Abstract
Purpose
Liver fibrosis is a major cause of morbidity and mortality and the outcome of various chronic liver diseases. Activation of hepatic stellate cells (HSCs) is the key event in liver fibrosis. Studies have confirmed that miR-140-3p plays a potential regulatory effect on HSC activation. However, whether miR-140-3p mediates the liver fibrosis remains unknown.
Materials and Methods
Expression of miR-140-3p was detected by real-time quantitative PCR (qPCR). Cell proliferation was measured by MTT, while cell apoptosis rate was determined via flow cytometry. Western blot assay was used to detect the expression of cleaved PARP. The fibrogenic effect was evaluated by expression of α-smooth muscle actin and desmin. Functional experiments were performed in transforming growth factor β1 (TGF-β1)-induced HSC-T6 cells with transfection of anti-miR-140-3p and/or siPTEN. Target binding between miR-140-3p and PTEN was predicted by the TargetScan database and identified using luciferase reporter assay and RNA immunoprecipitation.
Results
TGF-β1 induced the activation of HSC-T6 cells, and miR-140-3p expression varied according to HSC-T6 cell activation status. Knockdown of miR-140-3p reduced cell proliferation and the expressions of α-SMA and desmin, as well as increased apoptosis, in TGF-β1-induced HSC-T6 cells, which could be blocked by PTEN silencing. Additionally, inactivation of the AKT/mTOR signaling pathway stimulated by miR-140-3p knockdown was abolished when silencing PTEN expression. PTEN was negatively regulated by miR-140-3p via direct binding in HSC-T6 cells.
Conclusion
miR-140-3p is an important mediator in HSC-T6 cell activation, and miR-140-3p knockdown suppresses cell proliferation and fibrogenesis in TGF-β1-induced HSC-T6 cells, indicating that miR-140-3p may be a potential novel molecular target for liver fibrosis.
Keywords: miR-140-3p, PTEN, liver fibrosis, TGF-β1, hepatic stellate cells (HSCs)
INTRODUCTION
Liver fibrosis is a chronic wound-healing response represented by excessive extracellular matrix (ECM) deposition that occurs in most types of chronic liver diseases,1,2 including hepatocellular carcinoma and hepatitis B/C. Activation of hepatic stellate cells (HSCs), the major mesenchymal cells in the liver, are the critical cellular basis for liver fibrogenesis:3,4 activated HSCs are responsible for excess synthesis and deposition of ECM following a fibrotic stimulus.5,6 A variety of cytokines and growth factors induced in liver injury can activate HSCs to produce α-smooth muscle actin (α-SMA) and cause liver fibrosis,7,8 later leading to cirrhosis. Transforming growth factor-β1 (TGF-β1) is considered the main pro-fibrogenic mediator of the transdifferentiation of fibroblasts to myofibroblast-like cells.9 Despite fundamental advances in understanding of the pathophysiology of liver fibrosis, detail mechanisms of liver fibrosis remain elusive. Therefore, a comprehensive understanding of the molecular mechanisms involved in HSC activation and fibrogenesis will be helpful to providing novel therapeutic targets with which to inhibit, even mitigate, this devastating progression.
There has been an increasing number of studies on microRNAs (miRNAs) as key regulators in the development of liver fibrosis.10 miRNAs are endogenous, small, noncoding RNAs ranging in size from 20 to 25 nt, and deregulation of miRNAs affect various intracellular functional complexes,11 such as cell proliferation, apoptosis, and differentiation; ECM; cytoskeleton; tyrosine kinase; and G-protein signaling. With the use of in vitro cell activation and miRNA microarray hybridization, many differentially expressed miRNAs, among which miR-140 was upregulated, were identified in rat HSCs during activation.12 Serum miR-138 and miR-140 were highly detected in early fibrosis and late fibrosis, compared to healthy patients.13 Moreover, increasing expression thereof during the development of fibrosis of the liver and progressive liver fibrosis have been posited in the late stages of various chronic liver diseases. Research has demonstrated that miR-140-3p has a pro-fibrotic effect in the mammary glands14 and is deeply involved in liver disorders,12,15,16 including hepatic impact injury, non-alcoholic fatty liver disease, and hepatocellular carcinoma. Thus, we planned to investigate the role of miR-140-3p in HSC activation and its molecular signaling pathway.
Phosphatase and tensin homolog deleted on chromosome 10 (PTEN), a tumor suppressor, is a dual phosphatase, and its major function is to dephosphorylate phosphatidylinositol 3, 4, 5-triphosphate (PIP3) to phosphatidylinositol 4, 5-bisphosphate (PIP2), antagonizing PI3K/AKT signaling.17 Alteration of PTEN expression and activity has been recognized as being pervasive among tumor cells.18 Accumulating evidence has indicated that PTEN is dysregulated in liver diseases19,20 and has demonstrated decreased PTEN expression in fibrotic diseases of the lungs, kidneys, and skin.21,22,23 PTEN activity and expression are controlled by several mechanisms, including phosphorylation, acetylation, oxidation, ubiquitination, noncoding RNAs, and DNA methylation.24 Here, we sought to determine the role of miR-140-3p in HSC activation through PTEN. In this work, we studied the fibrogenic role of miR-140-3p in rat hepatic stellate HSC-T6 cells and its downstream regulation.
MATERIALS AND METHODS
Cells and cell culture
This study was approved by the Institutional Review Board of Puai Hospital, Tongji Medical College, Huazhong University of Science and Technology. The HSC-T6 cell line was obtained from the Cell Bank of Type Culture Collection (Chinese Academy of Sciences, Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (DMEM, Hyclone, Logan, UT, USA), 10% (v/v) fetal bovine serum (FBS, Hyclone), 100 units/mL of penicillin, and 100 µg/mL of streptomycin in a humidified atmosphere containing 5% (v/v) CO2 at 37℃.
Cell medium was refreshed every other day, and cells grown to subconfluence were pretreated with serum-free DMEM for 16 h. Then, cells were incubated with DMEM supplemented with platelet derived growth factor (PDGF)-BB (GF310; Merck; MO, USA) and TGF-β1 (T7039; Merck) for 48 h.
Cell transfection
In six-well plates (Corning, NY, USA), 10 ng/mL of TGF-β1 treated HSC-T6 cells were seeded at a density of 2×105 cells per well 24 h prior to the transfection. siRNA against PTEN (siPTEN)/scramble, pre-miR-140-3p/NC, and anti-miR-140-3p/NC were provided by GenePharma (Shanghai, China). Oligonucleotides were transfected into cells at a final concentration of 100 nM using Lipofectamine 2000 (Invitrogen, Shanghai, China) according to the manufacturer's instructions. Samples were collected after 48 h of transfection for further studies, such as RNA isolation and protein extraction.
Cell proliferation assay
Cell proliferation assay was determined by standard 3-(4, 5-dimethylthiazol-2-yl)-2, 4-diphenyl-tetrazolium bromide (MTT, Sigma-Aldrich, Louis, MO, USA) assay. After transfection, cells were implanted at a density of 2×104 cells per well in 96-well plates (Corning) for 24 h. Briefly, 5 mg/mL of MTT (Sigma-Aldrich) was added and incubated at 37℃ for another 4 h; thereafter, the medium was replaced and the formazan crystals were dissolved in 150 µL of dimethyl sulphoxide (DMSO, Dingguo, Beijing, China). The optical density was determined with a Thermomax microplate reader (Bio-Tek EL, VT, USA) at 490 nm wavelength. All experiments were performed in triplicate.
Cell apoptosis assay
Cell apoptosis assay was performed on flow cytometry using Cell Cycle and Apoptosis Analysis Kits (Beyotime, Shanghai, China). In brief, cells were collected and washed with cold phosphate buffer solution (PBS) three times after transfection. Then, cells were fixed with 70% ethanol, stained with 0.5 mL of propidium iodide (PI) staining buffer, which contained 200 mg/mL of RNase A and 50 µg/mL of PI, at 37℃ for 30 min in the dark. Analyses were performed on a BD LSR flow cytometer (BD Biosciences, Jiangsu, China), and apoptotic cells were calculated. The experiments were repeated three times.
Western blot assay
TGF-β1-treated HSC-T6 cells with translation or not were lysed in RIPA (Beyotime), and the protein concentration was determined by Bradford. 20 µg of sample were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). Prior to incubation of primary antibodies overnight at 4℃ with gentle shaking, the blots were blocked in 5% of nonfat milk. GAPDH on the same membrane was the loading control. After incubation of horseradish peroxidase conjugated secondary antibodies correspondingly, proteins were visualized with ECL Plus (Solarbio, Beijing, China), and ImageJ (National Institutes of Health, Bethesda, MD, USA) was utilized to analyze the gray intensity of the bands. Primary antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA) and Abcam (Cambridge, UK): α-SMA (#14968), COL1A1 (#84336), PCNA (#13110), cleaved caspase 3 (#9664), caspase 3 (#9665), p-AKT (#2965), p-mTOR (#2971), AKT (#9272), mTOR (#2972), GAPDH (#97166), and fibronectin (#6328).
Real-time quantitative PCR
Total RNA from cultured cells was isolated using TRIzol reagent (Thermo, Waltham, MA, USA) following the protocol, and 300 ng of total RNA was reverse transcribed to cDNA using Bestar QPCR RT Kit (DBI Bioscience, Ludwigshafen, Germany). Real-time quantitative PCR (qPCR) was performed with Bestar SYBR Green qPCR Master Mix (DBI Bioscience) on ABI 7500. The expression of miR-140-3p was calculated according to the comparative threshold cycle value (2−ΔΔCt) method, compared with U6 small nuclear RNA (U6). PCR primers were as follows: miR-140-3p, 5′-TCGGCAGGTAACACTG TCTGGT-3′ (sense) and 5′-CTCAACTGGTGTCGTGGA-3′ (antisense); U6, 5′-CTCGCTTCGGCAGCACA-3′ (sense) and 5′-AACGCTTCACGAATTTGCGT-3′ (antisense). All experiments were performed at least in triplicate.
Luciferase reporter assay
The potential binding sites of miR-140-3p on PTEN 3′UTR were mutated and cloned by PCR into plasmid pGL3-basic. Cells were plated in a 24-well plate (Corning) at 1×104 cells/well, followed by co-transfection with 20 nM of miR-140-3p mimic/NC and 20 ng of either PTEN wt/mut 3′UTR in TGF-β1 treated HSC-T6 cells for 48 h. Cells were collected to measure the relative luciferase using the Dual-luciferase Reporter Assay System (Promega, Madison, WI, USA). The ratio of Firefly to Renilla luciferase activity was used as the relative luciferase activity, and all experiments were carried out in triplicate.
RNA immunoprecipitation
TGF-β1-treated HSC-T6 cells were transfected with miR-140-3p/NC. RNA immunoprecipitation (RIP) was performed with cell extract, and Magna RIP™ RNA-binding protein immunoprecipitation kit (Millipore) was chosen to detect expression of PTEN from the samples bound to the Ago2 antibody or IgG. All procedures followed the standard protocol. Total RNA isolation was extracted from RIP-Ago2/IgG immunoprecipitation using TRIzol (Thermo). All operations were performed in triplicate.
Transwell assay
For migration assay, HSC-T6 cells with different processing were supplemented in 200 µL of serum-free medium and plated in the upper chamber with a non-coated membrane (Corning). The medium containing 10% FBS (Gibco) was used as a chemo-attractant and loaded in the lower chamber. The transwell system was maintained at 37℃ for 48 h. The migratory cells into the lower chambers were stained with 0.5% crystal violet and quantitated under a light microscope (DM2500M, Leica, Wetzlar, Germany) separately, and the ability of migration/invasion was assessed as the percentage of migratory/invasive cells compared with control group.
Statistical analysis
All statistical analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). Values given represent mean±SEM, and the differences between two groups were determined by two-tailed Student's t-test. p<0.05 was considered to be statistically different.
RESULTS
miR-140-3p is highly expressed upon HSC-T6 cell activation
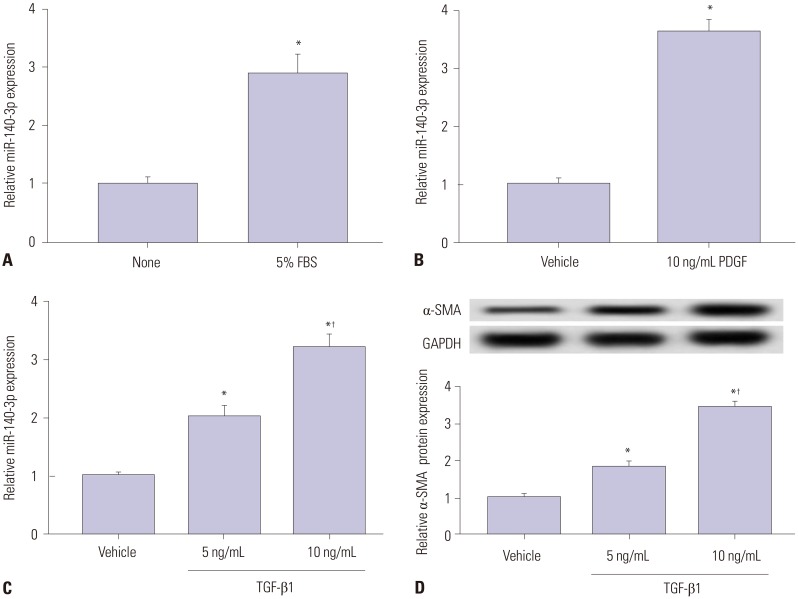
First and foremost, the expression of miR-140-5p was measured in activated HSC-T6 cells and compared with that in quiescent cells. 5% FBS, PDGF-BB (PDGF), and TGF-β1 were selected to stimulate cells independently. As was shown in Fig. 1A–C, miR-140-3p expression was upregulated over threefold in 5% FBS-, 10 ng/mL of PDGF-, and 10 ng/mL of TGF-β1-incubated HSC-T6 cells. Additionally, the levels of α-SMA, a marker of HSC activation, in TGF-β1-treated cells and relative α-SMA protein expression were elevated (Fig. 1D). To explore the role of miR-140-3p in HSC activation, HSC-T6 cells were exposed to 10 ng/mL of TGF-β1 for further experiments. This data indicated that miR-140-3p is highly expressed in activated HSCs.
Fig. 1. miR-140-3p expression according to HSC-T6 cell activation status. Expression of miR-140-5p was measured in HSC-T6 cells by qPCR. (A) Cells were incubated with 5% FBS for 48 h. The control was FBS-free cells. (B) Cells were treated with 10 ng/mL of PDGF-BB (PDGF) for 48 h. The control was no treatment cells. (C) Cells were exposed to TGF-β1 for 48 h. (D) Levels of the marker of hepatic stellate cell activation, α-SMA in TGF-β1 treated cells were detected using western blot assay. The control was no treatment cells. All experiments were performed in triplicate, and *p<0.05, †p<0.01. PDGF, platelet derived growth factor; α-SMA, α-smooth muscle actin; TGF-β1, transforming growth factor β1.
miR-140-3p knockdown reduces cell fibrosis in HSC-T6 cells
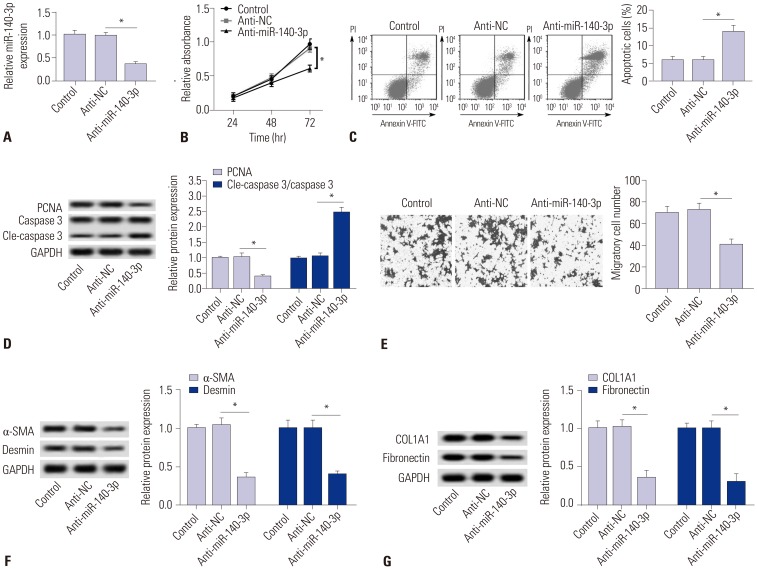
The role of miR-140-3p knockdown in cell fibrosis were measured in TGF-β1-induced HSC-T6 cells. miR-140-3p knockdown was obtained after transfection with anti-miR-140-3p (Fig. 2A). Cell proliferation was impaired by anti-miR-140-3p (Fig. 2B), and apoptosis rate and level of cleaved caspase 3 were higher in miR-140-3p knockdown cells (Fig. 2C and D); meanwhile, expression of the proliferation marker proliferating cell nuclear antigen (PCNA) was opposite to that of cleaved caspase 3. Additionally, migratory cells were decreased (Fig. 2E). Moreover, we observed that anti-miR-140-3p induced lower expression levels of markers of fibrosis α-SMA, desmin, COL1A1, and fibronectin (Fig. 2F and 2G). These results illuminated a certain cytotoxicity and fibrosis suppressor role of miR-140-3p knockdown in TGF-β1-induced HSC-T6 cells.
Fig. 2. miR-140-3p knockdown reduces cell fibrosis in HSC-T6 cells. miR-140-3p knockdown was obtained after transfection of anti-miR-140-3p into TGF-β1-induced HSC-T6 cells. (A) Relative expression levels of miR-140-3p. (B) Cell proliferation was measured with cell proliferation assay. (C) Cell apoptosis was measured on flow cytometry, and apoptosis rate was calculated. (D) Expressions of PCNA, caspase 3, and cleaved caspase 3 (cle-caspase 3) were detected using Western blot assay, and the gray intensity was analyzed. (E) Cell migration was determined using transwell assay (0.5% crystal violet, ×100). (F and G) Expressions of α-SMA, desmin, COL1A1, and fibronectin were detected using Western blot assay, and the gray intensity was analyzed. All experiments were compared with cells transfected of anti-NC and performed in triplicate, *p<0.05. TGF-β1, transforming growth factor β1; α-SMA, α-smooth muscle actin; anti-miR-140-3p, miR-140-3p inhibitor; PCNA, proliferating cell nuclear antigen; COL1A1, collagen type 1A1.
PTEN is negatively regulated by miR-140-3p target binding in HSC-T6 cells
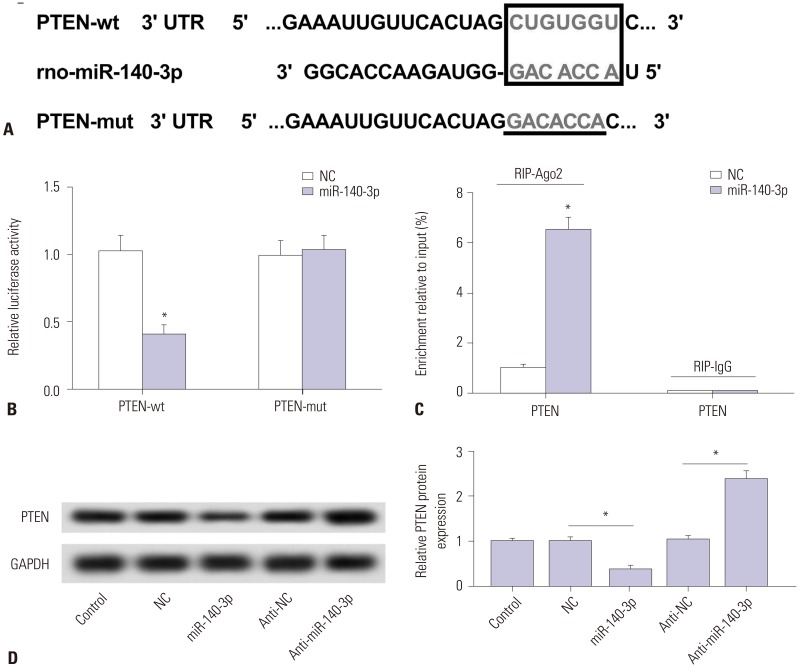
Potential target sites were predicted on TargetScan software (http://www.targetscan.org/) (Fig. 3A). The site on PTEN wide type (wt) 3′UTR was mutated (mut), and full-length PTEN-wt/mut 3′UTR was cloned into pGL3 vector. Luciferase reporter assay (Fig. 3B) and RIP (Fig. 3C) were performed to identify target binding in TGF-β1 treated HSC-T6 cells. As shown, luciferase activity relatively declined after co-transfection with PTEN-wt 3′UTR and miR-140-3p mimic, compared with cotransfected with PTEN-mut 3′UTR and miR-140-3p mimic. Besides, dramatically high expression of PTEN was obtained from Ago2 immunoprecipitation. We also observed that expression of PTEN protein was upregulated by anti-miR-140-3p and downregulated by miR-140-3p mimic (Fig. 3D). These outcomes indicated that PTEN was a downstream target gene of miR-140-3p in HSC-T6 cells.
Fig. 3. PTEN is negatively regulated by miR-140-3p target binding in HSC-T6 cells. (A) Potential target sites (red) on PTEN 3′UTR were predicted on TargetScan software and mutated. (B) Luciferase reporter assay was conducted to identify the binding sites. Relative luciferase activity was significantly reduced in cells transfected with PTEN wild type (wt) 3′UTR and miR-140-3p mimic (miR-140-3p). (C) RIP were performed. Dramatically high expression of PTEN were obtained from Ago2, separately. (D) Effect of miR-140-3p on expression of PTEN in cells. Relative expression levels of PTEN were evaluated using Western blotting, and the gray intensity was analyzed. All experiments were performed in triplicate, *p<0.05. RIP, RNA immunoprecipitation; PTEN, phosphatase and tensin homolog deleted on chromosome.
PTEN silencing blocks the effect of miR-140-3p knockdown in HSC-T6 cells
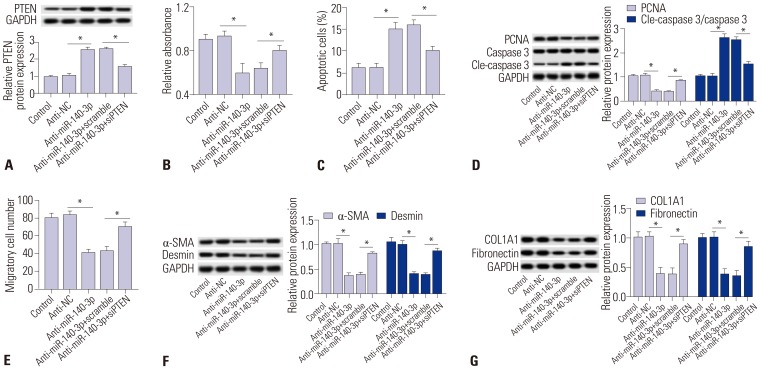
Further, the effects of PTEN silencing on miR-140-3p knockdown-induced inhibition of cell proliferation and fibrosis were measured in TGF-β1-induced HSC-T6 cells. Silencing expression of PTEN was obtained after transfection of specific siRNA against PTEN (siPTEN). Expression levels of PTEN were upregulated in cells transfected with anti-miR-140-3p and then reduced with siPTEN incubation (Fig. 4A). Cell proliferation was impaired by anti-miR-140-3p, which was improved by siPTEN (Fig. 4B). Apoptosis rate and levels of cleaved caspase 3 were higher in miR-140-3p knockdown cells, and were subsequently blocked with siPTEN incubation (Fig. 4C and 4D). PCNA expression was opposite that of cleaved caspase 3. In addition, the number of migratory cells declined (Fig. 4E). Next, we found that anti-miR-140-3p-induced lower expression levels of α-SMA, desmin, COL1A1, and fibronectin were rescued with treatment of siPTEN (Fig. 4F and G). These results showed that PTEN silencing blocks the inhibitory effects of miR-140-3p knockdown on cell proliferation and fibrosis.
Fig. 4. PTEN silencing blocks the effect of miR-140-3p knockdown in activated HSC-T6 cells. Silencing expression of PTEN was obtained after transfection with specific siRNA against PTEN (siPTEN) into TGF-β1-induced HSC-T6 cells. Effects of siPTEN on anti-miR-140-3p action were measured. (A) Expression levels of PTEN in cells transfected with anti-miR-140-3p and siPTEN or not. (B and C) Cell proliferation and apoptosis was analyzed in cells transfected with anti-miR-140-3p and siPTEN or not. (D) Expressions of PCNA, caspase 3, and cle-caspase 3 were analyzed in cells transfected with anti-miR-140-3p and siPTEN or not, and the gray intensity was calculated. (E) Cell migration was determined using transwell assay after transfection with anti-miR-140-3p and siPTEN or not. (F and G) Expressions of α-SMA, desmin, COL1A1, and fibronectin were analyzed in cells transfected with anti-miR-140-3p and siPTEN or not, and the gray intensity was calculated. All experiments were performed in triplicate, *p<0.05. TGF-β1, transforming growth factor β1; PTEN, phosphatase and tensin homolog deleted on chromosome 10; PCNA, proliferating cell nuclear antigen; α-SMA, α-smooth muscle actin; COL1A1, collagen type 1A1.
miR-140-3p knockdown inactivates AKT/mTOR signaling, which is abolished by PTEN silencing
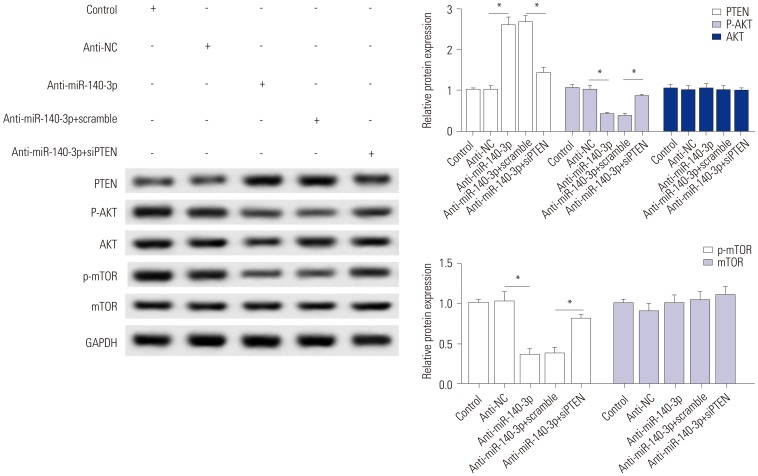
While we discovered that PTEN silencing blocks the effect of miR-140-3p knockdown in TGF-β1-induced HSC-T6 cells, the associated signaling pathway remains unknown. The effects of siPTEN on anti-miR-140-3p-inactivated signaling pathway were measured, and the expressions of p-AKT, p-mTOR, and PTEN were detected using Western blot assay (Fig. 5). As shown, anti-miR-140-3p significantly reduced the p-AKT and p-mTOR, while the expressions of total AKT and mTOR were not affected. Co-expression of anti-miR-140-3p and siPTEN improved p-AKT and p-mTOR expression. These data indicated that miR-140-3p knockdown inhibits AKT/mTOR signaling in HSC-T6 cells, which could be blocked by PTEN silencing.
Fig. 5. miR-140-3p knockdown inactivates the AKT/mTOR signaling pathway, which is abolished by PTEN silencing. Effects of siPTEN on anti-miR-140-3p-inactivated signaling pathway were measured. Expressions of p-AKT and p-mTOR and PTEN were detected using Western blot assay, and the gray intensity was calculated. All experiments were performed in triplicate. *p<0.05. PTEN, phosphatase and tensin homolog deleted on chromosome.
DISCUSSION
In this study, we discovered that miR-140-3p is highly expressed in TGF-β1-induced HSC-T6 cells, indicating the regulatory role of miR-140-3p in HSCs activation and fibrogenesis. Knockdown of miR-140-3p reduced cell proliferation and increased cell apoptosis and fibrosis through upregulating its target PTEN, thus inhibiting p-AKT and p-mTOR. Our work suggests a novel pro-fibrotic factor, miR-140-3p, that affects PTEN expression and PTEN-mediated AKT/mTOR signaling in rat HSCs.
miRNAs have become promising targets for liver fibrosis therapy. Targeting specific molecules to combat liver fibrosis is a growing challenge in translational medicine. Though significant progress has been made to elucidate the physiopathology related to liver fibrosis, there is still a lack of effectively established drugs for liver fibrosis prognosis and treatment. Also, with the central role of HSCs activation in the development of liver fibrosis,4,25 suppression of its activation is the most promising therapeutic approach. Inhibition of cytokine-mediated ECM synthesis26 and promotion of HSCs apoptosis27 are the predominant strategies of liver fibrosis. In this study, we found that a noncoding RNA, namely miR-140-3p, universally affected TGF-β1-induced HSC-T6 cell proliferation, apoptosis, and activation. In other research, the miR-29 family exhibited dramatic decreases in the two models of liver fibrosis, compared with nonfibrotic liver, as well as in patients with liver fibrosis, through inducing apoptosis of HSCs and reducing the accumulation of ECM.28 The miR-34 family29 has also been found to play promotion roles in liver fibrosis by target binding to acyl-CoA synthetase long-chain family member 1 (ACSL1) and PPARγ to deposit ECM proteins and metabolize fatty acids in HSCs: the accumulation of vitamin A-containing lipid droplets is characteristic of quiescent HSCs.
miR-140-3p has a pro-fibrosis effect. It has been demonstrated that miR-140-3p has a pro-fibrotic effect in the mammary gland14 and is deeply involved in liver disorders,13,15,16 including hepatic impact injury, non-alcoholic fatty liver disease, and hepatocellular carcinoma. Progressive liver fibrosis is observed in the late stages of various chronic liver diseases, and miR-140 has been predicted to be upregulated in rat HSCs during activation.12 Additionally, serum miR-138 and miR-140 have been highly detected in early fibrosis and late fibrosis, compared to the healthy patients, with increasing expression in late stages of liver fibrosis.13 Thus, we planned to investigate the role of miR-140-3p in HSC activation and its molecular signaling pathway. Herein, we hypothesized and verified the pro-fibrotic properties of miR-140-3p in TGF-β1-induced HSC-T6 cells. Because our results were from in vitro experiments depending on rat immortalized HSC line, primary HSCs30 and rat liver fibrosis models31 should be established to further identify the pro-fibrosis effect of miR-140-3p. Additionally, more investigations are desperately needed to provide evidence of the pro-fibrosis effect of miR-140-3p in fibrotic diseases involving the lungs, heart, and skin.
Decreased PTEN controlled by miRNAs in the liver is a mechanism of fibrogenesis. PI3K/AKT pathway is believed to regulate liver fibrosis,32,33 Its upstream natural inhibitor PTEN exhibits a tumor suppressor role and is dysregulated in liver diseases.19,34 PTEN has recently been identified as a target/biomarker for pharmacological therapy of liver fibrosis,35 and approaches to decrease PTEN expression would be promising strategies in liver injury.34,36 A growing number of miRNAs have been found to directly/indirectly target PTEN in fibrogenesis: miR-21 mediates PDGF-BB-induced LX-2 cell activation,37 and its target, PTEN-mediated PI3K/AKT pathway, mediates the fibrogenic effects of miR-21. Adiponectin-induced upregulation of miR-29b34 can suppress transcription of the DNA methylation protein DNMT3B in LX-2 cells, thus resulting in reduced methylation of PTEN CpG islands and ultimately suppressing the PI3K/AKT pathway. P53 and PTEN deficiency has been shown to be associated with chronic hepatitis B-induced liver fibrosis.36 PTEN regulates ECM protein expression, including collagen metabolism and α-SMA, as well as hepatic macrophage activation and function, in progression and reversal of liver fibrosis. In this study, miR-140-3p-stimulated PTEN silencing improved cell proliferation and expression of α-SMA and desmin, accompanied by less apoptosis, through facilitating p-AKT and p-mTOR levels.38
Recently published studies have clarified the role of miRNAs in liver fibrosis and miRNAs, and not surprisingly, both play an anti-fibrogenic and a pro-fibrogenic role, depending on the genes targeted and the nature of the stimulus. Here, we showed that miR-140-3p is upregulated according to HSC-T6 cell activation status. Moreover, miR-140-3p knockdown increases PTEN expression, while miR-140-3p overexpression decreases PTEN expression. PTEN silencing could partly block, even reverse, the effects of miR-140-3p knockdown on anti-proliferation, pro-apoptosis, and anti-fibrosis. Our data suggest the presence of a novel miR-140-3p/PTEN/AKT/mTOR signaling pathway underlying the pathogenesis of HSC activation and fibrogenesis.
ACKNOWLEDGEMENTS
This work was supported by the Applied Basic Research Program of Wuhan Municipal Bureau of Science and Technology (Grant No. NO 2017060201010154).
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Shi-Min Wu.
- Data curation: Tian-Hong Li.
- Formal analysis: Shi-Min Wu, Tian-Hong Li.
- Funding acquisition: Hao Yun.
- Investigation: Shi-Min Wu, Tian-Hong Li.
- Methodology: Hao Yun.
- Project administration: Shi-Min Wu.
- Resources: Hao Yun.
- Software: Hong-Wu Ai.
- Supervision: Tian-Hong Li.
- Validation: Hong-Wu Ai.
- Writing—original draft: Ke-Hui Zhang.
- Writing—review & editing: Shi-Min Wu, Tian-Hong Li.
References
- 1.Kocabayoglu P, Friedman SL. Cellular basis of hepatic fibrosis and its role in inflammation and cancer. Front Biosci (Schol Ed) 2013;5:217–230. doi: 10.2741/s368. [DOI] [PubMed] [Google Scholar]
- 2.Arriazu E, Ruiz de Galarreta M, Cubero FJ, Varela-Rey M, Pérez de Obanos MP, Leung TM, et al. Extracellular matrix and liver disease. Antioxid Redox Signal. 2014;21:1078–1097. doi: 10.1089/ars.2013.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee UE, Friedman SL. Mechanisms of hepatic fibrogenesis. Best Pract Res Clin Gastroenterol. 2011;25:195–206. doi: 10.1016/j.bpg.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mimche PN, Lee CM, Mimche SM, Thapa M, Grakoui A, Henkemeyer M, et al. EphB2 receptor tyrosine kinase promotes hepatic fibrogenesis in mice via activation of hepatic stellate cells. Sci Rep. 2018;8:2532. doi: 10.1038/s41598-018-20926-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dooley S, ten Dijke P. TGF-β in progression of liver disease. Cell Tissue Res. 2012;347:245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J, Luo Z, Pan Y, Zheng W, Li W, Zhang Z, et al. H19/miR-148a/USP4 axis facilitates liver fibrosis by enhancing TGF-β signaling in both hepatic stellate cells and hepatocytes. J Cell Physiol. 2018 Oct 26; doi: 10.1002/jcp.27656. [Epub] [DOI] [PubMed] [Google Scholar]
- 7.Preisser L, Miot C, Le Guillou-Guillemette H, Beaumont E, Foucher ED, Garo E, et al. IL-34 and macrophage colony-stimulating factor are overexpressed in hepatitis C virus fibrosis and induce profibrotic macrophages that promote collagen synthesis by hepatic stellate cells. Hepatology. 2014;60:1879–1890. doi: 10.1002/hep.27328. [DOI] [PubMed] [Google Scholar]
- 8.Breitkopf-Heinlein K, Meyer C, König C, Gaitantzi H, Addante A, Thomas M, et al. BMP-9 interferes with liver regeneration and promotes liver fibrosis. Gut. 2017;66:939–954. doi: 10.1136/gutjnl-2016-313314. [DOI] [PubMed] [Google Scholar]
- 9.Gressner AM, Weiskirchen R. Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF-beta as major players and therapeutic targets. J Cell Mol Med. 2006;10:76–99. doi: 10.1111/j.1582-4934.2006.tb00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambrecht J, Mannaerts I, van Grunsven LA. The role of miRNAs in stress-responsive hepatic stellate cells during liver fibrosis. Front Physiol. 2015;6:209. doi: 10.3389/fphys.2015.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar V, Mahato RI. Delivery and targeting of miRNAs for treating liver fibrosis. Pharm Res. 2015;32:341–361. doi: 10.1007/s11095-014-1497-x. [DOI] [PubMed] [Google Scholar]
- 12.Guo CJ, Pan Q, Cheng T, Jiang B, Chen GY, Li DG. Changes in microRNAs associated with hepatic stellate cell activation status identify signaling pathways. FEBS J. 2009;276:5163–5176. doi: 10.1111/j.1742-4658.2009.07213.x. [DOI] [PubMed] [Google Scholar]
- 13.El-Ahwany E, Nagy F, Zoheiry M, Shemis M, Nosseir M, Taleb HA, et al. Circulating miRNAs as predictor markers for activation of hepatic stellate cells and progression of HCV-induced liver fibrosis. Electron Physician. 2016;8:1804–1810. doi: 10.19082/1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfson B, Zhang Y, Gernapudi R, Duru N, Yao Y, Lo PK, et al. A high-fat diet promotes mammary gland myofibroblast differentiation through microRNA 140 downregulation. Mol Cell Biol. 2017;37:e00461-16. doi: 10.1128/MCB.00461-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakasugi H, Takahashi H, Niinuma T, Kitajima H, Oikawa R, Matsumoto N, et al. Dysregulation of miRNA in chronic hepatitis B is associated with hepatocellular carcinoma risk after nucleos(t)ide analogue treatment. Cancer Lett. 2018;434:91–100. doi: 10.1016/j.canlet.2018.07.019. [DOI] [PubMed] [Google Scholar]
- 16.Wolfson B, Lo PK, Yao Y, Li L, Wang H, Zhou Q. Impact of miR-140 deficiency on non-alcoholic fatty liver disease. Mol Nutr Food Res. 2018;62:e1800189. doi: 10.1002/mnfr.201800189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanco-Aparicio C, Renner O, Leal JF, Carnero A. PTEN, more than the AKT pathway. Carcinogenesis. 2007;28:1379–1386. doi: 10.1093/carcin/bgm052. [DOI] [PubMed] [Google Scholar]
- 18.Chen CY, Chen J, He L, Stiles BL. PTEN: tumor suppressor and metabolic regulator. Front Endocrinol (Lausanne) 2018;9:338. doi: 10.3389/fendo.2018.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takashima M, Parsons CJ, Ikejima K, Watanabe S, White ES, Rippe RA. The tumor suppressor protein PTEN inhibits rat hepatic stellate cell activation. J Gastroenterol. 2009;44:847–855. doi: 10.1007/s00535-009-0073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shearn CT, Orlicky DJ, McCullough RL, Jiang H, Maclean KN, Mercer KE, et al. Liver-specific deletion of phosphatase and tensin homolog deleted on chromosome 10 significantly ameliorates chronic EtOH-induced increases in hepatocellular damage. PLoS One. 2016;11:e0154152. doi: 10.1371/journal.pone.0154152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia H, Diebold D, Nho R, Perlman D, Kleidon J, Kahm J, et al. Pathological integrin signaling enhances proliferation of primary lung fibroblasts from patients with idiopathic pulmonary fibrosis. J Exp Med. 2008;205:1659–1672. doi: 10.1084/jem.20080001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parapuram SK, Shi-wen X, Elliott C, Welch ID, Jones H, Baron M, et al. Loss of PTEN expression by dermal fibroblasts causes skin fibrosis. J Invest Dermatol. 2011;131:1996–2003. doi: 10.1038/jid.2011.156. [DOI] [PubMed] [Google Scholar]
- 23.McClelland AD, Herman-Edelstein M, Komers R, Jha JC, Winbanks CE, Hagiwara S, et al. miR-21 promotes renal fibrosis in diabetic nephropathy by targeting PTEN and SMAD7. Clin Sci (Lond) 2015;129:1237–1249. doi: 10.1042/CS20150427. [DOI] [PubMed] [Google Scholar]
- 24.Ciuffreda L, Falcone I, Incani UC, Del Curatolo A, Conciatori F, Matteoni S, et al. PTEN expression and function in adult cancer stem cells and prospects for therapeutic targeting. Adv Biol Regul. 2014;56:66–80. doi: 10.1016/j.jbior.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Xu MY, Hu JJ, Shen J, Wang ML, Zhang QQ, Qu Y, et al. Stat3 signaling activation crosslinking of TGF-β1 in hepatic stellate cell exacerbates liver injury and fibrosis. Biochim Biophys Acta. 2014;1842:2237–2245. doi: 10.1016/j.bbadis.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 26.Roderfeld M, Weiskirchen R, Wagner S, Berres ML, Henkel C, Grötzinger J, et al. Inhibition of hepatic fibrogenesis by matrix metalloproteinase-9 mutants in mice. FASEB J. 2006;20:444–454. doi: 10.1096/fj.05-4828com. [DOI] [PubMed] [Google Scholar]
- 27.de Oliveira da Silva B, Ramos LF, Moraes KCM. Molecular interplays in hepatic stellate cells: apoptosis, senescence, and phenotype reversion as cellular connections that modulate liver fibrosis. Cell Biol Int. 2017;41:946–959. doi: 10.1002/cbin.10790. [DOI] [PubMed] [Google Scholar]
- 28.Jiang XP, Ai WB, Wan LY, Zhang YQ, Wu JF. The roles of microRNA families in hepatic fibrosis. Cell Biosci. 2017;7:34. doi: 10.1186/s13578-017-0161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li WQ, Chen C, Xu MD, Guo J, Li YM, Xia QM, et al. The rno-miR-34 family is upregulated and targets ACSL1 in dimethylnitrosamine-induced hepatic fibrosis in rats. FEBS J. 2011;278:1522–1532. doi: 10.1111/j.1742-4658.2011.08075.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhu J, Zhang Z, Zhang Y, Li W, Zheng W, Yu J, et al. MicroRNA-212 activates hepatic stellate cells and promotes liver fibrosis via targeting SMAD7. Biochem Biophys Res Commun. 2018;496:176–183. doi: 10.1016/j.bbrc.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 31.Diniz GP, Huang ZP, Liu J, Chen J, Ding J, Fonseca RI, et al. Loss of microRNA-22 prevents high-fat diet induced dyslipidemia and increases energy expenditure without affecting cardiac hypertrophy. Clin Sci (Lond) 2017;131:2885–2900. doi: 10.1042/CS20171368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Son MK, Ryu YL, Jung KH, Lee H, Lee HS, Yan HH, et al. HS-173, a novel PI3K inhibitor, attenuates the activation of hepatic stellate cells in liver fibrosis. Sci Rep. 2013;3:3470. doi: 10.1038/srep03470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Chu ES, Chen HY, Man K, Go MY, Huang XR, et al. microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget. 2015;6:7325–7338. doi: 10.18632/oncotarget.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar P, Raeman R, Chopyk DM, Smith T, Verma K, Liu Y, et al. Adiponectin inhibits hepatic stellate cell activation by targeting the PTEN/AKT pathway. Biochim Biophys Acta Mol Basis Dis. 2018;1864:3537–3545. doi: 10.1016/j.bbadis.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng J, Wu C, Xu Z, Xia P, Dong P, Chen B, et al. Hepatic stellate cell is activated by microRNA-181b via PTEN/Akt pathway. Mol Cell Biochem. 2015;398:1–9. doi: 10.1007/s11010-014-2199-8. [DOI] [PubMed] [Google Scholar]
- 36.Hu TH, Wang CC, Huang CC, Chen CL, Hung CH, Chen CH, et al. Down-regulation of tumor suppressor gene PTEN, overexpression of p53, plus high proliferating cell nuclear antigen index predict poor patient outcome of hepatocellular carcinoma after resection. Oncol Rep. 2007;18:1417–1426. [PubMed] [Google Scholar]
- 37.Wei J, Feng L, Li Z, Xu G, Fan X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed Pharmacother. 2013;67:387–392. doi: 10.1016/j.biopha.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Tsang CK, Wang S, Li XX, Yang Y, Fu L, et al. MAF1 suppresses AKT-mTOR signaling and liver cancer through activation of PTEN transcription. Hepatology. 2016;63:1928–1942. doi: 10.1002/hep.28507. [DOI] [PMC free article] [PubMed] [Google Scholar]