Abstract
The basic helix-loop-helix (bHLH) transcription factor family is crucial for plant development and stress responses. In this study, we identified 159 bHLH-encoding genes in the wheat (Triticum aestivum L.) genome and determined their roles in biotic and abiotic stress tolerance. Phylogenetic analyses showed that the TabHLH genes were classified into 19 groups, which shared similar gene structures and conserved motifs. A comprehensive transcriptome analysis revealed that bHLH genes were differentially expressed in diverse wheat tissues and were responsive to multiple abiotic and biotic stresses. A gene ontology analysis indicated that most bHLH proteins involved in DNA-binding activities and the gene expression regulation. Analyses of interaction networks suggested that TabHLHs mediate networks involved in multiple stress-signaling pathways. The findings of this study may help clarify the intricate transcriptional control of bHLH genes and identify putative stress-responsive genes relevant to the genetic improvement of wheat.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1742-4) contains supplementary material, which is available to authorized users.
Keywords: Stress, bHLH, Wheat, Gene expression, Interaction network
Introduction
The basic helix–loop–helix (bHLH) transcription factors, which contain a conserved bHLH domain, form a large superfamily widely distributed in plants, and are involved in multiple plant biological processes (Abe et al. 2003; Toledo-Ortiz et al. 2003). The conserved bHLH domain is composed of 60 amino acids, with two functional segments, the basic and the HLH regions. The basic region, which is located at the N-terminus, exhibits DNA-binding activity, whereas the HLH region, with two α-helices separated by a loop, functions in protein–protein interactions (Murre et al. 1989). Because their dual functions enable simultaneous interactions with DNA and proteins, bHLH transcription factors are important for diverse signaling networks, plant growth regulation and stress responses.
In plants, bHLH transcription factors help regulate many biological processes. Specific R2R3-MYB and bHLH transcription factors interact with WD repeat proteins to form MYB–bHLH–WD repeat complexes that contribute to the tight regulation of flavonoid biosynthesis (Xu et al. 2015). In tomato, a PRE-like atypical bHLH transcription factor, SlPRE2, affects plant morphology and negatively regulates the accumulation of fruit pigments (Zhu et al. 2017). Some bHLH genes are involved in biotic and abiotic stress responses (Fujita et al. 2006). A previous study revealed that AtbHLH115 positively regulates the Fe-deficiency response by binding to the promoters of the Fe-deficiency-responsive genes, bHLH38/39/100/101 and POPEYE, and interacting with E3 ligase in A. thaliana (Liang et al. 2017). Furthermore, bHLH transcription factors ILR3 and POPEYE contribute to multiple regulatory networks that control plant root responses to wounding by modulating glucosinolate (GLS) accumulation under Fe-deficiency conditions (Samira et al. 2018). The bHLH proteins also control brassinosteroid-signaling pathways and influence others (Kim et al. 2017). The overexpression of grapevine VvbHLH1 in A. thaliana can enhance drought and salt tolerance (Wang et al. 2016). The overexpression of a bHLH gene from Tamarix hispida in A. thaliana can improve salt and drought tolerance by enhancing the osmotic potential and decreasing the accumulation of reactive oxygen species (Ji et al. 2016). Overexpression of GhbHLH171 in cotton improves plant tolerance to the fungus V. dahlia by activating the JA synthesis (He et al. 2018). Thus, bHLH transcription factors appear to function as crucial regulators of plant development and stress responses.
To date, several plant bHLH proteins have been characterized in many species based on genome sequencing. There are 162 and 167 bHLHs in A. thaliana and rice, respectively (Bailey et al. 2003; Toledo-Ortiz et al. 2003; Li et al. 2006), while 155, 124, and 188 bHLHs have been identified in bean, potato and apple, respectively (Kavas et al. 2016; Mao et al. 2017; Wang et al. 2018). However, less is known about the bHLH family in wheat (Triticum aestivum L.), which is the second largest cereal crop. Because of the complex origin of the allohexaploid wheat species, which contains three homologous genomes (A, B, and D), systematic studies of wheat gene families have proceeded slowly. Owing to the economic and social importance of wheat and the critical roles of bHLHs in plants, investigations of wheat bHLH transcription factors are necessary to elucidate the biological processes underlying wheat development and stress responses.
In this study, we identified 159 bHLH-encoding genes in the wheat genome. We also analyzed their phylogenetic relationships, protein motifs, gene structures, expression patterns in diverse tissues, and responses to biotic and abiotic stresses. Furthermore, we investigated the TabHLH genes’ effects on biological processes, and characterized the bHLH interaction networks in response to stress. The data from this comprehensive study will increase our understanding of TabHLH functions associated with stress responses, and form a solid foundation for future evaluations of the utility of bHLH transcription factors in the genetic improvement of wheat.
Materials and methods
Identification of bHLH proteins and analyses of phylogenetic relationships
Wheat bHLH protein sequences were acquired from the Ensembl database (http://plants.ensembl.org/index.html). The bHLH protein sequences from rice and A. thaliana were downloaded from the RGAP (http://rice.plantbiology.msu.edu/) and TAIR (http://www.arabidopsis.org/) databases, respectively. The hidden Markov model profiles of the bHLH domain (PF00010) were used as queries to search for predicted bHLH proteins in the wheat dataset with the HMMER program (http://hmmer.wustl.edu/) (Eddy 2011). A BLAST search with rice and A. thaliana bHLHs as queries was used to identify predicted wheat bHLHs. The potential bHLHs were further examined with the Pfam database (http://pfam.sanger.ac.uk/) and Conserved Domain database (http://www.ncbi.nlm.nih.gov/cdd/). Additionally, bHLH protein sequences from wheat, rice, and A. thaliana were aligned with the Clustal X 2.0 program, and the resulting alignments were used to construct a phylogenetic tree with the MEGA 5.0 program (Tamura et al. 2011).
Analyses of TabHLH sequences and the properties of the encoded proteins
The ExPASy database (http://expasy.org/) was used to predict the molecular weight and isoelectric point of wheat bHLH transcription factors. The MEME (version 5.0.1) software (http://meme-suite.org/tools/meme) was used to analyze conserved protein motifs, while the GSDS database (http://gsds.cbi.pku.edu.cn/) was used to analyze gene structures.
Analysis of TabHLH expression profiles
Tissue-specific TabHLH expression levels were analyzed based on wheat RNA-sequencing (RNA-seq) datasets downloaded from the ENA database (https://www.ebi.ac.uk/ena,ERP004714). Additionally, the expression profiles induced by biotic and abiotic stress treatments were analyzed based on wheat RNA-seq datasets downloaded from the NCBI sequence read archive database (https://www.ncbi.nlm.nih.gov/sra; cold: SRR1460552; heat and drought: SRP045409; salt: SRR2306546; Fusarium pseudograminearum: SRP048912;), and ENA database (Stripe rust pathogen: ERP013983; Zymoseptoria tritici: ERP009837). Transcriptome mapping and assembly were completed with TopHat (version 2.0.10) and Cufflinks (Trapnell et al. 2012). Gene expression levels were calculated according to the fragments per kilobase of exon per million reads mapped (FPKM). The TM4 software was used to generate heat maps with log2-transformed FPKM values for the TabHLH gene expression levels induced by different stresses (Saeed et al. 2003).
Plant materials, treatments, and quantitative real-time PCR (qRT-PCR) analysis
Wheat cultivar ‘Chinese Spring’ plants were analyzed in this study. Plants were grown in soil under greenhouse conditions of 23 °C and a 16-h light/8-h dark photoperiod. We treated 10-day-old seedlings with 150 mM NaCl or 20% PEG6000 (w/v), 100 μM methyl jasmonate (MeJA), which simulated salt, drought stress conditions and pathogens infection, respectively, while seedlings were incubated at 4 °C to simulate cold stress conditions (Chartzoulakis and Loupassaki 1997; Li et al. 2011; Deng et al. 2013). Seedlings grown under normal conditions were used as controls. Leaves were collected from stress-treated and control seedlings at 0, 1, 3, 6, 12 and 24 h after treatments for the subsequent RNA isolation step (Liu et al. 2013; Wang et al. 2013). Additionally, the roots, stems, leaves, spikes and grains (10 days after pollination) were collected from flowering plants for the subsequent tissue-specific gene expression analysis.
Changes in TabHLH expression levels in response to drought, cold, salt stresses and MeJA treatment were assessed using qRT-PCR assays, which were conducted with SYBR® Premix Ex Taq™ (TaKaRa, Shiga, Japan) and the Real-Time Detection System (Bio-Rad, CFX96, USA). The specificity and efficiency levels of the qRT-PCR primers were verified by a melting curve analysis and agarose gel electrophoresis (additional data are given in Online Resource: Supplementary table S1). The β-actin gene was used as an internal reference for all the qRT-PCR analyses. The reaction conditions of qRT-PCR are described as follows: 95 °C for 10 min, followed by cycling for 40 cycles of denaturation at 95 °C for 10 s, annealing at 56 °C for 15 s and extension at 72 °C for 30 s. At the end of the reaction, a melting curve analysis of 65–95 °C was performed. Relative gene expression levels were calculated according to the 2−ΔΔCt method (Livark and Schmittgen 2001). All the samples were analyzed with three replicates.
Gene ontology (GO) enrichment and network interaction analyses
Information regarding TabHLH protein functional annotations was acquired from the wheat genome database, after which the Blast2GO and WEGO programs were used to complete a GO functional enrichment analysis (Conesa et al. 2005; Ye et al. 2006). We used the AraNet V2 (http//www.inetbio.org/aranet/) tool to investigate the TabHLH interaction network, which was based on the orthologous genes between wheat and A. thaliana. The network was drawn with the Cytoscape (version 3.6.1) software (Shannon et al. 2003).
Results
Identification of the wheat bHLH family and analyses of phylogenetic relationships
To identify wheat bHLH family members, a hidden Markov model was applied to search the wheat genome database, with the bHLH domain PF00010-related sequences as queries. After using the conserved domain database and Pfam database to verify the presence of known conserved bHLH domains, we identified 159 TabHLH proteins, renamed TabHLH1–159 based on the chromosomal locations of the corresponding genes. The bHLH proteins were predicted to comprise 115 (TabHLH58) to 618 (TabHLH59) amino acid residues, with relative molecular masses of 13.17–67.49 kDa and theoretical isoelectric points ranging from 4.62 (TabHLH50) to 11.62 (TabHLH61) (additional data are given in Online Resource: Supplementary table S2).
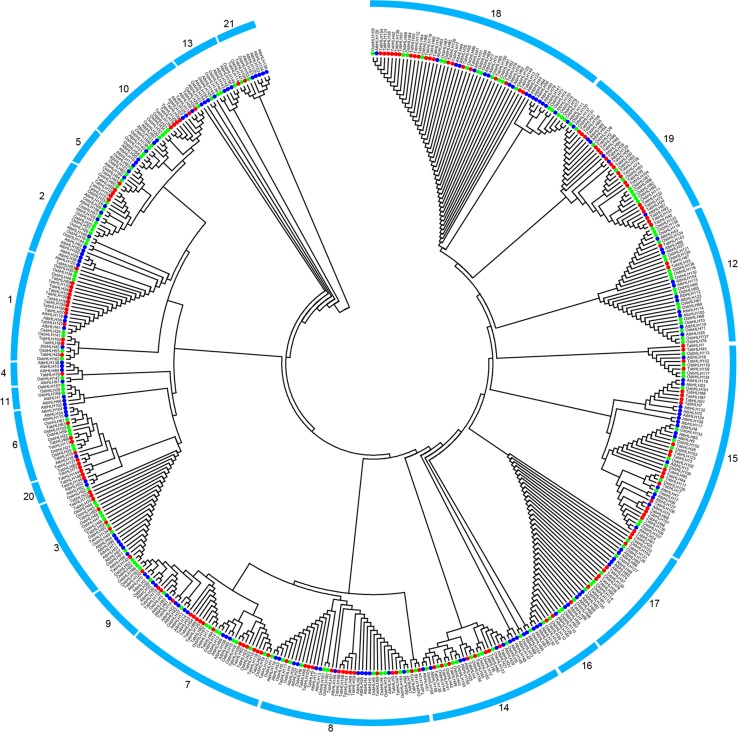
To investigate the evolutionary relationships within the bHLH family, a phylogenetic tree based on bHLH proteins from wheat, rice, and A. thaliana was constructed according to the maximum-likelihood method, because rice and A. thaliana are model monocot and dicot plants, respectively (Fig. 1). The A. thaliana bHLH family has 21 groups (Toledo-Ortiz et al. 2003), while TabHLH was divided into 19 groups, having no members corresponding to those of A. thaliana groups 11 and 13. The largest groups were 15 and 18, with more than 20 members, whereas groups 4, 5, 16, and 21 each had fewer than three bHLH members. Thus, the diverse wheat bHLH family members may have different functions. Some bHLH orthologs among wheat, rice, and A. thaliana were also identified based on the phylogenetic analysis, implying that some ancestral bHLH genes existed prior to the divergence of these plant species.
Fig. 1.
Phylogenetic analysis of bHLH proteins of wheat, rice and A. thaliana. The maximum-likelihood (ML) tree was constructed using Clustal X 2.0 and MEGA 5.0 softwares. Signs of different shapes represent bHLH proteins from wheat (red round, Ta), rice (green round, Os) and A. thaliana (blue round, At)
Analyses of conserved motifs and gene structures within the wheat bHLH family
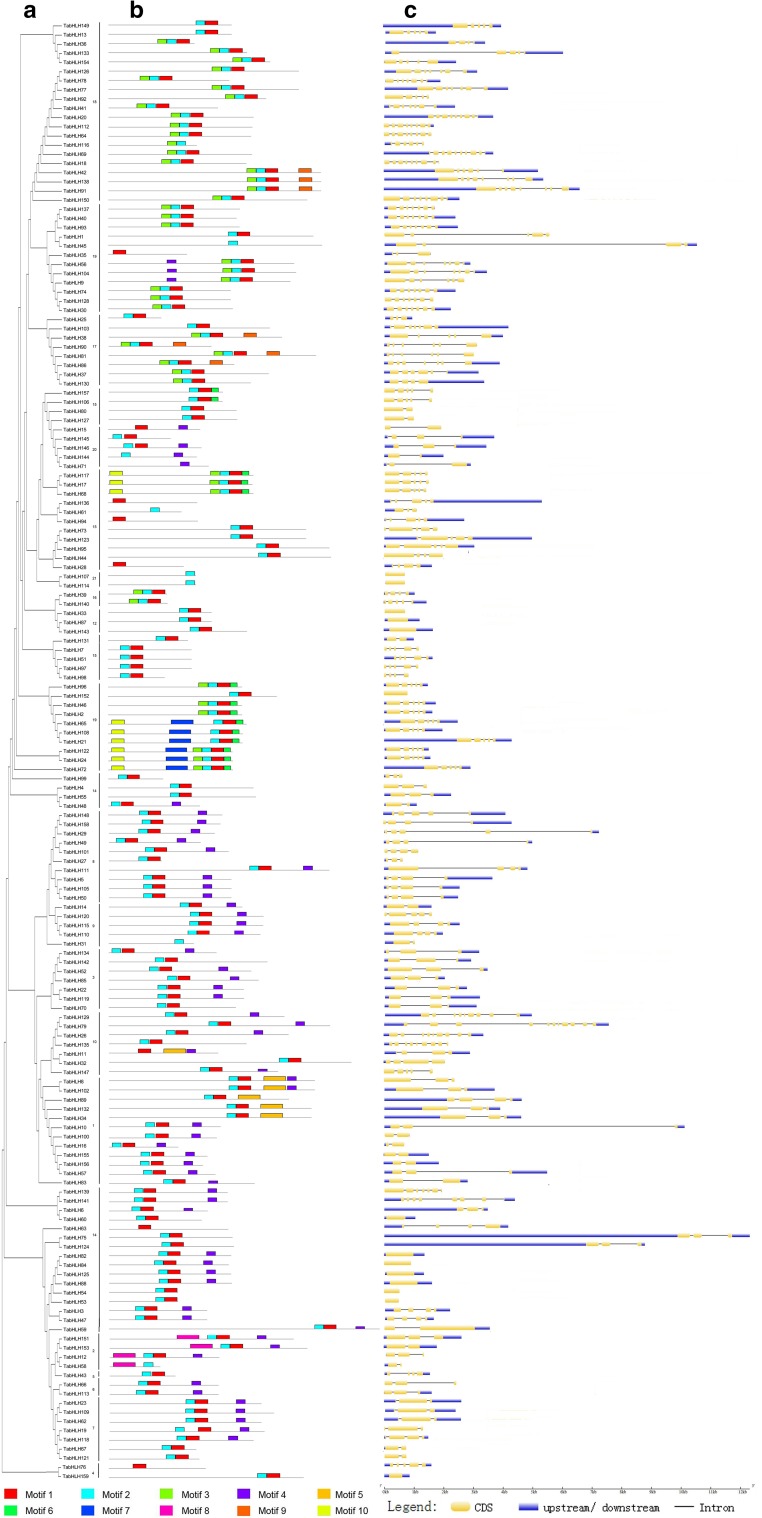
To clarify the evolution of bHLH genes, ten conserved motifs within bHLH proteins were identified using MEME software (Fig. 2b). We determined that all TabHLH proteins contained conserved bHLH motif 1 or 2. The TabHLH proteins in groups 15–19 also contained motif 3, while the proteins in the other groups contained motif 4. Motif analyses indicated that same group harbored similar conserved motifs. However, some motifs were present in some specific groups. For example, motif 7 was detected in only group 19, motif 5 was detected exclusively in group 1, and motif 8 was present only in group 2. Additionally, motif 10 was detected in groups 15 and 19. Most of the bHLH members in group 19 harbored motifs 3, 6, 7, and 10, while most of the bHLHs in group 2 shared motifs 4 and 8. Thus, the TabHLHs clustered in the same group shared similar amino acid sequences, which may be useful for characterizing the phylogenetic relationships among wheat bHLH genes.
Fig. 2.
The conserved motifs and gene structure analyses of wheat bHLH family. a The phylogenetic relationship of TabHLHs. b Conserved motifs analysis of TabHLHs. All motifs were identified by MEME database. c Gene structure analysis of TabHLHs. The gene structures were drawn using GSDS database. The blue boxes, yellow boxes, and the black lines indicate upstream/downstream, exons and introns, respectively
Exon–intron organizations within the bHLH genes were also investigated to gain insights into the evolution of their gene structures (Fig. 2c). The TabHLH genes in groups 12 and 21 contained only one exon, while the genes in groups 17, 18, and 19 contained 5–8 exons. In contrast, the genes in groups 1, 2, 5, 7, 15, and 20 had no more than five exons, and the genes in groups 3 and 6 included three exons. These observations suggested that similar exon–intron organizations exist in TabHLH genes from the same group, and that gene structures may have influenced the evolution of these genes.
Expression profiles of bHLH family genes in different wheat organs
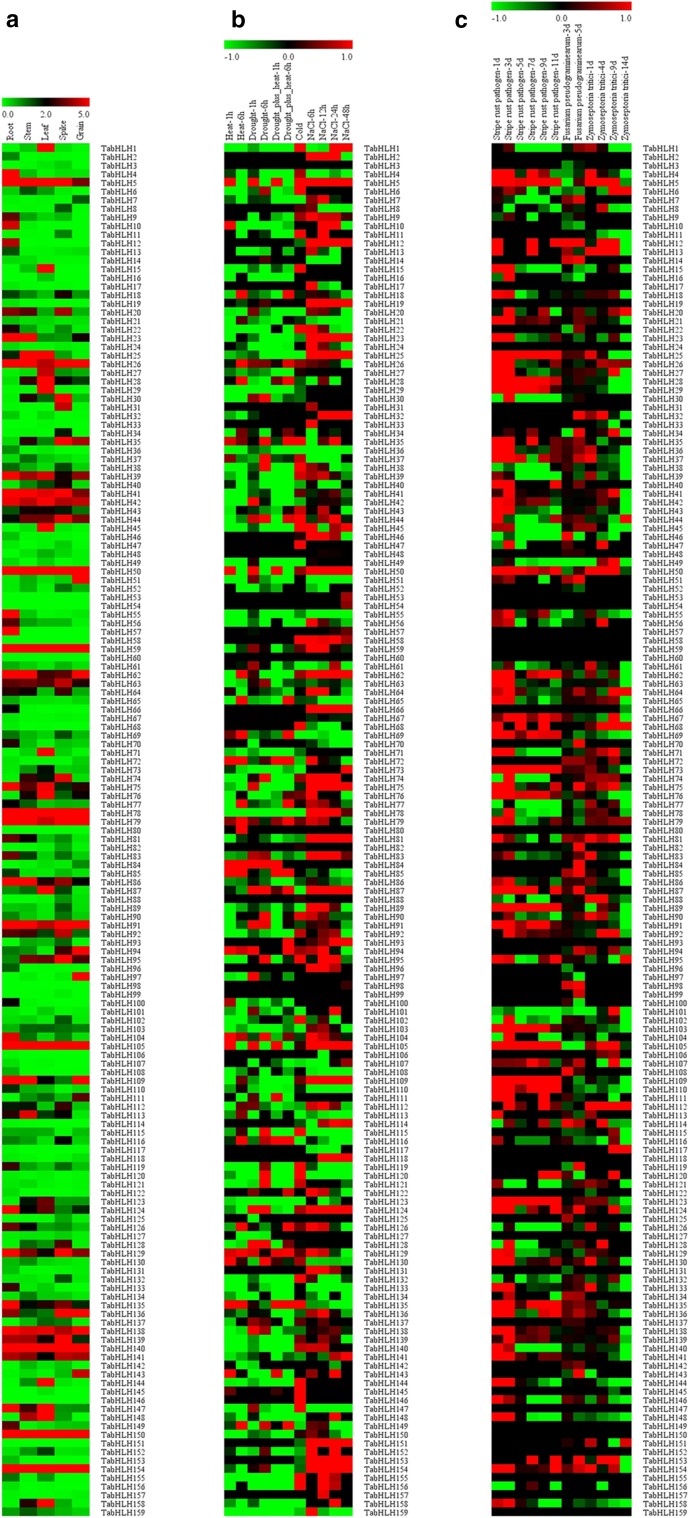
To examine the organ-specific bHLH gene expression profiles in wheat plants, we generated expression-level data for root, stem, leaf, spike and grain samples based on RNA-seq analyses (Fig. 3a; Supplementary table S3). Of the 159 TabHLH genes, 154 were expressed in at least one tested organ, and 146, 141, 147, 141, and 137 genes were expressed in the root, stem, leaf, spike, and grain tissues, respectively. Moreover, 16.4, 10.7, 13.2, 11.3, and 10.7% of the genes were highly expressed (value > 5) in the root, stem, leaf, spike, and grain tissues, respectively, indicating that bHLH genes have important functions related to wheat organ development.
Fig. 3.
Expression profiles of wheat bHLH gene family. a Expression profiles of TabHLHs in different organs of root, stem, leaf, spike and grain. The heat map was constructed according to the FPKM value of wheat RNA-seq data. b Expression profiles of TabHLHs in response to heat, drought, heat plus drought, cold and salt stresses. c Expression profiles of TabHLHs in response to stripe rust pathogen, Fusarium pseudograminearum and Zymoseptoria tritici infection. Log2-based FPKM value was used to create the heat map. Changes in gene expression are shown in color as the scale
A comparison of gene expression profiles among the five analyzed organs revealed that most genes were more highly expressed in the roots and leaves than in the stems and grains, suggesting that these genes are important for roots and leaves development. Additionally, 131 genes were expressed in all five organs, with seven of these genes (TabHLH-50, -59, -78, -105, -140, -150, -154) being highly expressed (value > 5). Thus, these genes are likely important for wheat organ development. Furthermore, regarding tissue-specific expression, five genes (TabHLH-4, -55, -57, -124, -135), 11 genes (TabHLH-1, -15, -28, -29, -45, -71, -76, -87, -144, -148, -158), two genes (TabHLH-30, -95) and three genes (TabHLH-51, -97, -94) were specifically expressed (value > 5) in the roots, leaves, spikes, and grains, respectively. The TabHLH62 and TabHLH109 genes were predominantly expressed (value > 5) in the roots, stems, and grains. Moreover, TabHLH25 was preferentially expressed (value > 5) in the stems and leaves, while TabHLH136 was preferentially expressed in the spikes and grains. These results implied that TabHLH functions differ in an organ-dependent manner. The organ-specific TabHLH expression patterns may provide valuable information for future investigations of wheat organ development and function.
Expression profiles of wheat bHLH genes under abiotic stress
The TabHLH expression patterns in response to heat, drought, cold, and salt stress conditions were investigated based on RNA-seq data to elucidate bHLH functions during exposures to abiotic stresses (Fig. 3b; Supplementary table S4). The expression levels of most of the 159 TabHLH genes were affected by these abiotic stresses, and 98.7% of the genes were responsive to more than one stress treatment. Additionally, the expression levels of 20.1%, 26.4%, 38.4%, and 47.8% of the genes were upregulated by heat, drought, cold, and salt stresses, respectively, of which, 9.4%, 10.7%, 15.7%, and 25.8% of the genes exhibited significantly upregulated expression levels (value > 1). In contrast, the expression levels of 57.2%, 50.9%, 34.6%, and 45.8% of the genes were downregulated by heat, drought, cold, and salt stresses, respectively, of which, 38.4%, 26.4%, 11.9%, and 25.8% of the genes exhibited significantly downregulated expression levels (value < − 1). Moreover, two genes (TabHLH72 and -85), two genes (TabHLH87 and -128), eight genes (TabHLH-6, -47, -119, -120, -144, -145, -146, -147), 21 genes (TabHLH-5, -25, -32, -50, -56, -58, -59, -62, -66, -74, -75, -87, -105, -124, -143, -148, -151, -152, -153, -154, -158) showed specific expression of significantly upregulated (value > 1) under heat, drought, cold and salt stress conditions, respectively. Meanwhile, the expression levels of three genes (TabHLH5, TabHLH35, and TabHLH95) were significantly upregulated in response to all the tested abiotic stresses, indicating that they may contribute to stress responses. Furthermore, eight genes (TabHLH-4, -11, -22, -29, -40, -55, -74, and -121), three genes (TabHLH-109, -143 and -154), one genes (TabHLH-25), 13 genes (TabHLH-2, -22, -46, -65, -69, -70, -85, -108, -110, -113, -115, -119 and -149) showed specific expression of significantly downregulated under heat, drought, cold and salt stress conditions, respectively. Thus, TabHLH proteins may help regulate wheat responses to diverse abiotic stresses.
Expression profiles of wheat bHLH genes under biotic stress
The TabHLH expression patterns in response to stripe rust pathogen, Fusarium pseudograminearum and Zymoseptoria tritici infection were investigated based on RNA-seq data to elucidate bHLH functions during exposures to biotic stresses (Fig. 3c; Supplementary table S5). The 86.7% of the genes were responsive to more than one biotic stress. Additionally, the expression levels of 53.5%, 45.9%, and 31.4% of the genes were upregulated by stripe rust pathogen, Fusarium pseudograminearum and Zymoseptoria tritici infection, respectively, of which, 32.7%, 10.1%, and 14.5% of the genes exhibited significantly upregulated expression (value > 1). In contrast, the expression levels of 30.8%, 34.0%, and 30.8% of the genes were downregulated by the tested biotic stress, respectively, of which, 12.6%, 4.4%, and 6.3% of the genes exhibited significantly downregulated expression (value < − 1). Moreover, nine genes (TabHLH-12, -13, -37, -50, -64, -75, -81, -124,-153) showed upregulated in response to all tested biotic stresses, indicating they may contribute to multiple biotic stress responses. These results implied that TabHLHs may help regulate wheat responses to diverse biotic stresses.
Validation of differentially expressed bHLH genes
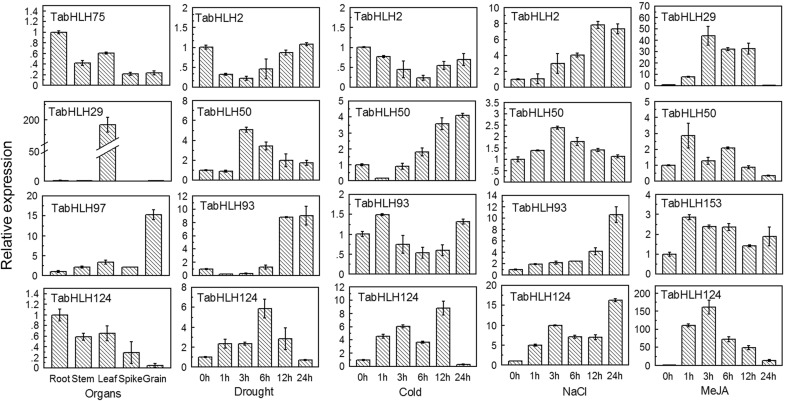
According to the RNA-seq data, TabHLH-29, -75, -97, and -124 showed organ-specific expression patterns; TabHLH-2, -50, -93 and -124 were upregulated by more than one abiotic stress; and TabHLH-29, -50, -124 and -153 were induced under tested biotic stress. These differentially expressed TabHLH genes were selected for a qRT-PCR analysis to validate the RNA-seq data (Fig. 4). After normalization, the majority of selected bHLH genes showed the same trend and consistent results between RNA-seq data and qRT-PCR data, such as TabHLH75 and TabHLH124 were relatively highly expressed in roots. TabHLH29 and TabHLH97 were relatively highly expressed in leaves and grain, respectively. Except for TabHLH2, the other tested genes were upregulated by all the tested biotic and abiotic stresses. These results indicated that the qRT-PCR data were consistent with the RNA-seq data, suggesting it is reasonable to use RNA-seq data to assess the expression patterns of wheat bHLH genes in different organs and in response to various stresses.
Fig. 4.
Validation of the expression level of TabHLHs by qRT-PCR analyses. The relative expression levels of selected TabHLHs in different organs, under drought, cold, salt and MeJA treatments. The mRNA fold difference was relative to that of root samples for organs special expression and untreated samples for different stress treatments expression used as calibrator. Data are mean ± SD of n = 3 biological replicates
GO enrichment among TabHLH transcription factor genes
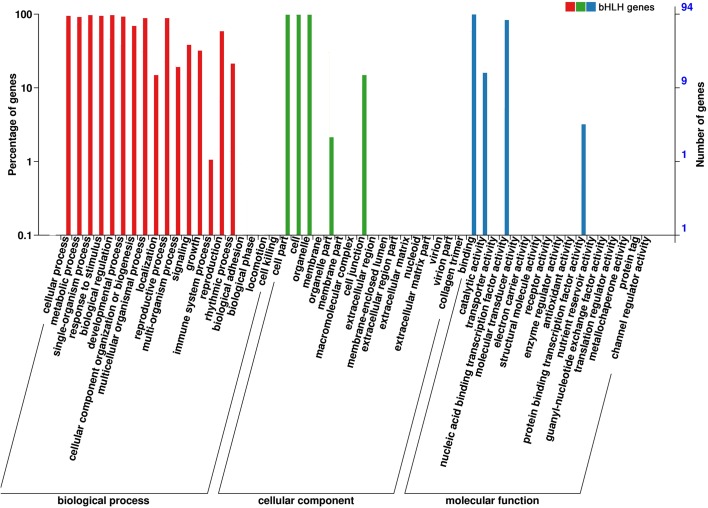
To characterize the functions of TabHLH transcription factors, a GO enrichment analysis was conducted using the Blast2GO and WEGO programs. A total of 94 annotated TabHLH genes were categorized into the three main GO categories (biological process, cellular component, and molecular function) (Fig. 5; Supplementary table S6). For the biological process category, 91 TabHLH genes were assigned to 16 of the 20 GO terms, with biological regulation and single-organism process representing the dominant categories (96.8%). Of the genes belonging to the cellular component category, 97.8% were assigned to cell and organelles. Within the molecular function category, binding activity (98.9%) and nucleic acid binding transcription factor activity (82.9%) were the most highly represented terms. These annotations implied that TabHLH genes are involved in extensive metabolic activities.
Fig. 5.
Gene ontology (GO) enrichment of the TabHLHs. The results are summarized under the three main GO categories: biological process, cellular component and molecular function. The right y-axis indicates the number of genes in each category. The left y-axis indicates the percentage of a specific category of genes in the corresponding GO category
Regulatory network of TabHLH transcription factors
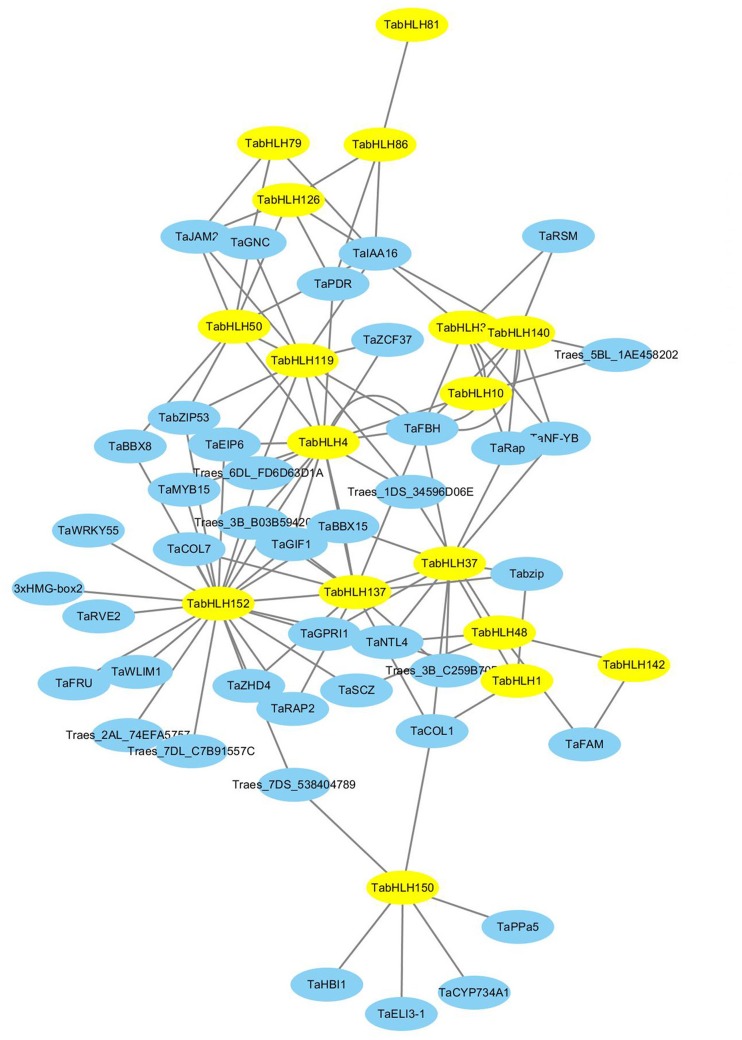
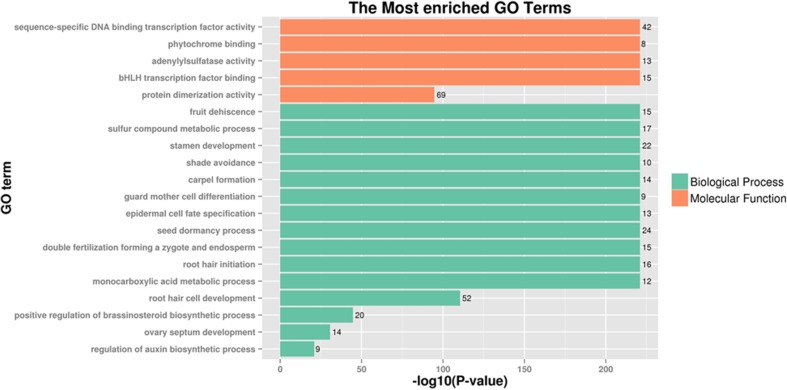
To decipher the interactions among TabHLH transcription factors and wheat genes, we predicted the associated regulatory network using an orthology-based method (Fig. 6). We detected 17 TabHLH genes with similarities to A. thaliana genes. Additionally, a wheat regulatory network involving 117 gene pairs was created, suggesting that the wheat bHLHs commonly function in networks regulating gene expression and metabolic processes (Supplementary tables S7 and S8). Moreover, TabHLH152 was orthologous to A. thaliana transcription factor bHLH87 (AT3G21330), and interacted with 24 wheat genes, including genes encoding transcription factors (bZIP53, WRKY55, RVE1, MYB15, and COL7) and other factors (GIF1). Furthermore, TabHLH137 and TabHLH119 interacted with 13 and 11 genes, respectively. The other TabHLHs interacted with fewer than ten genes. In addition, some TabHLHs interacted with each other, such as the interactive gene pairs TabHLH81–TabHLH86, TabHLH86–TabHLH126 and so on. A GO analysis of the genes involved in the predicted network revealed functions related to the regulation of transcription, leaf morphogenesis, root meristem, and seed growth (Fig. 7). Thus, TabHLH transcription factors may be crucial members of the regulatory networks that mediate wheat development and stress responses.
Fig. 6.
The interaction network of TabHLHs in wheat according to the orthologs in A. thaliana. The network was drawn using Cytoscape software database. The yellow nodes and blue nodes indicate TabHLHs and interacted proteins, respectively
Fig. 7.
Functional categories of proteins in TabHLH interaction networks
Discussion
The bHLH transcription factors play vital roles in regulating plant developmental processes and responses to environmental stresses. Characterizing the core underlying regulatory network may advance our understanding of bHLH functions in diverse biological processes. Although some bHLH genes have been studied in many plants, relatively little information is available regarding wheat bHLH genes. In this study, we identified 159 genes encoding bHLH transcription factors in the wheat genome. The phylogenetic, conserved motif, and gene structure analyses of wheat bHLH genes revealed that each group shared similar motifs and exon–intron organizations, which typical characteristics have also been observed in other plants (Sun et al. 2015; Kavas et al. 2016). The results suggested that TabHLH genes in the same group were relatively closely related during evolution.
Wheat is a vital crop worldwide, and plant development and stress responses are important factors influencing crop yield and quality. The bHLH transcription factors are reportedly important for plant development and stress responses in several species, including potato, carrot, and apple (Chen et al. 2015; Mao et al. 2017; Wang et al. 2018). In this study, TabHLH110 was specifically expressed in the roots. In A. thaliana, AtABS5 (AT1G68810), which is an ortholog of TabHLH110, is expressed in the roots, where it affects vascular cell division in the root apical meristem (KyokoOhashi-Ito et al. 2014). Our data also revealed that most bHLH genes were induced by tested abiotic and biotic stresses, suggesting that they have a regulatory role influencing responses to biotic and abiotic stresses. Similar phenomena have been observed in apple, potato, and carrot (Chen et al. 2015; Mao et al. 2017; Wang et al. 2018). Therefore, wheat bHLHs may be extensively involved in different wheat developmental processes and regulate multiple stress responses.
In plants, transcription factors, which regulate gene expression by interacting with the promoter regions of downstream target genes, are crucial for multiple biological processes. The plant bHLH–MYB complex helps regulate developmental activities, and metabolism, as well as gibberellin, jasmonate, and abscisic acid (ABA) signaling. In A. thaliana, the jasmonate ZIM-domain-containing proteins can interact with MYB–bHLH–WD repeat complexes, and activate downstream signal cascades to modulate jasmonate-mediated anthocyanin accumulation, trichome initiation, and gibberellin and jasmonate signaling (Qi et al. 2011, 2014). The bHLH–MYB complex can bind to the rd22 promoter region functioning as a cis-acting element to regulate the drought- and ABA-induced expression of rd22, thereby functioning as a transcriptional activator influencing abscisic acid (ABA) signaling (Abe et al. 2003). MYB–bHLH–TTG1 transcription factor complexes can regulate anthocyanin accumulation in developing A. thaliana seedlings (Appelhagen et al. 2011). Additionally, bHLHs form homodimers and heterodimers to regulate gene expression networks (Zhu et al. 2015). The auxin-regulated bHLH transcription factor TMO5 and its heterodimeric bHLH partner LHW form a complex that mediates vascular cell division and early vascular development (Vera-Sirera et al. 2015; Ohashi-Ito and Fukuda 2016). Moreover, bHLH proteins can also interact with other factors to form bHLH–PAS and bHLH–FBXO45 complexes (Salat et al. 2015; Fribourgh and Partch 2017). Although the A. thaliana bHLH family affects multiple biological processes, there are relatively few reports describing bHLH signaling in wheat. In this study, we constructed a wheat bHLH interaction network based on our orthological analyses (Fig. 6; Supplementary Tables 6, 7 and 8). We identified TabHLH dimer complexes, bHLH–MYB complexes, and some other complexes based on the network. These complexes may have vital functions in networks regulating wheat gene expression and metabolic processes. Furthermore, a GO functional enrichment analysis indicated that TabHLH genes are significantly enriched for functions related to the regulation of gene expression. The TabHLH target genes are also significantly enriched for functions related to cellular processes and pathways for organ development and stress responses (Fig. 7). From the interaction network and the expression profile analyses, some putative functional genes were identified, such as TabHLH152, which interacted with more than 20 wheat genes and was induced by drought, cold and NaCl treatments, indicating that it might have important roles in wheat responses to abiotic stress. The AtbHLH87, ortholog of TabHLH152 in A. thaliana, has clear functional connections to cold- and heat-stress responses (Rasmussen et al. 2013), and AtbHLH is involved in light stress response by association with some zinc finger proteins and phytochrome (Jiao et al. 2007). The interaction network suggested that TabHLH genes are critical for wheat development and external stress responses.
Conclusions
In summary, a genome-wide analysis identified 159 wheat bHLH genes, and their classifications and evolutionary relationships were clarified using phylogenetic, conserved motif, and gene structures analyses. The bHLH expression profiles revealed that these genes mediate wheat development and stress responses. GO enrichment and interaction network analyses confirmed that TabHLH genes are crucial for responses to stresses. As important transcription factors, TabHLHs function as pivotal regulators of the expression of downstream gene targets associated with specific cellular responses. In wheat, these transcription factors are activated by diverse external stimuli and interact with target genes to form a regulatory signaling network. The data presented herein will be valuable for future functional characterizations of bHLH genes and the breeding of improved wheat cultivars.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by grants from Henan key scientific and technological projects (182102110476) and civic key scientific and technological project (2017007/7.3). We thank Lesley Benyon, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author contributions
LW, ZX and BL conceived and designed the experiments. LX, JH performed the experiments. LW and ZX analyzed the data. LW and BL wrote the paper.
Compliance with ethical standards
Conflict of interest
All authors declare no competing interests.
Contributor Information
Zhaohui Xie, Email: xiezhaohui@hncj.edu.cn.
Bingbing Li, Email: hncj111@126.com.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelhagen I, Jahns O, Bartelniewoehner L, Sagasser M, Weisshaar B, Stracke R. Leucoanthocyanidin dioxygenase in Arabidopsis thaliana: characterization of mutant alleles and regulation by MYB-BHLH-TTG1 transcription factor complexes. Gene. 2011;484:61–68. doi: 10.1016/j.gene.2011.05.031. [DOI] [PubMed] [Google Scholar]
- Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Werber M, Weisshaar B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell. 2003;15:2497–2502. doi: 10.1105/tpc.151140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartzoulakis KS, Loupassaki MH. Effects of NaCl salinity on germination, growth, gas exchange and yield of greenhouse eggplant. Agric Water Manag. 1997;32:215–225. doi: 10.1016/S0378-3774(96)01276-0. [DOI] [Google Scholar]
- Chen Y, Li M, Wu X, Huang Y, Ma J, Xiong A. Genome-wide analysis of basic helix-loop-helix family transcription factors and their role in responses to abiotic stress in carrot. Mol Breed. 2015;35:1–12. doi: 10.1007/s11032-015-0202-z. [DOI] [Google Scholar]
- Conesa A, Götz S, Miguel J, García-Gómez Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Deng X, Zhou S, Hu W, Feng J, Zhang F, Chen L, Huang C, Luo Q, He Y, Yang G, He G. Ectopic expression of wheat TaCIPK14, encoding a calcineurin B-like protein-interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiol Plant. 2013;149:367–377. doi: 10.1111/ppl.12046. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourgh JL, Partch CL. Assembly and function of bHLH-PAS complexes. Proc Natl Acad Sci USA. 2017;114:5330–5332. doi: 10.1073/pnas.1705408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- He X, Zhu L, Wassan GM, Wang Y, Miao Y, Shaban M, Hu H, Sun H, Zhang X. GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171. Mol Plant Pathol. 2018;19:896–908. doi: 10.1111/mpp.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Nie X, Liu Y, Zheng L, Zhao H, Zhang B, Huo L, Wang Y. A bHLH gene from Tamarix hispida improves abiotic stress tolerance by enhancing osmotic potential and decreasing reactive oxygen species accumulation. Tree Physiol. 2016;36:193–207. doi: 10.1093/treephys/tpv139. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- Kavas M, Baloglu MC, Atabay ES, Ziplar UT, Dasgan HY, Ünver T. Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol Genet Genomics. 2016;291:129–143. doi: 10.1007/s00438-015-1095-6. [DOI] [PubMed] [Google Scholar]
- Kim Y, Song J-H, Park S-U, Jeong Y-S, Kim S-H. Brassinosteroid-induced transcriptional repression and dephosphorylation-dependent protein degradation negatively regulate BIN2-interacting AIF2 (a BR signaling-negative regulator) bHLH transcription factor. Plant Cell Physiol. 2017;58:227–239. doi: 10.1093/pcp/pcx003. [DOI] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H W, Zang BS, Deng XW, Wang XP. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta. 2011;234:1007–1018. doi: 10.1007/s00425-011-1458-0. [DOI] [PubMed] [Google Scholar]
- Liang G, Zhang H, Li X, Ai Q, Yu D. bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J Exp Bot. 2017;68:1743–1755. doi: 10.1093/jxb/erx043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang L, Xing X, Sun L, Pan J, Kong X, Zhang M, Li D. ZmLEA3, a multifunctional group 3 LEA protein from maize (Zea mays L.), is involved in biotic and abiotic stresses. Plant Cell Physiol. 2013;54:944–959. doi: 10.1093/pcp/pct047. [DOI] [PubMed] [Google Scholar]
- Livark K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mao K, Dong Q, Li C, Liu C, Ma F. Genome-wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front Plant Sci. 2017;8:480. doi: 10.3389/fpls.2017.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, Caw PSM, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-X. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Fukuda H. Functional mechanism of bHLH complexes during early vascular development. Curr Opin Plant Biol. 2016;33:42–47. doi: 10.1016/j.pbi.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 2011;23:1795–1814. doi: 10.1105/tpc.111.083261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Huang H, Wu D, Yan J, Qi Y, Song S, Xie D. Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell. 2014;26:1118–1133. doi: 10.1105/tpc.113.121731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, Bones AM, Nielsen HB, Mundy J. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 2013;161:1783–1794. doi: 10.1104/pp.112.210773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saegusa M, Ohashi-Ito K, Oda Y, Iwamoto K, Kojima M, Katayama H, Fukuda H, Sakakibara H. A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr Biol. 2014;24:2053–2058. doi: 10.1016/j.cub.2014.07.050. [DOI] [PubMed] [Google Scholar]
- Salat D, Winkler A, Urlaub H, Gessler M. Hey bHLH proteins interact with a FBXO45 containing SCF ubiquitin ligase complex and induce its translocation into the nucleus. PLoS One. 2015;10:e0130288. doi: 10.1371/journal.pone.0130288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samira R, Li B, Kliebenstein D, Li C, Davis E, Gillikin JW, Long TA. The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis. Plant Mol Biol. 2018;97:297–309. doi: 10.1007/s11103-018-0735-8. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Fan HJ, Ling HQ. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics. 2015;16:9. doi: 10.1186/s12864-014-1209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff LA, Pachter L. Differential gene and transcript expression analysis of RNA-Seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Sirera F, Rybe BD, Úrbez C, Kouklas E, Pesquera M, Álvarez-Mahecha JC, Tuominen H, Carbonell J, Borst JW, Weijers D, Blázquez AM. A bHLH-based feedback loop restricts vascular cell proliferation in plants. Dev Cell. 2015;35:432–443. doi: 10.1016/j.devcel.2015.10.022. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng P, Chen L, Wang X, Ma H, Hu W, Yao N, Feng Y, Chai R, Yang G, He G. A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS One. 2013;8:e65120. doi: 10.1371/journal.pone.0065120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhu H, Chen D, Li Z, Peng R, Yao Q. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Org. 2016;125:387–398. doi: 10.1007/s11240-016-0953-1. [DOI] [Google Scholar]
- Wang R, Zhao P, Kong N, Lu R, Pei Y, Huang C, Ma H, Chen Q. Genome-wide identification and characterization of the potato bHLH transcription factor family. Genes. 2018;9:54. doi: 10.3390/genes9010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–185. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu E, You C, Wang S, Cui J, Niu B, Wang Y, Qi J, Ma H, Chang F. The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. 2015;83:976–990. doi: 10.1111/tpj.12942. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chen G, Guo X, Yin W, Yu X, Hu J, Hu Z. Overexpression of SlPRE2, an atypical bHLH transcription factor, affects plant morphology and fruit pigment accumulation in tomato. Sci Rep. 2017;7:5786. doi: 10.1038/s41598-017-04092-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.