Abstract
Inflammation in tumor microenvironments is implicated in the pathogenesis of tumor development. In particular, inflammasomes, which modulate innate immune functions, are linked to tumor growth and anticancer responses. However, the role of the NLRC4 inflammasome in gliomas remains unclear. Here, we investigated whether the upregulation of the NLRC4 inflammasome is associated with the clinical prognosis of gliomas. We analyzed the protein expression and localization of NLRC4 in glioma tissues from 11 patients by immunohistochemistry. We examined the interaction between the expression of NLRC4 and clinical prognosis via a Kaplan-Meier survival analysis. The level of NLRC4 protein was increased in brain tissues, specifically, in astrocytes, from glioma patients. NLRC4 expression was associated with a poor prognosis in glioma patients, and the upregulation of NLRC4 in astrocytomas was associated with poor survival. Furthermore, hierarchical clustering of data from the Cancer Genome Atlas dataset showed that NLRC4 was highly expressed in gliomas relative to that in a normal healthy group. Our results suggest that the upregulation of the NLRC4 inflammasome contributes to a poor prognosis for gliomas and presents a potential therapeutic target and diagnostic marker.
Subject terms: CNS cancer, Cancer microenvironment
Introduction
Glioma represents the most prevalent primary tumor of the central nervous system, with high morbidity and mortality rates. The standard therapy for gliomas comprises tumor resection and subsequent radiotherapy and chemotherapy with temozolomide in the adjuvant setting1,2. However, the efficacy of this multimodal therapeutic strategy is limited as a result of the overall resistance to therapy and the high risk of recurrence3, for which there are limited therapies, including with nitrosoureas and bevacizumab, with minimal clinical efficacy4. Therefore, a novel approach is greatly needed to address the extremely poor therapeutic results for patients with gliomas.
Mounting evidence indicates that the inflammatory microenvironment plays an important role in tumor development5,6. However, innate immunity regulating the inflammatory reaction in tumor microenvironments remains poorly characterized. Inflammasomes are critical intracellular multiprotein complexes of innate inflammatory pathways that are responsible for the production of interleukin (IL)-1β and IL-187,8. Although inflammasomes are important for central nervous system diseases such as Alzheimer’s disease, multiple sclerosis, and traumatic injury, the role of inflammasomes in brain tumors remains unclear9–11.
Inflammasomes are formed by the assembly of cytosolic sensors (AIM2-like receptors and nucleotide-binding domain and leucine-rich-repeat-containing receptors, also known as NOD-like receptors [NLRs]), an adaptor protein apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC), and an effector caspase, procaspase-17. Two distinct signals are required for the activation of inflammasomes. The first involves pathogen- or danger-associated molecular patterns, including bacteria and viruses or reactive oxygen species, ATP, and DNA12,13, which are recognized by pattern recognition receptors such as Toll-like receptor or NOD family members on microglia, astrocytes, and macrophages in the central nervous system and prime the expression of pro-IL-1β and pro-IL-1814. The second signal involves tissue damage, metabolic dysregulation, ATP, and hyaluronan, which induce the cleavage of pro-IL-1β and pro-IL-18 to mature forms15,16.
Recent data indicate that inflammasomes play important roles in carcinogenesis and tumor progression17. Many endogenous and exogenous signals induce chronic inflammation and inflammasome activation in cancer18. We have previously reported that cellular glycolysis pathway, which plays an important role in cancer energy metabolism, are associated with NLRP3 inflammasome activation in macrophages under pro-inflammatory conditions19,20. However, the role of inflammasomes in tumors remains disputed.
In the present study, we comprehensively analyzed the immunohistological expression and distribution of inflammasome-associated components in tissues from glioma patients. We also performed a genome-wide analysis to assess the expression of inflammasome-associated genes and their association with the overall survival of patients diagnosed with gliomas included in the Cancer Genome Atlas (TCGA) database. Our data suggest that NLRC4 may represent not only a potential therapeutic target for gliomas, but also a biomarker for diagnosis and prognosis in brain cancer.
Results
Expression of the NLRC4 inflammasome in glioma patients
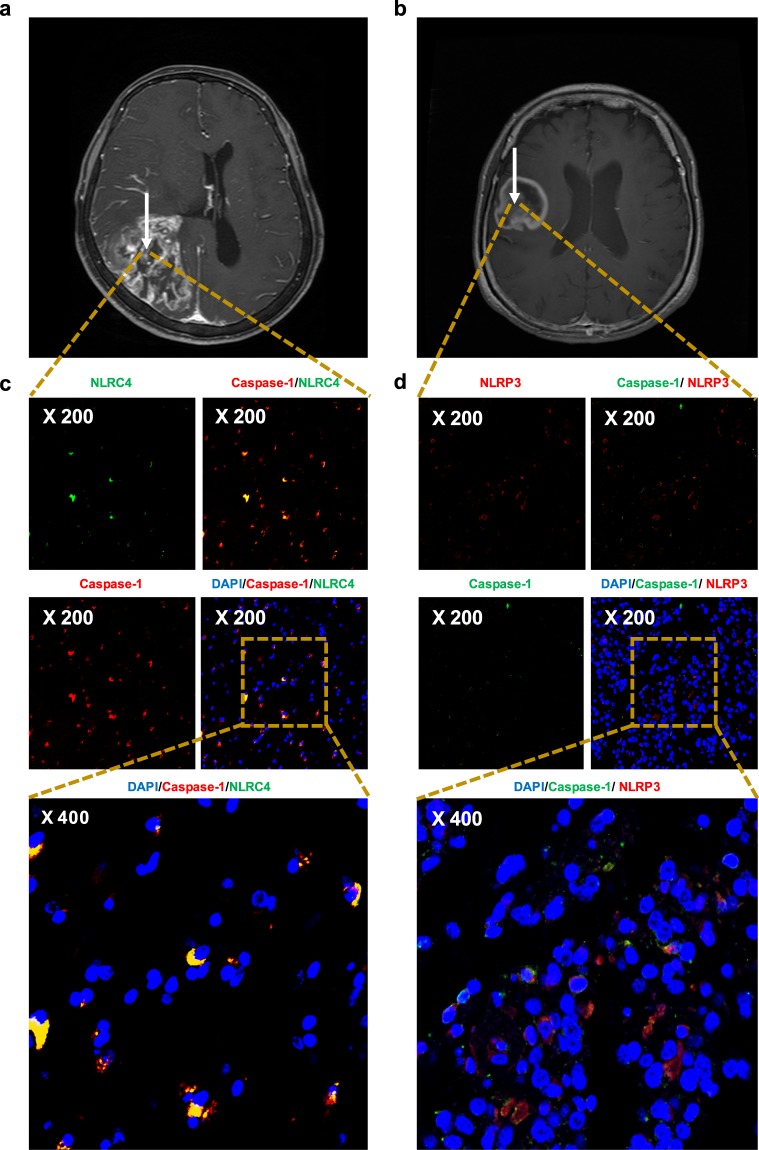
Representative MRI results from patients with GBM are shown in Fig. 1a,b. Consistent with a previous report showing that the NLRP3 inflammasome is activated in patient-derived cell lines and microglia17, we found that the mRNA levels of NLRC4 were significantly elevated in glioma patients (see Supplementary Fig. S1g). Thus, NLRC4 and NLRP3 were selected for immunohistochemical examination. Additionally, colocalization with caspase-1 was examined as a marker of inflammasome activation18. As shown in Fig. 1c,d (see also Supplementary Fig. S2), NLRC4 and NLRP3 were detected by immunostaining in the brains of glioma patients. Moreover, both proteins show colocalization with caspase-1.
Figure 1.
NLRC4 and NLRP3 inflammasomes are present and activated in brain tissue from glioma patients. Preoperative magnetic resonance imaging scans of 62-year-old (a) and 76-year-old (b) female patients with GBM. White arrows highlight tumor areas. (c) Human glioma specimens were analyzed by immunohistochemistry for the expression of NLRC4 (green) and caspase-1 (red). (d) Samples were stained for NLRP3 (red) and caspase-1 (green). Nuclei were stained with DAPI (blue).
Caspase-1 is present in the cytosol as an inactive zymogen19. In response to pathogen infection and tissue damage, pattern recognition receptors (PRRs) (NLRP3, NLRC4) oligomer is formed and it can recruit ASC. Then procaspase-1 can be recruited to the PRR-ASC complex by their CARD (caspase recruitment domain). This recruitment converts procaspase-1 into active caspase-1 via proximity-mediated cleavage20. Therefore, Fig. 1c,d (colocalization of NLR families and caspase-1) indicates that inflammasome were assembled and activated in the tumor tissues.
High NLRC4 expression is associated with poor overall survival in glioma patients
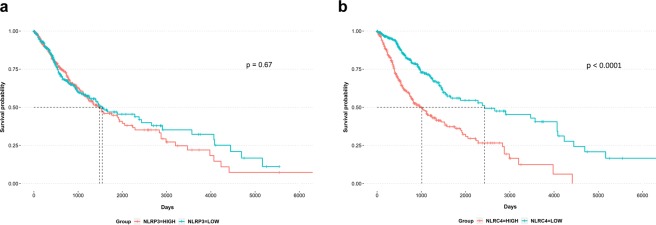
To investigate NLRC4 and NLRP3 inflammasomes as potential prognostic markers in glioma patients, we compared NLRP3 (Fig. 2a) or NLRC4 (Fig. 2b) expression with survival using TCGA data. Kaplan-Meier survival plots indicated that glioma patients with high NLRC4 expression had a significantly lower overall survival than those with low NLRC4 expression, whereas the expression of NLRP3 did not have a significant association with overall survival.
Figure 2.
High NLRC4 expression is associated with poor survival. Kaplan-Meier plots for comparing survival rates of glioma patients in relation to expression levels of NLRP3 (a) and NLRC4 (b) in the tumors based on the GBMLGG dataset from TCGA. Divided two groups by median value of each expression level.
Sutterwala et al. reported that NLRC4 acts independently of inflammasome activation to suppress melanoma tumor progression21. However, the role of inflammasome differ among cancer types. Figure 2b showed that the median survival time of glioma patients with highly expression NLRC4 was significantly decreased compared to patients with low NLRC4 expression. In addition, we provided expression patterns of genes which are involved to NLRC4-mediated inflammasome including IL1B which is one of final product of NLRC4 inflammasome (Fig. S1). IL1B was significantly up-regulated according to poor prognosis group. On the other hand, previous report demonstrated that secreted IL-1β promotes chronic inflammation and induces glioma progression17. Collectively, these results indicate that NLRC4 inflammasome promotes glioma progression.
High NLRC4 expression in astrocytes
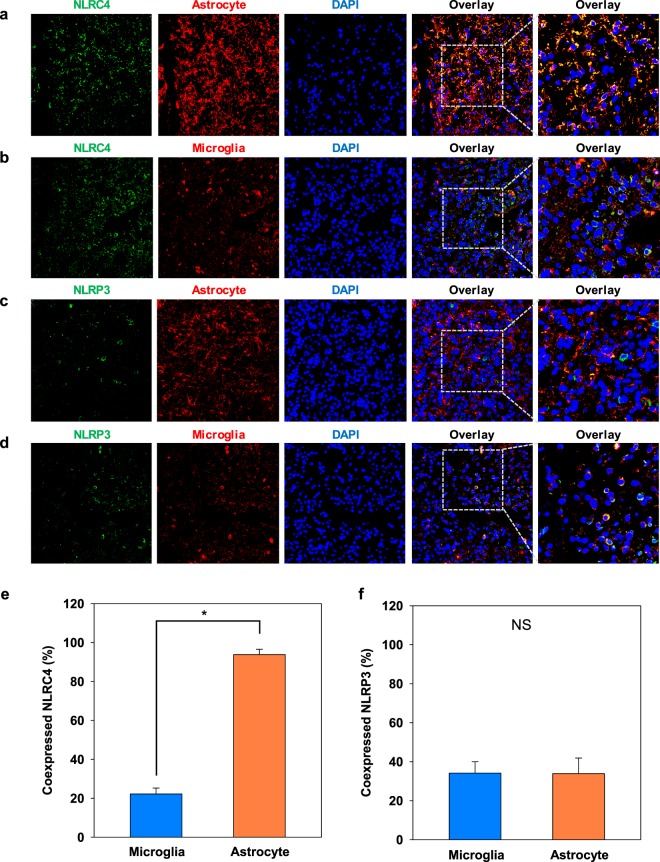
Despite the study of NLRC4 and NLRP3 inflammasomes in several neurological diseases (Alzheimer’s disease, Parkinson’s disease, stroke, and multiple sclerosis), their roles in gliomas remain undefined. The results in Fig. 1 indicated that NLRC4 and NLRP3 inflammasomes are activated in glioma patients. To better understand inflammasome function in the glial compartment, we compared their expression among different cell types, mainly microglia (identified by staining for Iba1) and astrocytes (identified by GFAP expression). Confocal images showed that both microglia and astrocytes expressed NLRC4 and NLRP3 components (Fig. 3a–d), with strong overlap between NLRC4 and GFAP, as shown by the yellow color in the merged image. Quantification via analyses with ImageJ confirmed this colocalization of NLRC4 in astrocytes (t-test, p-value < 0.001), whereas NLRP3 was expressed similarly by both astrocytes and microglia (Fig. 3e,f).
Figure 3.
NLRC4 is highly expressed in astrocytes from glioma patients. Astrocytes (a,c) and microglia (b,d) were analyzed by immunohistochemistry for the expression of NLRC4 and NLRP3, respectively. Nuclei were stained with DAPI (blue). Co-expression of NLRC4 (e) or NLRP3 (f) with markers for microglia and astrocytes was quantified by ImageJ software. The values are means ± SEMs from three replicates (n = 8). *T-test p-value < 0.001.
Expression of inflammasome-associated genes in glioma patients
As there is a report demonstrating that human malignant gliomas activate NLRP3 inflammasomes and express IL-1β17, we sought to further investigate the connection between inflammasomes and gliomas. Pairwise Pearson’s correlation matrix heat maps of the expression of genes in NOD like receptors signaling pathway (hsa04621) which is related to inflammasomes from the KEGG database are shown for normal solid tissue and LGG and GBM samples from the TCGA database (Supplementary Fig. S1a,c,e, respectively). We found that the genes necessary for the assembly of functional inflammasome complexes were positively correlated in glioma patients but not in normal individuals. Supplementary Fig. 2b,d,f show the nearest neighbors for components of the canonical inflammasome complex (CASP1 [encoding caspase-1], PYCARD [encoding ASC], and NLRP [encoding NLRP]) with strong positive correlations. Genes encoding numerous NLR family proteins (NLRC4, NOD1, NOD2, NLRP1, NLRP3, NLRP12, and NAIP) and proinflammatory caspases/inflammasome adaptor molecules (PYCARD, CASP1, CASP4, and CASP5) were positively correlated with inflammasome complex genes in both LGG and GBM groups (Supplementary Fig. S1b,d,f) but not in normal brain (e.g., NLRC4, NLRP3, PYCARD, and CASP1; Supplementary Fig. S1b). Additionally, genes encoding proinflammatory cytokines regulated by the inflammasome (e.g., IL1B) and transcription factors (e.g., NFKB1) that prime inflammasome oligomerization and activation were significantly associated with inflammasome complex expression in LGG and GBM TCGA datasets. Furthermore, the expression levels of genes encoding NLR proteins (NLRP12, NOD2, NOD1, NLRC4, and NAIP), inflammasome adaptor molecules (PYCARD), and proinflammatory caspases (CASP1, CASP4, and CASP5) were significantly higher in glioma patients than in normal controls (Supplementary Fig. S1g). Moreover, the expression of inflammasome-associated genes was higher in GBM than in LGG. These results indicate that the inflammasome-associated pathway is activated in glioma patients.
NLRC4 inflammasome in glioma patients
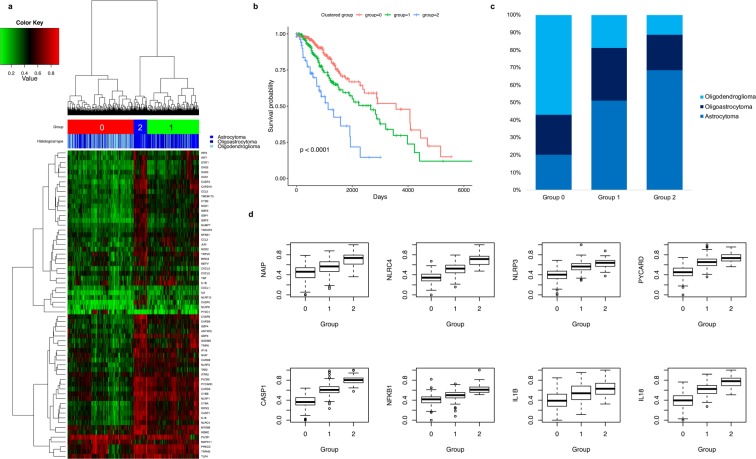
To identify further potential associations between groups of inflammasome-associated genes with similar profiles and histological subtype and patient survival, we performed unsupervised hierarchical clustering of LGG data from the TCGA database (Fig. 4a). Most of the genes that were expressed at low levels clustered into group 0, whereas those with medium levels of expression were clustered into group 1 (Fig. 4a). A stronger association was found among highly expressed genes, such as NLRC4, as well as NLRP3, PYCARD, CASP1, IL1B, and IL18, which clustered in group 2 (Fig. 4d, Supplementary Tables S2 and S3). The expression levels of these genes (Fig. 4d) were higher in the astrocytoma subtype than in oligodendroglioma and oligoastrocytoma subtypes (Fig. 4c).
Figure 4.
NLRC4 is highly expressed in patients with astrocytomas and associated with poor prognosis. (a) Cluster map depicting the 64 mRNAs that were significantly differently expressed according to agglomerative clustering for inflammasome-associated genes from the TCGA LGG dataset (n = 525). Red represents high expression, green represents low expression, and the color scale depicted above the heat map displays histological types. (b) Kaplan-Meier survival curves for groups in panel A. (c) Plot of proportions of histological types by group. (d) Boxplots of expression levels with inflammasome-related key proteins including NLRC4 and NLRP3. The expression levels, log2[x + 1] RNA-seq expectation maximization (RSEM) counts, were transformed to minimum-maximum (0–1) normalized value.
To investigate the prognosis of cancer with high expression of inflammasome-associated genes, overall survival of the patients was analyzed within the groups 0 to 2 (Fig. 4b). The best outcomes were associated with patients clustered in group 0, which exhibit low expression of these genes (Fig. 4d), whereas those with high levels of expression, in group 2, had the worst clinical outcomes (Pair-wise log-rank p-values < 0.01 after Benjamini-Hochberg corrections). These results indicate that high expression of inflammasome-related genes in glioma patients is associated with a poor prognosis (Fig. 4b,d and Tables S2 and S3). Especially, expression levels of the genes in NLRC4-mediated inflammasome pathway, including NAIP, NLRC4, PYCARD, CASP1, NFKB1, IL1B, and IL18, significantly increased according to the poor prognosis group (Fig. 4b,d).
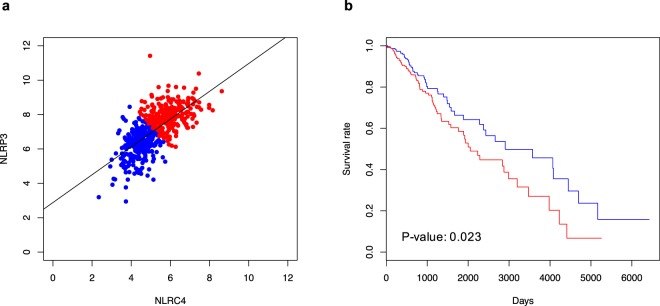
Although NLRC4 and NLRP3 typically form distinct inflammasome complexes22–24, recent reports demonstrate that the NLRC4 inflammasome recruits NLRP3 during Salmonella infection25, indicating that both are important for the activation of bacteria-mediated inflammasomes. To determine if this may also occur with inflammasomes in gliomas, we performed a correlation analysis using data from the TCGA dataset. NLRC4 expression was positively correlated with NLRP3 expression in GBM and LGG patients (correlation coefficient = 0.64, p < 0.001; Fig. 5a). In addition, a Kaplan-Meier survival analysis based on NLRP3 and NLRC4 expression demonstrated that patients with high expression of both had a significantly poorer prognosis (p < 0.023; Fig. 5b). Collectively, these data indicate that both NLRC4 and NLRP3 are upregulated in gliomas and suggest that they potentially associated with glioma progression.
Figure 5.
Correlated NLRC4 and NLRP3 expression in glioma patients (a) Dot plots from LGG dataset from TCGA for gliomas presenting correlations between NLRC4 and NLRP3. (b) Kaplan-Meier survival plot for the TCGA glioma samples according to the co-expression levels of NLRC4 and NLRP3.
Discussion
There is emerging evidence that chronic inflammation in the tumor microenvironment impacts all stages of carcinogenesis and, depending on the proinflammatory cytokines, is associated with a poor prognosis26. Such inflammation, which involves chemokines, cytokines, and the extracellular matrix, is considered the seventh hallmark of cancer5. Inflammasomes contribute to the initiation of cancer-related inflammation and have various functions in tumorigenesis27. However, the roles of inflammasome components differ among cancer types27,28, inhibiting tumorigenesis in colitis-associated colon cancer29,30 and promoting the development of epithelial skin cancer and breast cancer tumor growth and metastasis29,31. Feng et al.32 showed that high NLRP3 expression was associated with poor survival in patients with oral squamous cell carcinoma, whereas the survival of patients with breast cancer was correlated positively with NLRP3 and negatively with NLRC4 as reported by Kolb et al.28. By contrast, NLRC4 expression was found to show no correlation with the survival of patients with hepatocellular carcinoma by Wang et al.33. Recently, Xiao-Feng et al. reported that NLRP3 inflammsome promotes glioma by regulating cellular proliferation, apoptosis and metastasis34. However, the clinical role of the inflammasome in gliomas is not known. The results of this study reveal that NLRP3 and NLRC4 inflammasomes are expressed and activated in gliomas and that high expression of NLRC4 in particular is associated with poor overall survival.
To better understand the role of inflammasomes in gliomas, their composition and activation in inflammatory cells should be characterized. Whereas microglia, the resident macrophages, account for only 10–15% of all cells in brain35, astrocytes, which maintain ion homeostasis and support neurons, are the most abundant36. Gustin et al.37 showed that NLRP3 inflammasomes are present and activated by lipopolysaccharide in microglia but not astrocytes in mouse brain. However, Freeman et al.38 showed that NLRP3 and NLRC4 inflammasomes are active in both microglia and astrocytes, with stronger NLRC4 expression in astrocytes, in a model of multiple sclerosis, similar to what we observed in samples from glioma patients in this study. Remarkably, the expression of NLRC4 was higher in the astrocytoma subtype than in oligodendroglioma and oligoastrocytoma subtypes (Fig. 4c,d) and was associated with poor survival (Fig. 4b).
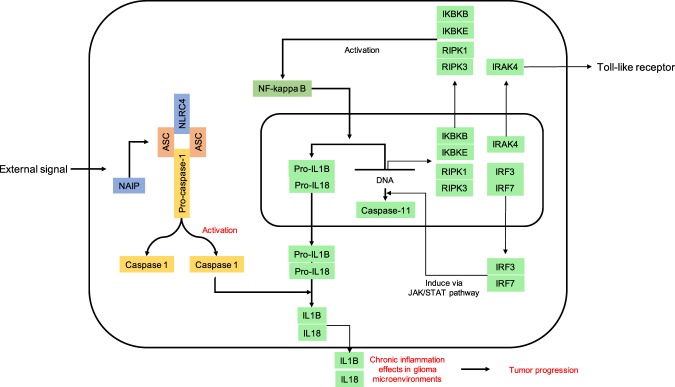
Although astrocytic NLRC4 has been implicated in Alzheimer’s disease and multiple sclerosis38,39, the present report using the TCGA glioma dataset is the first description of NLRC4 expression and its effects on the clinical prognosis in patients with astrocytomas (Fig. 4). We also provide the first characterization of inflammasome-associated transcripts in LGG and GBM patients. Overall, we observed positive correlations in glioma patients, but not normal controls, among genes encoding NLR family proteins, an adaptor molecule, and proinflammatory caspases, which are key components of the inflammasome (Fig. 6). Of note, patients with GBM showed high expression of many of these as well as of IKBKB and IKBKE (Supplementary Fig. S1g and Table S1), which encode kinases that relieve inhibition of NF-κB40. NF-κB regulates inflammasome activation by inducing the transcription of pro-IL-1β and NLRP341,42. Recently, Irrera et al.43 blocked the NF-κB and NLRP3 inflammasome pathways with pharmacological inhibition of IKK-β (encoded by IKBKB), demonstrating its importance in inflammasome activation. Patients with GBM also showed high expression of RIPK1 and RIPK3, which encode receptor-interacting serine/threonine protein kinases shown to drive NF-κB and inflammasome activation44,45, and IRAK4, which encodes IL-1 receptor-associated kinase 4, a protein kinase involved in innate immune response signaling from Toll-like receptors and associated with NLRP3 inflammasome activation46,47. Finally, our results show that tumor tissues in GBM patients highly express IRF3 and IRF7, encoding interferon regulatory factors that can lead to the transcription of caspase-11, which is important for noncanonical NLRP3 inflammasome activation, via activation of the JAK/STAT pathway48–50.
Figure 6.
Schematic illustration of NLRC4 inflammasome pathway in glioma.
In conclusion, the histological and bioinformatics analyses of tissue samples and TCGA data from glioma patients provide evidence that NLRC4 and NLRP3 inflammasomes are activated in gliomas. Moreover, high expression of NLRC4 was associated with poor patient survival. These findings contribute to the understanding of the role of the inflammasome in brain gliomas and suggest that NLRC4 is a potential therapeutic target and biomarker for diagnosis and prognosis in brain cancer.
Materials and Methods
Sample acquisition
Glioma specimens obtained during surgery from 11 patients (Table 1) at the Department of Neurosurgery, CHA Bundang Medical Center, CHA University (Republic of Korea), were stored at room temperature in neutral buffered saline. Among these patients, six had GBM (WHO grade IV), two had oligodendrogliomas (WHO grade II), one had a diffuse midline glioma (WHO grade IV), one had an anaplastic ganglioglioma (WHO grade III), and one had an anaplastic astrocytoma (WHO grade III). The median age of the patients was 51 years (range, 9–76 years).
Table 1.
Glioma patients: clinical synopsis.
| Patient no. | Gender/Age | Clinical diagnosis | IDH-1 mutant |
|---|---|---|---|
| 1 | F/76 | Glioblastoma | Negative |
| 2 | F/51 | Oligodendroglioma | Positive |
| 3 | M/28 | Oligodendroglioma | Negative |
| 4 | M/67 | Glioblastoma | Negative |
| 5 | F/9 | Diffuse midline glioma (grade IV) | Negative |
| 6 | F/10 | Anaplastic ganglioglioma (grade III) | Negative |
| 7 | F/59 | Anaplastic astrocytoma | Negative |
| 8 | F/62 | Glioblastoma | Negative |
| 9 | F/46 | Glioblastoma | Negative |
| 10 | M/50 | Glioblastoma | Negative |
| 11 | M/53 | Glioblastoma | Negative |
The Bundang CHA Hospital Institutional Review Board approval was obtained for this study (CHAMC 2016-04-012), and all patients signed informed consent forms. And all methods were performed in accordance with the relevant guidelines and regulations.
Immunohistochemistry
Formalin-fixed paraffin-embedded glioma blocks were serially sectioned at a 4 µm thickness and deparaffinized in xylene. The sections were rehydrated after immersing in ethanol and incubated with goat serum to block nonspecific binding. The sections were then incubated overnight at 4 °C with primary rabbit monoclonal antibodies against NLRP3 and CARD12 or with mouse monoclonal antibodies against ionized calcium-binding adaptor protein 1 (Iba1) or glial fibrillary acidic protein (GFAP) (Abcam, Cambridge, MA). Secondary antibodies were applied for 1 h at room temperature. Images were acquired using a confocal laser scanning microscope (Zeiss LSM 880; Carl Zeiss, Jena, Germany).
Description of TCGA data
Transcriptome sequencing and clinical data of GBM and low-grade glioma (LGG) were downloaded from the XenaTCGA database. The tumor statuses were LGG (n = 525) and GBMLGG (n = 702) dataset, and the sample types of GBMLGG dataset were recurrent tumor (n = 27), primary tumor (n = 670), and solid tissue normal (n = 5).
Correlation analysis and expression patterns of inflammasome-associated genes in the KEGG pathway
The genes associated with the NLR signaling pathway (KEGG entry number hsa04621), involving two prototypic NLRs that induce caspase-1 via inflammasomes, were identified from the KEGG database. Pairwise Pearson’s correlation analyses between the expression of each of the NLR signaling pathway-related genes were performed and visualized as a cluster map based on a hierarchical method with Euclidean distances for recurrent, primary tumor, and solid tissue normal groups. To analyze gene expression patterns, agglomerative clustering (using ward method and Euclidean distance) was performed with whole NLR signaling pathway-related genes which are expressed over the 1 and divided 2 groups in LGG patients (n = 525) of TCGA. We extracted 64 differentially expressed genes between the 2 groups by t-test with Bonferroni corrections and absolute difference their expression levels over the 0.5. After extracting 64 genes, we performed agglomerative clustering again with the 64 genes using ward method and Euclidean distance after minimum 0 to maximum 1 normalized value.
Statistical analysis
Pairwise log-rank tests with Benjamini-Hochberg corrections were performed to compare prognoses between the three groups which were divided by agglomerative clustering with 64 genes, and Kaplan-Meier plots were generated in all survival analyses. Two-dimensional K-means clustering of NLRC4 and NLRP3, the two major inflammasome-related genes in the NOD-like receptors signaling pathway in KEGG, was performed, and the survival rates were compared between the two clusters.
Supplementary information
Acknowledgements
This research was supported by grants of the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1D1A1B03029598), the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant no. HI16C1559), and the Industrial Technology Innovation Program through the Ministry of Trade, Industry and Energy (Korea), funded by the Ministry of Trade, Industry and Energy, Republic of Korea (Grant No. 10067378).
Author Contributions
J.J. Lim, M.J. Kim and Y. Park are co-first authors. J.J. Lim, M.J. Kim and Y. Park performed the majority of experiments and analyses. Y. Park and J.W. performed bioinformatic analyses for public data base. J.J. Lim and K.G. Cho provided surgical specimens. J.J. Lim and S.J. Hwang helped interpretation of clinical data. J.J. Lim, M.J. Kim, Y. Park and J.W. Ahn organized and processed specimens and genome data. J.J. Lim and M.J. Kim wrote the manuscript with the feedback from J.S. Moon, K.G. Cho, K. Kwack. K.G. Cho and K. Kwack designed and supervised the entire project.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jaejoon Lim, Min Jun Kim and YoungJoon Park contributed equally.
Contributor Information
Kyung Gi Cho, Email: sandori50@gmail.com.
KyuBum Kwack, Email: kbkwack@cha.ac.kr.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44261-9.
References
- 1.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, et al. Maintenance Therapy With Tumor-Treating Fields Plus Temozolomide vs Temozolomide Alone for Glioblastoma: A Randomized Clinical Trial. JAMA. 2015;314:2535–2543. doi: 10.1001/jama.2015.16669. [DOI] [PubMed] [Google Scholar]
- 3.Ammirati M, Galicich JH, Arbit E, Liao Y. Reoperation in the treatment of recurrent intracranial malignant gliomas. Neurosurgery. 1987;21:607–614. doi: 10.1227/00006123-198711000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Tosoni A, Franceschi E, Poggi R, Brandes AA. Relapsed Glioblastoma: Treatment Strategies for Initial and Subsequent Recurrences. Curr Treat Options Oncol. 2016;17:49. doi: 10.1007/s11864-016-0422-4. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rathinam VA, Fitzgerald KA. Inflammasome Complexes: Emerging Mechanisms and Effector Functions. Cell. 2016;165:792–800. doi: 10.1016/j.cell.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Halle A, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heneka MT, et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature. 2013;493:674–678. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adamczak SE, et al. Pyroptotic neuronal cell death mediated by the AIM2 inflammasome. J Cereb Blood Flow Metab. 2014;34:621–629. doi: 10.1038/jcbfm.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 13.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Bryant C, Fitzgerald KA. Molecular mechanisms involved in inflammasome activation. Trends Cell Biol. 2009;19:455–464. doi: 10.1016/j.tcb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Koizumi Y, et al. Inflammasome activation via intracellular NLRs triggered by bacterial infection. Cell Microbiol. 2012;14:149–154. doi: 10.1111/j.1462-5822.2011.01707.x. [DOI] [PubMed] [Google Scholar]
- 16.Lamkanfi M, Dixit VM. Inflammasomes: guardians of cytosolic sanctity. Immunol Rev. 2009;227:95–105. doi: 10.1111/j.1600-065X.2008.00730.x. [DOI] [PubMed] [Google Scholar]
- 17.Tarassishin L, Casper D, Lee SC. Aberrant expression of interleukin-1beta and inflammasome activation in human malignant gliomas. PLoS One. 2014;9:e103432. doi: 10.1371/journal.pone.0103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, et al. Activation of Nod-like receptor protein 3 inflammasomes turns on podocyte injury and glomerular sclerosis in hyperhomocysteinemia. Hypertension. 2012;60:154–162. doi: 10.1161/HYPERTENSIONAHA.111.189688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thornberry NA, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y. Caspase activation: revisiting the induced proximity model. Cell. 2004;117:855–858. doi: 10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Janowski AM, et al. NLRC4 suppresses melanoma tumor progression independently of inflammasome activation. J Clin Invest. 2016;126:3917–3928. doi: 10.1172/JCI86953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 23.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 25.Qu Y, et al. NLRP3 recruitment by NLRC4 during Salmonella infection. J Exp Med. 2016;213:877–885. doi: 10.1084/jem.20132234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, Grivennikov SI, Karin M. The unholy trinity: inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer Cell. 2011;19:429–431. doi: 10.1016/j.ccr.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zitvogel L, Kepp O, Galluzzi L, Kroemer G. Inflammasomes in carcinogenesis and anticancer immune responses. Nat Immunol. 2012;13:343–351. doi: 10.1038/ni.2224. [DOI] [PubMed] [Google Scholar]
- 28.Kolb R, et al. Obesity-associated NLRC4 inflammasome activation drives breast cancer progression. Nat Commun. 2016;7:13007. doi: 10.1038/ncomms13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Man SM, et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell. 2015;162:45–58. doi: 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dupaul-Chicoine J, et al. The Nlrp3 Inflammasome Suppresses Colorectal Cancer Metastatic Growth in the Liver by Promoting Natural Killer Cell Tumoricidal Activity. Immunity. 2015;43:751–763. doi: 10.1016/j.immuni.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Guo B, Fu S, Zhang J, Liu B, Li Z. Targeting inflammasome/IL-1 pathways for cancer immunotherapy. Sci Rep. 2016;6:36107. doi: 10.1038/srep36107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng X, et al. The role of NLRP3 inflammasome in 5-fluorouracil resistance of oral squamous cell carcinoma. J Exp Clin Cancer Res. 2017;36:81. doi: 10.1186/s13046-017-0553-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, et al. NLRC and NLRX gene family mRNA expression and prognostic value in hepatocellular carcinoma. Cancer Med. 2017;6:2660–2672. doi: 10.1002/cam4.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin XF, et al. NLRP3 in human glioma is correlated with increased WHO grade, and regulates cellular proliferation, apoptosis and metastasis via epithelial-mesenchymal transition and the PTEN/AKT signaling pathway. Int J Oncol. 2018;53:973–986. doi: 10.3892/ijo.2018.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filiano AJ, Gadani SP, Kipnis J. Interactions of innate and adaptive immunity in brain development and function. Brain Res. 2015;1617:18–27. doi: 10.1016/j.brainres.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gustin A, et al. NLRP3 Inflammasome Is Expressed and Functional in Mouse Brain Microglia but Not in Astrocytes. PLoS One. 2015;10:e0130624. doi: 10.1371/journal.pone.0130624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freeman L, et al. NLR members NLRC4 and NLRP3 mediate sterile inflammasome activation in microglia and astrocytes. J Exp Med. 2017;214:1351–1370. doi: 10.1084/jem.20150237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L, Chan C. IPAF inflammasome is involved in interleukin-1beta production from astrocytes, induced by palmitate; implications for Alzheimer’s Disease. Neurobiol Aging. 2014;35:309–321. doi: 10.1016/j.neurobiolaging.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boone DL, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 41.Sun SC. Non-canonical NF-kappaB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutterwala FS, Haasken S, Cassel SL. Mechanism of NLRP3 inflammasome activation. Ann N Y Acad Sci. 2014;1319:82–95. doi: 10.1111/nyas.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Irrera N, et al. BAY 11-7082 inhibits the NF-kappaB and NLRP3 inflammasome pathways and protects against IMQ-induced psoriasis. Clin Sci (Lond) 2017;131:487–498. doi: 10.1042/CS20160645. [DOI] [PubMed] [Google Scholar]
- 44.Moriwaki K, Chan FK. Necroptosis-independent signaling by the RIP kinases in inflammation. Cell Mol Life Sci. 2016;73:2325–2334. doi: 10.1007/s00018-016-2203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/S1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 46.Kang LL, et al. Cinnamaldehyde and allopurinol reduce fructose-induced cardiac inflammation and fibrosis by attenuating CD36-mediated TLR4/6-IRAK4/1 signaling to suppress NLRP3 inflammasome activation. Sci Rep. 2016;6:27460. doi: 10.1038/srep27460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandes-Alnemri T, et al. Cutting edge: TLR signaling licenses IRAK1 for rapid activation of the NLRP3 inflammasome. J Immunol. 2013;191:3995–3999. doi: 10.4049/jimmunol.1301681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aachoui Y, Sagulenko V, Miao EA, Stacey KJ. Inflammasome-mediated pyroptotic and apoptotic cell death, and defense against infection. Curr Opin Microbiol. 2013;16:319–326. doi: 10.1016/j.mib.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schauvliege R, Vanrobaeys J, Schotte P, Beyaert R. Caspase-11 gene expression in response to lipopolysaccharide and interferon-gamma requires nuclear factor-kappa B and signal transducer and activator of transcription (STAT) 1. J Biol Chem. 2002;277:41624–41630. doi: 10.1074/jbc.M207852200. [DOI] [PubMed] [Google Scholar]
- 50.Vigano E, Mortellaro A. Caspase-11: the driving factor for noncanonical inflammasomes. Eur J Immunol. 2013;43:2240–2245. doi: 10.1002/eji.201343800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.