Abstract
Quantitative assessment of MT1-MMP cell surface-associated proteolytic activity remains undefined. Presently, MT1-MMP was stably expressed and a cell-based FRET assay developed to quantify activity towards synthetic collagen-model triple-helices. To estimate the importance of cell surface localization and specific structural domains on MT1-MMP proteolysis, activity measurements were performed using a series of membrane-anchored MT1-MMP mutants and compared directly with those of soluble MT1-MMP. MT1-MMP activity (kcat/KM) on the cell surface was 4.8-fold lower compared with soluble MT1-MMP, with the effect largely manifested in kcat. Deletion of the MT1-MMP cytoplasmic tail enhanced cell surface activity, with both kcat and KM values affected, while deletion of the hemopexin-like domain negatively impacted KM and increased kcat. Overall, cell surface localization of MT1-MMP restricts substrate binding and protein coupled motions (based on changes in both kcat and KM) for catalysis. Comparison of soluble and cell surface-bound MT2-MMP revealed 12.9-fold lower activity on the cell surface. The cell-based assay was utilized for small molecule and triple-helical transition state analog MMP inhibitors, which were found to function similarly in solution and at the cell surface. These studies provide the first quantitative assessments of MT1-MMP activity and inhibition in the native cellular environment of the enzyme.
Keywords: Cell-based Assay, Protease Inhibitor, Collagenolysis, Matrix Metalloproteinase, Cell Surface Proteolysis
Graphical Abstract
MT1-MMP was stably expressed and a cell-based FRET assay developed to quantify activity towards synthetic collagen-model triple-helices. Activity measurements were performed using a series of membrane-anchored MT1-MMP mutants and compared directly with those of soluble MT1-MMP. Cell surface localization of MT1-MMP was found to restrict substrate binding and protein coupled motions for catalysis. Small molecule and triple-helical transition state analog MMP inhibitors were found to function similarly in solution and at the cell surface.
The quantification of cell surface-associated protease activity, along with the evaluation of inhibitor potency, is often performed using isolated enzyme and substrate. As a result, the contribution of the cell surface environment to the regulation of proteolytic activity is negated. Membrane type-1 matrix metalloproteinase (MT1-MMP) is a member of the matrix metalloproteinase (MMP) gene family, multidomain enzymes that are characterized by an N-terminal propeptide domain, a zinc-coordinating active site within the catalytic (CAT) domain, and a C-terminal hemopexin-like (HPX) domain (1). However, unlike secreted members of the MMP family, MT1-MMP is distinguished by the presence of a short transmembrane (TM) domain and a cytoplasmic tail (CT) that serve to localize the enzyme to discrete regions of the plasma membrane while providing access to the intracellular compartment (1).
Although MT1-MMP can hydrolyze a variety of substrates (2), one of its most important functions is the serve as a pericellular, interstitial collagenase that plays key roles in events ranging from mesenchymal stem cell differentiation and adipose tissue development to carcinoma cell proliferation, invasion, and metastasis (1, 3–6). MT1-MMP collagenolytic activity has also been implicated in facilitating secondary infections (7). Interestingly, even though MT1-MMP serves as an interstitial collagenase, in similar fashion to several secreted MMPs (i.e., MMP-1, MMP-8, and MMP-13), only MT1-MMP activity is critical for conferring cells with tissue-invasive properties (8–14). Hence, the plasma membrane environment appears to regulate MT1-MMP collagenolytic activity. While quantitative assessments of MT1-MMP catalytic activity at the cell surface should clarify the potential influences and effects that the membrane environment elicits on catalytic activity under native conditions, such determinations have remained problematic (see below).
Visualization of membrane-bound, active MT1-MMP has been achieved by fluorescence resonance energy transfer (FRET) imaging of surface-anchored sensors. An initial MT1-MMP sensor was created using the Cys-Pro-Lys-Glu-Ser-Cys-Asn-Leu-Phe-Val-Leu-Lys-Asp sequence, derived from the MT1-MMP cleavage site in proMMP-2 (15). The FRET pair was an Enhanced Cyan Fluorescence Protein (ECFP, the fluorophore) and a Yellow Fluorescence Protein variant (YPet, the quencher) (15). A second generation sensor was created using the sequence Cys-Arg-Pro-Ala-His-Leu-Arg-Asp-Ser-Gly and the FRET pair mOrange2 (fluorophore) and mCherry (quencher), yielding a biosensor that was largely insensitive to MMP-2- or MMP-9-dependent hydrolysis (16). To improve the biosensor’s selectivity, a pentapeptide library was screened, and the sensor CyPet-Ser-Leu-Ala-Pro-Leu-Gly-Leu-Gln-Arg-Arg-YPet (where Cyan Fluorescence Protein variant (CyPet) was the fluorophore) was found to be more selective for MT1-MMP compared with other MMPs, with the exception of MMP-9 (17). A further optimized MT-MMP probe was developed based on the sequence AHLR (Cys-Arg-Pro-Ala-His-Leu-Arg-Asp-Ser-Gly) with Gly-Gly-Ser-Gly-Gly-Thr linkers flanking each side of the sequence and ECFP and YPet as a FRET pair (18). MT1-MMP activity was favored over MMP-2 and MMP-9 as well as the membrane-anchored MMPs, MT2-MMP and MT3-MMP. However, the insufficient selectivity of sensors that are based on short linear peptides did not allow for the precise quantification of cellular MT1-MMP activity under native conditions, and the recorded changes in fluorescence were not sufficiently robust for routine analysis.
A more recent biosensor for MT1-MMP was created using a separate donor and acceptor that assembled in situ (19). Specifically, a monobody (PEbody) was developed to bind to R-phycoerythrin (R-PE) dye. The PEbody was fused with ECFP and also inserted into the cell membrane. An MT1-MMP labile sequence (Cys-Arg-Pro-Ala-His-Leu-Arg-Asp-Ser-Gly) was incorporated between the ECFP and the PEbody. MT1-MMP hydrolysis resulted in a decrease in FRET. Images were reported to be clearer than for the ECFP/YPet sensor (15). The ECFP-PEbody/R-PE biosensor was used to study the localization and mobility of MT1-MMP, but not to quantify activity. Interestingly, this study found that MT1-MMP mobility was restricted by inhibition partners (19).
Imaging of MT1-MMP activity on the surface of human mesenchymal stem cells was achieved using a three-dimensional PEG-hydrogel that incorporated the MMP substrate Dabcyl-Gly-Gly-Pro-Gln-Gly-Ile-Trp-Gly-Gln-Lys(fluorescein)-Ahx-Cys (20). The relative change in fluorescence was quantified, but no kinetic parameters were reported. The sequence used was not specific for MT1-MMP.
For the purpose of analyzing cell-surface proteolytic enzymes, one would ideally utilize substrates that correspond to the most prominent activity of a targeted protease. As such, synthetic triple-helical peptide (THP) substrates that model interstitial (types I-III) collagen have been developed for convenient, continuous activity-monitoring assays. FRET THPs (fTHPs) have typically used (7-methoxycoumarin-4-yl)-acetyl (Mca) as a fluorophore that, in turn, is efficiently quenched by 2,4-dinitrophenyl (Dnp) moieties (21, 22). These fTHPs have been employed to discriminate MMP family members in kinetic assays in vitro, as well as in transfer FRET assays in cultured cells. Additionally, we have described an in situ (non-transfer) MMP cell-based assay using FRET peptide substrates (23).
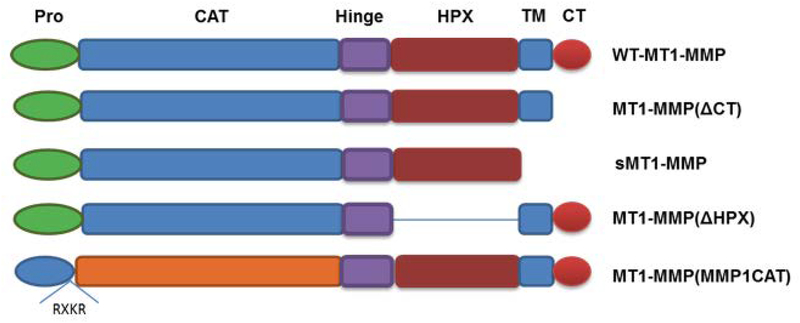
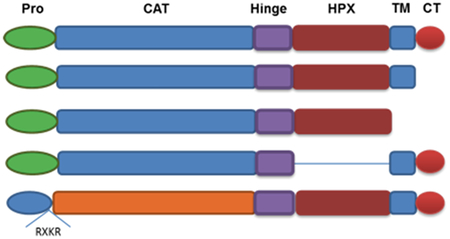
In the present study, MT1-MMP was stably expressed in cells and a cell-based FRET assay used to quantify cell surface-associated protease activity and its kinetic parameters. To determine the effect of the cell surface and the individual MT1-MMP domains on catalysis, activity comparisons were made using soluble (i.e., transmembrane-deleted) MT1-MMP and surface-bound MT1-MMP mutants. Given recent, and often contradictory, reports regarding the role of the MT1-MMP CT, CAT domain, and HPX domain in regulating proteolytic activity (14, 24–30), we also assessed the enzymatic properties of MT1-MMP following (i) deletion of the CT [MT1-MMP(ΔCT)], to determine if a lack of enzyme internalization, partitioning into lipid rafts, and/or CT posttranslation modification modulates activity, (ii) deletion of the HPX domain [MT1-MMP(ΔHPX)], to determine the role of the HPX domain in cell-surface collagenolysis, and (iii) replacement of the MT1-MMP CAT domain with the MMP-1 CAT domain [MT1-MMP(MMP-1 CAT)], to determine if the MT1-MMP CAT domain is optimal for cell-surface collagenolysis (Figure 1). Activity of the soluble and cell-bound forms of MT2-MMP were evaluated for comparison to MT1-MMP. Finally, the effect of two distinct classes of inhibitors on cell surface MT1-MMP proteolysis was examined.
Figure 1.
Schematic illustration of MT1-MMP constructs. Domains of MT1-MMP are propeptide (Pro) in green, catalytic (CAT) in blue, hinge (Hinge) in purple, hemopexin-like (HPX) in burgundy, transmembrane (TM) in blue, and cytoplasmic tail (CT) in red. Blue prodomain and orange CAT domain represent MMP-1.
1. Experimental Section
1.1. Methods and materials
Cell culture reagents were obtained from Invitrogen unless otherwise stated. Standard chemicals were of analytical or molecular biology grade and purchased from Fisher Scientific. Antibodies were purchased from EMD Millipore and Pierce. The triple-helical substrate fTHP-9 [(Gly-Pro-Hyp)5-Gly-Pro-Lys(Mca)-Gly-Pro-Gln-Gly~Cys(Mob)-Arg-Gly-Gln-Lys(Dnp)-Gly-Val-Arg-(Gly-Pro-Hyp)5-NH2] and the triple-helical peptide inhibitor GlyΨ{PO2H-CH2}Ile-Tyr THPI [(Gly-Pro-Hyp)4-Gly-mep-Flp-Gly-Pro-Gln-[GlyΨ(PO2H-CH2)Ile]-Tyr-Phe-Gln-Arg-Gly-Val-Arg-Gly-mep-Flp-(Gly-Pro-Hyp)4-Tyr-NH2, where mep = 4-methylproline and Flp = 4-fluoroproline] were synthesized in house using methods described previously (31–35). Marimastat, a nonselective inhibitor of MMPs (36, 37), was purchased from Sigma. Tissue inhibitor of metalloproteinase 2 (TIMP-2) was obtained from Abcam (catalog # ab39314).
1.2. Cell culture and transfection
COS-1 cells (CRL-1650) were obtained from ATCC. Human MCF-7 breast carcinoma cells that express low levels of MT1-MMP and negligible levels of MMP-8 were cultured as described previously in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum (FCS) (38, 39). The plasmid construct for producing human soluble MT1-MMP (MT1-MMP without its TM domain and CT, designated sMT1-MMP) was described previously (40, 41). The pCDNA 3.1 plasmids containing human wild-type MT1-MMP (WT-MT1-MMP), MT1-MMP with its cytoplasmic tail deleted [MT1-MMP(ΔCT)], MT1-MMP with the HPX domain deleted [MT1-MMP(ΔHPX)], and MT1-MMP in which the entire CAT domain was replaced with the CAT domain of human MMP-1 [MT1-MMP(MMP-1 CAT)] have also been described (14, 28). The MT1-MMP(MMP-1 CAT) construct was composed of MMP-1 Met1 to Tyr260 with the Gly-Leu-Ser-Ser-Aal-Arg-Asn-Arg-Gln-Lys-Arg sequence inserted between the Pro and CAT domains, and MT1-MMP Gly284 to Val582 (28, 42) The inserted sequence allows for furin activation of the resulting chimera (42). All pCDNA 3.1 plasmids containing WT-MT1-MMP and mutants were used for stable transfection of MCF-7 cells. Control cells were transfected with the original pCDNA3.1 plasmid. MCF-7 cells were stably transfected using X-tremeGENE 9 Reagent (Roche) followed by isolation of single colonies after 4–6 weeks of geneticin selection. Transfected cells were routinely grown in selective medium (DMEM supplemented with 10% FCS and 0.55 mg/mL geneticin).
The MT2-MMP construct (Met1 to Val669 with an HA tag (human influenza hemagglutinin residues 98–106) in the linker 2 region (between Glu584 and Pro585) has been described previously (13, 43).
1.3. Soluble protein production and detection
To generate soluble MT1-MMP, sMT1-MMP was transiently transfected in COS-1 cells. Transfected COS-1 cells were cultured for 56 h in serum-free OptiMEM. Conditioned medium was collected, concentrated 20-fold, and desalted using Ultracel®−30K centrifugal filters (Millipore, catalog # UFC903024). Concentrated samples were pooled and dialyzed against TSB buffer (50 mM Tris, 50 mM NaCl, 10 mM CaCl2, 0.05% Brij-35, pH 7.5) using the Slide-A-Lyzer Dialysis Cassette 20K (Thermo Scientific, product # 66102) overnight at 4 °C. Protein samples were resolved by reducing 12% SDS-polyacrylamide gel electrophoresis and transferred onto a PVDF membrane. The membrane was blocked with 5% non-fat dry milk in PBS plus 0.1% Tween (PBST) for 1 h at ambient temperature and then probed with an anti-MT1-MMP HPX domain mAb (Millipore, catalog # MAB3317) for 16 h at 4 °C. After extensive washing with PBST, the membrane was reprobed using goat anti-mouse IgG conjugated with horseradish peroxidase (HRP) (Millipore, catalog # AP124P) in PBST for 1 h at ambient temperature. The blot was developed in a SuperSignal® West Pico Chemiluminescent Substrate (Thermo Scientific).
To generate soluble MT2-MMP, MT2-MMP Met1 to Asn625 with an N-terminal His tag was stably transfected in MCF-7 cells as described (13, 43). FBS free media was collected, concentrated, and buffer exchanged to TSB buffer. The protein was then isolated using His trap resin, eluted with 100 mM imidazole and dialyzed overnight in TSB buffer.
MT1-MMP and MT2-MMP were activated by incubation of the proMT-MMP in TSB buffer with 0.1 μg/mL of rhTrypsin-3 for 1 h (proMT1-MMP) or 2 h (proMT2-MMP) at 37 °C (44). After MT-MMP activation, remaining trypsin-3 activity was quenched by addition of 1 mM AEBSF (R&D Systems) and incubation for 15 min at room temperature. Immediately after activation the enzyme was diluted in cold TSB buffer. Enzyme aliquots were kept on wet ice and used the same day.
1.4. Cell extract preparation from stably transfected cells
Stably transfected MCF-7 cells with WT-MT1-MMP or its mutants were cultured in selective medium. For Western blot analysis, the cultured cells were directly lysed on the 6-well tissue culture plates using 200 μL/well of RIPA buffer with protease inhibitor cocktail (Sigma, catalog # P8340). A tissue culture cell scraper was applied to each well and cell lysates were collected with a pipette and kept on ice. After centrifugation, the protein content of the supernatants was quantified by the Pierce BCA Protein Assay Kit (Thermo Scientific) carried out in triplicate in wells of a 96-well plate. MT1-MMP production was confirmed by Western blot as described above using mAbs against either the MT1-MMP CAT domain or the MT1-MMP hinge domain (Millipore, catalog # ab6005 and ab6004, respectively) and goat anti-rabbit IgG-HRP (Genescript, catalog # A00098) as a secondary antibody. β-actin mAb BA2R (Pierce, catalog #MA5–15739) was used as a loading control.
1.5. Soluble enzyme assay
Substrate stock solutions were prepared at various concentrations in TSB buffer containing 0.5% DMSO. MT1-MMP and MT2-MMP assays were conducted in TSB buffer by incubating a range of substrate concentrations (0.05–30 μM) with 7 and 9 nM enzyme, respectively, at 37 °C. Fluorescence was measured on a multiwell plate fluorimeter (Biotek Synergy H1) using λexcitation = 324 nm and λemission = 405 nm. The same kinetic assay was also run in OptiMEM media as opposed to TSB buffer and showed no deviation in the kinetic parameters (data not shown). Rates of hydrolysis were obtained from plots of fluorescence versus time, using data points from the linear portion of the hydrolysis curve alone. The slope from these plots was divided by the fluorescence change corresponding to complete hydrolysis and then multiplied by the substrate concentration to obtain rates of hydrolysis in units of μM/sec. The relationship between the rate of hydrolysis and substrate concentration for the MT-MMP/fTHP-9 pair for which individual kinetic parameters were determined was found to follow the Michaelis-Menten model. Kinetic parameters were evaluated by Lineweaver-Burk, Eadie-Hofstee, and Hanes-Woolf analyses. Data were additionally analysed using nonlinear regression, one-site hyperbolic binding model with GraphPad Prism 5 software. All the values reported are mean ± SD (n=3). MMP substrate cleavage sites were established by MALDI-TOF MS and found to be consistent with previously published data (31).
1.6. Cell surface bound enzyme assay
The in situ enzyme assays were run in wells of a 384-well tissue-culture treated opaque microplate (Greiner Bio-One, catalog # 781080). To minimize plate-based differences, the same plate type was used for both soluble and cell surface bound enzyme assays. Stably transfected cells with passage number 3 to 8 were used in the assay. Cells (6 × 103/well) were seeded in OptiMEM Medium and then incubated overnight at 37 °C in a CO2 incubator before the assay. The assays were carried out in serum-free OptiMEM with fTHP-9 dissolved in the same media with 0.5% DMSO in a total volume of 60 μL. Proteolytic activity was determined by calculating the percentage increase in fluorescence compared to the background signal provided by the corresponding dilution of the substrates with no cells using a multiwell plate fluorimeter (as described previously). Estimation of the active MT1-MMP and MT2-MMP enzyme levels was performed by TIMP-2 titration (31). Cells (6 × 103/well) were incubated with a 1.5–200 nM concentration range of TIMP-2. By accounting for active protease concentrations, enzyme kinetics will not be affected by different levels of protein expression. The rate of hydrolysis was calculated as described above. All the values reported are mean ± SD (n=3).
1.7. Inhibition assay
Stock solutions of GlyΨ{PO2H-CH2}Ile-Tyr THPI, marimastat, and fTHP-9 were prepared in OptiMEM media with 0.5% DMSO. Inhibitors were prepared over a 1 nM to 5 μM concentration range. Cells (6 × 103/well) were seeded in wells of a 384-well plate. Inhibitors were added to the wells and incubation proceeded for 45 min at 37 °C. After incubation, 15 μM of fTHP-9 was added to the wells and fluorescence was recorded for 30 min and an increase in relative fluorescence units (RFU) determined as described above. IC50 values were determined using GraphPad Prism 5 software.
2. Results
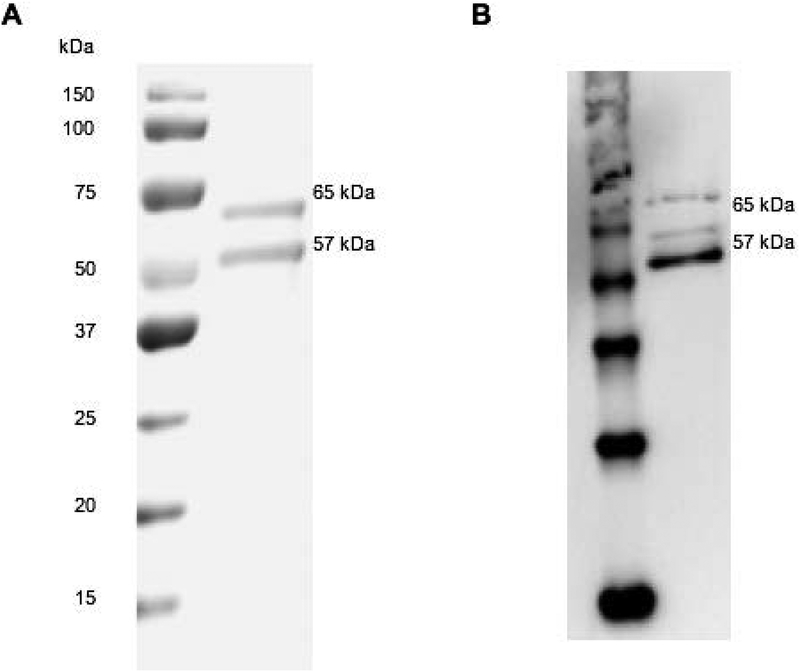
Soluble MT1-MMP (sMT1-MMP; Figure 1) was isolated from the media of COS-1 cells transiently transfected with the sMT1-MMP cDNA. As expected, sMT1-MMP was secreted as both proenzyme (65 kDa) and activated (57 kDa) forms (Figure 2) (40, 45, 46). The proenzyme form of sMT1-MMP was then activated using rhTrypsin-3 for 1 h, followed by trypsin-3 inactivation by addition of AEBSF. Trypsin activation of proMT1-MMP generates a single product at the N-terminus starting with Tyr112 (47). This cleavage site identical to that observed following the cellular activation of proMT1-MMP by the trans-Golgi-associated serine proteinase furin (40). Following TIMP-2 titration, 160 nM of the active enzyme was detected in the sMT1-MMP samples (40% of total protein) (Table 1). Initially, the single-stranded modified Knight substrate [Mca-Lys-Pro-Leu-Gly-Leu-Lys(Dnp)-Ala-Arg-NH2] served as a control to assess activity. Hydrolysis of the substrate by sMT1-MMP proceeded with kcat/KM = 38,560 M−1sec−1, KM = 21.3 μM, and kcat = 0.82 sec−1. The recorded kcat/KM value was 2.6–3.7-fold higher compared to prior kcat/KM values obtained with other sMT1-MMP samples (Tyr112-Glu523, expressed in Pichia pastoris) and similar FRET substrates (48).
Figure 2.
Protein expression, production, and purification of soluble MT1-MMP (sMT1-MMP). (A) SDS-PAGE analysis of purified protein. Protein was run on 12% SDS-PAGE gel under reducing conditions and (B) analyzed by Western blot with anti-MT1-MMP HPX domain mAb. Gel and Western blot analyses showed the proenzyme (65 kDa) and active (57 kDa) forms of sMT1-MMP.
Table 1:
TIMP-2 titration for active enzyme evaluation.
| MT-MMP Variant | MT-MMP (nM) per well |
|---|---|
| Mock cells | 0.96 ± 0.09a |
| WT-MT1-MMP | 2.28 ± 0.18a |
| MT1-MMP(ΔCT) | 4.46 ± 0.24a |
| MT1-MMP(ΔHPX) | 3.38 ± 0.13a |
| MT1-MMP(MMP-1 CAT) | 3.32 ± 0.15a |
| WT-MT2-MMP | 4.61 ± 0.23a |
Enzyme concentrations were determined as described in Experimental Section 1.6.
Kinetic parameters (KM, kcat, and kcat/KM) were next determined for hydrolysis of the triple-helical substrate fTHP-9 by sMT1-MMP using a 384-well plate format. For sMT1-MMP hydrolysis of fTHP-9, kcat/KM = 45,130 M−1sec−1, KM = 18.6 μM, and kcat = 0.9 sec−1 were recorded (Table 2). To quantify the catalytic activity of cellular MT1-MMP, MCF-7 cells that express little, if any, endogenous MT1-MMP, were engineered to stably express the wild-type proteinase. High levels of MT1-MMP were confirmed in transfected cells relative to mock-transfected cells as determined by Western blotting (Figure 3), wherein 63- and 42-kDa protein bands were detected, corresponding to the active enzyme and its autocatalytically-generated inactive fragment, respectively (49, 50). The levels of MT1-MMP stably expressed in the MCF-7 cells were similar to those observed for MDA-MB-231 cells (data not shown), the latter of which endogenously expresses MT1-MMP (51). Parameters such as optimal cell number and suitable media for the assay were subsequently established. When the cell number was varied between 1.25 × 103 to 1.0 × 104 MT1-MMP-transfectants/well and activity measured using fTHP-9, activity peaked at 5–6 × 103 cells/well (data not shown). For estimation of the levels of active enzyme mobilized to the cell surface, the hydrolytic activity of 6 × 103 cells/well was titrated with increasing concentrations of TIMP-2 (52). Under these conditions, WT-MT1-MMP transfectants were estimated to express ~2.28 nM active proteinase, whereas control cells expressed ~0.96 nM of active metalloproteinase activity, likely representing endogenous proteases produced by MCF-7 cells (e.g., MT2-MT6-MMPs) that are also inhibited by TIMP-2 (Table 1). With the endogenous activity treated as background, stable transfection of MCF-7 cells resulted in the expression of approximately 7.95 × 106 active MT1-MMP molecules/cell ((1.32 × 10−9 moles/L) × (60 × 10−6 L) × (6.022 × 1023 molecules/mole))/(6 × 103 cells)). This number is comparable to those reported in prior studies using imaging approaches. For example, using the MP-3653 reporter (a PEG-liposome possessing a hydroxamic acid for MMP targeting and carboxyfluorescein for imaging) for quantification, MT1-MMP transfection of MCF-7 cells resulted in ~1.1 × 106 molecules/cell (53). MT1-MMP concentrations on uterus carcinoma SiHa cell surfaces were determined using gold nanoclusters containing MT1-AF7p (sequence Cys-Cys-Tyr-His-Trp-Lys-His-Leu-His-Asn-Thr-Lys-Thr-Phe-Leu) (54), where MT1-AF7p binds to the MT1-MMP “MT-loop” region. MT1-MMP concentration was 5.24–12.47 × 10−18 moles per cell (3.16–7.51 × 106 MT1-MMP molecules/cell).
Table 2:
Kinetic parameters for fTHP-9 hydrolysis by secreted and membrane-anchored MT-MMP.
| MT-MMP Variant | KM (μM) | kcat/KM (M−1sec−1) | kcat (sec−1) |
|---|---|---|---|
| sMT1-MMP | 18.6 ± 1.4 | 45130 ± 6641** | 0.84 |
| WT-MT1-MMP | 15.1 ± 2.2 | 9315 ± 3262* | 0.14 |
| MT1-MMP(ΔCT) | 11.2 ± 0.79 | 19060 ± 1760** | 0.21 |
| MT1-MMP(ΔHPX) | 23.4 ± 2.9 | 8430 ± 608** | 0.2 |
| MT1-MMP(MMP-1 CAT) | 36.8 ± 4.4 | 2237 ± 367** | 0.08 |
| sMT2-MMP | 2.41 ± 1.0 | 43180 ± 51.1 | 0.1 ± 0.032 |
| WT-MT2-MMP | 6.95 ± 0.81 | 3336 ± 51.1 | 0.03 ± 0.003 |
Kinetic parameters were determined as described in Experimental Section 1.5 and 1.6. Results are presented as mean ± SD, as indicated. Statistical comparisons were performed with one-way analysis of variance (ANOVA) using SigmaPlot v12.5 (SPSS, Inc., Chicago, IL). Statistical significance was defined as
p < 0.05 and
p <0.001.
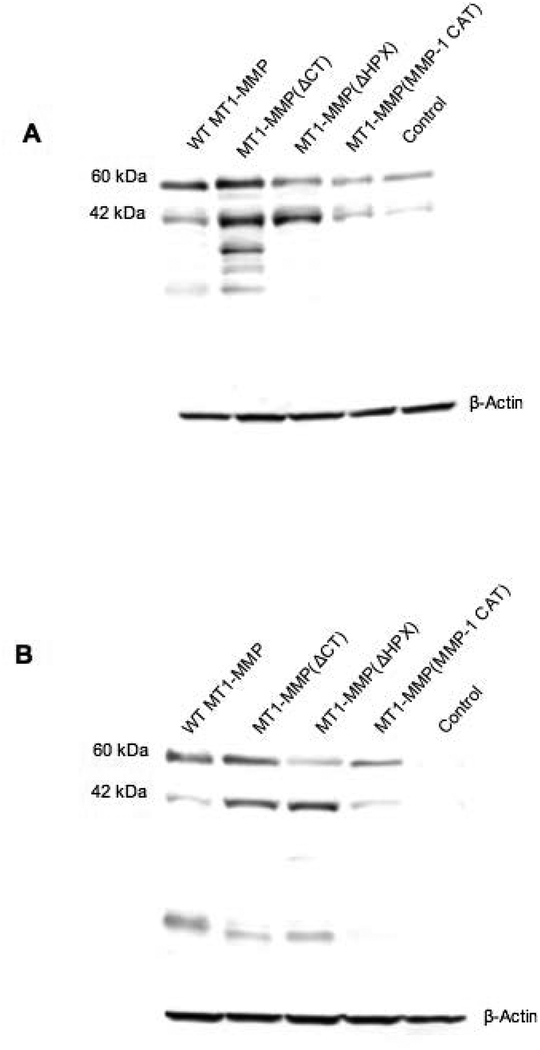
Figure 3.
Western blotting of transfected MCF-7 cells. MCF-7 cells stably transfected with the original pcDNA3.1 plasmid (Control) or WT-MT1-MMP, MT1-MMP(ΔCT), MT1-MMP(ΔHPX), or MT1-MMP(MMP-1 CAT) were lysed and samples identified by Western blotting with anti-MT1-MMP (A) CAT domain mAb or (B) hinge domain mAb. Western blot analysis showed the active (63 kDa) and autodegraded (42 kDa) forms of WT-MT1-MMP or MT1-MMP(ΔCT), as well as the active forms of MT1-MMP(MMP-1 CAT) (56 kDa) or MT1-MMP(ΔHPX) (42 kDa). In the case of MT1-MMP(ΔHPX), the protein observed at 63 kDa is natural production of WT-MT1-MMP by MCF-7 cells. The lowest MW degradation product in the WT-MT1-MMP and MT1-MMP(ΔCT) samples (~18 kDa) probably corresponds to the autocatalytically generated Tyr112-Ala255 MT1-MMP, which is inactive and does not bind TIMP-2 (101). The other degradation products observed in the MT1-MMP(ΔCT) sample (MW ~31–35 kDa) may be related to non-autocatalytic processing within the HPX domain (101). These fragments would be released from the cell surface (49). Results shown are representative of three or more experiments performed. As observed previously, WT-MT1-MMP, MT1-MMP(ΔCT), and MT1-MMP(MMP-1 CAT) exhibit similar mobility in Western blot analysis (28, 41). The loading control was β-actin.
Kinetic analyses demonstrated that WT-MT1-MMP displayed a KM = 15.1 μM (Table 2), a value similar to that observed with sMT1-MMP (i.e., 18.6 μM). Interestingly, the kcat/KM value for the membrane-tethered enzyme was 4.8-fold lower relative to that determined for the soluble enzyme (Table 2), based primarily on a kcat for WT-MT1-MMP that was decreased 6-fold relative to that of sMT1-MMP. These results suggest that, under similar experimental conditions, the membrane-anchored enzyme is less efficient collagenolytically compared with the soluble protease. As the effect is primarily a function of kcat, membrane-bound MT1-MMP may have limited conformational flexibility, a parameter that restricts the coupled motions required for catalysis (55).
To identify putative effects of the individual MT1-MMP domains on catalytic activity, MT1-MMP mutants (MT1-MMP(ΔCT), MT1-MMP(ΔHPX), and MT1-MMP(MMP-1 CAT)) (see Figure 1) were each stably expressed in MCF-7 cells and the active protease levels quantified by TIMP-2 titration. MT1-MMP(ΔCT) transfected cells displayed a 2.0-fold increase of the active enzyme at the cell surface relative to the wild-type construct (Table 1). These results are consistent with prior studies demonstrating that deletion of the CT delays endocytosis and consequently allows the mutant enzyme to accumulate at the cell surface, likely in the lipid raft compartment (56, 57), but contradict reports suggesting that the MT1-MMP CT plays a required role in regulating proteolytic activity at the cell surface (29, 58). MCF-7 cells expressing MT1-MMP(ΔHPX) also displayed higher levels of bound TIMP-2 when compared to cells expressing WT-MT1-MMP (Table 1), likely due to decreased MT1-MMP endocytosis, as the HPX domain can regulate MT1-MMP internalization as a function of its interactions with the tetraspanins (50). Interestingly, cells expressing MT1-MMP(MMP-1 CAT) also had higher levels of the active enzyme compared with WT-MT1-MMP cells (Table 1). This may be due to the reduced ability of the MMP-1 CAT domain to support the autocatalytic cleavage of the chimera from the cell surface compared with the native MT1-MMP construct (see Figure 3) (28, 49).
The active enzyme concentrations at the cell surface differed (Table 1), while Western blotting showed similar levels of protein production (Figure 3). However, the Western blots were for cell lysates, and thus included both cell surface bound and internalized enzyme. Biotinylation of cell surface proteins (50) followed by Western blot analysis revealed higher levels of cell surface MT1-MMP(ΔCT) protein compared with WT-MT1-MMP (data not shown).
The kinetic parameters were next determined for fTHP-9 hydrolysis by each of the MT1-MMP mutants (Table 2). Deletion of the CT resulted in a small decrease in the KM compared to the wild-type enzyme, whereas kcat was substantially increased. Hence, the MT1-MMP CT does not play a required role in controlling the enzyme’s catalytic activity. By contrast, in the absence of the HPX domain, the KM value increased compared with the wild-type enzyme, supporting an important role for this domain in the binding to the collagen triple-helix (25, 28, 45), while the kcat value increased significantly. Nevertheless, these results demonstrate that the HPX domain, while capable of modulating MT1-MMP activity, is not required for the expression of collagenolytic activity (28). When the CAT domain of MT1-MMP was replaced with that of MMP-1, fTHP-9 hydrolytic activity was likewise retained although with a 2-fold increase in KM and a 50% decrease in kcat (Table 2). Hence, though the structure of the chimeric enzyme may be negatively impacted by potential domain clashes between MT1-MMP and MMP-1, the membrane-anchored construct retains significant enzymatic activity against the triple-helical substrate, highlighting the fact that cell surface collagenolytic activity per se is not a unique characteristic of the MT1-MMP CAT domain (14, 28).
To examine the effects of the cell surface on another MMP, the activity of MT2-MMP in solution and in cells was compared. MT2-MMP was found previously to catalyze the hydrolysis of fTHP-9 (31). In this case, soluble MT2-MMP was found to have much greater activity towards fTHP-9 than cell surface-bound MT2-MMP (Table 2).
Finally, to compare the efficacy of synthetic inhibitors towards soluble versus membrane-bound MT1-MMP, we utilized two well-characterized MMP inhibitors. Marimastat is a small molecule hydroxamate that chelates the active site Zn2+ (59), whereas GlyΨ{PO2H-CH2}Ile-Tyr THPI is a transition state (phosphinate) analog that interacts with both the active site and secondary binding sites (exosites) of MT1-MMP (60). Marimastat exhibited an IC50 = 19 ± 3 nM for the soluble enzyme and an IC50 = 36 ± 8 nM for membrane-bound WT-MT1-MMP as determined in the cell-based assay. GlyΨ{PO2H-CH2}Ile-Tyr THPI had an IC50 = 20 ± 4 nM with the soluble enzyme and an IC50 = 36 ± 6 nM with the membrane-bound WT-MT1-MMP. Thus, the potencies of both inhibitors were decreased ~2-fold when comparing the cell-surface bound to the soluble form of MT1-MMP.
3. Discussion
Evaluating enzyme kinetics of proteinases tethered to the cell surface relative to those of soluble proteinases can provide insight into the roles of the membrane microenvironment in regulating catalytic activity (61). To this end, we used a series of membrane-anchored and soluble forms of MT1-MMP to define the role of the cell surface and individual protease domains on MT1-MMP activity and catalytic efficiency. Enzyme activity was measured via a cell-compatible FRET assay, whereby an increase in fluorescence upon hydrolysis allowed for the rapid kinetic evaluation of the MT1-MMP proteolytic activity in an intact cell system. In the past, THP substrate models of triple-helical collagen have allowed for significant advancements in the characterization of collagenolysis (60, 62–64). The use of fTHP-9 is based its high selectivity for MT1-MMP and MMP-8 compared with other collagenolytic MMPs as a consequence of the presence of Cys(Mob) in the P1’ subsite (31). Structural analyses of MMP S1’ binding pockets indicated that MT1-MMP can better accommodate large hydrophobic residues compared with MMP-1, based on the Arg214 residue in MMP-1 versus Leu in MT1-MMP (65, 66). MT1-MMP is thus considered to have a deep, tunnel-like S1’ pocket, while MMP-1 has a shallow S1’ pocket (67). Indeed, fTHP-9 was hydrolyzed at a rate 36-fold higher by MT1-MMP compared with MMP-1 (31). Further, in contrast to MT1-MMP, fTHP-9 is a very inefficient substrate for MMP-2 and MMP-9 (31). As MMP-2 and MMP-9 possess an intermediate-sized S1’ pocket (67), these proteinases may not efficiently accommodate the Cys(Mob) side chain. Based on these advantageous parameters, fTHP-9 was used as a preferred substrate to quantify the collagenolytic activity of various MT1-MMP forms in our current study.
Soluble MT1-MMP exhibited a higher activity than cell surface bound proteinase (Table 2), an expected result given that tethering of the proteinase to the cell surface restricts its diffusion to the soluble substrate (26). However, the difference was manifested almost entirely in kcat, suggesting that the cell surface modulates the coupled motions for catalysis (55). Protein motions and catalytic activity are likely linked in collagenolytic MMPs with prior studies supporting the existence of multiple coupled conformational states in MMP-1 (60, 63, 68).
Soluble MT2-MMP was found to process fTHP-9 at a similar kcat/KM value as MT1-MMP (Table 2). This differs from the collagenolytic activities of these enzymes, where soluble MT1-MMP has much greater activity towards type I collagen than MT2-MMP (69). The difference in activity between soluble and cell surface bound MT2-MMP was greater than the difference for these two MT1-MMP constructs (Table 2). In this case, the difference between the MT2-MMP constructs was manifested in both kcat and KM. The results suggest that MT2-MMP is a far less efficient collagenolytic enzyme than MT1-MMP when bound to the cell surface. This result is consistent with observations for type I collagen invasion by COS-1 cells transiently transfected with MT1-MMP or MT2-MMP (13).
The MT1-MMP constructs evaluated herein allowed for the examination of the roles of several individual domains on the overall collagenolytic activity of MT1-MMP. It has been shown previously that MT1-MMP undergoes clathrin-dependent internalization mediated via the CT (70–72). However, the MT1-MMP CT has also been shown to interact with intracellular binding partners, such as the FAK-p130Cas complex, and to undergo posttranslational modifications, including Tyr and Thr phosphorylation, Lys ubiquitination, and Cys palmitoylation (73–77). While the CT has been reported to negatively regulate cell-mediated MT1-MMP proteolytic activity (29, 58), this domain did not play a required role in directly regulating proteolytic activity and the measured KM values were not significantly different between WT-MT1-MMP and MT1-MMP(ΔCT) (Table 2). The enhanced activity of MT1-MMP(ΔCT) compared with WT-MT1-MMP may be the result of the internalization of the latter in a complex with substrates, reducing, as a result, the observed hydrolysis rate for the wild type enzyme. In agreement with these findings, CT deletion has previously been shown to enhance the type I collagen-invasive potential of epithelial as well as mesenchymal cell populations (14, 28, 78). In addition, replacement of the CT and the TM domain of MT1-MMP with the glycosylphosphatidylinositol anchor of MT6-MMP enhanced MDCK cell invasion in a three-dimensional type I collagen matrix (79).
The MT1-MMP HPX domain has been reported to play critical roles in a range of functions, including the formation of MT1-MMP homodimers, MT1-MMP•CD44 and MT1-MMP•tetraspanin heterodimerization, and MT1-MMP•type I collagen binding interactions (4, 25, 45, 80–83). However, deletion of the HPX domain only slightly reduced the kcat/KM value of the mutant enzyme relative to WT-MT1-MMP (Table 2). The reduction was entirely due to an increased KM value. In solution, deletion of the HPX domain only reduced MT1-MMP activity towards the triple-helical substrate fTHP-15 3.5-fold (i.e., kcat/KM = 26,700 M−1sec−1 for soluble MT1-MMP versus 7,660 M−1sec−1 for soluble MT1-MMP(ΔHPX)) (84). Interestingly, despite changes in catalytic activity, these results are consistent with previous studies demonstrating the ability of MT1-MMP(ΔHPX) to support collagenolytic and tissue-invasive activities when expressed in COS cells or fibroblasts (14, 28). Thus, our data support the contention that the triple-helicase and collagen-binding activities of the HPX domain play contributory, but not absolute, roles for MT1-MMP function in intact cell systems (28). Most likely, the HPX domain serves as a “modulator” of the CAT domain by enhancing activity through coupled motions (as discussed earlier) and/or dampening activity of hyperactive CAT domains (85). Additional support for this latter notion is provided by the observation that the MT1-MMP CAT domain alone exhibits a 5.0–7.5-fold higher catalytic activity towards single-stranded synthetic substrates compared with the soluble full-length enzyme (48, 86).
In our efforts to identify potentially unique properties associated with the MT1-MMP CAT domain, we replaced the wild-type CAT domain with that of secreted interstitial collagenase, MMP-1. MT1-MMP(MMP-1 CAT) had a ~4.2-fold reduced activity compared with WT-MT1-MMP (Table 2). This reduction was a result of both an increased KM value and a decreased kcat value. As previously reported, soluble MMP-1 had reduced activity towards fTHP-9 compared with soluble MT1-MMP (31). The magnitude of the difference was greater for the soluble enzymes, but the comparison is not straightforward, as the mutant we used included the MT1-MMP HPX domain (while the soluble MMP-1 has its native HPX domain). Though the collagenolytic activity of MT1-MMP(MMP-1 CAT) is about a half of that of WT-MT1-MMP (28), this result is consistent with previous studies demonstrating that the collagen-invasive activity of fibroblasts, though retained to a significant degree, was decreased when the MMP-1 CAT domain was substituted for the MT1-MMP CAT domain (14).
Contributions from secondary binding sites within the MT1-MMP CAT domain and/or other cell surface biomolecules (such as integrins) may also promote MT1-MMP collagenolysis (1, 87). The need for proper interaction of MT1-MMP with cell surface partners is suggested by a recent study of the 163–170 loop region within MT1-MMP CAT domain. This MT (membrane type)-loop region is present in the CAT domain of MT1-MT6-MMPs, but is absent in all other MMPs. Deletion of this loop has been reported to result in the mis-localization of MT1-MMP relative to β1 integrin adhesion complexes with subsequent decreases in collagenolytic activity (88). However, these results stand in contrast with earlier reports that the MT-loop region does not play a required role in MT1-MMP-dependent collagenolysis (28) and further studies are required to resolve these discrepancies.
Finally, the present approach allowed for the evaluation of the effectiveness of proteinase inhibitors at the cell surface. Both small molecule and mini-protein inhibitors were found to be active towards cell surface triple-helical peptidase activity, albeit with reduced efficiency compared with inhibition in solution. The inhibition of MMP cell surface activity has been quantified previously using fluorescein-conjugated gelatin (89). However, gelatin is a substrate for multiple proteases, and MMPs were expressed on the surface of yeast cells (89), which are quite different from mammalian cell surfaces. As several selective and/or secondary binding site (exosite) MT1-MMP inhibitors have been described (2, 90–92), the evaluation of activity at the cell surface will allow for examination of such inhibitors in a more native-like environment. This will also facilitate the development of inhibitors that may act indirectly on MT1-MMP activity. For example, MT1-MMP activity has been reported to be enhanced by tetraspanins (81, 93) and bilayer membranes (94, 95), and thus inhibitors may be designed to mitigate those interactions. MT1-MMP inhibitors have been described that disrupt the association of the enzyme with β integrin subunits on the cell surface (88, 96).
The present cell-based assay could potentially be improved by creating a cell-surface bound substrate. To achieve this goal, lipid-like alkyl chains can be attached to the N-terminus of the MT1-MMP substrate fTHP-9. We have previously used this approach to stabilize fTHPs (31, 32, 97). An MMP-12 FRET substrate has been anchored to the cell surface via palmitoylation (98). The previously discussed ECFP/Ypet, mOrange2/mCherry, and ECFP-PEbody/R-PE MT1-MMP biosensors all incorporated the transmembrane domain of platelet-derived growth factor receptor to promote cell surface anchoring (15, 16, 19). Quasi-irreversible insertion into membranes can also be achieved via peptide modification by cholesterol, whereby cholesteryl chloroformate or cholest-5-en-3-yl bromoacetate is used to attach cholesterol to the N-terminus of the peptide or the side chain of a Cys residue, respectively (99, 100). With a surface bound substrate, one can minimize diffusion effects and increase the local enzyme•substrate concentration. A potential drawback is that the surface bound substrate might disrupt interactions between the enzyme and native cell surface binding partners (see above). Future studies can be designed to evaluate soluble versus cell-surface bound substrates in the MT1-MMP cell-based assay described herein.
Acknowledgments
We are most appreciative of Dr. Stanley Zucker’s scientific input and review of the manuscript. This work was supported by the National Institutes of Health (CA098799 and NHLBI contract 268201000036C to G.B.F., CA088308 and CA071699 to S.J.W.), the James and Esther King Biomedical Research Program (to G.B.F.), the US-Israel Binational Science Foundation (BSF) (to G.B.F.), and the State of Florida, Executive Office of the Governor’s Department of Economic Opportunity.
Footnotes
Conflict of Interest
The authors have no conflicts of interest.
Contributor Information
Sonia Pahwa, Email: pahwa.sonia@gmail.com.
Manishabrata Bhowmick, Email: manishabrata.bhowmick@sial.com.
Sabrina Amar, Email: amar.sabrina@gmail.com.
Jian Cao, Email: jian.cao2@nih.gov.
Alex Y. Strongin, Email: strongin@SBPdiscovery.org.
Rafael Fridman, Email: rfridman@med.wayne.edu.
Stephen J. Weiss, Email: sjweiss@umich.edu.
References
- 1.Rowe RG, Weiss SJ (2009) Navigating ECM barriers at the invasive front: the cancer cell-stroma interface. Annu Rev Cell Dev Biol;25: 567–95. [DOI] [PubMed] [Google Scholar]
- 2.Pahwa S, Stawikowski MJ, Fields GB (2014) Monitoring and inhibiting MT1-MMP during cancer initiation and progression. Cancers;6: 416–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabova L, Chrysovergis K, Yamada SS, Holmbeck K (2007) MT1-MMP is required for efficient tumor dissemination in experimental metastatic disease. Oncogene;27: 3274–81. [DOI] [PubMed] [Google Scholar]
- 4.Zarrabi K, Dufour A, Li J, Kuscu C, Pulkoski-Gross A, Zhi J, et al. (2011) Inhibition of matrix metalloproteinase-14 (MMP-14)-mediated cancer cell migration. J Biol Chem;286: 33167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang Y, Rowe RG, Botvinick EL, Kurup A, Putnam AJ, Seiki M, et al. (2013) MT1-MMP-dependent control of skeletal stem cell commitment via a β1-integrin/YAP/TAZ signaling axis. Dev Cell;25: 402–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castro-Castro A, Marchesin V, Monteiro P, Lodillinsky C, Rossé C, Chavrier P (2016) Cellular and Molecular Mechanisms of MT1-MMP-Dependent Cancer Cell Invasion. Annu Rev Cell Dev Biol;32: 555–76. [DOI] [PubMed] [Google Scholar]
- 7.Talmi-Frank D, Altboum Z, Solomonov I, Udi Y, Jaitin DA, Klepfish M, et al. (2016) Extracellular Matrix Proteolysis by MT1-MMP Contributes to Influenza-Related Tissue Damage and Mortality. Cell Host Microbe;20: 458–70. [DOI] [PubMed] [Google Scholar]
- 8.Fisher KE, Sacharidou A, Stratman AN, Mayo AM, Fisher SB, Mahan RD, et al. (2009) MT1-MMP- and Cdc42-dependent signaling co-regulate cell invasion and tunnel formation in 3D collagen matrices. J Cell Sci;122: 4558–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, et al. (2007) Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol;9: 893–904. [DOI] [PubMed] [Google Scholar]
- 10.Koike T, Vernon RB, Hamner MA, Sadoun E, Reed MJ (2002) MT1-MMP, but not secreted MMPs, influences the migration of human microvascular endothelial cells in 3-dimensional collagen gels. J Cell Biochem;86: 748–58. [DOI] [PubMed] [Google Scholar]
- 11.Zhang W, Matrisian LM, Holmbeck K, Vick CC, Rosenthal EL (2006) Fibroblast-derived MT1-MMP promotes tumor progression in vitro and in vivo. BMC Cancer;6: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M, et al. (2004) Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol;167: 769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hotary K, Allen E, Punturieri A, Yana I, Weiss SJ (2000) Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J Cell Biol;149: 1309–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabeh F, Li X-Y, Saunders TL, Rowe RG, Weiss SJ (2009) Secreted versus membrane-anchored collagenases: Relative roles in fibroblast-dependent collagenolysis and invasion. J Biol Chem;284: 23001–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouyang M, Lu S, Li X-Y, Xu J, Seong J, Glepmans BNG, et al. (2008) Visualization of polarized membrane type 1 matrix metalloproteinase activity in live cells by fluorescence resonance energy transfer imaging. J Biol Chem;283: 17740–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouyang M, Huang H, Shaner NC, Remacle AG, Shiryaev SA, Strongin AY, et al. (2010) Simultaneous visualization of protumorigenic Src and MT1-MMP activities with fluorescence resonance energy transfer. Cancer Res;70: 2204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jabaiah A, Daugherty PS (2011) Directed evolution of protease beacons that enable sensitive detection of endogenous MT1-MMP activity in tumor cell lines. Chem Biol;18: 392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu S, Wang Y, Huang H, Pan Y, Chaney EJ, Boppart SA, et al. (2013) Quantitative FRET imaging to visualize the invasiveness of live breast cancer cells. PLoS One;8: e58569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Limsakul P, Peng Q, Wu Y, Allen ME, Liang J, Remacle AG, et al. (2018) Directed Evolution to Engineer Monobody for FRET Biosensor Assembly and Imaging at Live-Cell Surface. Cell Chem Biol;25: 370–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leight JL, Alge DL, Maier AJ, Anseth KS (2013) Direct measurement of matrix metalloproteinase activity in 3D cellular microenvironments using a fluorogenic peptide substrate. Biomaterials;34: 7344–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight CG, Willenbrock F, Murphy G (1992) A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett;296: 263–6. [DOI] [PubMed] [Google Scholar]
- 22.Nagase H, Fields CG, Fields GB (1994) Design and characterization of a fluorogenic substrate selectively hydrolyzed by stromelysin 1 (matrix metalloproteinase-3). J Biol Chem;269: 20952–7. [PubMed] [Google Scholar]
- 23.Giricz O, Lauer JL, Fields GB (2011) Comparison of Metalloproteinase Protein and Activity Profiling. Anal Biochem;409: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh Y, Takamura A, Ito N, Maru Y, Sato H, Suenaga N, et al. (2001) Homophilic complex formation of MT1-MMP facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. EMBO J;20: 4782–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam EM, Moore TR, Butler GS, Overall CM (2004) Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinase 2 and 14 (gelatinase A and MT1-MMP): The differential roles of the MMP hemopexin C domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J Biol Chem;279: 43336–44. [DOI] [PubMed] [Google Scholar]
- 26.Itoh Y, Ito N, Nagase H, Evans RD, Bird SA, Seiki M (2006) Cell surface collagenolysis requires homodimerization of the membrane-bound collagenase MT1-MMP. Mol Biol Cell;17: 5390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao J, Chiarelli C, Richman O, Zarrabi K, Kozarekar P, Zucker S (2008) Membrane type 1 matrix metalloproteinase induces epithelial-to-mesenchymal transition in prostate cancer. J Biol Chem;283: 6232–40. [DOI] [PubMed] [Google Scholar]
- 28.Li X-Y, Ota I, Yana I, Sabeh F, Weiss SJ (2008) Molecular dissection of the structural machinery underlying the tissue-invasive activity of membrane type-1 matrix metalloproteinase. Mol Biol Cell;19: 3221–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu X, Zech T, McDonald L, Gonzalez EG, Li A, Macpherson I, et al. (2012) N-WASP coordinates the delivery and F-actin-mediated capture of MT1-MMP at invasive pseudopods. J Cell Biol;199: 527–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weaver SA, Wolters B, Ito N, Woskowicz AM, Kaneko K, Shitomi Y, et al. (2014) Basal localization of MT1-MMP is essential for epithelial cell morphogenesis in 3D collagen matrix. J Cell Sci;127: 1203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, et al. (2006) The roles of substrate thermal stability and P2 and P1’ subsite identity on matrix metalloproteinase triple-helical peptidase activity and collagen specificity. J Biol Chem;281: 38302–13. [DOI] [PubMed] [Google Scholar]
- 32.Minond D, Lauer-Fields JL, Nagase H, Fields GB (2004) Matrix metalloproteinase triple-helical peptidase activities are differentially regulated by substrate stability. Biochemistry;43: 11474–81. [DOI] [PubMed] [Google Scholar]
- 33.Bhowmick M, Fields GB (2012) Synthesis of Fmoc-Gly-Ile Phosphinic Pseudodipeptide: Residue Specific Conditions for Construction of Matrix Metalloproteinase Inhibitor Building Blocks. Int J Pept Res Ther;18: 335–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauer-Fields JL, Brew K, Whitehead JK, Li S, Hammer RP, Fields GB (2007) Triple-helical transition-state analogs: A new class of selective matrix metalloproteinase inhibitors. J Am Chem Soc;129: 10408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhowmick M, Tokmina-Roszyk D, Onwuha-Ekpete L, Harmon K, Robichaud T, Fuerst R, et al. (2017) Second Generation Triple-Helical Peptide Transition State Analog Matrix Metalloproteinase Inhibitors. J Med Chem;60: 3814–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasmussen HS, McCann PP (1997) Matrix metalloproteinase inhibition as a novel anticancer strategy: A review with special focus on batimastat and marimastat. Pharmacol Ther;75: 69–75. [DOI] [PubMed] [Google Scholar]
- 37.Beckett RP, Whittaker M (1998) Matrix metalloproteinase inhibitors 1998. Exp Opin Ther Patents;8: 259–82. [Google Scholar]
- 38.Rozanov DV, Deryugina EI, Ratnikov BI, Monosov EZ, Marchenko GN, Quigley JP, et al. (2001) Mutation analysis of membrane type-1 matrix metalloproteinase (MT1-MMP). The role of the cytoplasmic tail Cys(574), the active site Glu(240), and furin cleavage motifs in oligomerization, processing, and self-proteolysis of MT1-MMP expressed in breast carcinoma cells. J Biol Chem;276: 25705–14. [DOI] [PubMed] [Google Scholar]
- 39.Köhrmann A, Kammerer U, Kapp M, Dietl J, Anacker J (2009) Expression of matrix metalloproteinases (MMPs) in primary human breast cancer and breast cancer cell lines: New findings and review of the literature. BMC Cancer;9: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pei D, Weiss SJ (1996) Transmembrane-deletion mutants of the membrane-type matrix metalloproteinase-1 process progelatinase A and express intrinsic matrix-degrading activity. J Biol Chem;271: 9135–40. [DOI] [PubMed] [Google Scholar]
- 41.Cao J, Kozarekar P, Pavlaki M, Chiarelli C, Bahou WF, Zucker S (2004) Distinct roles for the catalytic and hemopexin domains of membrane type 1-matrix metalloproteinase in substrate degradation and cell migration. J Biol Chem;279: 14129–39. [DOI] [PubMed] [Google Scholar]
- 42.Pei D, Weiss SJ (1995) Furin-dependent intracellular activation of the human stromelysin-3 zymogen. Nature;375: 244–7. [DOI] [PubMed] [Google Scholar]
- 43.Ota I, Li X-Y, Hu Y, Weiss SJ (2009) Induction of a MT1-MMP and MT2-MMP-dependent basement membrane transmigration program in cancer cells by Snail1. Proc Natl Acad Sci USA;106: 20318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu HL, Sohail A, Valiathan RR, Wasinski BD, Kumarasiri M, Mahasenan KV, et al. (2013) Shedding of discoidin domain receptor 1 by membrane-type matrix metalloproteinases. J Biol Chem;288: 12114–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.d’Ortho MP, Will H, Atkinson S, Butler G, Messent A, Gavrilovic J, et al. (1997) Membrane-type matrix metalloproteinases 1 and 2 exhibit broad-spectrum proteolytic capacities comparable to many matrix metalloproteinases. Eur J Biochem;250: 751–7. [DOI] [PubMed] [Google Scholar]
- 46.Roderfeld M, Büttner FH, Bartnik E, Tschesche H (2000) Expression of human membrane type 1 matrix metalloproteinase in Pichia pastoris. Protein Expr Purif;19: 369–74. [DOI] [PubMed] [Google Scholar]
- 47.Will H, Atkinson SJ, Butler GS, Smith B, Murphy G (1996) The soluble catalytic domain of membrane type 1 matrix metalloproteinase cleaves the propeptide of progelatinase a and initiates autoproteolytic activation. J Biol Chem;271: 17119–23. [DOI] [PubMed] [Google Scholar]
- 48.Hurst DR, Schwartz MA, Ghaffari MA, Jin Y, Tschesche H, Fields GB, et al. (2004) Catalytic- and ecto-domains of membrane type 1-matrix metalloproteinase have similar inhibition profiles but distinct endopeptidase activities. Biochem J;377: 775–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osenkowski P, Toth M, Fridman R (2004) Processing, shedding, and endocytosis of membrane type 1-matrix metalloproteinase (MT1-MMP). J Cell Physiol;200: 2–10. [DOI] [PubMed] [Google Scholar]
- 50.Schröder HM, Hoffman S, Hecker M, Korff T, Ludwig T (2013) The tetraspanin network modulates MT1-MMP cell surface trafficking. Int J Biochem Cell Biol;45: 1133–44. [DOI] [PubMed] [Google Scholar]
- 51.Lafleur MA, Drew AF, de Sousa EL, Blick T, Bills M, Walker EC, et al. (2005) Upregulation of matrix metalloproteinases (MMPs) in breast cancer xenografts: a major induction of stromal MMP-13. Int J Cancer;114: 544–54. [DOI] [PubMed] [Google Scholar]
- 52.Zucker S, Drews M, Conner C, Foda HD, DeClerck YA, Langley KE, et al. (1998) Tissue inhibitor of metalloproteinase-2 (TIMP-2) binds to the catalytic domain of the cell surface receptor, membrane type 1-matrix metalloproteinase (MT1-MMP). J Biol Chem;273: 1216–22. [DOI] [PubMed] [Google Scholar]
- 53.Remacle AG, Shiryaev SA, Golubkov VS, Freskos JN, Brown MA, Karwa AS, et al. (2013) Non-destructive and Selective Imaging of the Functionally Active, Pro-invasive Membrane Type-1 Matrix Metalloproteinase (MT1-MMP) Enzyme in Cancer Cells. J Biol Chem;288: 20568–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Liu R, Shu Q, Yuan Q, Xing G, Gao X (2018) Quantitative Analysis of Multiple Proteins of Different Invasive Tumor Cell Lines at the Same Single-Cell Level. Small;14: e1703684. [DOI] [PubMed] [Google Scholar]
- 55.Watt ED, Shimada H, Kovrigin E, Loria JP (2007) The mechanism of rate-limiting motions in enzyme function. Proc Natl Acad Sci USA;104: 11981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Remacle A, Murphy G, Roghi C (2003) Membrane type I-matrix metalloproteinase (MT1-MMP) is internalised by two different pathways and is recycled to the cell surface. J Cell Sci;116: 3905–16. [DOI] [PubMed] [Google Scholar]
- 57.Jiang A, Lehti K, Wang X, Weiss SJ, Keski-Oja J, Pei D (2001) Regulation of membrane-type matrix metalloproteinase 1 activity by dynamin-mediated endocytosis. Proc Natl Acad Sci USA;98: 13693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uekita T, Itoh Y, Yana I, Ohno H, Seiki M (2001) Cytoplasmic tail-dependent internalization of membrane-type 1 matrix metalloproteinase is important for its invasion-promoting activity. J Cell Biol;155: 1345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whittaker M, Floyd CD, Brown P, Gearing AJH (1999) Design and therapeutic application of matrix metalloproteinase inhibitors. Chem Rev;99: 2735–76. [DOI] [PubMed] [Google Scholar]
- 60.Lauer-Fields JL, Chalmers MJ, Busby SA, Minond D, Griffin PR, Fields GB (2009) Identification of Specific Hemopexin-like Domain Residues That Facilitate Matrix Metalloproteinase Collagenolytic Activity. J Biol Chem;284: 24017–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Black RA, Doedens JR, Mahimkar R, Johnson R, Guo L, Wallace A, et al. (2003) Substrate specificity and inducibility of TACE (tumour necrosis factor α-converting enzyme) revisited: the Ala-Val preference, and induced intrinsic activity. Biochem Soc Symp;70: 39–52. [DOI] [PubMed] [Google Scholar]
- 62.Arnold LH, Butt L, Prior SH, Read C, Fields GB, Pickford AR (2011) The Interface Between Catalytic and Hemopexin Domains in Matrix Metalloproteinase 1 Conceals a Collagen Binding Exosite. J Biol Chem;286: 45073–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bertini I, Fragai F, Luchinat C, Melikian M, Toccafondi M, Lauer JL, et al. (2012) Structural Basis for Matrix Metalloproteinase 1 Catalyzed Collagenolysis. J Am Chem Soc;134: 2100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manka SW, Carafoli F, Visse R, Bihan D, Raynal N, Farndale RW, et al. (2012) Structural insights into triple-helical collagen cleavage by matrix metalloproteinase 1. Proc Natl Acad Sci USA;109: 12461–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bode W, Fernandez-Catalan C, Tschesche H, Grams F, Nagase H, Maskos K (1999) Structural properties of matrix metalloproteinases. Cell Mol Life Sci;55: 639–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maskos K (2005) Crystal structures of MMPs in complex with physiological and pharmacological inhibitors. Biochimie;87: 249–63. [DOI] [PubMed] [Google Scholar]
- 67.de Oliveira CA, Zissen M, Mongon J, McCammon JA (2007) Molecular dynamics simulations of metalloproteinases types 2 and 3 reveal differences in the dynamic behavior of the S1’ binding pocket. Curr Pharm Design;13: 3471–5. [DOI] [PubMed] [Google Scholar]
- 68.Cerofolini L, Fields GB, Fragai M, Geraldes CFGC, Luchinat C, Parigi G, et al. (2013) Examination of matrix metalloproteinase-1 (MMP-1) in solution: A preference for the pre-collagenolysis state. J Biol Chem;288: 30659–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morrison CJ, Overall CM (2006) TIMP-independence of MMP-2 activation by MT2-MMP is determined by contributions of both the MT2-MMP catalytic and hemopexin domains. J Biol Chem;281: 26528–39. [DOI] [PubMed] [Google Scholar]
- 70.Lehti K, Valtanen H, Wickstrom SA, Lohi J, Keski-Oja J (2000) Regulation of membrane-type-1 matrix metalloproteinase activity by its cytoplasmic domain. J Biol Chem;275: 15006–13. [DOI] [PubMed] [Google Scholar]
- 71.Lehti K, Lohi J, Juntunen MM, Pei D, Keski-Oja J (2002) Oligomerization through hemopexin and cytoplasmic domains regulates the activity and turnover of membrane-type 1 matrix metalloproteinase. J Biol Chem;277: 8440–8. [DOI] [PubMed] [Google Scholar]
- 72.Itoh Y, Seiki M (2006) MT1-MMP: a potent modifier of pericellular microenvironment. J Cell Physiol;206: 1–8. [DOI] [PubMed] [Google Scholar]
- 73.Anilkumar N, Uekita T, Couchman JR, Nagase H, Seiki M, Itoh Y (2005) Palmitoylation at Cys574 is essential for MT1-MMP to promote cell migration. FASEB J;19: 1326–8. [DOI] [PubMed] [Google Scholar]
- 74.Moss NM, Wu YI, Liu Y, Munshi HG, Stack MS (2009) Modulation of the membrane type 1 matrix metalloproteinase cytoplasmic tail enhances tumor cell invasion and proliferation in three-dimensional collagen matrices. J Biol Chem;284: 19791–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nyalendo C, Michaud M, Beaulieu E, Roghi C, Murphy G, Gingras D, et al. (2007) Src-dependent phosphorylation of membrane type I matrix metalloproteinase on cytoplasmic tyrosine 573: role in endothelial and tumor cell migration. J Biol Chem;282: 15690–9. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y, McNiven MA (2012) Invasive matrix degradation at focal adhesions occurs via protease recruitment by a FAK-p130Cas complex. J Cell Biol;196: 375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eisenach PA, de Sampaio PC, Murphy G, Roghi C (2012) Membrane type 1 matrix metalloproteinase (MT1-MMP) ubiquitination at Lys581 increases cellular invasion through type I collagen. J Biol Chem;287: 11533–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang A, Pei D (2003) Distinct roles of catalytic and pexin-like domains in membrane-type matrix metalloproteinase (MMP)-mediated pro-MMP-2 activation and collagenolysis. J Biol Chem;278: 38765–71. [DOI] [PubMed] [Google Scholar]
- 79.Nie J, Pei J, Blumenthal M, Pei D (2007) Complete restoration of cell surface activity of transmembrane-truncated MT1-MMP by a glycosylphosphatidylinositol anchor. Implications for MT1-MMP-mediated prommp2 activation and collagenolysis in three-dimensions. J Biol Chem;282: 6438–43. [DOI] [PubMed] [Google Scholar]
- 80.Mori H, Tomari T, Koshifumi I, Sato H, Tojo H, Yana I, et al. (2002) CD44 directs membrane-type I matrix metalloproteinase to lamellipodia by associating with its hemopexin-like domain. EMBO J;21: 3949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yañez-Mó M, Barreiro O, Gonzalo P, Batista A, Megías D, Genís L, et al. (2008) MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood;112: 3217–26. [DOI] [PubMed] [Google Scholar]
- 82.Murphy G, Nagase H (2011) Localizing matrix metalloproteinase activities in the pericellular environment. FEBS J;278: 2–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tochowicz A, Goettig P, Evans R, Visse R, Shitomi Y, Palmisano R, et al. (2011) The dimer interface of the membrane type 1 matrix metalloproteinase hemopexin domain: crystal structure and biological functions. J Biol Chem;286: 7587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao Y, Marcink T, Gari RRS, Marsh BP, King GM, Stawikowska R, et al. (2015) Transient Collagen Triple Helix Binding to a Key Metalloproteinase in Invasion and Development. Structure;23: 257–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gioia M, Fasciglione GF, Marini S, D’Alessio S, De Sanctis G, Diekmann O, et al. (2002) Modulation of the catalytic activity of neutrophil collagenase MMP-8 on bovine collagen I. J Biol Chem;277: 23123–30. [DOI] [PubMed] [Google Scholar]
- 86.Neumann U, Kubota H, Frei K, Ganu V, Leppert D (2004) Characterization of Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2, a fluorogenic substrate with increased specificity constants for collagenases and tumor necrosis factor converting enzyme. Anal Biochem;328: 166–73. [DOI] [PubMed] [Google Scholar]
- 87.Fields GB (2013) Interstitial collagen catabolism. J Biol Chem;288: 8785–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Woskowicz AM, Weaver SA, Shitomi Y, Ito N, Itoh Y (2013) MT-LOOP-dependent localization of membrane type I matrix metalloproteinase (MT1-MMP) to the cell adhesion complexes promotes cancer cell invasion. J Biol Chem;288: 35126–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Diehl B, Hoffmann TM, Mueller NC, Burkhart JL, Kazmaier U, Schmitt MJ (2011) Novel yeast bioassay for high-throughput screening of matrix metalloproteinase inhibitors. Appl Environ Microbiol;77: 8573–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ling B, Watt K, Banerjee S, Newsted D, Truesdell P, Adams J, et al. (2017) A novel immunotherapy targeting MMP-14 limits hypoxia, immune suppression and metastasis in triple-negative breast cancer models. Oncotarget;8: 58372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Levin M, Udi Y, Solomonov I, Sagi I (2017) Next generation matrix metalloproteinase inhibitors - Novel strategies bring new prospects. Biochim Biophys Acta;1864: 1927–39. [DOI] [PubMed] [Google Scholar]
- 92.Santamaria S, de Groot R (2018) Monoclonal antibodies against metzincin targets. Br J Pharmacol;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lafleur MA, Xu D, Hemler ME (2009) Tetraspanin proteins regulate membrane type-1 matrix metalloproteinase-dependent pericellular proteolysis. Mol Biol Cell;20: 2030–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cerofolini L, Amar S, Lauer JL, Martelli T, Fragai M, Luchinat C, et al. (2016) Bilayer Membrane Modulation of Membrane Type 1 Matrix Metalloproteinase (MT1-MMP) Structure and Proteolytic Activity. Nat Sci Rep;6: 29511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marcink TC, Simoncic JA, An B, Knapinska AM, Fulcher YG, Akkaladevi N, et al. (2018) MT1-MMP Binds Membranes by Opposite Tips of its β-Propeller to Position it for Pericellular Proteolysis. Structure;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yosef G, Arkadash V, Papo N (2018) Targeting the MMP-14/MMP-2/integrin αvβ3 axis with multispecific N-TIMP2-based antagonists for cancer therapy. J Biol Chem;293: 13310–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lauer-Fields JL, Sritharan T, Stack MS, Nagase H, Fields GB (2003) Selective hydrolysis of triple-helical substrates by matrix metalloproteinase-2 and −9. J Biol Chem;278: 18140–5. [DOI] [PubMed] [Google Scholar]
- 98.Cobos-Correa A, Trojanek JB, Diemer S, Mall MA, Schultz C (2009) Membrane-bound FRET probe visualizes MMP12 activity in pulmonary inflammation. Nat Chem Biol;5: 628–30. [DOI] [PubMed] [Google Scholar]
- 99.Ingallinella P, Bianchi E, Ladwa NA, Wang YJ, Hrin R, Veneziano M, et al. (2009) Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc Natl Acad Sci USA;106: 5801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li CG, Tang W, Chi XJ, Dong ZM, Wang XX, Wang XJ (2013) A cholesterol tag at the N terminus of the relatively broad-spectrum fusion inhibitory peptide targets an earlier stage of fusion glycoprotein activation and increases the peptide’s antiviral potency in vivo. J Virol;87: 9223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Toth M, Hernandez-Barrantes S, Osenkowski P, Bernardo MM, Gervasi DC, Shimura Y, et al. (2002) Complex pattern of membrane type I matrix metalloproteinase shedding. J Biol Chem;277: 26340–50. [DOI] [PubMed] [Google Scholar]