Abstract
In recent years, with the increase in cancer mortality caused by metastasis, and with the development of individualized and precise medical treatment, early diagnosis with precision becomes the key to decrease the death rate. Since detecting tumour biomarkers in body fluids is the most non‐invasive way to identify the status of tumour development, it has been widely investigated for the usage in clinic. These biomarkers include different expression or mutation in microRNAs (miRNAs), circulating tumour DNAs (ctDNAs), proteins, exosomes and circulating tumour cells (CTCs). In the present article, we summarized and discussed some updated research on these biomarkers. We overviewed their biological functions and evaluated their multiple roles in human and small animal clinical treatment, including diagnosis of cancers, classification of cancers, prognostic and predictive values for therapy response, monitors for therapy efficacy, and anti‐cancer therapeutics. Biomarkers including different expression or mutation in miRNAs, ctDNAs, proteins, exosomes and CTCs provide more choice for early diagnosis of tumour detection at early stage before metastasis. Combination detection of these tumour biomarkers may provide higher accuracy at the lowest molecule combination number for tumour early detection. Moreover, tumour biomarkers can provide valuable suggestions for clinical anti‐cancer treatment and execute monitoring of treatment efficiency.
Keywords: CTCs, ctDNAs, exosomes, miRNAs, proteins, tumour biomarkers
1. INTRODUCTION
Tumour biomarkers are molecules produced by tumour cells, which can indicate the biological status of tumour and can be used to evaluate the disease status and the efficiency of therapeutic interventions.
To survive and adapt in human and animal body, tumour cells have inherited genetic instability that leads to genetics alteration, including cancer‐specific mutations or changes in gene expression. These genetic alterations not only promote tumour development but provide researchers with a chance to chase the disease status at the same time. Although the term “tumour biomarker” now covers any molecular, biochemical, physiological, or anatomical property that reflects tumour's presence and status which can be quantified or measured, an ideal tumour biomarker is preferred to be collected non‐invasively from body fluids, such as the blood. These biomarkers include microRNAs, ctDNAs, proteins, exosomes and CTCs released by the tumour and circulating in the body fluids.
Generally, tumour biomarkers are not expected to simply show the status of tumour, but to exhibit important functions for tumour's survival, growth and metastasis. Based on this fact, tumour biomarkers are recently regarded as treatment targets. Moreover, tumour biomarkers get an emerging role to direct the treatment of anti‐tumour drugs. In 2017, Food and Drug Administration (FDA) accelerated the approval of Keytruda (pembrolizumab), an antibody drug targeting PD‐1(programmed death 1), for the treatment of adult and paediatric patients with unresectable or metastatic solid tumours that have been identified as having a specific genetic feature (or tumour biomarker) referred to as microsatellite instability‐high (MSI‐H) or mismatch repair deficient (dMMR). Doctor Richard Pazdur, the acting director of the Office of Hematology and Oncology Products in the FDA's Center for Drug Evaluation and Research and director of the FDA's Oncology Center of Excellence, recommended this work as “this is an important first for the cancer community,” he said, “Until now, the FDA has approved cancer treatments based on where in the body the cancer started‐for example, lung or breast cancers. We have now approved a drug based on a tumor's biomarker without regard to the tumor's original location.” [https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm560167.htm]. In this review, we will overview some current tumour biomarkers, discuss their biological functions, evaluate their roles in clinical treatment and compare the strength and limitations between different detected markers (Table 1 ), which may provide a prospect for the clinic applications of these markers during different stages of tumour development and anti‐cancer treatment (Figure 1 ).
Table 1.
Comparison of different tumour biomarker detection methods for clinical applications
| Biomarker | Modality | Strengths | Limitations | Ref. |
|---|---|---|---|---|
| Imaging‐based methods | ||||
| CT, MRI, PET, etc | High accuracy, displaying solid tumour visually | High ionizing radiation, unable to detect minimal tumours | 183 | |
| Solid biopsy | ||||
| IHC staining, etc | Reflecting histological situations | Invasive detection methods, cannot cover all heterogeneity | 124 | |
| Body fluids biopsy | ||||
| miRNAs | Altered level of tumour‐specific miRNAs, such as miR‐21 and miR‐155. | Non‐invasive, high sensitivity, allowing for early detection | Unstable, limited by individual difference | 50‐52 |
| ctDNAs | Tumour‐specific mutations, such as EGFR and BRAF. | Non‐invasive, high sensitivity, reflect individual difference, allowing for early detection | Lack of functional studies | 199‐122 |
| DNA methylations, such as ALX4. | ||||
| Proteins | Elevated level of proteins, such as AFP and CA‐125. | Non‐invasive, high sensitivity, allowing for early detection | Limited by individual difference | 116 |
| Different expression profiles, such as ER, PR, HER2, etc | ||||
| Exosomes | Increased exosome number | Non‐invasive, relatively stable in exosome, allowing for early detection | Limited isolation efficiency, lack of large scale studies | 166‐168 |
| Different exosomal nucleotides and proteins | ||||
| CTCs | Increased CTC number | Non‐invasive, reflecting the evolutions of tumour cells timely during tumour development and treatment | Affected by isolation and selection methods, lack of large scale studies, can only be detectable during metastasis but can hardly be detected at an early stage | 194‐196 |
| Altered nucleotides and proteins in CTCs | ||||
The table shows the classification of currently used tumour biomarker detection methods and compared their strengths and limitations considering whether it is less harmful to patients, convenient to detect, with a high accuracy, high stability, can be detectable at an early stage, reflecting individual difference and indicating tumour evolution during development and treatment.
CT, computed tomography; IHC, immunohistochemistry; MRI, magnetic resonance imaging; PET, positron emission tomography.
Figure 1.
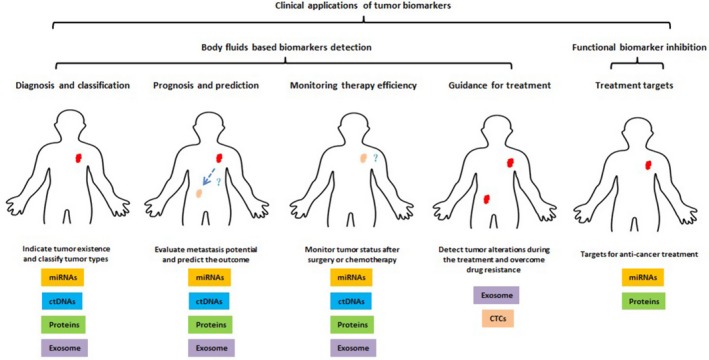
Clinical applications of tumour biomarker in different stage during cancer development and anti‐cancer treatment
2. THE MOLECULAR FUNCTIONS AND CLINICAL USE OF MIRNAS AS TUMOUR BIOMARKERS
2.1. The discovery of miRNAs as biomarker
miRNAs are small non‐coding RNAs (ncRNAs) that target corresponding messenger RNAs (mRNAs) to post‐transcriptionally downregulate certain gene expression. miRNAs were first identified in Caenorhabditis elegans in 1993,1 and extracellular miRNAs were first discovered in plants in 1996.2 Until now, over 2500 human miRNAs have been identified.3
2.2. Cancer‐related molecular functions of circulating miRNAs
The first study linking miRNA with cancer was published in 2002.4 After that, many groups focused their research on miRNA regulating cancer process and found that it involves in all hallmarks of cancer as defined by Hanahan and Weinberg.5 The functions of miRNAs can either be tumour supportive or tumour suppressive, often depending on the genes they targeted. For example, some best‐characterized cancer‐related microRNAs were listed below.
2.2.1. Let‐7 family
The Let‐7 family include 13 different members and have been reported to be related with many types of cancer, and it was recognized as a tumour suppressor generally. Let‐7 regulates cancer cell cycle and proliferation by targetingRAS genes,6, 7 HMGA2,8, 9 STAT3,10 UHRF211 and MYC,12, 13, 14 and additionally, it can regulate cell apoptosis by targeting CASP3.15
2.2.2. miR‐15/16
miR‐15/16 is also an important tumour‐suppressing miRNA during various types of tumour progression. It can regulate apoptosis through targeting FEAT/METTL13,16 RPS6KB1, IGF1R,17 CCND1,18 BCL2,19 RECK and/or SOX6.20 It is a regulator of cell cycle process by targeting FG2F, CCNE1 and E2F1,21, 22, 23 and it is also involved in cell autophagy and metastasis by targeting mTORC2 and SOX5.24, 25
2.2.3. miR‐21
The function of miR‐21 is mainly tumour promoting, since it targets many genes that are important tumour suppressors. These targeted genes mainly related to cell apoptosis, growth, invasion and tumour migration, such as BCL2,26 PTEN,27, 28 TP53, TGFB1,29 RECK,30 RHOB,31 TPM132and PDCD4.33, 34
2.2.4. The miR‐29 Family
Members of miR‐29 family usually act as tumour suppressors, and their downregulation always related to many types of cancer. They directly target cell cycle gene CDK6,35, 36, 37 apoptosis genes MCL1, BCL2 and FHIT,35, 38, 39 and migration and invasion genes LAMC1 and CDC42.40, 41
2.2.5. The miR‐34 Family
ThemiR‐34 family are well known to regulate cell cycle, senescence, apoptosis and invasiveness in cancer. They target at genes that encode factors required for G1/S transition such as MYC, E2F, CDK4 and CDK6. They also target anti‐apoptotic genes such as BCL2, SIRT1 and genes involved in tumour cell invasion such as MET.42
2.2.6. miR‐155
The genes targeted by miR‐155 are involved in multiple pathways related to multiple cancer‐related processes. For example, SMAD5 regulates the epithelial‐mesenchymal transition (EMT) process, while SOCS1, INPP5D and CSF1R regulate cell proliferation, and CASP3, FADD, APAF1 and FOXO3A regulate cell apoptosis.43, 44, 45, 46, 47, 48, 49
Currently, there are about thousands of studies about miRNAs as tumour biomarkers, including numerous reviews that have summarized the detail information about the history, classification and functions of tumour‐related miRNAs. Since tumour is of highly heterogeneity, different cancer types have different regulating molecule mechanisms, so some reviews also summarized the miRNA clinical usage by cancer types.50, 51, 52
2.3. Present applications of circulating miRNAs in clinic
In cancer patients, cancer‐related miRNAs will get some changes in expression or mutations and resulted in abnormal functions that facilitate cancer progression. The high‐throughput sequencing was applied to analyse the expression and mutation of miRNA genes and identified a series of aberrant expression profiles in many human cancers types, such as lymphoma,53 breast cancer,54 colorectal cancer,55 prostate cancer56 and glioma.57 These miRNAs change can be reflected in blood or other body fluid and FFPE tissues, and is even detectable in exosome or CTCs.
2.3.1. Circulating miRNAs for the diagnosis of cancers
In plasma, the combination of miR‐21, miR‐145 and miR‐155 could help distinguish lung cancer patients with 69.4% sensitivity and 78.3% specificity.58 Combination of miR‐148b, miR‐409‐3p and miR‐801 could significantly distinguish breast cancer cases and healthy controls.59 In other body fluids, such as sputum, the combination of miR‐205, miR‐210 and miR‐708 distinguished lung squamous cell carcinoma patients with 73% sensitivity and 96% specificity.60
2.3.2. Circulating miRNAs for the classification of cancers
The reason why miRNAs can be used for the classification of cancers is that different tissues have different miRNAs expression pattern, and miRNAs can reflect the origin of a specific type of tumour or even cellular subsets. In a blind study including 22 different tumour types, classifying tumours according to tissue of origin, the miRNA expression signatures can reach accuracy higher than 90%.61 Recent studies showed that distinct miRNA expression signatures could indicate different cellular subsets in acute myeloid leukaemia (AML)62 and prostate cancer.63 Since we are entering the era of personalized medicine, the anti‐cancer treatment for each patient increasingly depends on molecular analyses, which means establishing a classification according to miRNAs molecular functions that can direct clinic therapy is urgently in need.
2.3.3. The prognostic and predictive values of circulating miRNA for therapy response
Since miRNAs have important regulatory functions during cancer development, their levels can reflect tumour status to some extent and thus could predict the outcome of therapy response. For example, low level of let‐7, a tumour suppressor miRNA, is correlated with poor prognosis including tumour size, overall survival and early recurrence. Moreover, the expression of miR‐21, a tumour‐promoting miRNA, is negatively correlated with relapse‐free survival of diffuse large B‐cell lymphoma (DLBCL) patients.64 In a study of 391 patients with advanced NSCLC, Wang et. al. found that high expression of miR‐16 was obviously associated with better survival.65
2.3.4. Circulating miRNAs as monitors for therapy efficacy
In chronic myeloid leukaemia (CML), the level of cells with the BCR‐ABL rearrangement is widely used to characterize the disease progression, and decreases after imatinib treatment. It was reported that miR‐451 levels negatively correlate with BCR‐ABL levels and can monitor the therapy effect of imatinib at both the time of diagnosis and after treatment.66
2.3.5. Circulating miRNA as targets for anti‐cancer therapeutics
The first microRNA‐based anti‐cancer therapy is MRX34, a synthetic miR‐34a mimic that is loaded into liposomal nanoparticles,67 which acts as a tumour suppressor miRNA downstream of p53. Another example is Miravirsen, a modified sequence complementary to miR‐122. Miravirsen was used for hepatitis C therapy and showed reduction in viral RNA with no evidence of resistance.68 Despite the different therapy effects of different miRNAs targets, the remaining problem is drug resistance, so developing a proper drug combination is one way to have better therapy outcomes.
3. THE MOLECULAR FUNCTIONS AND CLINICAL USE OF CTDNAS AS TUMOUR BIOMARKERS
3.1. The discovery of ctDNAs as biomarker
Cell‐free DNA (cfDNA) is small pieces of DNAs released into blood by various mechanisms mainly including cell apoptosis and necrosis. It was first identified in 1948 by Mandel and Metais in the blood of healthy people.69 Under normal conditions, cell‐free DNA levels are relatively low since apoptotic and necrotic cells are cleared by infiltrating phagocytes. For cancer patients, the cell‐free DNA fraction is often tumour cells derived, which is called circulating tumour DNA (ctDNA). ctDNAs are usually at a higher level with cancer patients and contain some genetic alterations specific for tumour cells.
3.2. Cancer‐associated genetic alterations of ctDNAs
ctDNAs were initially used to identify the presence of tumour in 1994, when Vasioukhin et al detected tumour‐specific RAS mutations in the plasma of cancer patients.70 Generally, ctDNA carries genomic and epigenomic information different from normal cfDNAs, such as point mutations, changed integrity, rearranged sequences, copy number variation (CNV), loss of heterozygosity (LOH), microsatellite instability (MSI) and DNA methylation.71
3.2.1. Tumour‐specific genetic alterations
Abundant mutations have been detected in the ctDNAs of patients with various types of cancer. For example, PIK3CA mutations,72 HER273 and ESR174 higher amplification were detected in breast cancers patients. In colorectal cancers, tumour‐specific gene alterations of EGFR, BRAF, ALK, KIT, PDGFR, HER2 and KRAS75, 76, 77 were detected via ctDNA‐based assays. In the cases of lung cancers, EGFR mutations and ALK rearrangements were also identified.78, 79, 80 ctDNA concentration was significantly increased in other types of cancers such as periampullary cancer,81 oesophageal cancer,82 head and neck cancer,83 renal cancer,84 melanoma85 and prostate cancer.86 Besides, high LOH frequencies, particularly the observed CCND2 loss, were associated with the aggressiveness of breast cancer.87
3.2.2. DNA methylation in ctDNA
DNA methylation plays important regulatory roles in gene expression and genome stability. For example, high levels of 5‐methylcytosine at the promoter region always result in gene transcriptionally silence. And methylation at the promoter region or non‐coding sequences is often dysregulated in many types of tumour and is associated with tumour initiation, progression, dissemination and metastasis.88 Some detectable ctDNA methylation in cancer patients consists of MLH1, CDKN2A (INK4A), ALX4, CDH4, NGFR, RUNX3, SEPT9, TMEFF289, 90, 91, 92, 93, 94, 95and so on.
3.3. Present applications of ctDNAs in clinic
It is believed that cancers are results of gene mutation accumulation. These oncogenic genetic alterations can not only facilitate tumour progression and metastasis, but also be closely correlated with acquired treatment resistance.
3.3.1. ctDNAs for the diagnosis of cancers
Present studies for ctDNAs used in diagnosis are lack of large scale study, and the specificity is not ideal enough for early diagnosis of certain types of cancers. This is partially because the detection methods at present are limited to detect the very low amount of ctDNAs in early stage of cancer patients, and some mutations such as KRAS are not specific for certain types of tumour but exist in many tumour types.96
Recently, Dennis Lo group use plasma Epstein‐Barr virus DNA to screen for nasopharyngeal cancer. In the study, a total of 20,174 participants underwent nasopharyngeal cancer screening, and the sensitivity and specificity were 97.1% and 98.6%, respectively.97 This study may provide us a new sight about detecting cell‐free DNAs, not simply limited by tumour secreted factors but also include those factors that cause the cancer.
3.3.2. The prognostic and predictive values of ctDNAs
Some patients cured by surgery still receive adjuvant chemotherapy in case of tumour relapse. Studies showed that detecting ctDNAs before and after surgical resection can identify individuals with residual disease,98 and predict disease recurrence.83, 84 For example, the high concentration of ctDNA is positively correlated with a poorer survival in metastatic colorectal cancers with detectable KRAS ctDNA.99 Another example is that detecting the methylation of MGMT promoter region on ctDNAs in glioblastoma multiforme patients can also direct whether it is necessary to have adjuvant treatment after surgery.100, 101 Nowadays, the prognostic and predictive value of ctDNA has been extended to different type of cancers, such as cervical cancer,102 colorectal cancer,103, 104 pancreatic cancer,105, 106, 107 melanoma108, 109 and breast cancer,110, 111 in which the increased levels of ctDNA are related to poor overall survival.
3.3.3. ctDNAs as monitors for therapy efficacy
The level of ctDNA is closely correlated with tumour burden and therapeutic responses. It has been reported that its levels increased rapidly with disease progression and declined correspondingly after successful treatment in melanoma,109, 112 breast,110 ovarian113 and colon cancers.114, 115
3.3.4. ctDNAs as guidance for treatments
Recently, there are many new methods for the detection of ctDNA to monitor emerging resistant mutations during anti‐cancer treatment, which allow us to choose appropriate treatment based on specific mutations detected in the drug‐resistant tumour for each individual. For example, in colorectal cancer patients undergoing anti‐EGFR treatment, detecting KRAS mutations in ctDNAs of patients with anti‐EGFR therapies can identify relapse10 months before radiographic documentation of disease progression.116 Similar situations also include BRAF L597 mutation in cutaneous melanoma with MEK inhibitor and PIK3CA mutation in solid tumours with PIK3CA inhibitors.117, 118
There are many more reports which introduce ctDNAs as cancer biomarkers at different aspects; a selection of reviews119, 120, 121, 122may also serve as a starting point for readers outside the field.
4. THE MOLECULAR FUNCTIONS AND CLINICAL USE OF PROTEINS AS TUMOUR BIOMARKERS
Compared with other types of tumour biomarkers, cancer‐related proteins are earlier and more widely used in the clinic. Until now, numerous proteins have been identified to be upregulated with tumour burden, which can either be detectable in tumour tissues or in patients’ blood.
4.1. Cancer‐associated protein markers
4.1.1. Present protein markers in clinic
At present, American National Cancer Institute lists the protein tumour markers that are now used in clinic, for example, alpha‐fetoprotein (AFP) for liver cancer and germ cell tumours, CA15‐3 for breast cancer, CA19‐9 for pancreatic cancer and gastric cancer, CA‐125 for ovarian cancer, carcinoembryonic antigen (CEA) for colorectal cancer and some other cancers, and so on. Other protein biomarkers also include calcitonin for medullary thyroid cancer, CD20 for non‐Hodgkin lymphoma, chromogranin A (CgA) for neuroendocrine tumours, beta‐2‐microglobulin (B2M) for multiple myeloma, and so on.
4.1.2. Other potential markers to be used in clinic
In breast cancer, there is a clear molecular subtype based on some important protein such as ER, PR and HER2, whose function is correlated with breast cancer progression. And the combination of several genes was also commercially used to predict the clinical outcome of breast cancer patients.123 Another widely used marker is EGFR mutations for lung cancer, whose function is closely related to tumour progression through important signalling pathways such as MAPK and AKT/PI3K, and there are also many clinical drugs targeting EGFR.124
4.2. Present applications of cancer biomarker proteins in clinic
4.2.1. Protein markers for the diagnosis of cancers
At present, the accuracy of a single protein biomarker can only discriminate cancer patients and healthy individuals, which is certainly not enough for early diagnosis and further clinical use. Currently, only few body fluid‐based protein markers were approved by FDA, and none of them have high accuracy for early clinical diagnosis. One promising way to increase the accuracy for disease diagnosis is to combine several protein markers. For example, the OVA1 test for ovarian cancer identified five protein markers in serum including CA125, transthyretin, apolipoprotein A‐I (APOA1), β2‐microglobulin and transferrin125, 126; the combination of these five proteins has a ROC AUC of 0.90 and predicts 91.4% ovarian malignancy in the cases of early‐stage disease. This result shows a dramatic accuracy improvement compared with 65.7% for CA125 alone.127, 128
4.2.2. The prognostic and predictive values of protein markers
In breast cancer patients, 21 proteins were identified from an antibody microarray containing 135 antibody fragments, whose functions related to the development of metastasis. The combination of these 21 proteins could distinguish patients at high or low risk for developing metastasis, with an ROC AUC of 0.85.129 What is more, this 21 proteins combination also provided an added value to clinic. That is when combined with conventional clinical parameters, which the ROC AUC is 0.66, the ROC AUC could increase to 0.90 for prediction of recurrence.129
4.2.3. Protein markers as monitors for therapy efficacy
Another function that is widely used in clinic is to monitor the therapy efficacy. Besides some traditional tumour biomarkers such as CA125, many studies work to identify other new tumour biomarkers in different cancer types to improve the present situation. For example, the level of a newly identified tumour biomarker Hsp90α in patients’ plasma showed significant correlation with therapy efficacy in lung cancer.130
5. THE MOLECULAR FUNCTIONS AND CLINICAL USE OF EXOSOMES AS TUMOUR BIOMARKER
5.1. The discovery of tumour‐derived exosomes as tumour biomarkers
Exosomes are 30‐100 nm small vesicles secreted by cells to the extracellular matrix or body fluids. It was first discovered in 1980s by Johnstone group who declared that transferrin receptor could be selectively released in circulating vesicles, which was later named exosomes.131, 132, 133 In 1990s, people recognized exosomes to be related with immune system functions.134 In 2010s, researchers found that exosomes contain RNAs, DNAs, proteins and metabolites.135 And in recent years, exosomes were found to have an important role in cell‐cell communication and signalling transduction.
5.2. Functions of exosomes during cancer development
Exosomes play an important role in cell‐cell communication, since exosomes can package certain RNAs, DNAs, proteins and other metabolism from the donor cells. There are emerging evidences that tumour cells secret exosomes to facilitate cancer growth, angiogenesis, invasion, metastasis, immunity and even drug resistance acquirement.
5.2.1. Exosomal Nucleic Acids
The nucleic acids in exosomes include RNAs such as miRNAs, mRNAs, tRNAs, lncRNAs136, 137, 138, 139 and DNAs including single stranded and double stranded.135, 140 Among these nucleic acids, exosomal miRNAs draw most of the attention.141, 142, 143 It has been reported that pro‐angiogenic miRNAs within tumour‐derived exosomes can induce angiogenesis.144 Furthermore, miRNAs, such as miR21 and miR29a that are highly expressed in tumour cells, can be transported by exosomes and bind to toll‐like receptors to trigger the inflammatory response, which will facilitate tumour growth and metastasis.145 Cancer cells can also gain miRNAs information from other cancer‐associated cells. For example, miR21 could help to suppress ovarian cancer apoptosis and bind to apoptotic protease‐activating factor‐1 (APAF1) to confer drug resistance to paclitaxel.146
5.2.2. Exosomal Proteins
Tumour‐derived exosomes can transport many proteins to establish a complex metastatic microenvironment. For example, HSP70, HSP90 and survivin can inhibit apoptosis and promote cellular proliferation.147, 148 VEGF, FGF and TGF‐β were reported to facilitate angiogenesis.149 Tumour‐derived exosomes were enriched with MMPs (such as MMP‐1 and MMP‐19),150 which could degrade ECM components to facilitate cancer invasion.151 Recently, exosomes were found to inhibit immune system to promote tumour development by increasing the immune suppressive cells, decreasing NK and T cells proliferation and cytotoxicity, inhibiting antigen‐presenting cell number and function.152, 153, 154 There are emerging reports that tumour‐derived exosome also mediates the acquirement of drug resistance. One example is that exosomes of HER2‐overexpressed breast cancer cells also contain HER2 molecules, which can be combined with the HER2 antibody drug trastuzumab, thus prevent the drug from binding to tumour cells and inhibit the anti‐tumour effects.155
5.3. Present applications of tumour‐derived exosomes in clinic
5.3.1. Tumour‐derived exosomes for the diagnosis of cancers
One advantage for exosomal markers in clinical detection is its stability to avoid from enzyme‐based degradation compared with circulating markers, which may increase the accuracy of the detection. For example, miRNA‐1246 shows a sensitivity of 71.3% and a specificity of 73.9% for the diagnosis of oesophageal squamous cell cancer (ESCC), and its level is also correlated with the tumour metastasis and poor survival.156 Actually, many circulating cell‐free biomarkers can also be detected in exosomes in many types of cancer. For example, oncogene EGFR also exists in exosomes from prostate cancer patients.157 miRNAs such as miR‐21 and miR‐141, which was previously known as diagnostic markers for ovarian cancer, were also present in exosomes from ovarian cancer patients.141 KRAS and p53 mutations in exosomal DNA of pancreatic cancer also could predict the treatment option and therapy resistance.158
5.3.2. The prognostic and predictive values of tumour‐derived exosomes
Similar to the diagnosis property, exosomal biomarker also shows values in prognosis and prediction. For example, in tongue squamous cell carcinoma (TSCC), the higher level of caveolin‐1 (CAV1) in exosomes is negatively correlated with recurrence and survival.159 In nasopharyngeal carcinoma (NPC), the level of miR‐24‐3p was higher in exosomes from patients compared with healthy people and was correlated with lower disease‐free survival.154
5.3.3. Tumour‐derived exosomes as monitors for therapy efficacy
It was reported that the amount of cisplatin in exosomes released from cisplatin‐resistant cells is 2.6 times higher than that from cisplatin‐sensitive cells after treatment with cisplatin.160
5.3.4. Tumour‐derived exosomes in anti‐cancer therapy
Since exosomes play an important role in cancer growth, metastasis and drug resistance, some drugs target to inhibit the secretion of exosomes or just remove exosomes from blood circulation.161, 162 Since exosomes can protect its contents from degradation by enzymes, they are ideal drug delivery vehicles, especially for delivering some suppressor miRNAs such as miR‐143.163, 164 Because of the role of exosomes in the immune system, exosome could represent new antigens to the immune cells to evoke the immune system and finally overcome the immune escape of tumour cells.165
Although many challenges exist, it is still a promising field of biomarker, and interested readers are referred to some excellent reviews.166, 167, 168
6. THE MOLECULAR FUNCTIONS AND CLINICAL USE OF CTCS AS TUMOUR BIOMARKERS
6.1. The discovery of CTCs as tumour biomarkers
CTCs are tumour cells from a primary tumour that circulate in the blood around the body, and act as seeds for subsequent secondary metastatic tumour at distant organs. The amount of CTCs in blood is very low at about 1‐10 CTCs per mL of whole blood in metastatic patients.169 CTCs were first identified in 1869.170 Recent years, with the development of CTCs isolation and detection techniques, CTCs have been investigated as promising clinical tumour biomarkers in numerous types of cancer.
6.2. Functions of CTCs during cancer development
Since studies have recognized that CTCs are heterogeneous, which means a CTC cluster may not only contain different sizes or components of CTCs, but also include tumour‐associated stromal cells. These certain different characteristics have distinct biological functions and higher metastatic potential,171, 172, 173 and CTC clusters have higher metastatic potential (near 100‐fold) compared to individual CTCs.174 For example, the formation of CTC clusters requires protein expression such as plakoglobin and keratin 14, which are related to tumour metastases.171, 175 Some factors in the circulation microenvironment also participate in CTC metastasis ability such as pro‐inflammatory cytokines system.176 The presence of stromal cells such as endothelial cell and platelets also facilitates CTC cluster metastasis through different mechanisms.177, 178 What is more, bigger size CTC clusters are under more hypoxia conditions and are more potent to metastasis.179
6.3. Present applications of CTCs in clinic
6.3.1. CTCs for the diagnosis of cancers
The origin and function of CTCs determine that it can be detected in patients already undergoing metastasis. But for early‐stage non‐metastasis patients, it can rarely be detected in the circulation, which may limit its sensitivity and specificity for cancer diagnosis. However, in some cases, it can be used to distinguish lung cancer from benign lesions in patients at CTC count over 25.180 It can also be used for cancer screening, in tobacco‐induced chronic obstructive pulmonary disease, which are at high risk of developing lung cancer, only patients with detectable CTCs were diagnosed lung cancer later.181
6.3.2. The prognostic and predictive values of CTCs
The numbers and characteristics of CTCs are getting widely studied for the use of survival prognosis or therapy response prediction. For example, in metastasis breast cancer, 46.9% of the patients had higher CTC level (≥ 5 CTCs/7.5 mL), which meant lower progression‐free survival and overall survival compared to patients with lower CTC number (<5 CTCs/7.5 mL).182 What is more, CTCs that are undergoing EMT with the expression of EMT marker plastin‐3 could also predict therapy outcome.183
6.3.3. CTCs as monitors for therapy efficacy
Patients with lower CTC level after certain treatment exhibit better survival compared to those patients remain high CTC level. For example, in metastatic breast cancer, after one cycle of chemotherapy, patients with decreasing CTC levels have a better prognosis than patients with persistently high CTC levels.184 Similar results were also reported in colon cancer,185 castration‐resistant prostate cancer,186, 187 rectal cancer188, 189 and small cell lung cancer.190 In ovarian cancer, monitoring CTC has an even higher accuracy than protein marker CA125 for predicting chemotherapy response and cancer relapse.191
6.3.4. CTCs as guidance for treatments
The response of tumour cells to therapy can be dynamic; thus, measuring CTCs during the course of therapy may reveal tumour changes timely and provide us guides for further treatment. However, until now, only limited studies showed CTC‐based treatment direction applicable. For example, breast cancer patients with HER2‐negative setting but HER2‐positive CTCs were treated with trastuzumab or observation. The results showed that 27 of 36 patients treated with trastuzumab became CK19 mRNA‐negative compared to 7 of 39 observation patients, and trastuzumab treatment also decreased the risk of disease recurrence and prolonged disease‐free survival.192 In a recent clinical trial investigating the clinical utility of CTC numbers in ER‐positive metastatic breast cancer patients, patients with low CTC numbers are given hormone therapy, while patients with high CTC numbers are treated by first‐line chemotherapy. After one cycle of chemotherapy, patients remaining high CTC numbers (>5 CTCs/7.5 mL) were possibly switched to second‐line chemotherapy at earlier time. However, until now, this study was reported as negative, since an early switch of chemotherapy did not improve overall survival of these patients.193 Many reasons may cause this result; however, based on the property of CTCs, it is still believed to be the future direction for guiding treatment timely, and new effective treatment is also urgently needed after identifying the CTC status.
Some reviews that summarized the advantages and challenges in this field also listed here for readers who are interested.194, 195, 196
7. CONCLUSIONS
Large portions of cancer death are caused by metastasis, so early diagnosis becomes the key to decrease the death rate. Since detecting tumour biomarkers in body fluids is the most non‐invasive way to identify the status of tumour development, it has been widely investigated for the use in clinic. These biomarkers include different expression or mutation in miRNAs, ctDNAs, proteins, exosomes and CTCs. For the use of early diagnosis, which requires the detection of tumour markers at early stage before metastasis, high sensitivity and specificity are very important. Due to the characteristic of CTCs, which are often detected when metastasis already happened, the sensitivity and specificity for early diagnosis are not high enough to be ideal prospective marker for diagnosis. Other molecular markers such as miRNAs, ctDNAs and proteins can be secreted by tumour cells at very early stage to facilitate tumour development and metastasis, thus could be detected at an early stage (Figure 1). However, at present, the sensitivity and specificity of these molecular markers are not high enough, which may partially due to the present limited detection methods, limited stability or may be limited by the different molecular function of different patients. So, one promising way to solve this problem is to combine these biomarkers and achieve a highest accuracy at the lowest molecule combination number for tumour early detection. Recently, exosomes are identified as an emerging hot spot in the field of diagnostic tumour biomarker, because circulating exosomes shows important functions in long distance message transport between different cells, and detecting miRNAs, ctDNAs and proteins in exosomes can avoid enzyme‐based degradation of these molecules, which allow an elevated accuracy (Figure 1). However, methods for detection of isolate exosomes and molecular marker combinations still need further study, especially considering their biological functions.
After the diagnosis of cancer, patients usually need certain anti‐cancer treatment. Another important function for tumour biomarkers is to direct the determination of therapeutic regimen. Many oncogenic miRNAs, ctDNAs and proteins have functions to facilitate cancer progression and metastasis, so they usually correlate with poor prognosis (Figure 1). CTCs, which often occurred just before metastasis, possess ideal accuracy for prognosis and treatment efficiency prediction. These may provide some information for choosing certain anti‐cancer treatment. Further studies should also focus on biomarkers‐targeted treatments based on their molecular functions, which could have a more precise direction for drug treatment.
Another important clinic use for tumour biomarker is to monitor the treatment efficiency. Since the tumour markers could reflect the status of tumour development, these levels could evaluate whether the treatment is effective to inhibit tumour development. Meanwhile, it can monitor whether there is a relapse after a period of remission (Figure 1). However, the accuracy for this use is limited and required more efficient alternative treatments if the first‐line treatment is not effective.
During the anti‐cancer treatment, tumour cells may evolve new properties to gain drug resistance. These properties can be reflected in the genomic change of metastasis tumour cells, and the genetic information in CTCs provides an ideal way to identify these changes (Figure 1). At present, the study of this application is only at a very beginning stage, and the clinical results for CTCs directed treatment changes cannot improve their overall survival of these drug resistance patients. It may because that the alternative treatment is not effective. And the method for isolating CTCs is not mature, since different CTCs have different surface markers, and present method may not isolate all kinds of CTCs in the circulation.
At present, traditional tumour biomarkers widely used in clinic are still at protein level. However, due to their limited sensitivity and specificity, novel serum biomarkers such as CTCs and nucleic acids will have a great advantage in the future. Although, novel biomarkers have their own technical limitation, and protein markers may not soon be replaced, it is a trend that using different biomarkers in combination to increase the sensitivity and specificity. In the era of individualized medical treatment and precise medical treatment, the diagnosis and treatment decision relied much on the information provided by tumour biomarkers. One important event for this era is that in this year, FDA first approved a drug based on a tumour's biomarker without regard to the tumour's original location. And we believe that researchers will pay more attention on the molecular functions and the underlying mechanisms of these tumour biomarkers, to have a more precise use in the clinics in the future.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
JHL and LM wrote the manuscript and created the figures. DZ and JFG collected the related paper. YPJ, ZHH and DGL provided guidance and revised this manuscript. All authors approved the final manuscript.
Lin J, Ma L, Zhang D, et al. Tumour biomarkers—Tracing the molecular function and clinical implication. Cell Prolif. 2019;52:e12589 10.1111/cpr.12589
Jiahao Lin and Lie Ma contributed equally as first authors.
Funding information
We greatly acknowledge financial supports from the National Nature Science Foundation of China (Grant No. 31502108), Fundamental Research Funds for the Central Universities (Grant No. 2017QC076).
Contributor Information
Yipeng Jin, Email: yipengjin@sina.com.
Zhihai Han, Email: hanzhihai@hotmail.com.
Degui Lin, Email: csamalin@sina.com.
REFERENCES
- 1. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell. 1993;75(5):843‐854. [DOI] [PubMed] [Google Scholar]
- 2. Baulcombe DC. RNA as a target and an initiator of post‐transcriptional gene silencing in transgenic plants. Plant Mol Biol. 1996;32(1–2):79‐88. [DOI] [PubMed] [Google Scholar]
- 3. Griffiths‐Jones S, Grocock RJ, vanDongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down‐regulation of micro‐ RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 6. Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let‐7 microRNA family. Cell. 2005;120(5):635–647. [DOI] [PubMed] [Google Scholar]
- 7. Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non‐small cell lung tumor development by the let‐7 microRNA family. Proc Natl Acad Sci USA. 2008;105(10):3903–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee YS, Dutta A. The tumor suppressor microRNA let‐7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let‐7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y, Lu Y, Toh ST, et al. Lethal‐7 is down‐regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J hepatol. 2010;53(1):57–66. [DOI] [PubMed] [Google Scholar]
- 11. He X, Duan C, Chen J, et al. Let‐7a elevates p21(WAF1) levels by targeting of NIRF and suppresses the growth of A549 lung cancer cells. FEBS Lett. 2009;583(21):3501–3507. [DOI] [PubMed] [Google Scholar]
- 12. Sampson VB, Rong NH, Han J, et al. MicroRNA let‐7a down‐regulates MYC and reverts MYC‐induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67(20):9762–9770. [DOI] [PubMed] [Google Scholar]
- 13. Akao Y, Nakagawa Y, Naoe T. let‐7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29(5):903–906. [DOI] [PubMed] [Google Scholar]
- 14. Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let‐7/RISC to repress c‐Myc expression. Genes Dev. 2009;23(15):1743–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsang WP, Kwok TT. Let‐7a microRNA suppresses therapeutics‐induced cancer cell death by targeting caspase‐3. Apoptosis. 2008;13(10):1215–1222. [DOI] [PubMed] [Google Scholar]
- 16. Liang H, Fu Z, Jiang X, et al. miR‐16 promotes the apoptosis of human cancer cells by targeting FEAT. BMC Cancer. 2015;15:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen L, Wang Q, Wang GD, et al. miR‐16 inhibits cell proliferation by targeting IGF1R and the Raf1‐MEK1/2‐ERK1/2 pathway in osteosarcoma. FEBS Lett. 2013;587(9):1366–1372. [DOI] [PubMed] [Google Scholar]
- 18. Cai CK, Zhao GY, Tian LY, et al. miR‐15a and miR‐16‐1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol Rep. 2012;28(5):1764–1770. [DOI] [PubMed] [Google Scholar]
- 19. Yang TQ, Lu XJ, Wu TF, et al. MicroRNA‐16 inhibits glioma cell growth and invasion through suppression of BCL2 and the nuclear factor‐kappaB1/MMP9 signaling pathway. Cancer Sci. 2014;105(3):265–271. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Zhu Y, Xia Y, Niu H, Chen Y. MiR‐16 induced the suppression of cell apoptosis while promote proliferation in esophageal squamous cell carcinoma. Cell Physiol Biochem. 2014;33(5):1340–1348. [DOI] [PubMed] [Google Scholar]
- 21. He Q, Ren X, Chen J, et al. miR‐16 targets fibroblast growth factor 2 to inhibit NPC cell proliferation and invasion via PI3K/AKT and MAPK signaling pathways. Oncotarget. 2016;7(3):3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zubillaga‐Guerrero MI, Alarcon‐Romero Ldel C, Illades‐Aguiar B, et al. MicroRNA miR‐16‐1 regulates CCNE1 (cyclin E1) gene expression in human cervical cancer cells. Int J Clin Exp Med. 2015;8(9):15999–16006. [PMC free article] [PubMed] [Google Scholar]
- 23. Ofir M, Hacohen D, Ginsberg D. MiR‐15 and miR‐16 are direct transcriptional targets of E2F1 that limit E2F‐induced proliferation by targeting cyclin E. Mol Cancer Res. 2011;9(4):440–447. [DOI] [PubMed] [Google Scholar]
- 24. Huang N, Wu J, Qiu W, et al. MiR‐15a and miR‐16 induce autophagy and enhance chemosensitivity of Camptothecin. Cancer Biol Ther. 2015;16(6):941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Renjie W, Haiqian L. MiR‐132, miR‐15a and miR‐16 synergistically inhibit pituitary tumor cell proliferation, invasion and migration by targeting Sox5. Cancer Lett. 2015;356(2 Pt B):568–578. [DOI] [PubMed] [Google Scholar]
- 26. Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR‐21 expression and increases miR‐21 target gene expression in MCF‐7 breast cancer cells. Nucleic Acids Res. 2009;37(8):2584–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meng F, Henson R, Wehbe‐Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA‐21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He C, Dong X, Zhai B, et al. MiR‐21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;6(30):28867–28881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA‐21 targets a network of key tumor‐suppressive pathways in glioblastoma cells. Cancer Res. 2008;68(19):8164–8172. [DOI] [PubMed] [Google Scholar]
- 30. Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28(17):5369–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sabatel C, Malvaux L, Bovy N, et al. MicroRNA‐21 exhibits antiangiogenic function by targeting RhoB expression in endothelial cells. PLoS ONE. 2011;6(2):e16979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu S, Si ML, Wu H, Mo YY. MicroRNA‐21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282(19):14328–14336. [DOI] [PubMed] [Google Scholar]
- 33. Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005;65(14):6034–6041. [DOI] [PubMed] [Google Scholar]
- 34. Leupold JH, Yang HS, Colburn NH, Asangani I, Post S, Allgayer H. Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u‐PAR) gene expression via Sp‐transcription factors. Oncogene. 2007;26(31):4550–4562. [DOI] [PubMed] [Google Scholar]
- 35. Inoue A, Yamamoto H, Uemura M, et al. MicroRNA‐29b is a Novel Prognostic Marker in Colorectal Cancer. Ann Surg Oncol. 2015;22(Suppl 3):S1410–S1418. [DOI] [PubMed] [Google Scholar]
- 36. Li Y, Wang F, Xu J, et al. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV‐related target genes for miR‐29. J Pathol. 2011;224(4):484–495. [DOI] [PubMed] [Google Scholar]
- 37. Zhao JJ, Lin J, Lwin T, et al. microRNA expression profile and identification of miR‐29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115(13):2630–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu L, Xu Y, Jing Z, et al. Altered expression pattern of miR‐29a, miR‐29b and the target genes in myeloid leukemia. Exp Hematol Oncol. 2014;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu DW, Hsu NY, Wang YC, et al. c‐Myc suppresses microRNA‐29b to promote tumor aggressiveness and poor outcomes in non‐small cell lung cancer by targeting FHIT. Oncogene. 2015;34(16):2072–2082. [DOI] [PubMed] [Google Scholar]
- 40. Nishikawa R, Goto Y, Kojima S, et al. Tumor‐suppressive microRNA‐29s inhibit cancer cell migration and invasion via targeting LAMC1 in prostate cancer. Int J oncol. 2014;45(1):401–410. [DOI] [PubMed] [Google Scholar]
- 41. Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR‐29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16(1):23–29. [DOI] [PubMed] [Google Scholar]
- 42. Navarro F, Lieberman J. miR‐34 and p53: New Insights into a Complex Functional Relationship. PLoS ONE. 2015;10(7):e0132767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. George J, Lewis MG, Renne R, Mattapallil JJ. Suppression of transforming growth factor beta receptor 2 and Smad5 is associated with high levels of microRNA miR‐155 in the oral mucosa during chronic simian immunodeficiency virus infection. J Virol. 2015;89(5):2972–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xue H, Hua LM, Guo M, Luo JM. SHIP1 is targeted by miR‐155 in acute myeloid leukemia. Oncology Rep. 2014;32(5):2253–2259. [DOI] [PubMed] [Google Scholar]
- 45. Salemi D, Cammarata G, Agueli C, et al. miR‐155 regulative network in FLT3 mutated acute myeloid leukemia. Leukemia Res. 2015;39(8):883–896. [DOI] [PubMed] [Google Scholar]
- 46. Wei Y, Zhu M, Corbalan‐Campos J, Heyll K, Weber C, Schober A. Regulation of Csf1r and Bcl6 in macrophages mediates the stage‐specific effects of microRNA‐155 on atherosclerosis. Arterioscler Thromb Vasc Biol. 2015;35(4):796–803. [DOI] [PubMed] [Google Scholar]
- 47. De Santis R, Liepelt A, Mossanen JC, et al. miR‐155 targets Caspase‐3 mRNA in activated macrophages. RNA Biol. 2016;13(1):43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zang YS, Zhong YF, Fang Z, Li B, An J. MiR‐155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of Apaf‐1 expression. Cancer Gene Ther. 2012;19(11):773–778. [DOI] [PubMed] [Google Scholar]
- 49. Ling N, Gu J, Lei Z, et al. microRNA‐155 regulates cell proliferation and invasion by targeting FOXO3a in glioma. Oncology Rep. 2013;30(5):2111–2118. [DOI] [PubMed] [Google Scholar]
- 50. Tiberio P, Callari M, Angeloni V, Daidone MG, Appierto V. Challenges in using circulating miRNAs as cancer biomarkers. BioMed Res Int. 2015;2015:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. BioMed Res Int. 2015;2015:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Larrea E, Sole C, Manterola L, et al. New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies. Int J Mol Sci. 2016;17(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mazan‐Mamczarz K, Gartenhaus RB. Role of microRNA deregulation in the pathogenesis of diffuse large B‐cell lymphoma (DLBCL). Leukemia Res. 2013;37(11):1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ouyang M, Li Y, Ye S, et al. MicroRNA profiling implies new markers of chemoresistance of triple‐negative breast cancer. PLoS ONE. 2014;9(5):e96228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dong Y, Wu WK, Wu CW, Sung JJ, Yu J, Ng SS. MicroRNA dysregulation in colorectal cancer: a clinical perspective. British J Cancer. 2011;104(6):893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Maugeri‐Sacca M, Coppola V, Bonci D, De Maria R. MicroRNAs and prostate cancer: from preclinical research to translational oncology. Cancer J. 2012;18(3):253–261. [DOI] [PubMed] [Google Scholar]
- 57. Tumilson CA, Lea RW, Alder JE, Shaw L. Circulating microRNA biomarkers for glioma and predicting response to therapy. Mol Neurobiol. 2014;50(2):545–558. [DOI] [PubMed] [Google Scholar]
- 58. Tang D, Shen Y, Wang M, et al. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev. 2013;22(6):540–548. [DOI] [PubMed] [Google Scholar]
- 59. Cuk K, Zucknick M, Heil J, et al. Circulating microRNAs in plasma as early detection markers for breast cancer. Int J Cancer. 2013;132(7):1602–1612. [DOI] [PubMed] [Google Scholar]
- 60. Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Modern Pathol. 2010;23(8):1157–1164. [DOI] [PubMed] [Google Scholar]
- 61. Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nature Biotechnol. 2008;26(4):462–469. [DOI] [PubMed] [Google Scholar]
- 62. Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111(6):3183–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu C, Kelnar K, Vlassov AV, Brown D, Wang J, Tang DG. Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor‐suppressive functions of let‐7. Cancer Res. 2012;72(13):3393–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour‐associated microRNAs in serum of patients with diffuse large B‐cell lymphoma. British J Haematol. 2008;141(5):672–675. [DOI] [PubMed] [Google Scholar]
- 65. Wang Y, Gu J, Roth JA, et al. Pathway‐based serum microRNA profiling and survival in patients with advanced stage non‐small cell lung cancer. Cancer Res. 2013;73(15):4801–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scholl V, Hassan R, Zalcberg IR. miRNA‐451: A putative predictor marker of Imatinib therapy response in chronic myeloid leukemia. Leukemia Res. 2012;36(1):119–121. [DOI] [PubMed] [Google Scholar]
- 67. Bouchie A. First microRNA mimic enters clinic. Nature Biotechnol. 2013;31(7):577. [DOI] [PubMed] [Google Scholar]
- 68. Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. New England J Med. 2013;368(18):1685–1694. [DOI] [PubMed] [Google Scholar]
- 69. Mandel P. Les acides nucleiques du plasma sanguin chez l'homme. CR Acad Sci Paris. 1948;142:241–243. [PubMed] [Google Scholar]
- 70. Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. Point mutations of the N‐ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. British J Haematol. 1994;86(4):774–779. [DOI] [PubMed] [Google Scholar]
- 71. Marzese DM, Hirose H, Hoon DS. Diagnostic and prognostic value of circulating tumor‐related DNA in cancer patients. Expert Rev Mol Diagn. 2013;13(8):827–844. [DOI] [PubMed] [Google Scholar]
- 72. Higgins MJ, Jelovac D, Barnathan E, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clinical Cancer Res. 2012;18(12):3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gevensleben H, Garcia‐Murillas I, Graeser MK, et al. Noninvasive detection of HER2 amplification with plasma DNA digital PCR. Clinical Cancer Res. 2013;19(12):3276–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chu D, Paoletti C, Gersch C, et al. ESR1 Mutations in Circulating Plasma Tumor DNA from Metastatic Breast Cancer Patients. Clinical Cancer Res. 2016;22(4):993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Siravegna G, Bardelli A. Blood circulating tumor DNA for non‐invasive genotyping of colon cancer patients. Mol Oncol. 2016;10(3):475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Diaz LA Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nature Med. 2015;21(7):795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang Z, Lee JC, Lin L, et al. Activation of the AXL kinase causes resistance to EGFR‐targeted therapy in lung cancer. Nat Genet. 2012;44(8):852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bai H, Wang Z, Chen K, et al. Influence of chemotherapy on EGFR mutation status among patients with non‐small‐cell lung cancer. J Clin Oncol. 2012;30(25):3077–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Akca H, Demiray A, Yaren A, et al. Utility of serum DNA and pyrosequencing for the detection of EGFR mutations in non‐small cell lung cancer. Cancer Genet. 2013;206(3):73–80. [DOI] [PubMed] [Google Scholar]
- 81. Umetani N, Kim J, Hiramatsu S, et al. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: direct quantitative PCR for ALU repeats. Clin Chem. 2006;52(6):1062–1069. [DOI] [PubMed] [Google Scholar]
- 82. Tomita H, Ichikawa D, Ikoma D, et al. Quantification of circulating plasma DNA fragments as tumor markers in patients with esophageal cancer. Anticancer Res. 2007;27(4C):2737–2741. [PubMed] [Google Scholar]
- 83. Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nature Med. 1996;2(9):1035–1037. [DOI] [PubMed] [Google Scholar]
- 84. Gang F, Guorong L, An Z, Anne GP, Christian G, Jacques T. Prediction of clear cell renal cell carcinoma by integrity of cell‐free DNA in serum. Urology. 2010;75(2):262–265. [DOI] [PubMed] [Google Scholar]
- 85. Pinzani P, Salvianti F, Zaccara S, et al. Circulating cell‐free DNA in plasma of melanoma patients: qualitative and quantitative considerations. Clin Chim Acta. 2011;412(23–24):2141–2145. [DOI] [PubMed] [Google Scholar]
- 86. Hanley R, Rieger‐Christ KM, Canes D, et al. DNA integrity assay: a plasma‐based screening tool for the detection of prostate cancer. Clinical Cancer Res. 2006;12(15):4569–4574. [DOI] [PubMed] [Google Scholar]
- 87. Schwarzenbach H, Eichelser C, Kropidlowski J, Janni W, Rack B, Pantel K. Loss of heterozygosity at tumor suppressor genes detectable on fractionated circulating cell‐free tumor DNA as indicator of breast cancer progression. Clin Cancer Res. 2012;18(20):5719–5730. [DOI] [PubMed] [Google Scholar]
- 88. Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13(10):679–692. [DOI] [PubMed] [Google Scholar]
- 89. Miotto E, Sabbioni S, Veronese A, et al. Frequent aberrant methylation of the CDH4 gene promoter in human colorectal and gastric cancer. Cancer Res. 2004;64(22):8156–8159. [DOI] [PubMed] [Google Scholar]
- 90. Lofton‐Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood‐based colorectal cancer screening. Clin Chem. 2008;54(2):414–423. [DOI] [PubMed] [Google Scholar]
- 91. deVos T, Tetzner R, Model F, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009;55(7):1337–1346. [DOI] [PubMed] [Google Scholar]
- 92. Grady WM, Rajput A, Lutterbaugh JD, Markowitz SD. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res. 2001;61(3):900–902. [PubMed] [Google Scholar]
- 93. Silva JM, Dominguez G, Villanueva MJ, et al. Aberrant DNA methylation of the p16INK4a gene in plasma DNA of breast cancer patients. British J Cancer. 1999;80(8):1262–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lecomte T, Berger A, Zinzindohoue F, et al. Detection of free‐circulating tumor‐associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J cancer. 2002;100(5):542–548. [DOI] [PubMed] [Google Scholar]
- 95. Ebert MP, Model F, Mooney S, et al. Aristaless‐like homeobox‐4 gene methylation is a potential marker for colorectal adenocarcinomas. Gastroenterology. 2006;131(5):1418–1430. [DOI] [PubMed] [Google Scholar]
- 96. Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72(10):2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Chan K, Woo J, King A, et al. Analysis of Plasma Epstein‐Barr Virus DNA to Screen for Nasopharyngeal Cancer. New England J Med. 2017;377(6):513–522. [DOI] [PubMed] [Google Scholar]
- 98. Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single‐copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994;3(1):67–71. [PubMed] [Google Scholar]
- 99. Speicher MR, Pantel K. Tumor signatures in the blood. Nature Biotechnol. 2014;32(5):441–443. [DOI] [PubMed] [Google Scholar]
- 100. Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O‐6‐methylguanine‐DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clinical Cancer Res. 2004;10(6):1871–1874. [DOI] [PubMed] [Google Scholar]
- 101. Herrlinger U, Rieger J, Koch D, et al. Phase II trial of lomustine plus temozolomide chemotherapy in addition to radiotherapy in newly diagnosed glioblastoma: UKT‐03. J Clin Oncol. 2006;24(27):4412–4417. [DOI] [PubMed] [Google Scholar]
- 102. Campitelli M, Jeannot E, Peter M, et al. Human papillomavirus mutational insertion: specific marker of circulating tumor DNA in cervical cancer patients. PLoS ONE. 2012;7(8):e43393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. da Silva Filho BF, Gurgel AP, Neto MA, et al. Circulating cell‐free DNA in serum as a biomarker of colorectal cancer. J Clin Pathol. 2013;66(9):775–778. [DOI] [PubMed] [Google Scholar]
- 104. Wang JY, Hsieh JS, Chang MY, et al. Molecular detection of APC, K‐ ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J Surg. 2004;28(7):721–726. [DOI] [PubMed] [Google Scholar]
- 105. Chen H, Tu H, Meng ZQ, Chen Z, Wang P, Liu LM. K‐ras mutational status predicts poor prognosis in unresectable pancreatic cancer. Eur J Surg Oncol. 2010;36(7):657–662. [DOI] [PubMed] [Google Scholar]
- 106. Dabritz J, Preston R, Hanfler J, Oettle H. K‐ras mutations in the plasma correspond to computed tomographic findings in patients with pancreatic cancer. Pancreas. 2012;41(2):323–325. [DOI] [PubMed] [Google Scholar]
- 107. Kinugasa H, Nouso K, Miyahara K, et al. Detection of K‐ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;121(13):2271–2280. [DOI] [PubMed] [Google Scholar]
- 108. Santiago‐Walker A, Gagnon R, Mazumdar J, et al. Correlation of BRAF mutation status in circulating‐free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials. Clin Cancer Res. 2016;22(3):567–574. [DOI] [PubMed] [Google Scholar]
- 109. Shinozaki M, O'Day SJ, Kitago M, et al. Utility of circulating B‐RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res. 2007;13(7):2068–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. New England J Med. 2013;368(13):1199–1209. [DOI] [PubMed] [Google Scholar]
- 111. Murtaza M, Dawson SJ, Tsui DW, et al. Non‐invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. [DOI] [PubMed] [Google Scholar]
- 112. Bidard FC, Madic J, Mariani P, et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int J Cancer. 2014;134(5):1207–1213. [DOI] [PubMed] [Google Scholar]
- 113. Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra168. [DOI] [PubMed] [Google Scholar]
- 114. Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nature Med. 2008;14(9):985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26(8):1715–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti‐EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Teufel M, Van Laethem J‐L, et al. Abstract 5239: KRAS wild‐type status as detected by circulating tumor DNA analysis may be a prognostic or predictive factor for clinical benefit in patients with unresectable, locally advanced or metastatic pancreatic cancer (PC) treated with the MEK inhibitor refametin. Cancer Res. 2015;75(15 Supplement):5239–5239. [Google Scholar]
- 118. Janku F, Tsimberidou AM, Garrido‐Laguna I, et al. <em>PIK3CA</em> Mutations in Patients with Advanced Cancers Treated with PI3K/AKT/mTOR Axis Inhibitor. Mol Cancer Ther. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Han X, Wang J, Sun Y. Circulating tumor DNA as biomarkers for cancer detection. Genomics, Proteomics & Bioinformatics. 2017;15(2):59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early‐ and late‐stage human malignancies. Sci Transl Med. 2014;6(224):224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. He WS, Bishop KS. The potential use of cell‐free‐circulating‐tumor DNA as a biomarker for prostate cancer. Expert Rev Mol Diagn. 2016;16(8):839–852. [DOI] [PubMed] [Google Scholar]
- 122. Rapisuwon S, Vietsch EE, Wellstein A. Circulating biomarkers to monitor cancer progression and treatment. Comput Struct Biotechnol J. 2016;14:211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Dai X, Li T, Bai Z, et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5(10):2929–2943. [PMC free article] [PubMed] [Google Scholar]
- 124. Cancer Genome Atlas Research N . Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zhang Z, Bast RC Jr, Yu Y, et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64(16):5882–5890. [DOI] [PubMed] [Google Scholar]
- 126. Bast RC Jr, Feeney M, Lazarus H, Nadler LM, Colvin RB, Knapp RC. Reactivity of a monoclonal antibody with human ovarian carcinoma. J Clin Invest. 1981;68(5):1331–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zhang Z, Chan DW. The road from discovery to clinical diagnostics: lessons learned from the first FDA‐cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev. 2010;19(12):2995–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bristow RE, Smith A, Zhang Z, et al. Ovarian malignancy risk stratification of the adnexal mass using a multivariate index assay. Gynecologic Oncol. 2013;128(2):252–259. [DOI] [PubMed] [Google Scholar]
- 129. Carlsson A, Wingren C, Kristensson M, et al. Molecular serum portraits in patients with primary breast cancer predict the development of distant metastases. Proc Natl Acad Sci. 2011;108(34):14252–14257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Shi Y, Liu X, Lou J, et al. Plasma Levels of Heat Shock Protein 90 Alpha Associated with Lung Cancer Development and Treatment Responses. Clin Cancer Res. 2014;20(23):6016–6022. [DOI] [PubMed] [Google Scholar]
- 131. Johnstone RM, Mathew A, Mason AB, Teng K. Exosome formation during maturation of mammalian and avian reticulocytes: evidence that exosome release is a major route for externalization of obsolete membrane proteins. Journal Cell Physiol. 1991;147(1):27–36. [DOI] [PubMed] [Google Scholar]
- 132. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–978. [DOI] [PubMed] [Google Scholar]
- 133. Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101(3):942–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen‐presenting vesicles. J Exp Med. 1996;183(3):1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nature Commun. 2011;2:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biol. 2007;9(6):654–659. [DOI] [PubMed] [Google Scholar]
- 137. Ratajczak J, Wysoczynski M, Hayek F, Janowska‐Wieczorek A, Ratajczak MZ. Membrane‐derived microvesicles: important and underappreciated mediators of cell‐to‐cell communication. Leukemia. 2006;20(9):1487–1495. [DOI] [PubMed] [Google Scholar]
- 138. Gusachenko ON, Zenkova MA, Vlassov VV. Nucleic acids in exosomes: disease markers and intercellular communication molecules. Biochemistry (Mosc). 2013;78(1):1–7. [DOI] [PubMed] [Google Scholar]
- 139. Bullock M, Silva A, Kanlikilicer‐Unaldi P, et al. Exosomal Non‐Coding RNAs: Diagnostic, Prognostic and Therapeutic Applications in Cancer. Non‐Coding RNA. 2015;1(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kahlert C, Melo SA, Protopopov A, et al. Identification of double‐stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289(7):3869–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Taylor DD, Gercel‐Taylor C. MicroRNA signatures of tumor‐derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. [DOI] [PubMed] [Google Scholar]
- 142. Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood‐based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hunter MP, Ismail N, Zhang X, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3(11):e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Liu Y, Luo F, Wang B, et al. STAT3‐regulated exosomal miR‐21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016;370(1):125–135. [DOI] [PubMed] [Google Scholar]
- 145. Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll‐like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109(31):E2110–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Au Yeung CL, Co NN, Tsuruga T, et al. Exosomal transfer of stroma‐derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nature Commun. 2016;7:11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Graner MW, Cumming RI, Bigner DD. The heat shock response and chaperones/heat shock proteins in brain tumors: surface expression, release, and possible immune consequences. J Neurosci. 2007;27(42):11214–11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Khan S, Jutzy JM, Aspe JR, McGregor DW, Neidigh JW, Wall NR. Survivin is released from cancer cells via exosomes. Apoptosis. 2011;16(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Konstantinell A, Bruun JA, Olsen R, et al. Secretomic analysis of extracellular vesicles originating from polyomavirus‐negative and polyomavirus‐positive Merkel cell carcinoma cell lines. Proteomics. 2016;16(19):2587–2591. [DOI] [PubMed] [Google Scholar]
- 150. Tauro BJ, Mathias RA, Greening DW, et al. Oncogenic H‐ras reprograms Madin‐Darby canine kidney (MDCK) cell‐derived exosomal proteins following epithelial‐mesenchymal transition. Molecular Cell Proteomics. 2013;12(8):2148–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Endres M, Kneitz S, Orth MF, Perera RK, Zernecke A, Butt E. Regulation of matrix metalloproteinases (MMPs) expression and secretion in MDA‐MB‐231 breast cancer cells by LIM and SH3 protein 1 (LASP1). Oncotarget. 2016;7(39):64244–64259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Berchem G, Noman MZ, Bosseler M, et al. Hypoxic tumor‐derived microvesicles negatively regulate NK cell function by a mechanism involving TGF‐beta and miR23a transfer. Oncoimmunology. 2016;5(4):e1062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Rong L, Li R, Li S, Luo R. Immunosuppression of breast cancer cells mediated by transforming growth factor‐beta in exosomes from cancer cells. Oncology Lett. 2016;11(1):500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Ye SB, Zhang H, Cai TT, et al. Exosomal miR‐24‐3p impedes T‐cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma. J Pathol. 2016;240(3):329–340. [DOI] [PubMed] [Google Scholar]
- 155. Ciravolo V, Huber V, Ghedini GC, et al. Potential role of HER2‐overexpressing exosomes in countering trastuzumab‐based therapy. Journal Cell Physiol. 2012;227(2):658–667. [DOI] [PubMed] [Google Scholar]
- 156. Takeshita N, Hoshino I, Mori M, et al. Serum microRNA expression profile: miR‐1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. British J Cancer. 2013;108(3):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Kharmate G, Hosseini‐Beheshti E, Caradec J, Chin MY, Tomlinson Guns ES. Epidermal Growth Factor Receptor in Prostate Cancer Derived Exosomes. PLoS ONE. 2016;11(5):e0154967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Thakur BK, Zhang H, Becker A, et al. Double‐stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Vered M, Lehtonen M, Hotakainen L, et al. Caveolin‐1 accumulation in the tongue cancer tumor microenvironment is significantly associated with poor prognosis: an in‐vivo and in‐vitro study. BMC Cancer. 2015;15:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Chen KG, Valencia JC, Lai B, et al. Melanosomal sequestration of cytotoxic drugs contributes to the intractability of malignant melanomas. Proc Natl Acad Sci USA. 2006;103(26):9903–9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Gamperl H, Plattfaut C, Freund A, Quecke T, Theophil F, Gieseler F. Extracellular vesicles from malignant effusions induce tumor cell migration: inhibitory effect of LMWH tinzaparin. Cell Biol Int. 2016;40(10):1050–1061. [DOI] [PubMed] [Google Scholar]
- 162. Logozzi M, De Milito A, Lugini L, et al. High levels of exosomes expressing CD63 and caveolin‐1 in plasma of melanoma patients. PLoS ONE. 2009;4(4):e5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Kosaka N, Iguchi H, Yoshioka Y, Hagiwara K, Takeshita F, Ochiya T. Competitive interactions of cancer cells and normal cells via secretory microRNAs. J Biol Chem. 2012;287(2):1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Ohno S, Takanashi M, Sudo K, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Koyama Y, Ito T, Hasegawa A, et al. Exosomes derived from tumor cells genetically modified to express Mycobacterium tuberculosis antigen: a novel vaccine for cancer therapy. Biotechnol Lett. 2016;38(11):1857–1866. [DOI] [PubMed] [Google Scholar]
- 166. Wang Z, Chen JQ, Liu JL, Tian L. Exosomes in tumor microenvironment: novel transporters and biomarkers. J Transl Med. 2016;14(1):297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Nawaz M, Fatima F, Nazarenko I, et al. Extracellular vesicles in ovarian cancer: applications to tumor biology, immunotherapy and biomarker discovery. Expert Rev Proteomics. 2016;13(4):395–409. [DOI] [PubMed] [Google Scholar]
- 168. Soung YH, Ford S, Zhang V, Chung J. Exosomes in Cancer Diagnostics. Cancers. 2017;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Miller MC, Doyle GV, Terstappen LW. Significance of Circulating Tumor Cells Detected by the Cell Search System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J Oncol. 2010;2010:617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J. 1869;14:146. [Google Scholar]
- 171. Aceto N, Bardia A, Miyamoto DT, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158(5):1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Chen JF, Ho H, Lichterman J, et al. Subclassification of prostate cancer circulating tumor cells by nuclear size reveals very small nuclear circulating tumor cells in patients with visceral metastases. Cancer. 2015;121(18):3240–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. de Wit S, van Dalum G, Lenferink AT, et al. The detection of EpCAM(+) and EpCAM(‐) circulating tumor cells. Scientific Rep. 2015;5:12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Au SH, Storey BD, Moore JC, et al. Clusters of circulating tumor cells traverse capillary‐sized vessels. Proc Natl Acad Sci USA. 2016;113(18):4947–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Cheung KJ, Padmanaban V, Silvestri V, et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14‐expressing tumor cell clusters. Proc Natl Acad Sci USA. 2016;113(7):E854‐E863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Geng Y, Chandrasekaran S, Hsu JW, Gidwani M, Hughes AD, King MR. Phenotypic switch in blood: effects of pro‐inflammatory cytokines on breast cancer cell aggregation and adhesion. PLoS ONE. 2013;8(1):e54959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Upreti M, Jamshidi‐Parsian A, Koonce NA, et al. Tumor‐endothelial cell three‐dimensional spheroids: new aspects to enhance radiation and drug therapeutics. Transl Oncol. 2011;4(6):365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Sharma D, Brummel‐Ziedins KE, Bouchard BA, Holmes CE. Platelets in tumor progression: a host factor that offers multiple potential targets in the treatment of cancer. J Cell Physiol. 2014;229(8):1005–1015. [DOI] [PubMed] [Google Scholar]
- 179. Denes V, Lakk M, Makarovskiy A, et al. Metastasis blood test by flow cytometry: in vivo cancer spheroids and the role of hypoxia. Int J Cancer. 2015;136(7):1528–1536. [DOI] [PubMed] [Google Scholar]
- 180. Fiorelli A, Accardo M, Carelli E, Angioletti D, Santini M, Di Domenico M. Circulating Tumor Cells in Diagnosing Lung Cancer: Clinical and Morphologic Analysis. Ann Thorac Surg. 2015;99(6):1899–1905. [DOI] [PubMed] [Google Scholar]
- 181. Ilie M, Hofman V, Long‐Mira E, et al. "Sentinel" circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS ONE. 2014;9(10):e111597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Bidard FC, Peeters DJ, Fehm T, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15(4):406–414. [DOI] [PubMed] [Google Scholar]
- 183. Yokobori T, Iinuma H, Shimamura T, et al. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial‐mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res. 2013;73(7):2059–2069. [DOI] [PubMed] [Google Scholar]
- 184. Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. New England J Med. 2004;351(8):781–791. [DOI] [PubMed] [Google Scholar]
- 185. Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression‐free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(19):3213–3221. [DOI] [PubMed] [Google Scholar]
- 186. de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration‐resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302–6309. [DOI] [PubMed] [Google Scholar]
- 187. Goldkorn A, Ely B, Quinn DI, et al. Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: a phase III trial of docetaxel with or without atrasentan for metastatic castration‐resistant prostate cancer. J Clin Oncol. 2014;32(11):1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Zitt M, Zitt M, Muller HM, et al. Disseminated tumor cells in peripheral blood: a novel marker for therapy response in locally advanced rectal cancer patients undergoing preoperative chemoradiation. Dis Colon Rectum. 2006;49(10):1484–1491. [DOI] [PubMed] [Google Scholar]
- 189. Kienle P, Koch M, Autschbach F, et al. Decreased detection rate of disseminated tumor cells of rectal cancer patients after preoperative chemoradiation: a first step towards a molecular surrogate marker for neoadjuvant treatment in colorectal cancer. Ann Surg. 2003;238(3): 324–330; discussion 330–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small‐cell lung cancer. J Clin Oncol. 2012;30(5):525–532. [DOI] [PubMed] [Google Scholar]
- 191. Pearl ML, Dong H, Tulley S, et al. Treatment monitoring of patients with epithelial ovarian cancer using invasive circulating tumor cells (iCTCs). Gynecologic Oncol. 2015;137(2):229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Georgoulias V, Bozionelou V, Agelaki S, et al. Trastuzumab decreases the incidence of clinical relapses in patients with early breast cancer presenting chemotherapy‐resistant CK‐19mRNA‐positive circulating tumor cells: results of a randomized phase II study. Ann Oncol. 2012;23(7):1744–1750. [DOI] [PubMed] [Google Scholar]
- 193. Bidard FC, Pierga JY. Clinical utility of circulating tumor cells in metastatic breast cancer. J Clin Oncol. 2015;33(14):1622. [DOI] [PubMed] [Google Scholar]
- 194. Cabel L, Proudhon C, Gortais H, et al. Circulating tumor cells: clinical validity and utility. Int J Clin Oncol. 2017;22(3):421–430. [DOI] [PubMed] [Google Scholar]
- 195. Yap TA, Lorente D, Omlin A, Olmos D, de Bono JS. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res. 2014;20(10):2553–2568. [DOI] [PubMed] [Google Scholar]
- 196. Hong Y, Fang F, Zhang Q. Circulating tumor cell clusters: What we know and what we expect (Review). Int J Oncol. 2016;49(6):2206–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]


