Abstract
Background:
Advanced basal cell carcinomas (BCCs) suffer from a scarcity of effective treatment options. Previously, we found that the targetable histone methyltransferase EZH2 was upregulated in aggressive BCC subtypes, suggesting that epigenetics may play a role in BCC progression. The purpose of this study was to determine whether EZH2-associated proteins may be employed for the stratification of BCC histologic subtypes.
Methods:
Sixty-two specimens (from 61 patients), representing more or less aggressive BCC histologic subtypes and matching non-malignant epidermal cells, were included in this study. Immunohistochemistry of H3K27me3, 5hmC, NSD2, MOF and JARID1B was performed to assess their putative associations with BCC histologic subtypes, as well as with EZH2 and Ki67 expression levels.
Results:
We found that H3K27me3 and 5hmC upregulation was positively correlated with the occurrence of a less aggressive BCC histology. The modifications were also positively corelated with each other. Interestingly, we found that they were negatively correlated with the expression of EZH2, a marker for an aggressive BCC histology. The levels of NSD2, MOF, H3K27me3 and 5hmC were found to be universally upregulated in BCCs versus non-malignant epidermal cells.
Conclusions:
Our data reveal an EZH2-associated epigenetic marker profile that correlates with histologic signs of BCC aggressiveness. Our findings may have diagnostic and therapeutic implications, and indicate that epigenetic markers may be shared even with relatively less aggressive tumor types, thereby suggesting universal themes.
1. Introduction
Basal cell carcinoma (BCC) is diagnosed in 2.8–4 million USA citizens annually [1]. While usually considered “low-grade” and “easy” to treat, some BCC subtypes, such as locally advanced BCCs and basal cell nevus syndrome, can be difficult to control surgically [1]. For these tumors, hedgehog pathway inhibitors (vismodegib, sonidegib) constitute the only FDA-approved medical treatment option [1, 2]. However, these inhibitors are associated with only 15–58% response rates and, conversely, 14–30% serious adverse event rates [1, 2]. The identification of markers for an aggressive BCC histology could provide tools for predicting and assessing treatment responses, or even identifying actionable targets.
We recently identified immunohistochemical markers associated with an aggressive BCC histology, i.e., the proliferation-associated antigen Ki67 and the targetable histone methyltransferase EZH2 [1]. Here, we aimed to extend these observations by assessing whether additional EZH2-associated alterations may serve as BCC markers and/or provide additional pathophysiological and therapeutic insights. Using immunohistochemistry, we assessed the presence of H3K27me3 [3, 4], a highly-EZH2 correlated histone mark, the chromatin repressive complex binding partner JARID1B [5, 6], the EZH2-downstream methyltransferase NSD2/MMSET/WHSC1 [7], the acetyltransferase MOF [8] and the DNA hydroxymethyl mark 5hmC [9]. Our results revealed an epigenetic marker profile that correlates well with histologic signs of tumor aggressiveness.
2. Materials and methods
We conducted an IRB-approved (University of Michigan HUM #82579 and #40783) record review to identify 62 specimens from 61 patients with BCC, as well as matching non-malignant normal epidermal cells. Specifically, these include 60 BCC specimens from 59 patients in our tissue repository, which were included in separate analyses from our previous report [1], as well as 2 additional BCC specimens from 2 patients. The histologic subtypes were determined by a dermatopathologist (M.C.) and classified as more (morpheaform, infiltrative, micronodular) or less (nodular) aggressive. H3K27me3, 5hmC, NSD2, MOF and JARID1B immunohistochemical (IHC) stains were performed using an EnVision FLEX+ detection system (Dako) in conjunction with antibodies and conditions listed in Supplemental Table 1. Staining was scored by the weighted sum of intensity (0 = absent, 1 = moderate, 2 = strong) multiplied by the percentage of cells with each intensity (H-score, 0–2). For instance, for a given IHC stain in a single specimen with 10% tumor cells without staining, 60% tumor cells with moderate staining and 30% tumor cells with strong staining, the H-score = (0 × 10/100) + (1 × 60/100) + (2 × 30/100) = 1.2 [1]. Two-sided Student’s t-tests comparing BCC versus normal (paired samples) and more aggressive versus less aggressive (independent samples) were performed in R (v3.4.0). Pearson’s correlation was used to measure associations. P-values < 0.05 were considered statistically significant.
3. Results and discussion
Basal cell carcinomas (BCCs) are considered “less dangerous” malignancies because of the rarity of metastatic disease and the usually predictable form of local invasion. Therefore, research on the biology of BCC has been relatively limited. Yet, BCC is the most common malignancy in the developed world, which leads to a significant public health burden. In order to better understand the role of epigenetic regulation in driving BCC behavior, and building on our previously published findings [1], we set out to assess the presence of NSD2, MOF, H3K27me3, 5hmC and JARID1B via immunohistochemistry (IHC) in 32 cases with a less aggressive histologic subtype (nodular) and in 30 cases with more aggressive histologic subtypes (morpheaform, infiltrative, micronodular), as well as in normal epidermal cells. The H-scores for each IHC stain are listed in Table 1. NSD2, MOF, H3K27me3 and 5hmC were found to be preferentially present in BCC cells compared to normal epidermal cells (Table 1; Supplementary Fig. 1), whereas JARID1B was preferentially expressed in non-malignant epidermal cells compared to BCC cells (JARID1B: 0.82 ± 0.52 versus 0.71 ± 0.52, p = 0.007, Table 1). The levels of H3K27me3 and 5hmC were found to be positively correlated (r = 0.64, p < 0.0001), and preferentially enriched in less aggressive compared to more aggressive histological subtypes (Fig. 1) (H3K27me3: 1.82 ± 0.26 versus 1.41 ± 0.45, p < 0.0001; 5hmC: 1.53 ± 0.43 versus 1.03 ± 0.41, p < 0.0001, Table 1). These results are consistent with the reported loss of these epigenetic marks in other aggressive cancers [3, 10], suggesting that BCC serves as a model for studying the role of these modifications in regulating cancer cell behavior.
Table 1.
Expression by Histological Subtype.
| NSD2 | |||
| Data Summary | |||
| Specimen Type | n | mean | sd |
| Tumor More Aggressive | 32 | 0.838 | 0.461 |
| Tumor Less Aggressive | 30 | 0.740 | 0.425 |
| Tumor Overall | 62 | 0.790 | 0.443 |
| Tumor-Normal Paired | 59 | 0.776 | 0.424 |
| Normal Overall | 59 | 0.178 | 0.093 |
| Difference (Tumor-Normal) | 59 | 0.598 | 0.401 |
| Inference | |||
| Comparison | t | df | p |
| Tumor: More vs Less Aggr | 0.867 | 60 | 0.389 |
| Tumor vs Normal | 11.471 | 58 | 0.000 |
| H3K27me3 | |||
| Data Summary | |||
| Specimen Type | n | mean | sd |
| Tumor More Aggressive | 32 | 1.406 | 0.446 |
| Tumor Less Aggressive | 30 | 1.823 | 0.257 |
| Tumor Overall | 62 | 1.608 | 0.420 |
| Tumor-Normal Paired | 57 | 1.623 | 0.426 |
| Normal Overall | 57 | 1.363 | 0.446 |
| Difference (Tumor-Normal) | 57 | 0.260 | 0.365 |
| Inference | |||
| Comparison | t | df | p |
| Tumor: More vs Less Aggr | −4.549 | 50 | 0 |
| Tumor vs Normal | 5.372 | 56 | 0 |
| JARID1B | |||
| Data Summary | |||
| Specimen Type | n | mean | sd |
| Tumor More Aggressive | 32 | 0.762 | 0.588 |
| Tumor Less Aggressive | 30 | 0.650 | 0.435 |
| Tumor Overall | 62 | 0.708 | 0.518 |
| Tumor-Normal Paired | 58 | 0.678 | 0.500 |
| Normal Overall | 58 | 0.821 | 0.520 |
| Difference (Tumor-Normal) | 58 | −0.143 | 0.390 |
| Inference | |||
| Comparison | t | df | p |
| Tumor: Agg vs Cir | 0.860 | 57 | 0.393 |
| Tumor vs Normal | −2.796 | 57 | 0.007 |
| MOF | |||
| Data Summary | |||
| Specimen Type | n | mean | sd |
| Tumor More Aggressive | 32 | 1.419 | 0.521 |
| Tumor Circumscribed | 30 | 1.580 | 0.436 |
| Tumor Overall | 62 | 1.497 | 0.485 |
| Tumor-Normal Paired | 59 | 1.488 | 0.490 |
| Normal Overall | 59 | 1.175 | 0.472 |
| Difference (Tumor-Normal) | 59 | 0.314 | 0.410 |
| Inference | |||
| Comparison | t | df | p |
| Tumor: More vs Less Aggr | −1.324 | 59 | 0.191 |
| Tumor vs Normal | 5.881 | 58 | 0.000 |
| 5hmC | |||
| Data Summary | |||
| Specimen Type | n | mean | sd |
| Tumor More Aggressive | 32 | 1.031 | 0.405 |
| Tumor Circumscribed | 30 | 1.533 | 0.426 |
| Tumor Overall | 62 | 1.274 | 0.483 |
| Tumor-Normal Paired | 59 | 1.276 | 0.491 |
| Normal Overall | 59 | 0.793 | 0.433 |
| Difference (Tumor-Normal) | 59 | 0.483 | 0.448 |
| Inference | |||
| Comparison | t | df | p |
| Tumor: More vs Less Aggr | −4.748 | 59 | 0 |
| Tumor vs Normal | 8.281 | 58 | 0 |
The table presents sample sizes, means, and standard deviations for H-scores of five epigenetic marks. Also given are the t-statistics (t), degrees of freedom (df), and p-values (p) of Student’s t-tests that were used to compare mean expression between more and less aggressive subtypes and between tumor and matched non-malignant cells.
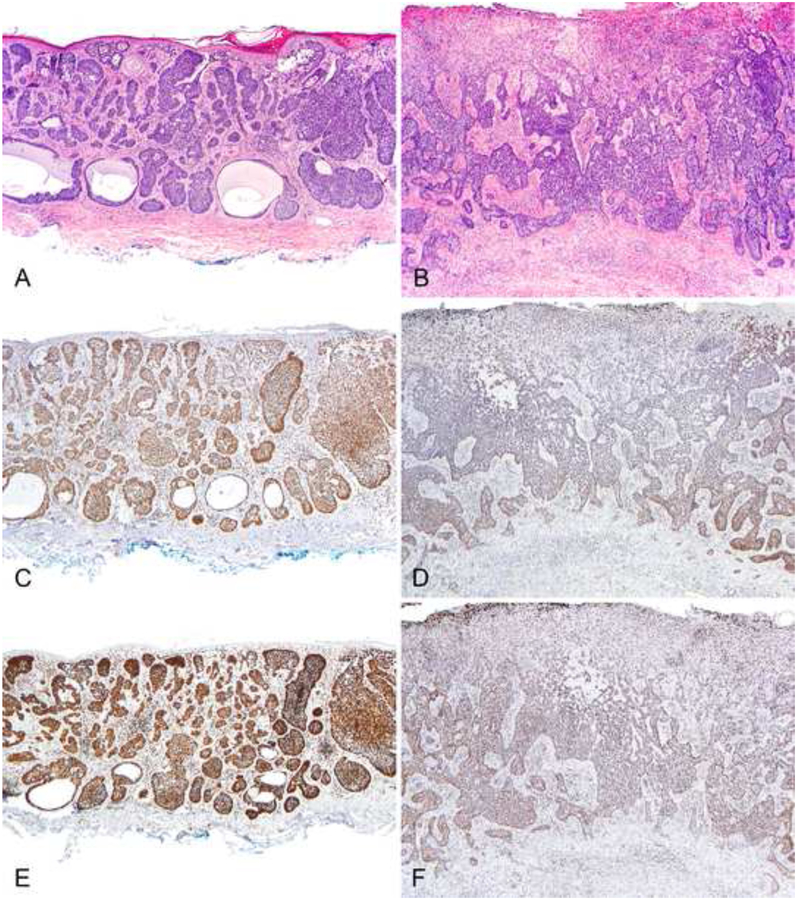
Figure 1. H3K27me3 and 5hmC staining in BCC.
A non-aggressive basal cell carcinoma (A, H&E) showing a strong 5hmC (C) and H3K27me3 (E) staining and an aggressive basal cell carcinoma (B, H&E) showing a weak 5hmC (D) and H3K27me3 staining (F). Original magnification: 40x.
We previously reported that the proliferation marker Ki67 and the histone methyltransferase EZH2 may serve as IHC markers for aggressive BCC histologic subtypes [1]. Since in this study the same BCC specimens were used, we wondered whether the presence of H3K27me3, 5hmC, NSD2, MOF and JARID1B might correlate with Ki67 and/or EZH2 expression. We found that Ki67 negatively correlated with 5hmC (r = −0.31, p = 0.016, Table 2) and that EZH2 negatively correlated with H3K27me3 (r = −0.29, p = 0.024) and 5hmC (r = −0.48, p < 0.0001). In contrast, we surprisingly found that EZH2 positively correlated with JARID1B (r = 0.39, p = 0.002) expression. JARID1B expression was lowest in the less aggressive BCCs, trending upward in both normal tissues at one hand, and more aggressive BCCs at the other hand. Taken together, these data reveal a dynamic epigenetic profile that links BCC tumor histology with proliferation and EZH2 status.
Table 2.
Correlations between Ki67, EZH2, and Epigenetic Marks
| CORRELATIONS | ||
| Ki67 | EZH2 | |
| Ki67 | 1.00 | 0.60 |
| EZH2 | 0.60 | 1.00 |
| NSD2 | 0.25 | 0.24 |
| H3K27me3 | −0.23 | −0.29 |
| JARID1B | 0.20 | 0.39 |
| MOF | −0.01 | −0.09 |
| 5hmC | −0.31 | −0.48 |
| PVALUES | ||
| Ki67 | EZH2 | |
| Ki67 | NA | 0.000 |
| EZH2 | 0.000 | NA |
| NSD2 | 0.055 | 0.063 |
| H3K27me3 | 0.076 | 0.024 |
| JARID1B | 0.126 | 0.002 |
| MOF | 0.942 | 0.489 |
| 5hmC | 0.016 | 0.000 |
The table presents Pearson’s correlations and associated p-values comparing Ki67 and EZH2, previously described aggressive BCC markers, with five epigenetic marks, using H-scores from analyzed aggressive and less aggressive BCCs (Table 1).
Other significant pairwise correlations (Supplementary Fig. 2) observed included: NSD2 and JARID1B (r = 0.37, p = 0.004), EZH2 and JARID1B (r = 0.39, p = 0.002), MOF and JARID1B (r = 0.33, p = 0.01) and MOF and 5hmC (r = 0.33, p = 0.003). EZH2 is an aggressive BCC marker, and while higher JARID1B expression trends towards aggressive versus less aggressive tumors (H-score: 0.762 versus 0.650), respectively, the difference was not found to be statistically significant (p = 0.39). Analysis of a larger number of BCC cases may reveal a statistically significant association between JARID1B expression and histological type, as has been observed in other tumors such as triple-negative breast cancer [11]. Our data suggest associations between certain histone methylases, demethylases, acetyltransferases and DNA hydroxymethyl marks. While EZH2 is known to associate with JARID1B in a complex linked to transcriptional repression [6], the other enzymes are known to catalyze unrelated modifications (i.e., none of them hydroxymethylates DNA) and, as yet, are not known to bind to each other. Further studies are required to determine whether the associations observed here bear causal and/or mechanistic significances.
The observation that EZH2 and H3K27me3 are negatively correlated in BCCs seems paradoxical since EZH2 catalyzes H3K27me3 formation [3, 4]. Such a pattern has, however, also been observed in other cancers and may relate to H3K27me3 dilution in highly proliferative cells of aggressive tumors [1, 3]. An alternative explanation may be that EZH2 has functions independent of histone H3 methylation [12]. Yet another possibility may be that an H3K27me3 demethylase, such as UTX or JMJD3, is differentially active in aggressive cancers, including BCCs [13]. Future studies should be aimed at tracing the presence of these demethylases in BCCs in order to account for this apparently paradoxical negative correlation, as well as at correlating findings in BCCs to other more aggressive tumor types.
The histological relationship between EZH2 and H3K27me3 observed here and in other cancers [3] highlights a caveat that is relevant to current trials that employ EZH2 inhibitors for the treatment of lymphomas and solid tumors [14]. Loss of H3K27me3 from cells in the epidermis has been proposed as a surrogate readout for pharmacologic EZH2 inhibition in clinical trials (http://www.epizyme.com/wp-content/uploads/2015/11/EZH2-Skin-McDonald-Final.pdf; http://www.epizyme.com/wp-content/uploads/2016/06/Ribrag-ASH-Lymphoma-final-06212016_2.pdf). While our findings do not support a causal link, our results raise the possibility that loss of H3K27me3 could be related to BCC progression. One may speculate that the absence of H3K27me3 may correlate with “poised” promoters related to proliferation and mesenchymal behavior. Further studies are needed to assess whether H3K27me3 loss by EZH2 inhibition increases the risk of BCC progression.
Increased NSD2 and MOF and decreased JARID1B expression levels in BCC cells versus non-malignant epidermal cells suggests that these histone modifying enzymes may regulate BCC tumorigenesis, again revealing similarities with other cancer types [5, 7, 8]. Similar to histone (de)methylases and acetyltransferases, these epigenetic BCC markers may represent therapeutic opportunities. The application of JARID1B and NSD2 inhibitors in animal models may reveal whether their antagonism promotes or inhibits BCC growth. JARID1B and NSD2 inhibitors are currently in clinical development for multiple myeloma and other cancers [5, 7, 15, 16]. Understanding their biology in the context of BCC may facilitate the optimization of their therapeutic potential. Our data suggest that BCC may serve as a model for studying fundamental biological processes that are shared among different cancers.
EZH2 has been found to methylate H3K27 upstream of NSD2 and MOF [4, 7, 8]. EZH2 and JARID1B are members of the same chromatin repressive complex and have both been found to be upregulated in prostate cancer [5–7]. Finally, EZH2 has been found to co-localize with 5hmC on chromatin, and the EZH2-containing polycomb repressive complex 2 has been found to recruit an enzyme responsible for 5hmC, i.e., Tet1 [9]. Our data support the notion that EZH2 occupies a central position within an epigenetic network intimately linked to BCC progression. Future studies are required to determine whether this network plays a causal role in BCC progression and/or predicts clinical responses to surgical and anti-hedgehog-based therapies, alone or in combination.
Supplementary Material
Acknowledgements
Supported in part by the National Institutes of Health (P30CA046592) VISORB clinical trial (ClinicalTrials.gov Identifier: NCT02436408; A.K. principal investigator) with support from Genentech, Inc. A.K. is supported by the Forbes Cancer Research Institute and the A. Alfred Taubman Medical Research Institute. A.K. is recipient of a Clinician-Scientist Award from Research to Prevent Blindness (RPB), and R.C.R. is a recipient of an RPB Career Development Award. R.C.R. is supported by Career Development Awards from the National Institutes of Health (K08EY026654) and the A. Alfred Taubman Medical Research Institute, where he is the Leslie and Abigail Wexner Emerging Scholar. R.C.R. received funding from the Leonard G. Miller Ophthalmic Research Fund at the Kellogg Eye Center, Grossman Research Fund, Barbara Dunn Research Fund, Roz Greenspoon Research Fund, Beatrice & Reymont Paul Foundation and March Hoops to Beat Blindness. The authors acknowledge generous support from an unrestricted research grant from RPB to the Department of Ophthalmology and Visual Sciences at the University of Michigan. None of the sponsors participated in the design and conduct of the study, collection, management, analysis and interpretation of the data, preparation, review or approval of the manuscript; and the decision to submit the manuscript for publication.
Footnotes
Conflict of interest: The authors report no other conflicts of interest.
R.C.R. and A.K. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
References (please include titles; journal issue numbers in bold)
- 1.Rao RC, Chan MP, Andrews CA and Kahana A, EZH2, proliferation rate, and aggressive tumor subtypes in cutaneous basal cell carcinoma. JAMA Oncol 2, 962–963 (2016) doi: 10.1001/jamaoncol.2016.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekulic A and Von Hoff D, Hedgehog pathway inhibition. Cell 164, 831 (2016) doi: 10.1016/j.cell.2016.02.021 [DOI] [PubMed] [Google Scholar]
- 3.Holm K, Grabau D, Lovgren K, Aradottir S, Gruvberger-Saal S, Howlin J, Saal LH, Ethier SP, Bendahl PO, Stal O, Malmstrom P, Ferno M, Ryden L, Hegardt C, Borg A and Ringner M, Global H3K27 trimethylation and EZH2 abundance in breast tumor subtypes. Mol Oncol 6, 494–506 (2012) doi: 10.1016/j.molonc.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD and Reinberg D, Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32, 503–518 (2008) doi: 10.1016/j.molcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP and Chen CD, JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci U S A 104, 19226–19231 (2007) doi: 10.1073/pnas.0700735104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banck MS, Li S, Nishio H, Wang C, Beutler AS and Walsh MJ, The ZNF217 oncogene is a candidate organizer of repressive histone modifiers. Epigenetics 4, 100–106 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asangani IA, Ateeq B, Cao Q, Dodson L, Pandhi M, Kunju LP, Mehra R, Lonigro RJ, Siddiqui J, Palanisamy N, Wu YM, Cao X, Kim JH, Zhao M, Qin ZS, Iyer MK, Maher CA, Kumar-Sinha C, Varambally S and Chinnaiyan AM, Characterization of the EZH2- MMSET histone methyltransferase regulatory axis in cancer. Mol Cell 49, 80–93 (2013) doi: 10.1016/j.molcel.2012.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, Sun H, Shu Y, Zou X, Zhao Y and Ge C, hMOF (human males absent on the first), an oncogenic protein of human oral tongue squamous cell carcinoma, targeting EZH2 (enhancer of zeste homolog 2). Cell Prolif 48, 436–442 (2015) doi: 10.1111/cpr.12177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neri F, Incarnato D, Krepelova A, Rapelli S, Pagnani A, Zecchina R, Parlato C and Oliviero S, Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells Genome Biol 14, R91 (2013) doi: 10.1186/gb-2013-14-8-r91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q and Pfeifer GP, 5-hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res 71, 7360–7365 (2011) doi: 10.1158/0008-5472.CAN-11-2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bamodu OA, Huang WC, Lee WH, Wu A, Wang LS, Hsiao M, Yeh CT and Chao TY, Aberrant KDM5B expression promotes aggressive breast cancer through MALAT1 overexpression and downregulation of hsa-miR-448. BMC Cancer 16, 160 (2016) doi: 10.1186/s12885-016-2108-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, Xu H, Cato L, Thornton JE, Gregory RI, Morrissey C, Vessella RL, Montironi R, Magi-Galluzzi C, Kantoff PW, Balk SP, Liu XS and Brown M, EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338, 1465–1469 (2012) doi: 10.1126/science.1227604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swigut T and Wysocka J, H3K27 demethylases, at long last. Cell 131, 29–32 (2007) doi: 10.1016/j.cell.2007.09.026 [DOI] [PubMed] [Google Scholar]
- 14.Kim KH and Roberts CW, Targeting EZH2 in cancer. Nat Med 22, 128–134 (2016) doi: 10.1038/nm.4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.di Luccio E, Inhibition of nuclear receptor binding SET domain 2/multiple myeloma SET domain by LEM-06 implication for epigenetic cancer therapies. J Cancer Prev 20, 113–120 (2015) doi: 10.15430/JCP.2015.20.2.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tumber A, Nuzzi A, Hookway ES, Hatch SB, Velupillai S, Johansson C, Kawamura A, Savitsky P, Yapp C, Szykowska A, Wu N, Bountra C, Strain-Damerell C, Burgess-Brown NA, Ruda GF, Fedorov O, Munro S, England KS, Nowak RP, Schofield CJ, La Thangue NB, Pawlyn C, Davies F, Morgan G, Athanasou N, Muller S, Oppermann U and Brennan PE, Potent and selective KDM5 inhibitor stops cellular demethylation of H3K4me3 at transcription start sites and proliferation of MM1S myeloma cells. Cell Chem Biol 24, 371–380 (2017) doi: 10.1016/j.chembiol.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.