of the priors and then generates a mean brain template by averaging the registered priors. This resulting template, which includes both normal and diseased brain states, has very high signal-to-noise ratio (SNR) and image contrast, making it useful as an “intermediate space” for registration between the priors and input images (Artaechevarria et al., 2008). The library of prior atlases was registered to an input target volume by first pre-computing and storing the warps from each prior subject to the mean brain template. The target volume was then diffeomorphically registered to the template using ANTs. The full transformation from each prior to the input target was formed by combing the pre-computed prior subject’s warp to the template with the inverse of the target-to-template warp using ComposeMultiTransform in ANTS. Crucially, the use of an intermediate template enabled the use of pre-computed warps and cut the number of nonlinear registration steps from 20 to 1, dramatically reducing the registration time from 60 hours to 3 hours on an 8-core dual Xeon processor Dell desktop.
b. Cropping.
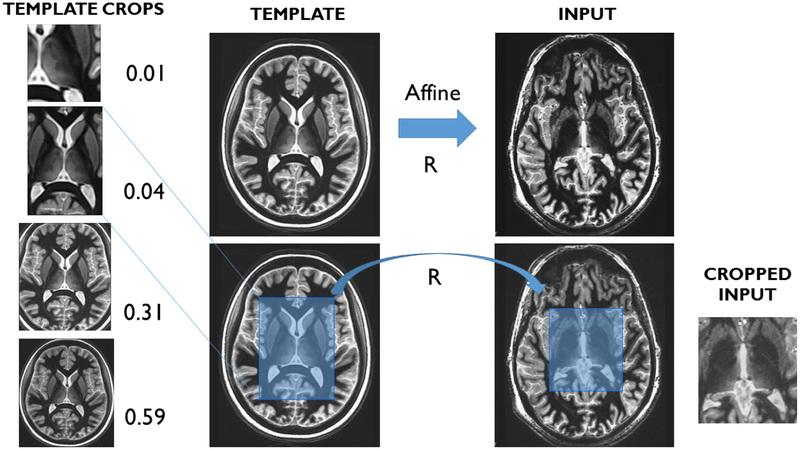
To further reduce processing times, we used a cropped version of both the template and the input target image. Planar cropping was performed manually on the template to generate a cropped template and a mask. This only needed to be performed once. To automatically crop the input image, we first linearly registered the full uncropped input and template images, warped the crop mask from template space to the input space, and finally cropped the input image with the desired crop mask. The use of cropping eliminates the need for pre-processing steps such as skull stripping, often used when processing whole volumes. We experimented with various crop sizes ranging from the full template size (256×256×440) to a tight crop encompassing either one (61×91×62) or both thalami (93×187×68), systematically spanning the sizes in all dimensions, with the crop ratio (defined as ratio of total pixels in the cropped template to the full uncropped template) ranging from 0.01 to 1. A small validation study was performed using 5 subjects to evaluate accuracy and processing times as a function of crop size to select a crop size that achieves the best combination of accuracy and processing times.
c. Label fusion.
After the cropping and registration steps, the 20 sets of candidate labels generated, one from each prior in the library, were fused to create a single set of labels for the target input image, which is the desired thalamic segmentation. We used PICSL MALF (Wang et al., 2013) for label fusion which has been shown to be superior to simplistic but fast approaches such as majority voting (Rohlfing et al., 2004). PICSL MALF has tunable hyper-parameters which were optimized for the task of thalamic segmentation from WMn MP-RAGE data. For each label, we implemented a grid search over a range of reasonable values to determine the best choice. This was run with leave-one-out cross-validation over the 20 priors, producing 20 cases for each hyper-parameter choice as described in (Wang et al., 2013). Our reward function was the Dice coefficient trimean over the validation cases, an efficient estimator of average segmentation performance. We searched over a 5×6×8 grid with ranges rp=[1, 5], rs=[0, 5], and β=[0.1, 10], making 39,000 executions.
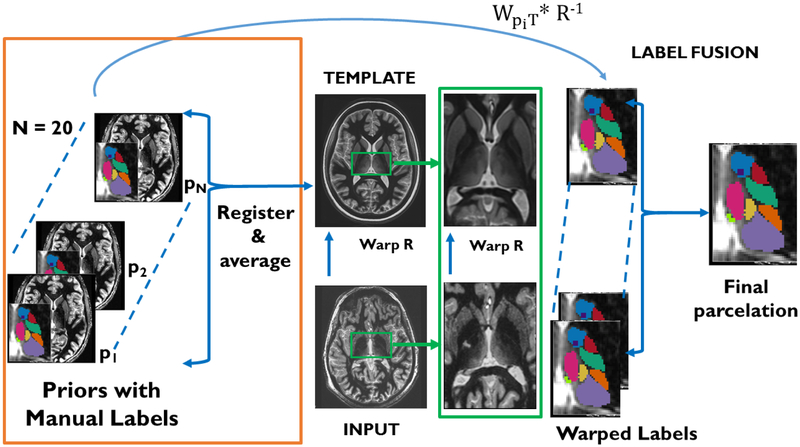
Figure 1 shows a flowchart of THOMAS. The automated cropping step for THOMAS is shown in Figure 2 along with four example crop sizes for the template.
Figure 1.
THOMAS workflow- The input image is nonlinearly registered to the average brain template. Precomputed warps from each of the 20 priors to the template is then combined with the template-input warp (R−1) to generate 20 candidate labels, which are then fused to get the final parcellation.
Figure 2.
Automated cropping scheme for THOMAS- the mean template is cropped manually with a desired crop mask (ranging in size from unilateral and bilateral thalamus to the whole brain). An affine registration between the template and the input is performed and the crop mask warped to the input space to crop generate a cropped input image.
2.2. MRI acquisition and datasets
2.2.1. White-matter-nulled MP-RAGE sequence
The inversion time of an MP-RAGE sequence can, in theory, be adjusted to null any one of three species-WM, GM, or CSF. Conventional MP-RAGE uses CSF suppression which results in good WM and GM signal but poor intra-thalamic contrast. On the other hand, WM nulling produces the best intra-thalamic (i.e. inter-nuclear) as well as thalamic-WM contrast (Tourdias et al., 2014a) with excellent signal-to-noise characteristics. Hence, WM nulling was chosen for our acquisition sequence. The WMn MP-RAGE sequence was further optimized to maximize SNR efficiency and contrast while minimizing blurring as described in (Tourdias et al., 2014a). Table 1 lists the optimized WMn MP-RAGE sequence parameters used at different field strengths.
Table 1.
White-matter-nulled MP-RAGE sequence parameters
| 1.5T (GE) | 3T (GE) | 3T (Siemens) | 7T (GE) | |
|---|---|---|---|---|
| TR/TS (ms) | 10.4/3000 | 10/4500 | 10/4000 | 10/6000 |
| TI (ms) | 350 | 500 | 500 | 680 |
| Flip (deg) | 10 | 9 | 7 | 4 |
| Matrix | 224×224×124 | 180×180×200 | 256×256×160 | 180×180×200 |
| Slice thickness (mm) | 1.2 | 1 | 1.1 | 1 |
| Parallel imaging | None | None | 2×1 | 1.5×1.5 |
| Scan time (min) | 7.5 | 10 | 5.5 | |
| Coil | 8-channel GE | 8-channel GE | 24-channel Nova | 32-channel Nova |
The datasets used to create the template and the atlas, as well as the data sets used for training and testing, were all acquired on a GE 7T MRI scanner (Discovery MR950, GE Healthcare) using a 32 channel head coil (Nova Medical), after obtaining informed consent. The 20 subjects that make up the prior library consisted of 9 healthy volunteers and 11 MS patients, chosen randomly from a larger set of 28 cases. We refer to this group as the training dataset. The remaining 8 subjects (4 healthy volunteers, 4 MS patients) were manually segmented and reserved for independent testing of our segmentation method. We refer to this group as the test dataset. The datasets were apportioned to roughly balance the number of controls and patients in each group. For volumetric analyses comparing MS patients and control subjects, we used 7 additional control subjects recruited to improve the balance between these two groups, resulting in a total of 15 MS patients and 20 healthy control subjects. We used multiple linear regression (MLR) analysis to determine whether there was a significant association between volume and disease state for each of the 26 volumes (12 thalamic nuclei plus whole thalamus on each of right and left sides), and report those significant associations that survived Bonferroni correction for 26 multiple comparisons.
The patient undergoing FUS thalamotomy was scanned pre-operatively on a GE 7T MRI scanner using a 32 receive-channel head coil (Nova Medical) and then post-operatively on a GE 3T MRI scanner (Discovery MR750, GE Healthcare) using an 8 receive-channel head coil. Patients undergoing DBS electrode placement were first scanned pre-operatively on a 3T Siemens MRI scanner (Skyra, Siemens Healthcare) using a 24 receive-channel head coil. Following the DBS surgery, these patients were scanned on a 1.5T GE MRI scanner using a 8 receive-channel head coil. The sequence used for thalamic segmentation was WMn MP-RAGE, although conventional T1 and T2 sequences were also acquired for lead visualization and routine clinical evaluation.
2.3. Image metrics
Accuracy was evaluated using two metrics: Dice coefficient and volume similarity index (VSI). They are defined as follows-
| (1) |
where X and Y refer to the two segmentation labels compared, with one being the ground truth and ∣X∣ and ∣Y∣ refers to the number of pixels in X and Y respectively.
To evaluate the performance of THOMAS, we first used images from the prior library as training cases by performing a leave-one-out cross validation. Each of the 20 prior images was segmented using THOMAS and compared with the ground truth manual segmentations. In this cross-validation, only 19 prior images (20 minus the prior image being analyzed) were used for label fusion for each prior being segmented, to eliminate bias.
Imaging data from eight subjects (test dataset) which were not part of the template or the library of twenty priors (training dataset) were then used as an unbiased test sample to assess the performance of THOMAS, also by evaluating against manual segmentation ground truth. A subset of 5 cases from this test dataset was used to compare the performance of THOMAS with the recently released thalamic segmentation module of FreeSurfer (Iglesias et al., 2018). Conventional T1 MP-RAGE data as well as both T1 MP-RAGE and WMn MP-RAGE were used as inputs to the thalamic segmentation module of FreeSurfer to generate two sets of thalamic nuclei labels; these labels were then compared both qualitatively and quantitatively to the corresponding labels produced by THOMAS.
2.4. Applications to FUS ablation and DBS planning
To illustrate the utility of THOMAS, we used it on a patient who was scheduled for FUS ablation of the Vim nucleus for treatment of essential tremor. This patient was scanned pre-operatively on a 7T MRI scanner and post-operatively after the FUS ablation procedure on a 3T MRI scanner, including the WMn MP-RAGE sequence in both exams. Targeting was based on standard indirect coordinates, adjusted at the time of surgery based on intraoperative test lesioning. The pre- and post-operative WMn MP-RAGE images were registered using affine registration while masking out the ablated region. The Vim label was then transferred from pre-op 7T to post-op 3T images and overlaid to visualize the location of the Vim relative to the ablated region.
To illustrate another application of THOMAS, we evaluated its performance on a patient who was scheduled for DBS implantation in the Vim nucleus for treatment of essential tremor. This patient had DBS electrodes implanted using a standard stereotactic frame and initial indirect coordinates, adjusted using awake microelectrode recording and physiologic mapping, followed by post-operative MRI. THOMAS was first used to segment the Vim nucleus from the pre-operative 3T WMn MP-RAGE images. The pre-operative and post-operative WMn MP-RAGE images were then registered using ANTS nonlinear registration to minimize the effect of distortion caused by the metal artifacts from the electrode. The Vim label was then warped to the post-operative image space and overlaid on T2 Cube to minimize the effects of dephasing from metal, prominent on gradient echo based sequences like MP-RAGE. The DBS electrode was manually segmented on post-operative T2 Cube showing the final lead position relative to the Vim segmentation.
2.5. Processing
All experiments were conducted on a dual-CPU 4-core 3 GHz Intel Xeon E5-2623 v3 Dell workstation with the exception of template creation which was on a 12-core 2.66 GHz Intel Xeon Mac workstation and hyperparameter optimization, which was performed on the Stanford Sherlock High Performance Cluster (HPC). A Wilcoxon signed-rank test was used to determine statistical significance. ANTs was used for all image registration using the Symmetric Normalization option for diffeomorphic nonlinear registration and affine option for linear registration. For multi-atlas label fusion, Python code from Wang et al. (Wang et al., 2013) was used. All pipelines were coded using Python. Brainlab was used for visualization of the DBS electrode on post-operative scans for the DBS cases.
3. Results
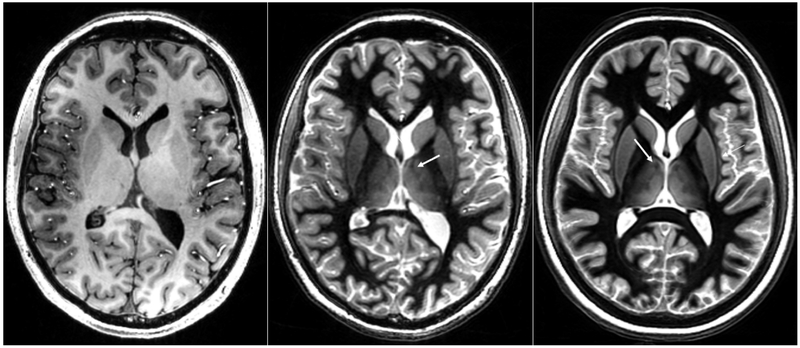
Figure 3b is a representative WMn MP-RAGE image showing the excellent inter-nuclear contrast achieved using the optimized pulse sequence parameters. Note also the clear depiction of the boundaries of the whole thalamus, which is not seen on conventional MP-RAGE imaging where CSF is nulled (Fig. 3a). The averaged mean brain template is shown in Figure 3c. The significantly improved SNR and contrast as well as the conspicuity of small features like the mammillo-thalamic tract (MTT) can be easily appreciated (white arrow), attesting to the quality of the registration steps involved in the template creation.
Figure 3.
Comparable slices from a conventional CSF-nulled MP-RAGE sequence (left), a white-matter-nulled MP-RAGE sequence (middle) and the mean brain template (right) generated from the white-matter-nulled MP-RAGE prior datasets. Note the lack of boundaries and poor inter nuclear contrast in the conventional MP-RAGE compared to the white-matter-nulled MP-RAGE sequence. Note also the improved SNR of the mean template as well as preservation of fine structures such as the MTT (white arrows).
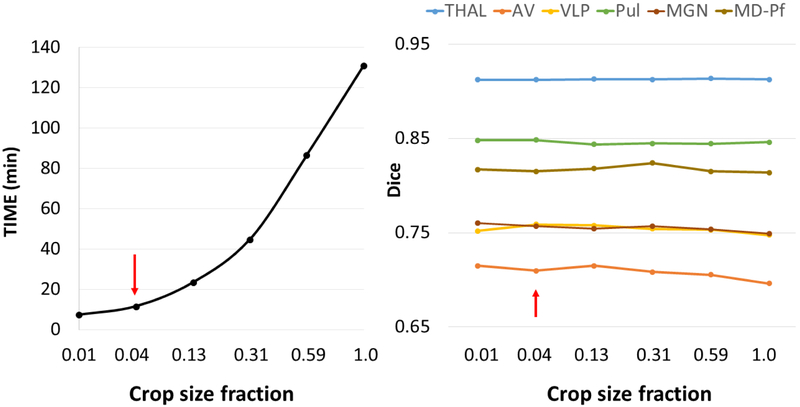
Figure 4 shows the effect of crop size fraction on mean processing times (a) and mean Dice coefficients (b) in comparison with the manual segmentation ground truth, shown for a subset of thalamic nuclei spanning a range of sizes from small nuclei like AV to the whole thalamus (THALAMUS) and averaged over 5 subjects. It can be seen that the mean Dice coefficients are fairly constant while the mean processing times increase exponentially with increasing crop size fraction, reaching its maximum for no cropping. The two smallest crop sizes (0.01 and 0.04) which represent unilateral and bilateral thalamic cropping both perform under 15 minutes and are ideal for minimizing processing times for applications such as DBS targeting. Since we are mostly interested in bilateral thalamic segmentation and since there is not a significant increase in processing time from unilateral to bilateral segmentation, the bilateral crop size 93×187×68 (red arrows) was chosen as the crop size for all subsequent experiments using THOMAS.
Figure 4.
Processing times and Dice coefficients as a function of crop size fraction for THOMAS obtained from a small subset of 5 datasets. While the crop size fractions of 0.01 and 0.04 are both ideal from a processing time standpoint, 0.04 corresponds to a bilateral thalamus crop and was chosen. See Figure 2 for the visualization. Note the relative insensitivity of Dice to crop fraction while the processing time increases exponentially with size.
Table 2 shows mean Dice coefficients for the whole thalamus and 12 thalamic nuclei including the MTT segmented using THOMAS compared against manual segmentation ground truth on the 20 priors processed using leave-one-out cross validation (training dataset) as well as the test dataset (8 subjects that were not part of the 20 priors training dataset). The nuclei are listed in descending size order. It can be seen that there is virtually no loss of accuracy while the processing time reduces by factor of 10 when going from full size to the cropped THOMAS. Note the high Dice values of 0.7 or higher for most nuclei including very small structures like the Hb and CM and reaching 0.85 or higher for larger nuclei such as the Pul and MD-Pf for both the training and test datasets. For the test dataset, the VSI ranges from a low of 0.82 (MGN) to a high of 0.98 (THALAMUS), indicating excellent accuracy in volume estimation. There were no statistically significant differences between the Dice of control subjects and MS patients for any of the thalamic nuclei, attesting to the ability of THOMAS to perform equally well on both healthy subjects and MS patients. Volumetric analyses using multiple linear regression which allowed us to adjust for age, sex and ICV, revealed significantly reduced volumes associated with MS for the following nuclei: Right AV, Right VPL, Left Pulvinar and Right Pulvinar (p<0.0019 prior to Bonferroni correction meaning these findings were significant even after conservative correction for multiple comparisons).
Table 2.
Comparison of Dice coefficients for THOMAS with no cropping and THOMAS (bilateral thalamus crop) for the whole thalamus and 12 thalamic nuclei for the training dataset computed using leave-one-out cross validation (n=20) as well as for the test dataset (n=8). The last column shows VSI for the test dataset. For each case, Dice coefficients were computed by comparison with the manually segmented ground truth.
| Method and Dataset |
THOMAS no cropping n=20 training set |
THOMAS cropped n=20 training set |
THOMAS cropped n=8 test set |
THOMAS cropped n=8 test set |
|
|---|---|---|---|---|---|
| Metric | Volume | Dice | Dice | Dice | VSI |
| THAL | 6064.00 | 0.92 | 0.92 | 0.93 | 0.98 |
| Pul | 1700.00 | 0.86 | 0.86 | 0.86 | 0.96 |
| VLP | 915.00 | 0.77 | 0.79 | 0.79 | 0.97 |
| MD-Pf | 743.00 | 0.85 | 0.85 | 0.89 | 0.93 |
| VPL | 372.00 | 0.68 | 0.69 | 0.71 | 0.92 |
| VA | 315.00 | 0.71 | 0.71 | 0.70 | 0.91 |
| AV | 180.00 | 0.79 | 0.80 | 0.77 | 0.89 |
| CM | 147.00 | 0.75 | 0.75 | 0.78 | 0.88 |
| LGN | 142.00 | 0.74 | 0.75 | 0.70 | 0.91 |
| VLa | 135.00 | 0.68 | 0.67 | 0.64 | 0.83 |
| MGN | 102.00 | 0.72 | 0.74 | 0.71 | 0.82 |
| MTT | 55.00 | 0.69 | 0.70 | 0.69 | 0.92 |
| Hb | 32.00 | 0.73 | 0.73 | 0.79 | 0.93 |
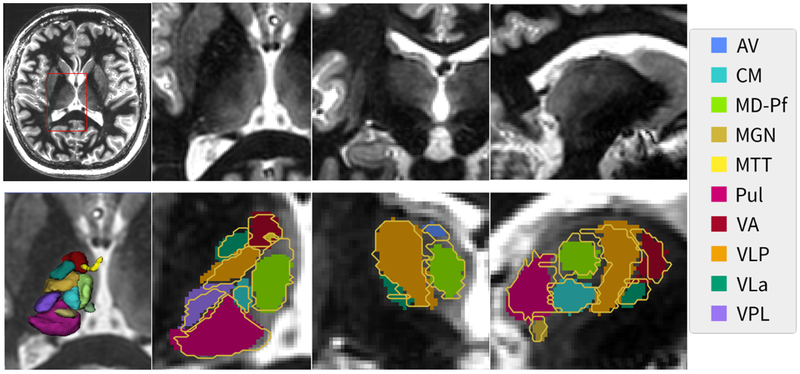
Figure 5 shows an example of the segmentation results from a 7T WMn MP-RAGE dataset processed using THOMAS. An axial slice is shown for reference in the top left and zoomed insets of the three orthogonal planes are shown on the top right with the segmented nuclei (solid color) overlaid on the zoomed insets in the bottom right panels. The manual segmentation ground truth data is depicted as yellow outlines. The close matching of the yellow boundaries to the automatic segmentation can be appreciated even for small nuclei despite the errors caused by digitization of the manual segmentation ground truth. A 3D rendering of all the segmented nuclei is also shown on the bottom left panel.
Figure 5.
A representative slice from a WMn MP-RAGE dataset (top left) along with the zoomed insets in three orthogonal planes (top right panels). THOMAS segmentation results (solid color) are shown in the bottom right panels with the manual segmentation shown as yellow outlines. A 3D rendering of the segmentation is shown in the bottom left panel. Note that even small structures like the MTT (yellow) are clearly visualized.
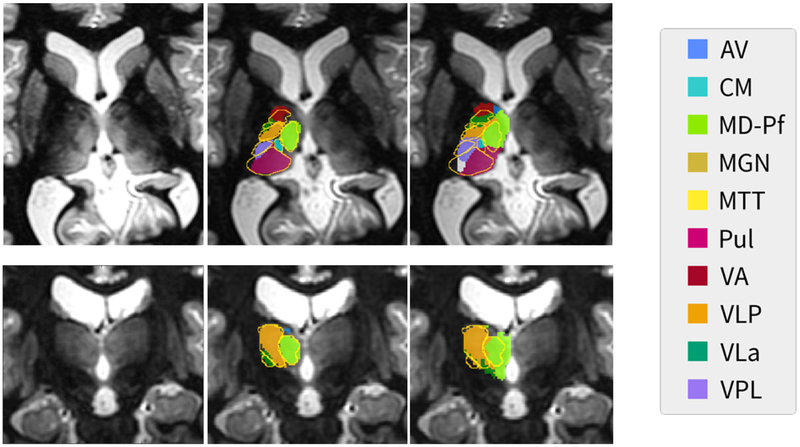
Table 3 shows a comparison of the performance of THOMAS and FreeSurfer using Dice and VSI metrics, evaluated against manual segmentation ground truth. It can be seen that THOMAS performs consistently well even in small nuclei. Figure 6 shows, on an example case, a comparison of the two segmentation schemes. Unlabeled WMn MP-RAGE images in axial (top) and coronal (bottom) planes are shown in the left column while the middle and right columns show overlays of thalamic nuclei produced by THOMAS and FreeSurfer respectively, with the ground truth manual segmentations showing in gold color in both cases. It can be seen that THOMAS follows the contours of the WMn MPRAGE contrast more closely as seen in the pulvinar and mediodorsal nuclei, and better matches the gold standard manual segmentations.
Table 3.
Comparison of Dice coefficients and VSI for THOMAS, FreeSurfer using just T1 MP-RAGE, and FreeSurfer using T1 MP-RAGE and WMn MP-RAGE data, evaluated by comparison with the manual segmentation ground truth for representative nuclei and the whole thalalamus.
| THOMAS Dice |
FS (T1) Dice |
FS (T1+WMn) Dice |
THOMAS VSI |
FS (T1) VSI |
FS (T1+WMn) VSI |
|
|---|---|---|---|---|---|---|
| THAL | 0.92 | 0.75 | 0.78 | 0.98 | 0.90 | 0.93 |
| Pul | 0.85 | 0.55 | 0.55 | 0.97 | 0.78 | 0.84 |
| VLP | 0.79 | 0.37 | 0.46 | 0.96 | 0.88 | 0.88 |
| MD-Pf | 0.86 | 0.52 | 0.55 | 0.92 | 0.87 | 0.81 |
| VPL | 0.71 | 0.27 | 0.37 | 0.92 | 0.62 | 0.60 |
| VA | 0.67 | 0.34 | 0.35 | 0.94 | 0.80 | 0.90 |
| AV | 0.75 | 0.26 | 0.18 | 0.83 | 0.76 | 0.61 |
| CM | 0.77 | 0.28 | 0.43 | 0.92 | 0.71 | 0.76 |
Figure 6.
Comparison of THOMAS (middle column) and FreeSurfer (right column) segmentation results on a representative slice shown in axial (top row) and coronal (bottom row) planes with the manual segmentation based on the Morel atlas overlaid as gold outline. Native WMn MP-RAGE images are shown in the left column. Note that the THOMAS segmentation clearly follows the thalamic boundaries demarcated in the WMn MP-RAGE images. FreeSurfer used both conventional and WMn MP-RAGE images for this segmentation.
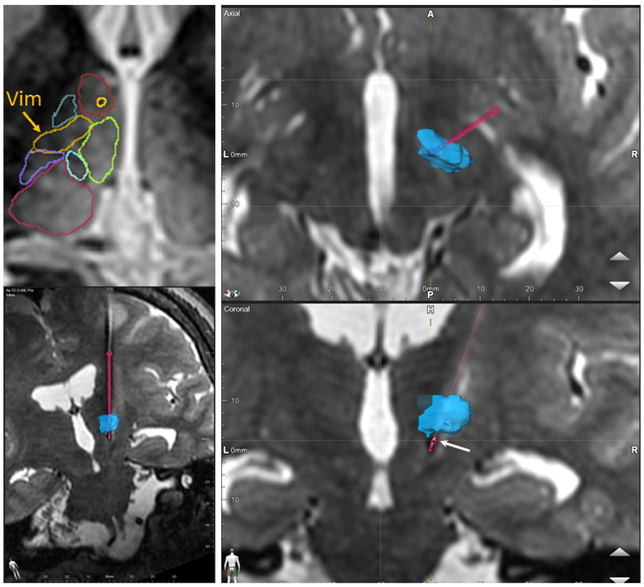
Figure 7 shows orthogonal planes of thalamic anatomy segmented from pre-operative 7T WMn MP-RAGE images (top panel) overlaid on top of post-op 3T WMn-MP-RAGE images (bottom panel). Note that the segmentation boundaries have been interpolated for display purposes. The dotted line is the AC-PC plane and the red cross marks the FUS ablation target inside the Vim nucleus (orange outline). The colored circles are additional targeting adjustments points that were explored during treatment before reaching the final treatment location at the cross. We can see that the target falls well within the segmentation of Vim. Note that the Vim was generated as the inferior half of the VLP nucleus.
Figure 7.
Application of THOMAS to focused ultrasound thalamotomy- the top 3 panels show orthogonal planes of post-operative 3T WMn MP-RAGE images obtained on a patient undergoing ablation of the Vim nucleus for treatment of essential tremor. Pre-operative 7T WMn MP-RAGE data acquired on the patient was segmented using THOMAS and warped to the post-operative space along with the labels. The lower panels show zoomed insets with thalamic nuclei boundaries overlaid on top. The ablation region (white edematous region) is clearly within the Vim nucleus (yellow). The red cross is the targeted ablation hot spot and the dotted line is the AC-PC axis.
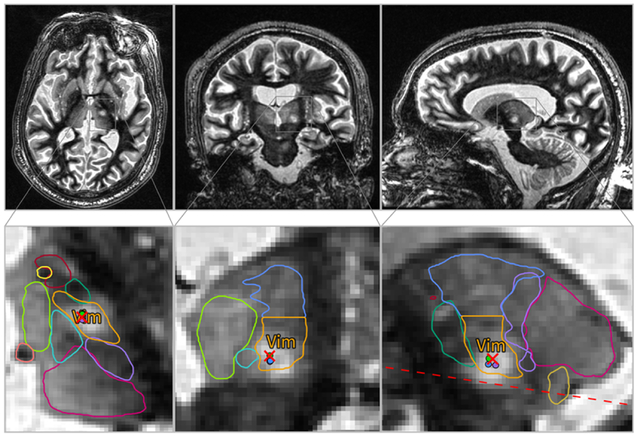
Figure 8 shows an example of Vim targeting on a patient treated for essential tremor using DBS electrode placement. The top left panel shows a pre-operative 3T WMn MP-RAGE axial slice overlaid with THOMAS segmented thalamic nuclei (Vim nucleus in ochre). An “electrode” view showing the path of the electrode is shown in the bottom left panel for reference. Axial and coronal views depicting the DBS electrode (pink) and a 3D volume rendering of the THOMAS segmented Vim nucleus (blue) overlaid on the post-operative T2 Cube data are depicted in the top and bottom right panels. The active contact point (second from the end) determined from post-surgical clinical follow-up is consistent with its location in the inferior edge of the Vim nucleus (white arrow).
Figure 8.
Application to DBS electrode placement- pre-operative 3T WMn MP-RAGE images acquired on a patient who underwent traditional DBS surgery using awake physiologic guidance were segmented using THOMAS (top left). The right top and bottom panels show axial and coronal planes of post-operative 1.5T T2 Cube images with the volume rendered Vim label overlaid after registration (blue). The segmented DBS electrode is shown in pink with the bottom left image showing the full path (“electrode” view). The active contact point (second from the end) is depicted using the white arrow and is at the inferior margin of the Vim.
4. Discussion
We have developed and validated a fully automated method for segmentation of 12 thalamic nuclei including the MTT in addition to the whole thalamus based entirely on anatomic imaging using the WMn MP-RAGE sequence. This is the first work to demonstrate multi-atlas-based thalamic nuclei segmentation based solely on structural MRI images. We rigorously evaluated the accuracy of our segmentation by comparison with manual ground truth segmentation based on the Morel atlas performed by an expert neuroradiologist on data acquired from healthy subjects as well as patients with MS. This comparison is missing in most published methods, especially those using DTI based methods. A high degree of accuracy was achieved even for small nuclei such as Hb and CM as evidenced by the high Dice coefficient values (>0.7) evaluating against manually segmented ground truth. The VSI indices were also very high, making the method potentially useful in assessment of small localized volumetric changes associated with neurological or neuropsychiatric diseases. Even on a small cohort of 15 MS patients and 20 healthy control subjects and using a very conservative Bonferroni correction, THOMAS was able to detect atrophy in specific nuclei in MS patients, adjusting for age, sex, and intracranial volumes.
Segmentation of the entire thalamus was achieved in approximately 10 minutes, opening the door for use in intra-operative procedures that involve direct MRI-based targeting such as Vim for treatment of essential tremor and ultrasound based thalamotomy. We have illustrated two example applications of our segmentation methods for targeting the Vim nucleus for treatment of essential tremor. The first application uses 7T pre-operative data to retrospectively delineate the Vim nucleus on 3T post-operative images on a patient undergoing FUS thalamotomy. The second uses 3T pre-operative data to retrospectively delineate the Vim nucleus in 1.5T post-operative images on a patient who underwent awake DBS lead placement using electrophysiological guidance. This shows the feasibility of using a combination of pre-operative 3T WMN MP-RAGE imaging and THOMAS combined with intra-operative 1.5T MRI to perform accurate, fast, direct and patient-specific targeting. While these results are qualitative, they demonstrate initial proof-of-concept that our methods can be of use to the neurosurgeon in prospective targeting of thalamic nuclei in near realtime even at clinical strength 3T MRI.
Our implementation of WMn MP-RAGE uses a radial fan beam k-space ordering scheme (Saranathan et al., 2015) which is inherently robust to motion due to the smearing of motion artifacts similar to other radial k-space MRI schemes and also helps achieve improved image contrast. However, our segmentation could also work with more easily available vendor supplied sequential or centric k-space encoding variants of MP-RAGE. In these cases, the use of navigator gating could further reduce the effect of motion and improve image quality as demonstrated for conventional MP-RAGE (van der Kouwe et al., 2008).
Liu et al. (Liu et al., 2015) segmented thalamic nuclei from 3T images using shape models estimated from 7T. They used conventional CSF-nulled MP-RAGE, which has poor thalamic contrast, and observed poor shape fidelity when performing label fusion from 7T to 3T. Our use of white-matter nulled MP-RAGE provided excellent boundaries, obviating the need for complicated shape models and enabling accurate label fusion and transfer learning from 7T prior data.
Most methods developed to date have used a DTI-based acquisition which inherits the limitations of EPI i.e. image distortion and limited spatial resolution (typically 2×2×2 mm^3). Battistella et al. (Battistella et al., 2017) reported a new clustering method based on the orientation distribution function (ODF) instead of angular direction, which seems to be much more stable and reliable compared to methods relying on diffusion direction orientation or angular direction. However, they used a data-driven method involving 5000 iterations of seeding to perform the clustering, making it time consuming. Furthermore their method produced 7 clusters, 6 of which correlated to histologically segmented nuclei and the seventh a composite. In future, methods like THOMAS (or even the average template we have created) may also be used to provide good initial conditions for seeding clusters to speed up the process. The whole thalamus mask used for many methods involve complicated preprocessing and minutes to hours of processing time (e.g. Battistella uses FreeSurfer reconstruction which takes 3-5 hours on desktop PC) which can be significantly shortened using the whole thalamus mask from our segmentation method.
Very recently, Iglesias et al. (Iglesias et al., 2018) introduced a Bayesian probabilistic atlas based segmentation technique (additional module in FreeSurfer) that works primarily on conventional MP-RAGE images to generate thalamic nuclei labels. This technique allows for provision of additional contrast images to help improve the thalamic segmentation. In our preliminary comparison study, THOMAS outperformed FreeSurfer using Dice as a metric especially for smaller nuclei like CM and VLa. While some of the performance can be attributed to the fact that THOMAS was developed using manual segmentations of WMn MP-RAGE data, guided by the Morel atlas, we believe that the main reason for THOMAS’s superior performance is the improved intra-thalamic contrast provided by the WMn MP-RAGE sequence, combined with the multi-atlas derived from those WMn volumes and the label fusion method used for segmentation. While the addition of WMn MP-RAGE to conventional MP-RAGE as an input to the thalamic segmentation module of FreeSurfer improved the Dice in most nuclei (Table 3), the performance of FreeSurfer is still largely determined by conventional MP-RAGE and is limited by the poor intrathalamic contrast of that sequence, an observation also reported by Iglesias et al. (Iglesias et al., 2018)
While we used single-echo WMN MP-RAGE magnitude images for segmentation, the use of susceptibility maps from a multi-echo variant of this sequence could further improve the accuracy and robustness of this technique similar to what has been reported for conventional MP-RAGE (van der Kouwe et al., 2008). Future work could improve the accuracy and speed of the label fusion step by using better models to combine the registered prior labels such as convolutional neural networks (CNN) as demonstrated by Yang et al. (arXiv https://arxiv.org/abs/1709.09641) for segmentation of cardiac data. We believe that the presence of sharp boundaries in WMn MP-RAGE will also help these methods converge well compared to conventional T1 or T2 imaging, where these boundaries are blurry or often non-existent.
One limitation of our study is the lack of direct comparisons to DTI based methods. Such direct comparisons were not possible due to the fact that DTI data was not acquired as part of our study. Comparing against Dice coefficients for thalamic nuclei segmentation reported in two state of the art methods reported in literature, our method performs comparably for the larger nuclei and significantly better for smaller nuclei like AV, MGN, and LGN. The first method (Liu et al., 2015) used shape models derived from 7T MP-RAGE and SWI data applied to 3T MP-RAGE data acquired with 1mm isotropic resolution while the second method (Glaister et al., 2016) used multi modal DTI data acquired at 3T with 2.2 mm isotropic resolution.
Another limitation common to all registration-based methods is suboptimal performance or even failure in the presence of large lesions or artifacts due to metal electrodes. This can be mitigated by masking those regions prior to the registration process, although this is usually tedious as it has to be done manually on a slice-by-slice basis. The composition of our library of prior images is limited to healthy subjects and multiple sclerosis patients. Other pathologies which can potentially affect the thalamus should be added to this library to provide more robust segmentation. The paucity of manually segmented prior datasets is a limitation for both our method and CNN-based methods. The combination of manual and automatically segmented data in the prior library as proposed by Pipitone et al. (Pipitone et al., 2014) could be used to widen the prior library to multiple pathologies. Finally, we used a mean brain template as an intermediate to achieve 20x speed-up. While this method has been shown to perform well (Artaechevarria et al., 2008), it can also be subject to registration errors. A method which directly registers each of the priors to the input would presumably be more accurate. The speed-up provided by cropping, as detailed in this paper, could reduce the computation burden significantly, allowing practical multi-atlas label-fusion segmentation that directly registers each of the priors to the input. Future improvements could incorporate 3D CNNs to perform segmentation and label fusion directly on the input data, significantly improving computational speeds as well as reducing errors due to the obviation of direct registration. However, the performance of these algorithms will be limited by the amount of available manually segmented prior data.
In conclusion, we have presented a fast fully automated segmentation method that uses structural 7T MRI data to segment 12 thalamic nuclei, requiring on the order of 10 minutes computation time on a relatively inexpensive desktop computer. THOMAS provides the highest accuracy for thalamic nuclear segmentation and volume estimation demonstrated to date. This method can be used to provide accurate volumes of nuclei which could be significant in understanding disease progression in Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, and other pathologies. It can also be used for MRI guided procedures such as FUS and DBS for essential tremor, where fast, accurate, and direct targeting and segmentation is critical.
Acknowledgments
We wish to acknowledge research support from the National Institutes of Health (NIH R21AA023582-02, NIH P41 EB015891) and from GE Healthcare. One of the authors (XX) received a grant from the French Agence Nationale de la Recherche within the context of the Investments for the Future program (ANR-10-LABX-57) and named “TRAIL” from ARSEP foundation.
Abbreviations
- AV
Anterior ventral nucleus
- CM
Centromedian nucleus
- CSF
Cerebrospinal fluid
- CSFn
Cerebrospinal fluid nulled
- DBS
Deep brain stimulation
- DTI
Diffusion tensor imaging
- EPI
Echo planar imaging
- FUS
Focused ultrasound
- GM
Grey matter
- Hb
Habenular nucleus
- LGN
Lateral geniculate nucleus
- MD
Mediodorsal nucleus
- MGN
Medial geniculate nucleus
- MP-RAGE
Magnetization-prepared rapidly acquired gradient echo
- MTT
Mammillothalamic tract
- Pul
Pulvinar nucleus
- SNR
Signal-to-noise ratio
- VA
Ventral anterior nucleus
- Vim
Ventralis intermedius nucleus
- VLa
Ventral lateral anterior nucleus
- VLp
Ventral lateral posterior nucleus
- VPL
Ventral posterior lateral nucleus
- WM
White matter
- WMn
White matter nulled
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abosch A, Yacoub E, Ugurbil K, Harel N, 2010. An assessment of current brain targets for deep brain stimulation surgery with susceptibility-weighted imaging at 7 tesla. Neurosurgery 67, 1745–1756; discussion 1756. 10.1227/NEU.0b013e3181f74105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen NC, 1997. The role of the thalamus in schizophrenia. Can J Psychiatry 42, 27–33. 10.1177/070674379704200104 [DOI] [PubMed] [Google Scholar]
- Artaechevarria X, Muñoz-Barrutia A, Ortiz-de-Solórzano C, 2008. Efficient classifier generation and weighted voting for atlas-based segmentation: two small steps faster and closer to the combination oracle, in: Reinhardt JM, Pluim JPW (Eds.), . p. 69141W 10.1117/12.769401 [DOI] [Google Scholar]
- Arts NJ, Walvoort SJ, Kessels RP, 2017. Korsakoff’s syndrome: a critical review. Neuropsychiatr Dis Treat 13, 2875–2890. 10.2147/NDT.S130078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistella G, Najdenovska E, Maeder P, Ghazaleh N, Daducci A, Thiran J-P, Jacquemont S, Tuleasca C, Levivier M, Bach Cuadra M, Fornari E, 2017. Robust thalamic nuclei segmentation method based on local diffusion magnetic resonance properties. Brain Struct Funct 222, 2203–2216. 10.1007/s00429-016-1336-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TEJ, Johansen-Berg H, Woolrich MW, Smith SM, Wheeler-Kingshott C. a. M., Boulby PA, Barker GJ, Sillery EL, Sheehan K, Ciccarelli O, Thompson AJ, Brady JM, Matthews PM, 2003. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat. Neurosci 6, 750–757. 10.1038/nn1075 [DOI] [PubMed] [Google Scholar]
- Benabid AL, Pollak P, Gao D, Hoffmann D, Limousin P, Gay E, Payen I, Benazzouz A, 1996. Chronic electrical stimulation of the ventralis intermedius nucleus of the thalamus as a treatment of movement disorders. J. Neurosurg 84, 203–214. 10.3171/jns.1996.84.2.0203 [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E, 1991. Alzheimer’s disease affects limbic nuclei of the thalamus. Acta Neuropathol. 81, 261–268. [DOI] [PubMed] [Google Scholar]
- Coscia DM, Narr KL, Robinson DG, Hamilton LS, Sevy S, Burdick KE, Gunduz-Bruce H, McCormack J, Bilder RM, Szeszko PR, 2009. Volumetric and shape analysis of the thalamus in first-episode schizophrenia. Hum Brain Mapp 30, 1236–1245. 10.1002/hbm.20595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deoni SCL, Rutt BK, Parrent AG, Peters TM, 2007. Segmentation of thalamic nuclei using a modified k-means clustering algorithm and high-resolution quantitative magnetic resonance imaging at 1.5 T. Neuroimage 34, 117–126. 10.1016/j.neuroimage.2006.09.016 [DOI] [PubMed] [Google Scholar]
- Elias WJ, Huss D, Voss T, Loomba J, Khaled M, Zadicario E, Frysinger RC, Sperling SA, Wylie S, Monteith SJ, Druzgal J, Shah BB, Harrison M, Wintermark M, 2013. A Pilot Study of Focused Ultrasound Thalamotomy for Essential Tremor. New England Journal of Medicine 369, 640–648. 10.1056/NEJMoa1300962 [DOI] [PubMed] [Google Scholar]
- Fama R, Rosenbloom MJ, Sassoon SA, Rohlfing T, Pfefferbaum A, Sullivan EV, 2014. Thalamic volume deficit contributes to procedural and explicit memory impairment in HIV infection with primary alcoholism comorbidity. Brain Imaging Behav 8, 611–620. 10.1007/s11682-013-9286-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R, 2012. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 30, 1323–1341. 10.1016/j.mri.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaister J, Carass A, Stough JV, Calabresi PA, Prince JL, 2016. Thalamus parcellation using multimodal feature classification and thalamic nuclei priors. Proc SPIE Int Soc Opt Eng 9784 10.1117/12.2216987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson JL, Suh JW, Nacewicz BM, Sutterer MJ, Cayo AA, Stodola DE, Burghy CA, Wang H, Avants BB, Yushkevich PA, Essex MJ, Pollak SD, Davidson RJ, 2012. Robust Automated Amygdala Segmentation via Multi-Atlas Diffeomorphic Registration. Front Neurosci 6 10.3389/fnins.2012.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias JE, Insausti R, Lerma-Usabiaga G, Bocchetta M, Van Leemput K, Greve DN, van der Kouwe A, Alzheimer’s Disease Neuroimaging Initiative, Fischl B, Caballero-Gaudes C, Paz-Alonso PM, 2018. A probabilistic atlas of the human thalamic nuclei combining ex vivo MRI and histology. Neuroimage 183, 314–326. 10.1016/j.neuroimage.2018.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TEJ, Sillery E, Ciccarelli O, Thompson AJ, Smith SM, Matthews PM, 2005. Functional-anatomical validation and individual variation of diffusion tractography-based segmentation of the human thalamus. Cereb. Cortex 15, 31–39. 10.1093/cercor/bhh105 [DOI] [PubMed] [Google Scholar]
- Kumar V, Mang S, Grodd W, 2015. Direct diffusion-based parcellation of the human thalamus. Brain Struct Funct 220, 1619–1635. 10.1007/s00429-014-0748-2 [DOI] [PubMed] [Google Scholar]
- Lee Jee-Young, Yoon EJ, Lee WW, Kim YK, Lee Jun-Young, Jeon B, 2016. Lateral geniculate atrophy in Parkinson’s with visual hallucination: A trans-synaptic degeneration? Mov. Disord 31, 547–554. 10.1002/mds.26533 [DOI] [PubMed] [Google Scholar]
- Liu Y, D’Haese P-F, Newton AT, Dawant BM, 2015. Thalamic nuclei segmentation in clinical 3T T1-weighted Images using high-resolution 7T shape models, in: Webster RJ, Yaniv ZR (Eds.), . p. 94150E 10.1117/12.2081660 [DOI] [Google Scholar]
- Maggioni E, Crespo-Facorro B, Nenadic I, Benedetti F, Gaser C, Sauer H, Roiz-Santiañez R, Poletti S, Marinelli V, Bellani M, Perlini C, Ruggeri M, Altamura AC, Diwadkar VA, Brambilla P, ENPACT group, 2017. Common and distinct structural features of schizophrenia and bipolar disorder: The European Network on Psychosis, Affective disorders and Cognitive Trajectory (ENPACT) study. PLoS ONE 12, e0188000 10.1371/journal.pone.0188000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnotta VA, Gold S, Andreasen NC, Ehrhardt JC, Yuh WT, 2000. Visualization of subthalamic nuclei with cortex attenuated inversion recovery MR imaging. Neuroimage 11, 341–346. 10.1006/nimg.2000.0552 [DOI] [PubMed] [Google Scholar]
- Mang SC, Busza A, Reiterer S, Grodd W, Klose AU, 2012. Thalamus segmentation based on the local diffusion direction: a group study. Magn Reson Med 67, 118–126. 10.1002/mrm.22996 [DOI] [PubMed] [Google Scholar]
- Minagar A, Barnett MH, Benedict RHB, Pelletier D, Pirko I, Sahraian MA, Frohman E, Zivadinov R, 2013. The thalamus and multiple sclerosis: modern views on pathologic, imaging, and clinical aspects. Neurology 80, 210–219. 10.1212/WNL.0b013e31827b910b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel A, Magnin M, Jeanmonod D, 1997. Multiarchitectonic and stereotactic atlas of the human thalamus. J. Comp. Neurol 387, 588–630. [DOI] [PubMed] [Google Scholar]
- Niemann K, Naujokat C, Pohl G, Wollner C, von Keyserlingk D, 1994. Verification of the Schaltenbrand and Wahren stereotactic atlas. Acta Neurochir (Wien) 129, 72–81. [DOI] [PubMed] [Google Scholar]
- Parnaudeau S, Bolkan SS, Kellendonk C, 2018. The Mediodorsal Thalamus: An Essential Partner of the Prefrontal Cortex for Cognition. Biol. Psychiatry 83, 648–656. 10.1016/j.biopsych.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipitone J, Park MTM, Winterburn J, Lett TA, Lerch JP, Pruessner JC, Lepage M, Voineskos AN, Chakravarty MM, Alzheimer’s Disease Neuroimaging Initiative, 2014. Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage 101, 494–512. 10.1016/j.neuroimage.2014.04.054 [DOI] [PubMed] [Google Scholar]
- Rohlfing T, Brandt R, Menzel R, Maurer CR, 2004. Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage 21, 1428–1442. 10.1016/j.neuroimage.2003.11.010 [DOI] [PubMed] [Google Scholar]
- Saranathan M, Tourdias T, Bayram E, Ghanouni P, Rutt BK, 2015. Optimization of white-matter-nulled magnetization prepared rapid gradient echo (MP-RAGE) imaging. Magn Reson Med 73, 1786–1794. 10.1002/mrm.25298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaier J, Anthofer J, Steib K, Fellner C, Rothenfusser E, Brawanski A, Lange M, 2015. Deep brain stimulation for essential tremor: targeting the dentato-rubro-thalamic tract? Neuromodulation 18, 105–112. 10.1111/ner.12238 [DOI] [PubMed] [Google Scholar]
- Sedrak M, Gorgulho A, Frew A, Behnke E, DeSalles A, Pouratian N, 2011. Diffusion tensor imaging and colored fractional anisotropy mapping of the ventralis intermedius nucleus of the thalamus. Neurosurgery 69, 1124–1129; discussion 1129-1130. 10.1227/NEU.0b013e3182296a42 [DOI] [PubMed] [Google Scholar]
- Spiegelmann R, Nissim O, Daniels D, Ocherashvilli A, Mardor Y, 2006. Stereotactic targeting of the ventrointermediate nucleus of the thalamus by direct visualization with high-field MRI. Stereotact Funct Neurosurg 84, 19–23. 10.1159/000092683 [DOI] [PubMed] [Google Scholar]
- Stough JV, Ye C, Ying SH, Prince JL, 2013. THALAMIC PARCELLATION FROM MULTI-MODAL DATA USING RANDOM FOREST LEARNING. Proc IEEE Int Symp Biomed Imaging 852–855. 10.1109/ISBI.2013.6556609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhyadhom A, Haq IU, Foote KD, Okun MS, Bova FJ, 2009. A high resolution and high contrast MRI for differentiation of subcortical structures for DBS targeting: the Fast Gray Matter Acquisition T1 Inversion Recovery (FGATIR). Neuroimage 47 Suppl 2, T44–52. 10.1016/j.neuroimage.2009.04.018 [DOI] [PubMed] [Google Scholar]
- Tourdias T, Saranathan M, Levesque IR, Su J, Rutt BK, 2014a. Visualization of intra-thalamic nuclei with optimized white-matter-nulled MPRAGE at 7T. Neuroimage 84, 534–545. 10.1016/j.neuroimage.2013.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourdias T, Saranathan M, Levesque IR, Su J, Rutt BK, 2014b. Visualization of intra-thalamic nuclei with optimized white-matter-nulled MPRAGE at 7T. NeuroImage 84, 534–545. 10.1016/j.neuroimage.2013.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynor CR, Barker GJ, Crum WR, Williams SCR, Richardson MP, 2011. Segmentation of the thalamus in MRI based on T1 and T2. Neuroimage 56, 939–950. 10.1016/j.neuroimage.2011.01.083 [DOI] [PubMed] [Google Scholar]
- van der Kouwe AJW, Benner T, Salat DH, Fischl B, 2008. Brain morphometry with multiecho MPRAGE. Neuroimage 40, 559–569. 10.1016/j.neuroimage.2007.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassal F, Coste J, Derost P, Mendes V, Gabrillargues J, Nuti C, Durif F, Lemaire J-J, 2012. Direct stereotactic targeting of the ventrointermediate nucleus of the thalamus based on anatomic 1.5-T MRI mapping with a white matter attenuated inversion recovery (WAIR) sequence. Brain Stimul 5, 625–633. 10.1016/j.brs.2011.10.007 [DOI] [PubMed] [Google Scholar]
- Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA, 2013. Multi-Atlas Segmentation with Joint Label Fusion. IEEE Trans Pattern Anal Mach Intell 35, 611–623. 10.1109/TPAMI.2012.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegell MR, Tuch DS, Larsson HBW, Wedeen VJ, 2003. Automatic segmentation of thalamic nuclei from diffusion tensor magnetic resonance imaging. Neuroimage 19, 391–401. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Zitella LM, Duchin Y, Teplitzky BA, Kastl D, Adriany G, Yacoub E, Harel N, Johnson MD, 2016. Multimodal 7T Imaging of Thalamic Nuclei for Preclinical Deep Brain Stimulation Applications. Front Neurosci 10, 264 10.3389/fnins.2016.00264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Akazawa K, Yuen S, Goto M, Matsushima S, Takahata A, Nakagawa M, Mineura K, Nishimura T, 2010. MR imaging of ventral thalamic nuclei. AJNR Am J Neuroradiol 31, 732–735. 10.3174/ajnr.A1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Wang H, Pluta J, Das SR, Craige C, Avants BB, Weiner MW, Mueller S, 2010. Nearly automatic segmentation of hippocampal subfields in in vivo focal T2-weighted MRI. Neuroimage 53, 1208–1224. 10.1016/j.neuroimage.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziyan U, Tuch D, Westin C-F, 2006. Segmentation of thalamic nuclei from DTI using spectral clustering. Med Image Comput Comput Assist Interv 9, 807–814. [DOI] [PubMed] [Google Scholar]