Abstract
Lactic acid bacteria (LAB) are capable of converting carbohydrate substrates into organic acids (mainly lactic acid) and producing a wide range of metabolites. Due to their interesting beneficial properties, LAB are widely used as starter cultures, as probiotics, and as microbial cell factories. Exploring LAB present in unknown niches may lead to the isolation of unique species or strains with relevant technological properties. Autochthonous rather than allochthonous starter cultures are preferred in the current industry of fermented food products, due to better adaptation and performance of autochthonous strains to the matrix they originate from. In this work, the lactic microbiota of eight different wild tropical types of fruits and four types of flowers were studied. The ability of the isolated strains to produce metabolites of interest to the food industry was evaluated. The presence of 21 species belonging to the genera Enterococcus, Fructobacillus, Lactobacillus, Lactococcus, Leuconostoc, and Weissella was evidenced by using culture-dependent techniques. The isolated LAB corresponded to 95 genotypically differentiated strains by applying rep-PCR and sequencing of the 16S rRNA gene; subsequently, representative strains of the different isolated species were studied for technological properties, such as fast growth rate and acidifying capacity; pectinolytic and cinnamoyl esterase activities, and absence of biogenic amine biosynthesis. Additionally, the strains' capacity to produce ethyl esters as well as mannitol was evaluated. The isolated fruit- and flower-origin LAB displayed functional properties that validate their potential use in the manufacture of fermented fruit-based products setting the background for the design of novel functional foods.
Keywords: lactic acid bacteria, fructophilic lactic acid bacteria, Fructobacillus, tropical fruits, microbial diversity, functional properties, mannitol, esterases
Introduction
Lactic acid bacteria (LAB) constitute an ubiquitous bacterial group that is widespread in nature in niches of dairy (fermented), meat and vegetable origin, the gastrointestinal and urogenital tracts of humans and animals, and soil and water (Liu et al., 2014). These microorganisms are well known for their ability to produce lactic acid as the main end-product of their anaerobic metabolism and for synthesizing a wide range of metabolites that beneficially affect the nutritional, sensorial, and technological properties of fermented food products. For these reasons, LAB have been extensively used (i) as starter cultures; (ii) as probiotics; and (iii) in the production of interesting compounds (i.e., nutraceuticals), due to their versatile metabolism (Naeem et al., 2012; Emerenini et al., 2013; Ruiz Rodríguez et al., 2017a).
Studies on the microbial diversity of unexplored niches and environments have led to the isolation of an endless number of novel bacterial species, which may display special or unique technological and/or health-promoting properties (Di Cagno et al., 2009, 2013; Endo and Salminen, 2013; Olofsson et al., 2014). Among the sparsely explored sources of LAB, flowers, fruits, and raw vegetables constitute a remarkable niche, due to their daily contact with man. These raw materials possess high carbohydrate but low protein contents and a slightly acidic pH, providing a suitable niche to several microorganisms (Naeem et al., 2012). However, the microbial composition in these environments is fluctuating and depends on intrinsic (physical and nutritional conditions) and extrinsic (environmental and harvesting conditions) parameters of the plant matrix (Naeem et al., 2012; Di Cagno et al., 2013; Garcia et al., 2016).
In general, the microbial population of vegetables and fruits is between 105 and 107 CFU/g; among which yeasts are the dominant group (102-106 CFU/g); LAB represent only a small part of the microbiota, ranging between 102 and 104 CFU/g (Di Cagno et al., 2013). To date, the LAB diversity present on fruits and flowers has been scarcely studied (Bae et al., 2006; Chambel et al., 2006; Nyanga et al., 2007; Yanagida et al., 2008; Chen et al., 2010; Neveling et al., 2012). Species belonging to the genera Weissella, Lactobacillus, Lactococcus, Leuconostoc, Fructobacillus, Enterococcus, Pediococcus, and Streptococcus have been found, among which W. cibaria, W. confusa, Lb. brevis, Lb. plantarum, Lb. rossiae, Leuc. mesenteroides, Leuc. pseudomesenteroides, Lc. lactis, Ec. faecalis, and Ec. durans have been reported as the most frequent species (Endo et al., 2009; Di Cagno et al., 2010; Askari et al., 2012; Naeem et al., 2012; Ong et al., 2012; Emerenini et al., 2013; Leong et al., 2014). Finally, fructophilic LAB (FLAB) species, such as Lb. kunkeei, Lb. florum, F. fructosus, F. ficulneus, F. pseudoficulneus, F. durionis and F. tropaeoli, have been detected in fruits, flowers, and vegetables (Edwards et al., 1998; Endo and Okada, 2008; Endo et al., 2009, 2010, 2011b, 2018). The origin of flower and fruit associated-microbiota remains still uncertain. It has been claimed that the microorganisms found in these niches may come from the environment, from pollinators visiting fruits and flowers when both coinciding in the plant at the same time, from birds fed with fruits, or from insects. Although the bacterial community present in the nectar of flowers may be affected by the atmosphere or by animals as dispersion vectors, the environmental and geographical factors, which shape microbial communities in the nectar, are still unknown (Alvarez-Perez et al., 2012; Fridman et al., 2012; Samuni-Blank et al., 2014). Samuni-Blank et al. (2014) found similar bacterial communities in the nectar of flowers and the surface of insects that visited the flowers indicating that dispersion of bacteria present in the nectar is not only formed by those present in the air and nectar consumers but also by other vectors such as insects. Anderson et al. (2013) found that many bacteria prevalent in beebread and the crop were also present in floral nectar suggesting frequent horizontal transmission. Also, a symbiotic LAB microbiota within the honey crop of honeybees has been reported (Vásquez et al., 2012). Recently, it was also suggested that floral microbes can mediate plant-bumblebee communication, going their potential beyond microbial effects on nectar chemistry (Russell and Ashman, 2019). On the other hand, Filannino et al. (2018) stated that although reports on endophyte populations of LAB in plants are scarce, advances on plant–microbe interactions have highlighted their importance as a new class of plant growth promoting microbes. To date, Lb. plantarum was the only endophytic LAB identified throughout the life cycle of the oregano and wheat plants (Pontonio et al., 2018).
It has been shown that the use of autochthonous LAB strains compared to allochthonous ones as starter cultures is advantageous to enhance the nutritional, sensorial and rheological properties of fermented food products as well as to ensure a prolonged shelf life. LAB selection can be based on pro-technological, sensory, and/or nutritional criteria (Di Cagno et al., 2013, 2015). Additionally, LAB strains belonging to particular niches may present specific metabolic traits as a result of environment adaptation (Siezen and Bachmann, 2008; Endo, 2012). In this regards, plant-associated LAB possess specific enzymes, such as levansucrase, tannase and phytase, and the common feature of producing high amounts of organic acids, such as lactic acid and acetic acid (Tyler et al., 2016). Furthermore, the production of other industrially interesting metabolites, such us aroma compounds, γ-aminobutyric acid, polyols, etc., may also be relevant (Mozzi et al., 2006; Hebert et al., 2008; Abeijón Mukdsi et al., 2009; Dhakal et al., 2012; Quinto et al., 2014; Ruiz Rodríguez et al., 2017a). For instance, mannitol, a compound widely applied in the cosmetic, food, and pharmaceutical industries, is highly produced by certain heterofermentative LAB by reduction of fructose, one of the main sugars present in fruits and vegetables (Endo et al., 2009; Patra et al., 2009, 2011; Carvalheiro et al., 2011; Saha and Racine, 2011; Ortiz et al., 2013; Tyler et al., 2016; Ruiz Rodríguez et al., 2017b).
Several studies on LAB isolation reported a polyphasic approach to achieve a precise microbial identification. However, the choice of appropriate identification methods may depend on certain factors, such as the origin of the sample (clinical, environmental, or food isolates), the number of isolates, and staff qualifications (Moraes et al., 2013). In general, with a few exceptions, a phenotypic test could be proper enough for clinical isolate identification, whereas for food isolates a molecular approach is probably the most sensitive and reliable method (Emerenini et al., 2013; Moraes et al., 2013). Molecular typing has been shown to be useful to group isolates from vegetables and fruits into several clusters for subsequent identification (Papalexandratou et al., 2011a; Di Cagno et al., 2013). One of the most suitable and widely used bacterial identification methods is 16S rRNA gene sequencing. These conserved genes present enough variability to be considered as excellent phylogenetic markers for genus and species level identification (Naeem et al., 2012; Emerenini et al., 2013; Moraes et al., 2013).
The tropical and subtropical areas of the Northern region of Argentina have a large diversity of fruit trees that may be considered as an interesting and rich source of LAB. In this work, we aimed to explore the LAB diversity present on diverse wild fresh fruits and flowers from Northern Argentina as well as to study their technological properties and their ability to produce industrially interesting metabolites.
Materials and Methods
Sample Collection
Several units of different fruits (9) and flowers (4) widespread in the Tucumán (27° 00′ 00′′ S, 65° 30′ 00′′ W) province in Northern Argentina were aseptically picked. Ripe wild fruits of guava (pink and yellow varieties), papaya, passion fruit, custard apple, medlar, mulberry, fig, and khaki, and flowers of medlar, passion fruit, custard apple, and papaya (Table 1) were aseptically collected with gloves, put into sterile stomacher bags, and immediately transported to the laboratory for analysis. Sampling was carried out according to the Southern hemisphere seasonal fruit production in the period between April 2013 and April 2014.
Table 1.
Tropical fruits and flowers from Northern Argentina studied in this work.
| Sample N° | Common name | Scientific name | Key | Collection season | Month (year) |
|---|---|---|---|---|---|
| 1 | Guava | Psidium guajava | G | Autumn | April (2013) |
| 2 | Papaya | Carica papaya | P | Autumn, winter | June, July (2013) |
| 3 | Papaya flowers | Carica papaya | FP | winter | June, July (2013) |
| 4 | Passion fruit | Passiflora edulis | My | Winter | July, August (2013) |
| 5 | Passion fruit flowers | Passiflora edulis | FMy | Autumn | April (2013) |
| 6 | Custard apple | Annona cherimola | Ch | Autumn | April (2013) |
| 7 | Custard apple flowers | Annona cherimola | FCh | Autumn | April (2013) |
| 8 | Meddlar | Eriobotrya japonica | N | Spring | September, October (2013) |
| 9 | Meddlar flowers | Eriobotrya japonica | FN | Autumn | April (2014) |
| 10 | Mulberries | Morus nigra | Mr | Spring | October (2013) |
| 11 | Fig | Ficus carica | H | Summer | January (2014) |
| 12 | Khaki | Diospyros kaki | Cq | Autumn | April (2014) |
Microbiological Analysis
Sample Processing and Isolation of LAB and FLAB From Fruits and Flowers
All samples were processed according to the characteristics of each fruit and flower. For large- and medium-size fruits, such as guava, papaya, passion fruit, custard apple, fig and khaki, separate pools of small portions of the surface and pulp of each unit of fruit were randomly taken. In the case of smaller fruits, such as meddlar and blackberries, as well as flowers, complete units were used. To analyze the LAB and FLAB microbiota present in the samples by direct plating; 90 or 45 mL of peptone water [0.1% (w/v) bacteriological peptone] were added to 10 g of fruit or to 5 g of flower, respectively, and homogenized for 1 min using a stomacher (Stomacher® 400; Seward, Worthing, UK). Appropriate serial dilutions of each suspension were plated onto MRS agar (De Man et al., 1960) for LAB isolation and MRSf agar [MRS agar containing 2% (w/v) of fructose instead of glucose] for FLAB isolation, both supplemented with 0.1 g/L of cycloheximide and 0.1 g/L of sodium azide to inhibit fungi and yeasts, and Gram negative microorganisms, respectively. The total microbial count and coliforms were determined for each sample using plate count agar (PCA; Oxoid Ltd., Basingstoke, Hampshire, UK) and Mac Conkey agar (Britania, Buenos Aires, Argentina), respectively.
Isolation of FLAB was done by culture enrichment according to Endo et al. (2009), with minor modifications. Briefly, 5 mL of FYP broth were added to small pieces of each fruit sample and incubated at 30°C for 24 h. After incubation, 100 μL of the enriched cultures were inoculated into fresh FYP broth and further incubated at 30°C until visible growth detection. Subsequently, serial dilutions of the cultures were plated onto FYP agar containing 5 g/L of CaCO3 (this component facilitates LAB detection by formation of a clearance zone around the colonies due to CaCO3 hydrolysis by lactic acid).
All plates were incubated at 30°C. MRS and MRSf plates were incubated anaerobically (Anaerobic System AnaeroGen™, Oxoid Ltd.) for 24 to 72 h. FYP and PCA plates were incubated aerobically for 24 to 72 h.
Colonies, randomly selected according to morphological differences (colony size and shape), were picked and purified by streaking on the suitable agar media and further characterized. Representative numbers (30%) of colonies from agar media containing between 30 and 300 CFU/g were picked; when the total colony count was <30, all colonies were analyzed.
Overnight cultures of the isolates were preliminarily assayed for Gram staining, microscopic morphology, and catalase activity. The catalase test was done by suspending bacterial cells in a 3% (v/v) hydrogen peroxide droplet. Gram-positive and catalase-negative cocci and rods were selected as presumptive LAB/FLAB. LAB were stored in a medium containing (g/L): skim milk, 100; yeast extract, 5; glucose, 10; and 10% (v/v) glycerol, while FLAB were stored in nutrient broth containing 20% (v/v) glycerol, both at −20°C.
Genotypic Characterization
Bacterial DNA Extraction
For genomic DNA extraction, two different protocols were used. DNA was either obtained using the commercial DNA extraction kit NucleoSpin®96 Tissue (MACHEREY-NAGEL GmbH & Co. KG, Germany) or extracted according to Pospiech and Neumann (1995) with some amendments. For the latter, three milliliters of stationary phase cultures were centrifuged at 10,000 rpm in an Eppendorf bench top centrifuge for 5 min. Cells were washed with 500 μL of TE buffer [10 mM Tris-HCl (pH 7.5), 10 mM EDTA] and resuspended in 400 mL of STET-lysozyme (15 mg/mL). After holding at 37°C for 2 h, 40 μL (1/10 vol) of 10% sodium dodecyl sulfate (SDS) and 5 μL of proteinase K (15 mg/mL) were added, and the mixture was incubated at 55°C for 2 h. Then, 170 μL (1/3 vol) of 5 M NaCl and 1 vol of chloroform: isoamyl alcohol (24:1) were added to the mixture maintaining it at room temperature for 30 min. After centrifugation at 13,000 rpm for 10 min, the aqueous phase was transferred to another tube, and the DNA was precipitated with isopropanol (1:1 v/v). The precipitate was washed with 500 μL of 70% (v/v) ethanol and centrifuged for 5 min at 13,000 rpm. DNA was dried by evaporating the alcohol and then resuspended in 30 μL of MilliQ water. DNA concentration and purity were spectrophotometrically determined by measuring the optical density (OD) at 260 and 280 nm and determining the OD260/OD280 ratio (Brown, 1995).
Molecular Dereplication of Isolates by rep-PCR Genomic Fingerprinting
To differentiate strains among isolates, microbial dereplication was achieved by rep-PCR fingerprinting (amplification of repetitive bacterial DNA elements through the polymerase chain reaction) of their genomic DNA with the single oligonucleotide primer (GTG)5 (5′-GTGGTGGTGGTGGTG-3′) (Versalovic et al., 1994; Gevers et al., 2001), referred to as (GTG)5-PCR fingerprinting. The PCR assay mixture (25 μL) consisted of 5 μL buffer [5X Green GoTaq® reaction buffer (Promega, WI, USA)], 1 μL of primer (63 mM), 7.35 μL nuclease-free water, 2.6 μL MgCl2 (50 mM), 4 μL bovine serum albumin (BSA; 1 mg/mL), 2.5 μL dimethyl sulfoxide [DMS, 100% (v/v)], 1.25 μL mixture of deoxyribonucleoside triphosphates (dNTPs; dATP, dTTP, dCTP and dGTP, 25 mM), 1 μL template DNA (50 ng/μL) and 0.3 μL Taq DNA polymerase (Promega, USA). PCR amplifications were performed with a My Cycler™ thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using the following program: 95°C for 5 min, 30 cycles of 94°C for 1 min, 40°C for 1 min, and 65°C for 8 min, and a final extension step at 65°C for 16 min.
The PCR products were electrophoresed in a 1.5% (w/v) agarose gel (15 × 20 cm) for 16 h at a constant voltage of 55 V in 1x TAE buffer [40 mM Tris-Acetate, 1 mM EDTA (pH 8.0)]. The rep-PCR profiles were visualized after staining with GelRed™ Nucleic Acid Gel Stain (Biotium, Hayward, CA, USA) under UV trans-illuminator (Syngene, Cambridge, UK), and digital image documentation was done using a CCD camera (Canon, Tokyo, Japan). The fingerprints were analyzed by the BioNumerics V4.0 software package (Applied Maths, Sint-Martens- Latem, Belgium). The similarity among digitized profiles was calculated using the Pearson correlation, and an average linkage (UPGMA, unweighted pair group method with arithmetic averages) dendrogram was derived from the profiles obtained.
Identification of LAB and FLAB by 16S rRNA Gene Sequencing
Representative LAB and FLAB isolates of all different (GTG)5-PCR fingerprint clusters were subjected to sequencing of the variable V1 region of the 16S ribosomal RNA gene using the PLB16 (5 ′AGA GTT TGA TCC TGG CTC AG 3′) and MLB16 (5′TGC GGC GTT TGG GTA CAC AG 3′) primers according to the protocol described by Hebert et al. (2000). The PCR assay mixture (50 μL) consisted of 5 μL 10 × buffer [20 mM Tris-HCl (pH 8.4), 500 mM KCl], 3 μL MgCl2 [50 mM], 2 μL of a mixture of dNTPs (dATP, dTTP, dCTP and dGTP, 5 mM), 1 U Taq polymerase (Inbio Highway, Buenos Aires, Argentina), 5 μL of each primer [10 μM], 28.7 μL nuclease-free water, and 1 μL of the purified chromosomal DNA (50 ng/μL) as template. PCR amplifications were performed with a My Cycler™ thermal cycler (Bio-Rad Laboratories, Inc.) using the following program: 94°C for 3 min, 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 45 s, and a final extension step at 72°C for 10 min.
PCR products were electrophoresed in a 1.0% (w/v) agarose gel at 100 V for 45 min in 1x TAE buffer, stained and visualized as described above. The size of DNA fragments (~500 bp) were estimated using a standard 1 kb DNA ladder (1 Kb Plus DNA Ladder, Invitrogen™, Carlsbad, CA, USA). Amplicons were purified by polyethylene glycol precipitation (protocol available at http://gator.biol.sc.edu/ http://labs.mcdb.lsa.umich.edu/labs/olsen/files/PCR.pdf). Nucleotide sequences of purified PCR products were determined at the CERELA-CONICET sequencing facility with an ABI 3130 DNA sequencer (Applied Biosystems, Foster, CA, USA).
Sequence Alignments
Identification at species level was achieved by using the BLAST (basic local alignment search tool) program (http://www.ncbi.nlm.nih.gov/BLAST) to compare the obtained rRNA gene sequences with those available at the GenBank database of the National Collection for Biotechnological Information (NCBI; http://www.ncbi.nlm.nih.gov/GenBank/) or Ribosomal Database Project (RDP; http://rdp.cme.msu.edu/) and estimate sequences similarities. For species assignation, a threshold of at least 97% identity with the reference strain in the databases was considered.
Analysis of Metabolic Properties
Mannitol and Organic Acids (Lactic Acid and Acetic Acid) Production
Thirty-eight selected strains belonging to different genera were grown in a formulated fruit simulation medium (FSM) (Ruiz Rodríguez et al., 2017b) with the following composition (g/L): glucose, 10.0; fructose 10.0; sorbitol, 5.0; malic acid, 2.0; MgSO4.7H2O, 0.2; MnSO4.H2O, 0.05; K2HPO4, 1.0; EDTA, 0.1; ammonium citrate, 2.0; vegetable peptone, 10.0; and Tween 80, 1 mL; pH 6.10. Cell-free supernatants (CFS) of 24-h cultures grown in FSM were analyzed for carbohydrate, organic acid, and mannitol content.
The consumption of glucose and fructose and mannitol production by selected strains were determined in the CFS by high-performance anion exchange chromatography (HPAEC) with pulsed amperometric detection, as described by Camu et al. (2007).
Production of lactic acid and acetic acid was determined by high-performance liquid chromatography (HPLC) according to Lefeber et al. (2010), except that an equal volume (500 μL) of acetonitrile was added to the CFS to remove proteins; the mixture was then centrifuged (16,060 × g for 15 min) and filtered (0.2 μm Minisart RC4 filters; Sartorius AG) before analysis.
All determinations were performed in triplicate and the mean values and standard deviations of each sample were calculated.
Aroma Compounds
Diacetyl Production
Overnight cultures of the selected strains were inoculated (1%, w/v) in 5 mL of FSM and incubated at 30°C for 48 h. After growth, 2 mL of the culture was supplemented with 1 mL of α-naphthol solution (4%, w/v) and 1 mL of KOH (30%, w/v) and incubated at 30°C for 30 min. Formation of a red/pink ring in the upper part of the cultures indicated diacetyl production (King, 1948). The results were qualitatively defined as negative (-), weak (+), medium (++) or strong (+++) according to the intensity of the color. The strain Lb. rhamnosus ATCC 7469 was used as positive control.
Ethyl Ester Production
Esterase enzymes, capable of both hydrolyzing and synthesizing esters, play an important role in food flavor development; the balance between synthesis and hydrolysis processes depends on several factors and the specific final product produced in each matrix confers differential organoleptic characteristics to fermented foods (Liu et al., 2004). Fruity ethyl esters can be synthesized by esterification when a fatty acid molecule reacts with ethanol to form an ester and water; this reaction is catalyzed by esterases and its activity is known as reverse esterase activity (REA). First, we evaluated the presence of esterase enzymes in the strains studied by determining the ability to hydrolyze esters. Then, the ability to synthesize esters with particular focus on the fruity ethyl esters was determined in esterase-positive strains.
Preparation of Cell-Free Extracts and Esterase Activity
Cells were harvested from 10 mL of FSM cultures after 16 h-incubation at 30°C by centrifugation (8,000 × g for 10 min) and washed three times with cold 50 mM potassium phosphate buffer (pH 7.0). The wet cell pellets were mixed with glass beads (150–212 μm, Sigma-Aldrich Chemical Co, MO, USA) in a 1:4:1 (mg cells:μL buffer:mg beads) ratio and subsequently disrupted using a Mini Bed Beater-8 (Biospec Products) for 10 min (with 2 min disruptions in ice every 2 min) at maximum speed. Cell debris and glass beads were removed by centrifugation (12,700 × g for 8 min, 4°C), and supernatants were immediately used as cell-free extracts (CFE) for esterase activity determination. Protein content (mg) of CFE were determined by Bradford (Bradford, 1976) with BSA as standard.
Esterase activity (EAh) of each CFE was determined on α-naphthyl (α-NA) derivatives (C2, C3, C4, C8, C10, and C12 of carbon atoms) as substrates (Sigma-Aldrich) according to Taboada et al. (2014) with minor changes. Briefly, the reaction mixture contained 18 μL of 100 mM sodium phosphate buffer (pH 7.0), 2 μL of α-NA substrate (10 mM in ethanol), and 20 μL of CFE. After incubation at 30°C for 1 h, the corresponding color was developed by adding 160 μL of Fast Garnet GBC (Sigma-Aldrich) preparation [5 g/L in 100 g/L SDS] and further incubation at room temperature for 15 min. The OD at 560 nm was measured by using a tuneable microplate reader (Versamax TM, Molecular Devices, Sunnyvale, CA, USA). Two standard curves were prepared using α-naphthol. One unit of esterase activity was defined as the amount of α-naphthol released by 1 mL of CFE/min. Specific EAh was defined as units per milligram of protein (U/mg) (Taboada et al., 2014).
Ester-Synthesizing Activity
The synthesis of fruity ethyl esters by esterification (also defined as REA) of 2 to 10 C atoms from butanoic acid and hexanoic acid, separately, in the presence of ethanol was studied. Specifically, the esters studied were: acetate-, propionate-, butanoate-, isovalerate-, caproate-, caprylate-, and ethyl caprate. Esters were detected by GC with a flame ionization detector (FID). Ester quantification was done using regression curves of the corresponding standards (R2 > 98%). These values were subtracted from the non-enzymatic production and the endogenous ester production, obtained from the substrate and CFE blanks, respectively. Afterwards, the results obtained were corrected by applying the recovery efficiency factor calculated previously, contemplating the product not recovered during the extraction procedure. One enzyme unit (U) was defined as the nmoles of esters formed per mL of enzyme extract per mL of reaction produced in 24 h. The specific activity was expressed as U per mg of total proteins.
Fruit and flower strains representing different LAB genera exhibiting EAh were selected for studying their ester-synthesizing capability. CFE were prepared from 300 mL cultures grown in MRS at 30°C for 16 h, harvested by centrifugation, washed twice with 100 mM phosphate buffer (pH 7.0), and resuspended at 40–60% (w/v) of the same buffer. Cell suspensions were disrupted by three successive passes through a French pressure cell (Thermo Spectronic model FA-078, NJ, USA) at 1,000 psi. Cell debris were removed and the supernatant was used as CFE. The protein content was determined by the Bradford method. Ester-synthesizing activity by esterification was determined by incubation of CFE in an assay mixture containing 100 mM sodium phosphate buffer (pH 7.0), 100 mM ethanol, and 10 mM free fatty acids (butanoic acid or hexanoic acid). After incubation at 30°C for 24 h, esters were extracted with ethyl ether from 1 mL of sample (2:1) and determined by GC [Agilent 6890N (CA, USA), Column HP5 (30 m, 0.32 mm d.i., 0.25 μm), carrier gas (nitrogen gas), detector, (FID)]. Controls lacking substrates or lacking CFE were included. One unit of ester-synthesizing activity was defined as nmoles of ethyl esters formed by 1 mL of CFE in 24 h. Specific ester-synthesizing activity was defined as units per milligram of protein (U/mg) (Abeijón Mukdsi et al., 2009).
Technological Properties
Acidifying Capacity and Growth Rate
Acidification rate and growth kinetics of 80 selected strains were evaluated. Active cultures (16–18 h) of the strains were inoculated [2% (v/v) inoculum] in FSM. After growth, cells were harvested by centrifugation (8,000 × g for 10 min at 4°C) and washed twice with 0.1 M sodium phosphate buffer (pH 7.0), resuspended in the same buffer, and the cell suspensions obtained were used as inoculum to evaluate the acidifying capacity (by measuring pH as a function of time), and bacterial growth by turbidimetry (optical density at 600 nm, OD600). For both assays, the OD600 of cell suspensions was measured and adjusted (Biotraza, model 722, Huida Medical Instruments Co., Jiangsu, China) to obtain microbial cultures with an initial OD600 of approximately 0.1.
Acidifying Capacity
pH values of culture samples were determined after 0, 2, 4, 6, 8, and 24 h of incubation (pH meter PT-10, Sartorius AG, Gottingen, Germany). Based on the pH curves obtained, the following data were calculated:
ΔpH8 (pH decrease after 8 h of incubation) = pH (0 h)–pH (8 h);
ΔpH24 (pH decrease after 24 h of incubation) = pH (0 h) − pH (24 h);
maximum acidification rate (Vmax, speed of the pH decrease): slope of the curve where the pH decrease is linear and maximum: .
Bacterial Growth
Bacterial growth was evaluated by measuring the OD600 (Versamax™ Microplate reader, Molecular Devices, CA, USA) in FSM every 30 min for 16 h. Growth curves were plotted (ln OD600 vs. time) and the maximum growth rates (μmax) were calculated as: .
The results of each parameter were analyzed individually; the Di Rienzo, Guzmán and Casanoves (DGC) test (Di Rienzo et al., 2002) was applied to evaluate statistical significant differences. All results were subjected to principal component analyses (PCA) using the RStudio software (RStudio-Team, 2016).
Pectinolytic Activity
Pectinolytic activity was qualitatively examined by pectin depolymerisation in pectin-containing agar media. Pure active cultures were inoculated with 5 μL spots on MRS agar (2%, w/v) without glucose and supplemented with 1% (w/v) citric pectin (Citromax S.A.C.I., Tucumán, Argentina) as the sole added carbon source. Plates were incubated at 30°C for 24 to 72 h. Then, iodine solution was added to detect a clearance zone due to pectin depolymerization after 15 min of staining followed by 15 min of washing with distilled water. The protocol was developed based on the works of Soares et al. (1999); Janani et al. (2011); Varghese et al. (2013); Vidhyasagar et al. (2013).
Cinnamoyl Esterase Activity
Cinnamoyl esterase activity was qualitatively evaluated by agar plate assays according to Donaghy et al. (1998) with modifications (Dr. Abeijón Mukdsi, personal communication, CERELA). Ethyl ferulate [EtFA, 2% (v/v) stock solution in methanol] was added to MRS agar media without glucose to a final concentration of 1 g/L. Overnight cultures were inoculated [2% (w/v)] in 5 mL of FSM and incubated at 30°C for 16 h. Grown cultures were centrifuged, washed twice with sodium phosphate buffer (0.1 M, pH 7.0), and resuspended in 5 mL of the same buffer. 50 μL of each suspension was transferred onto 1-cm diameter wells made on EtFA-supplemented agar media. Then, plates were incubated at 30°C for 24–72 h. The strain Lb. fermentum ATCC 14932 was used as positive control and the above mentioned buffer was used as negative control. The development of a clear zone around inoculated wells indicated breakdown of EtFA by the corresponding strain.
Biogenic Amine Production
Biogenic amine (BA) production was qualitatively evaluated according to Bover-Cid and Holzapfel (1999) with modifications. All strains under study, except for those of the Enterococcus species, were used. One strain of Ec. faecalis was used as positive control for tyramine production. From an active culture grown in MRS, two successive passages were done in MRSf containing 0.1% (w/v) of L-tyrosine, L-histidine, L-ornithine or L-lysine, to evaluate the production of tyramine, histamine, putrescine or cadaverine, respectively, by induction of the corresponding decarboxylase enzyme. Pyridoxal hydrochloride [0.005% (w/v)] was added as precursor of pyridoxal phosphate, the cofactor of the decarboxylase enzyme. Two successive passages of each active bacterial culture were done in MRSf without the addition of amino acids and were used as negative control. All cultures were incubated at 30°C for 16 h. BA production was determined in solid medium containing 1% (w/v) of each amino acid (tyrosine, histidine, ornithine, or lysine), separately, 0.005% (w/v) pyridoxal hydrochloride, and agar (1.5%, w/v), and with no addition of thiamin. Agar media lacking the presence of amino acids were used as negative control. Each culture was streaked on the five different amino acid-containing agar media and incubated at 30°C for 4 days. The production of BA was monitored by formation of purple color around the producing colonies due to the alkaline nature of BA in the presence of bromocresol purple as indicator.
Statistical Analysis
All assays were done in triplicate and average values with corresponding standard deviations (SD) are provided. Data were statistically analyzed using the Infostat Statistical Software (Universidad Nacional de Córdoba, Córdoba, Argentina). One-way analysis of variance (ANOVA) with the post hoc Tukey's test and DGC test were used to evaluate significant differences among samples. Principal component analysis was performed by using the tools for multivariate data analysis “ade4 package” for the RStudio software.
Results
Microbiological Analyses and Isolation of LAB
Selected weather conditions occurring during the fruit and flower sampling months are summarized in Table S1. Data were reported by the Tucumán Aerodrome meteorological station (ICAO: 871210–SANT). The majority of the samples were picked during the year 2013, which was particularly dry, registering 61.2% average annual relative humidity and 83 days of rain.
The total microbial counts were between 104 and 109 CFU/g on the fruits assayed and between 104 and 106 CFU/g on the flowers. Coliforms were present in counts of 104-108 CFU/g on the fruits and 104-106 CFU/g on the flowers, whereas LAB were found in lower numbers in the majority of the samples. LAB counts (directly isolated) were in the range of <102-104 CFU/g on the fruits or were not detectable (as in guava, custard apple, meddlar, and mulberries). Conversely, fig and papaya samples presented the highest LAB counts with 105 and 107 CFU/g, respectively. LAB counts in flower samples were in the order of 103 CFU/g with less variable count values than for fruits, the latter ones being strongly dependent on the samples (Table 2).
Table 2.
Viable colony counts of total bacteria (PCA agar), enterobacteria (Mac Conkey agar), and LAB (MRS and MRSf agar) in fruit and flower samples.
| log CFU/g | ||||||
|---|---|---|---|---|---|---|
| Sample | PCA (Total microbial count) | Mac Conkey (Enterobacteriaceae) | MRS (LAB) | MRSf (FLAB) | Picked colonies | Putative LAB |
| Guava | 9.0 | 8.0 | 3.0 | – | 135 | 84 |
| Papaya | 7.7 | 7.7 | 5.0 | – | 120 | 61 |
| Passion fruit | 5.7 | 4.7 | <2.0–4.7 | 4.7 | 265 | 85 |
| Passion fruit flowers | 5.7 | 4.7 | 3.7 | – | 30 | 25 |
| Custard apple | 4.7 | 4.0 | <2.0 | – | 78 | 24 |
| Custard apple flowers | 4.0 | 4.0 | 3.0 | – | 8 | 5 |
| Medlar | 7.0 | 6.0 | <2.0 | – | 89 | 68 |
| Medlar flowers | >6.0 | 6.0 | 3.7 | 3.7 | 106 | 81 |
| Mulberries | 4.0 | 4.0 | <2.0 | – | 72 | 44 |
| Fig | 7.7 | 4.0 | 6.7 | 5.7 | 317 | 245 |
| Khaki | 6.0 | 6.0 | 3.0 | 3.0 | 105 | 103 |
| Total | 1325 | 825 | ||||
A total of 1,325 colonies were picked from MRS, MRSf, and FYP agar media, derived from all fruit and flower samples; from these, 402 isolates could not be further recovered. Thus, 923 pure isolates were subjected to Gram staining and the catalase test, from which 825 Gram (+) and catalase (–) isolates were selected as putative LAB. The number of presumed LAB was variable among samples; figs being the fruits displaying the highest value (245); in contrast, only 5 isolates were obtained from custard apple flowers (Table 3).
Table 3.
Distribution of LAB isolates according to their species and the fruit and flower samples.
| Genus | Specie | Source | N° different strains/N° of isolates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | P | My | FMy | Ch | FCh | N | FN | Mr | H | Cq | |||
| Enterococcus | casseliflavus | 16 | 21 | 1 | 1 | 9 | 4/48 | ||||||
| casseliflavus/gallinarum | 1 | 27 | 4 | 3/32 | |||||||||
| durans | 3 | 2 | 2 | 1/7 | |||||||||
| faecalis | 1 | 1/1 | |||||||||||
| faecium | 4 | 3 | 2 | 3/9 | |||||||||
| hirae | 32 | 2 | 12 | 1 | 4/47 | ||||||||
| mundtii | 1 | 1/1 | |||||||||||
| Fructobacillus | durionis | 2 | 11 | 2/13 | |||||||||
| fructosus | 1 | 1/1 | |||||||||||
| pseudoficulneus | 1 | 1/1 | |||||||||||
| tropaeoli | 1 | 129 | 10 | 6/140 | |||||||||
| Lactobacillus | brevis | 13 | 1 | 4/14 | |||||||||
| plantarum | 1 | 1/1 | |||||||||||
| rhamnosus | 43 | 2/43 | |||||||||||
| Lactococcus | lactis | 13 | 5 | 3/18 | |||||||||
| lactis subsp. cremoris | 1 | 1/1 | |||||||||||
| lactis subsp. lactis | 8 | 5 | 4 | 5/17 | |||||||||
| Leuconostoc | citreum | 13 | 2/13 | ||||||||||
| mesenteroides | 3 | 10 | 26 | 1 | 6/40 | ||||||||
| mesenteroides subsp. dextranicum/mesenteroides | 1 | 30 | 3/31 | ||||||||||
| mesenteroides subsp. mesenteroides | 1 | 2 | 5 | 6/8 | |||||||||
| pseudomesenteroides | 27 | 1 | 3 | 2 | 52 | 9 | 1 | 60 | 24/155 | ||||
| Weissella | cibaria | 8 | 10 | 4/18 | |||||||||
| fabalis | 2 | 1/2 | |||||||||||
| minor | 12 | 6/12 | |||||||||||
| 56 | 56 | 49 | 24 | 19 | 2 | 60 | 79 | 44 | 193 | 91 | 95/673 | ||
Each fruit or flower sample were assigned with different colors.
Genotypical Analysis. Clustering of Isolates With Rep-PCR, and Identification of Representative Isolates
Putative LAB isolates were subjected to genotyping, using the rep-PCR technique to group those isolates corresponding to clones of the same strain. Once the (GTG)5-PCR assays were performed for each isolate, amplified bands were separated by electrophoresis, revealing a wide variety of band profiles among the different samples. Afterwards, (GTG)5-PCR fingerprints of the 825 putative LAB isolates were clustered into dendrograms (Figures S1, S2) to have an overview of the diversity of LAB species and strains present on the fruits and flowers assayed. One or more representative isolates of each profile group, representing genotypically different strains of LAB, were subjected to molecular identification. From the total putative LAB, 673 (81.6%) isolates were identified as LAB, whereas 152 (18.4%) isolates belonged to other bacterial groups or were environmental contaminants, such as acetic acid bacteria, staphylococci, etc. From the total LAB isolates, 44.7% (301) were obtained by direct plating of the samples, whereas enrichment cultures allowed the isolation of the remaining 55.3% (372). In this regard, it should be noted that the enrichment steps enabled the isolation of LAB strains from samples for which direct isolation failed, or very few isolates were recovered as in the case of guava, papaya, meddlar, and mulberries (data not shown). Papaya flowers were the only samples from which LAB failed to be isolated.
According to the band profiles obtained by (GTG)5-PCR, LAB isolates were distributed into 95 clusters, each representing different LAB strains. Representative isolates from each cluster were identified by comparing their 16S rRNA gene sequences with the available data in the NCBI or RDP databases. The fruits and flowers isolated strains belonged to six LAB genera, namely Enterococcus, Fructobacillus, Lactobacillus, Lactococcus, Leuconostoc and Weissella, and 21 different species were identified (Table 3). The largest cluster was constituted by 155 isolates belonging to Leuc. pseudomesenteroides, which were widely distributed among all samples assayed, except for papaya fruit, passion fruit, and custard apple flowers. The second largest group was formed by 140 fingerprints identified as F. tropaeoli; however, this species was only found in 3 different fruits, namely custard apple, fig, and khaki. Analysis of all LAB species showed that the genera Enterococcus and Leuconostoc were the most widely distributed among the samples studied; nine different Enterococcus species were found being this the most diverse genus present on fruits and flowers. On the other hand, Fructobacillus, Lactobacillus, and Weissella were the least spread genera among all samples. When studying the LAB species diversity present on each type of fruit or flower, it was found that figs and khakis harbored the highest numbers, with 8 and 7 different species, respectively (Table 3).
In those samples where LAB counting was possible, a microbial load analysis of each species was conducted (Table 4). The sample count estimation in MRS and MRSf (when possible) was similar, whereas the species load differed among the samples. For instance, Ec. casseliflavus was present in the order of 102 CFU/g on passion fruit flowers and custard apple flowers, 103 CFU/g on papaya and meddlar flowers, and 104 CFU/g on passion fruit. Further, Leuc. pseudomesenteroides counts were about 102 CFU/g on passion fruit; 102-104 CFU/g on meddlar flowers, and 104 CFU/g on khaki. Thus, the LAB species count was dependent on the fruit or floral matrix studied.
Table 4.
Microbial load of each LAB species present in the fruits and flowers assayed, as grown in MRS and MRSf incubated at 30°C for 48 h.
| CFU/g sample | |||
|---|---|---|---|
| Sample | Species | MRS | MRSf |
| GUAVA | |||
| G2 | Lb. brevis | 5.93 102 | – |
| PASSION FRUIT FLOWER | |||
| FMy1 | W. cibaria | 2.92 103 | – |
| Leuc. mesenteroides subsp. mesenteroides | 2.92 103 | – | |
| Lc. lactis subsp. lactis | 7.30 103 | – | |
| Ec. faecalis | 1.46 103 | – | |
| FMy2 | W. cibaria | 1.78 104 | – |
| Lc. lactis subsp. lactis | 8.88 103 | – | |
| FMy3 | Ec. gallinarum/casseliflavus | 1.28 103 | – |
| Ec. casseliflavus | 3.20 102 | – | |
| PASSION FRUIT | |||
| My3 | Ec. casseliflavus | 2.04 104 | 3.67 104 |
| My4 | Ec. casseliflavus | 1.33 104 | – |
| 2.33 103 | – | ||
| My11 | Leuc. pseudomesenteroides | – | 1.00 102 |
| Ec. casseliflavus | – | 4.00 102 | |
| CUSTARD APPLE FLOWER | |||
| FCh3 | Lb. Brevis | 4.37 102 | – |
| Ec. casseliflavus | 4.37 102 | – | |
| PAPAYA | |||
| P1 | Ec. casseliflavus | 1.00 103 | – |
| P3 | Leuc. mesenteroides subsp. dextranicum/mesenteroides | 1.00 102 | – |
| MEDLAR FLOWER | |||
| FN2 | Leuc. pseudomesenteroides | 1.39 104 | 1.18 104 |
| Lc. lactis | – | 6.20 103 | |
| FN3 | Leuc. pseudomesenteroides | 2.00 102 | 5.33 102 |
| Lc. lactis | 4.00 102 | 8.00 102 | |
| 1.00 103 | – | ||
| Ec. casseliflavus | 8.00 102 | – | |
| 1.00 103 | – | ||
| FIG | |||
| H1 | F. tropaeoli | 5.00 102 | 1.29 103 |
| F. durionis | 2.00 102 | 2.87 102 | |
| 1.00 103 | – | ||
| H2 | F. tropaeoli | 6.00 105 | 2.10 107 |
| H3 | F. tropaeoli | – | 3.00 105 |
| F. durionis | 6.88 105 | – | |
| 1.07 106 | – | ||
| Lc. lactis | – | 3.00 105 | |
| – | 3.31 105 | ||
| Lb. rhamnosus | 3.85 106 | 1.16 106 | |
| KHAKI | |||
| Cq1 | W. fabalis | 2.00 102 | – |
| Leuc. pseudomesenteroides | 1.00 104 | 5.00 103 | |
| 2.10 103 | 2.20 103 | ||
| Leuc. citreum | 1.00 102 | – | |
| Ec. hirae | 1.00 102 | – | |
| F. tropaeoli | – | 4.00 102 | |
Metabolite Target Analysis
Representative strains of the lactic microbiota present on wild fruits of guava, papaya, passion fruit, custard apple, meddlar, mulberry, fig and khaki, as well as on flowers of medlar, passion fruit, and custard apple from Tucumán were evaluated for their capacity to produce enzymes and compounds of biotechnological interest. Therefore, the metabolites mannitol, lactic acid, acetic acid, and diacetyl, as well as the enzymes cinnamoyl esterase, pectinase, and esterases were determined. These production properties correspond to the criteria normally used for the selection of functional starter cultures to be applied in fruit matrices.
Organic Acids and Mannitol Production
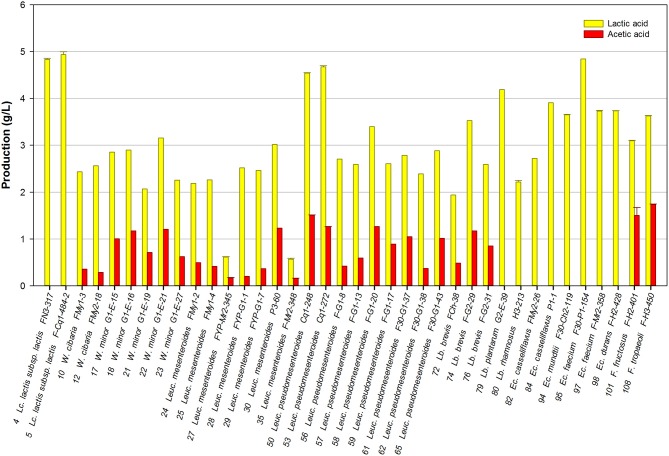
Thirty-eight representative strains of the isolated LAB species were selected and their ability to produce mannitol was evaluated in the FSM medium. Lactic acid and acetic acid production was simultaneously determined as end-fermentation products. As expected, lactic acid was the main organic acid produced by the strains studied, its concentration being in the range of 0.6–4.9 g/L. The maximal lactic acid production was achieved by strains of the homofermentative species Lc. lactis, followed by two Leuc. pseudomesenteroides, and one Enterococcus strain. Additionally, 28 strains produced acetic acid, though at lower concentrations (0.1–1.7 g/L), among which the Fructobacillus and Leuc. pseudomesenteroides strains produced the highest amounts (Figure 1). Lactic acid and acetic acid production was variable among the LAB isolates studied.
Figure 1.
Lactic acid and acetic acid production by selected LAB grown in FSM at 30°C for 24 h.
Mannitol is a relevant metabolite produced by certain heterofermentative LAB when growing on fructose, one of the most important carbohydrates present in fruits; so, mannitol production was studied in particular. Most of the strains studied were able to consume both monosaccharides present in the culture medium. Only the strains Lc. lactis FN3-317, Leuc. mesenteroides FYP-My2-345 and FYP-My2-348, Ec. faecium FYP-My2-38, and Ec. durans FYP-H2-428 grew at the expense of glucose solely, consuming very little fructose (0.22, 0.48, 0, 25, 0.62, and 0.55 g/L, respectively). The rest of the strains consumed equal amounts of both monosaccharides or mainly fructose, which was used for bacterial growth and mannitol production (Figure 2).
Figure 2.
Consumption of sugars and mannitol production by selected LAB grown in FSM at 30°C for 24 h.
Six strains belonging to the heterofermentative genera Leuconostoc and Fructobacillus produced high concentrations of mannitol (5.2–9.5 g/L). In particular, two strains, namely F. tropaeoli FYP-H3-450 (CRL2430, CERELA Culture Collection, Tucumán, Argentina) (Ruiz Rodríguez et al., 2017b) and F. fructosus FYP-H2-401, used fructose only as an alternative external electron acceptor, showing 100% mannitol yield in the presence of 10 g/L of fructose. The mannitol-producing phenotype was strain-dependent in most cases (Figure 2).
Aroma Compounds
Diacetyl Production
Fruits and vegetables are fermentable matrices wherein citrate is usually present. Some LAB species can ferment citrate, leading to the biosynthesis of aroma compounds, such as diacetyl, which positively impact the flavor of fermented food products (Smid and Kleerebezem, 2014). Under these considerations, the biosynthesis of 2,3-butanedione in FSM was tested for selected LAB isolates, among which only 23 strains were able to produce diacetyl. According to the intensity of the color, 10 strains of enterococci (5 Ec. casseliflavus, 1 Ec. faecalis, 1 Ec. hirae, 2 Ec. faecium, and 1 Ec. durans), 1 Lactobacillus (Lb. plantarum G2-E-39), 3 lactococci (Lc. lactis FMy2-21-2, FYP -Cq1-484-2, and FYP P-134-2), 1 Leuconostoc (Leuc. mesenteroides FYP30-P1-181), and 1 Fructobacillus (F. durionis H1-167) were weak (+) diacetyl producers, while Lc. lactis FN3-308 and FN3-317, Leuc. pseudomesenteroides FN3-319 and FN3F-306, and Ec. faecalis FMy1-8 showed moderate production (++); F. fructosus FYP-H2-401 and Lb. rhamnosus H3-213 and H3F-210 displayed strong pink rings (+++), indicating a positive diacetyl reaction (Table 5). Among all diacetyl-producing strains, disregarding the intensity of the fuchsia color generated, the majority corresponded to the genus Enterococcus and in second order to Lactococcus.
Table 5.
Technological properties of LAB strains isolated from fruits and flowers from Northern Argentina.
| Microorganism | Strain | Acidifing and kinetic parameters FSM medium | Biogenic amine production | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔpH8 ± SD | ΔpH24 ± SD | Vmax ± SD | μmax ± SD | Diacetyl production | Pectinolytic activity | Cinnamoyl esterase activity | T | H | P | C | |||
| Lactococcus lactis subsp. lactis | 1 | FN3-308 | 1.92 ± 0.21ª | 2.11 ± 0.15ª | −0.55 ± 0.24ª | 0.96 ± 0.02d | ++ | ++ | + | – | – | – | – |
| 2 | FMy2-21-2 | 1.94 ± 0.18ª | 2.17 ± 0.16ª | −0.54 ± 0.18ª | 0.97 ± 0.02d | + | ++ | + | – | – | – | – | |
| 4 | FN3-317 | 1.91 ± 0.16ª | 2.13 ± 0.15ª | −0.56 ± 0.10ª | 0.77 ± 0.01e | ++ | ++ | + | – | – | – | – | |
| 5 | F-Cq1-484-2 | 1.98 ± 0.16ª | 2.22 ± 0.16ª | −0.55 ± 0.03ª | 0.90 ± 0.00d | + | + | ++ | – | – | – | – | |
| Lc. lactis subsp. cremoris | 6 | F-P-134-2 | 1.68 ± 0.24ª | 1.91 ± 0.21ª | −0.43 ± 0.08b | 1.25 ± 0.04b | + | ++ | – | + | – | – | – |
| Lc. lactis | 9 | FN2F-266 | 1.66 ± 0.24ª | 2.03 ± 0.29ª | −0.27 ± 0.16b | 0.46 ± 0.00h | – | + | – | – | – | – | – |
| Weissella cibaria | 10 | FMy1-3 | 1.50 ± 0.02ª | 1.60 ± 0.19ª | −0.33 ± 0.08b | 0.95 ± 0.01d | – | + | – | – | – | – | – |
| 11 | FMy2-21-1 | 1.44 ± 0.08ª | 1.55 ± 0.27ª | −0.27 ± 0.10b | 0.97 ± 0.06d | – | + | – | – | – | – | – | |
| 12 | FMy2-18 | 1.45 ± 0.02ª | 1.56 ± 0.23ª | −0.26 ± 0.09b | 0.85 ± 0.03d | – | ++ | – | – | – | – | – | |
| W. fabalis | 14 | Cq1-277 | 0.17 ± 0.00c | 1.08 ± 0.50b | −0.10 ± 0.01b | 0.67 ± 0.03e | – | – | – | – | – | – | – |
| W. minor | 16 | G1-E-14 | 0.62 ± 0.16b | 1.70 ± 0.32ª | −0.12 ± 0.03b | 0.24 ± 0.02h | – | – | – | – | – | – | – |
| 17 | G1-E-15 | 0.72 ± 0.02b | 1.88 ± 0.23ª | −0.15 ± 0.03b | 0.49 ± 0.02g | – | – | – | – | – | – | – | |
| 18 | G1-E-16 | 0.62 ± 0.10b | 1.74 ± 0.08ª | −0.12 ± 0.01b | 0.55 ± 0.05f | – | – | – | – | – | – | – | |
| 21 | G1-E-19 | 1.64 ± 0.38ª | 1.92 ± 0.15ª | −0.32 ± 0.07b | 0.64 ± 0.03f | – | – | – | – | – | – | – | |
| 22 | G1-E-21 | 0.80 ± 0.18b | 1.70 ± 0.22ª | −0.18 ± 0.04b | 1.29 ± 0.15b | – | + | – | – | - | – | – | |
| 23 | G1-E-27 | 0.59 ± 0.04b | 1.58 ± 0.42ª | −0.15 ± 0.00b | 0.63 ± 0.07f | – | – | – | – | – | – | – | |
| Leuconostoc mesenteroides subsp. mesenteroides | 24 | FMy1-2 | 1.85 ± 0.11ª | 1.95 ± 0.19ª | −0.41 ± 0.00b | 0.64 ± 0.04f | – | – | – | – | – | – | – |
| 25 | FMy1-4 | 1.53 ± 0.00a | 1.93 ± 0.24ª | −0.28 ± 0.19b | 0.92 ± 0.06d | – | – | + | – | – | – | – | |
| 26 | F30-P1-181 | 1.46 ± 0.13ª | 1.61 ± 0.30ª | −0.35 ± 0.01b | 1.37 ± 0.08b | + | + | – | + | – | – | – | |
| 27 | F-Mr2-345 | 1.02 ± 0.15b | 1.92 ± 0.28ª | −0.21 ± 0.02b | 1.03 ± 0.12c | – | – | – | – | – | – | – | |
| 28 | F-G1-1 | 1.17 ± 1.04b | 1.95 ± 0.16ª | −0.18 ± 0.03b | 0.32 ± 0.04h | – | – | – | – | – | – | – | |
| 29 | F-G1-7 | 1.49 ± 0.07ª | 1.89 ± 0.11ª | −0.35 ± 0.02b | 0.62 ± 0.05f | – | – | – | – | – | – | – | |
| Leuc. mesenteroides | 30 | P3-60 | 1.81 ± 0.13ª | 1.89 ± 0.02ª | −0.40 ± 0.00b | 0.87 ± 0.05d | – | – | – | – | – | – | – |
| 34 | F-Cq1-489 | 1.92 ± 0.23ª | 2.09 ± 0.09ª | −0.39 ± 0.03b | 0.79 ± 0.00e | – | – | – | – | – | – | – | |
| 35 | F-Mr2-348 | 1.01 ± 0.28b | 2.08 ± 0.22ª | −0.20 ± 0.02b | 0.75 ± 0.03e | – | – | – | – | – | – | – | |
| 37 | F-Mr2-343 | 1.20 ± 0.13b | 2.05 ± 0.11ª | −0.26 ± 0.03b | 0.51 ± 0.01g | – | – | – | – | – | – | – | |
| 38 | F-Mr2-338 | 1.28 ± 0.19b | 1.87 ± 0.37ª | −0.22 ± 0.03b | 0.72 ± 0.03e | – | – | – | – | – | – | – | |
| Leuc. pseudomesenteroides | 39 | Cq1-260 | 1.43 ± 0.78ª | 2.00 ± 0.18ª | −0.27 ± 0.21b | 0.63 ± 0.03f | – | – | – | – | – | – | – |
| 41 | F-Mr1-297 | 1.44 ± 0.36ª | 2.02 ± 0.15ª | −0.25 ± 0.07b | 0.52 ± 0.01g | – | + | – | – | – | – | – | |
| 42 | F-N1-275 | 1.61 ± 0.08b | 1.70 ± 0.13ª | −0.37 ± 0.04b | 0.70 ± 0.01e | – | + | – | + | – | – | – | |
| 43 | FN2-284 | 2.04 ± 0.36a | 2.18 ± 0.33ª | −0.39 ± 0.12b | 0.60 ± 0.01f | – | + | – | – | – | – | – | |
| 44 | Cq1-245 | 1.72 ± 0.30a | 1.94 ± 0.23ª | −0.37 ± 0.09b | 0.46 ± 0.01g | – | – | – | – | – | – | – | |
| 45 | FN2-296 | 1.85 ± 0.21ª | 2.03 ± 0.13ª | −0.34 ± 0.02b | 0.56 ± 0.01f | – | – | – | – | – | – | – | |
| 46 | FN3-319 | 1.36 ± 0.01b | 1.66 ± 0.03ª | −0.37 ± 0.00b | 0.75 ± 0.01c | ++ | – | + | – | – | – | – | |
| 47 | FN3F-306 | 1.35 ± 0.05b | 1.67 ± 0.03ª | −0.36 ± 0.00b | 0.72 ± 0.01c | ++ | – | + | – | – | – | – | |
| 48 | Cq1-270 | 1.28 ± 0.41b | 2.05 ± 0.13ª | −0.37 ± 0.08b | 0.65 ± 0.00f | – | – | – | – | – | – | – | |
| 49 | Cq1F-218 | 1.55 ± 0.20ª | 1.89 ± 0.11ª | −0.34 ± 0.02b | 0.50 ± 0.01g | – | + | – | – | – | – | – | |
| 50 | Cq1-248 | 1.59 ± 0.06ª | 1.91 ± 0.08ª | −0.38 ± 0.05b | 0.65 ± 0.02f | – | – | – | – | – | – | – | |
| 51 | Cq1-256 | 1.56 ± 0.06ª | 1.89 ± 0.06ª | −0.38 ± 0.02b | 0.75 ± 0.02e | – | + | – | – | – | – | – | |
| 53 | Cq1-272 | 1.51 ± 0.01ª | 1.89 ± 0.11ª | −0.35 ± 0.01b | 0.61 ± 0.10f | – | + | – | – | – | – | – | |
| 56 | F-G1-8 | 1.61 ± 0.02ª | 1.91 ± 0.01ª | −0.36 ± 0.01b | 0.58 ± 0.01f | – | + | – | – | – | – | – | |
| 57 | F-G1-13 | 1.35 ± 0.34b | 1.77 ± 0.03ª | −0.24 ± 0.15b | 1.58 ± 0.01ª | – | + | – | – | – | – | – | |
| 58 | F-G1-20 | 1.03 ± 0.00b | 1.77 ± 0.00a | −0.31 ± 0.04b | 0.71 ± 0.01e | – | + | – | – | – | – | – | |
| 59 | F-G1-17 | 1.25 ± 0.01b | 1.75 ± 0.14ª | −0.29 ± 0.00b | 0.58 ± 0.01f | – | + | – | – | – | – | – | |
| 60 | F-G1-19 | 0.63 ± 0.00b | 1.86 ± 0.00a | −0.17 ± 0.02b | 0.86 ± 0.05d | – | + | – | – | – | – | – | |
| 61 | F30-G1-37 | 1.57 ± 0.05ª | 1.85 ± 0.17ª | −0.31 ± 0.04b | 1.04 ± 0.02c | – | ++ | – | – | – | – | – | |
| 62 | F30-G1-38 | 1.10 ± 0.54ª | 1.73 ± 0.09ª | −0.43 ± 0.10b | 0.71 ± 0.03e | – | + | – | – | – | – | – | |
| 65 | F30-G1-43 | 0.62 ± 0.25b | 1.70 ± 0.16ª | −0.18 ± 0.27b | 0.52 ± 0.01g | – | + | – | – | – | – | – | |
| 69 | F-G2-25 | 0.69 ± 0.13b | 1.86 ± 0.07ª | −0.28 ± 0.01b | 0.52 ± 0.01g | – | – | – | – | – | – | – | |
| Leuc. citreum | 70 | F-Cq1-501 | 1.85 ± 0.04ª | 1.95 ± 0.03ª | −0.33 ± 0.01b | 0.79 ± 0.00e | – | – | – | – | – | – | – |
| 71 | F-Cq1-496 | 1.77 ± 0.12ª | 1.86 ± 0.08ª | −0.37 ± 0.03b | 0.79 ± 0.00e | – | – | – | – | – | – | – | |
| Lactobacillus brevis | 72 | FCh3-38 | 0.83 ± 0.34b | 1.77 ± 0.07ª | −0.16 ± 0.02b | 0.34 ± 0.01h | – | + | – | – | – | – | – |
| 73 | G2-E-50 | 1.58 ± 0.45ª | 2.35 ± 0.06ª | −0.31 ± 0.06b | 0.40 ± 0.00g | - | – | – | – | – | – | – | |
| 74 | F-G2-29 | 0.90 ± 0.04b | 1.82 ± 0.23ª | −0.19 ± 0.01b | 0.42 ± 0.01g | – | + | – | – | – | – | – | |
| 76 | F-G2-31 | 0.97 ± 0.35b | 1.94 ± 0.11ª | −0.18 ± 0.02b | 0.46 ± 0.01g | – | + | – | – | – | – | – | |
| Lb. plantarum | 79 | G2-E-39 | 0.73 ± 0.45b | 2.18 ± 0.18ª | −0.21 ± 0.11b | 0.54 ± 0.02g | + | – | + | – | – | – | – |
| Lb. rhamnosus | 80 | H3-213 | 0.67 ± 0.19b | 1.96 ± 0.27ª | −0.16 ± 0.03b | 0.44 ± 0.08g | +++ | + | – | – | – | – | – |
| 81 | H3F-210 | 0.59 ± 0.18b | 2.14 ± 0.15ª | −0.15 ± 0.04b | 0.41 ± 0.04g | +++ | + | – | – | – | – | – | |
| Fructobacillus durionis | 100 | H1-167 | 1.50 ± 0.01ª | 1.62 ± 0.13ª | −0.28 ± 0.07b | 0.77 ± 0.02e | + | – | – | – | – | – | – |
| F. fructosus | 101 | F-H2-401 | 1.55 ± 0.18ª | 1.60 ± 0.10ª | −0.37 ± 0.05b | 0.68 ± 0.07e | +++ | – | – | – | – | – | – |
| F. pseudoficulneus | 102 | F-H3-468 | 1.59 ± 0.20ª | 1.64 ± 0.12ª | −0.45 ± 0.12b | 0.86 ± 0.04d | – | – | – | – | – | – | – |
| F. tropaeoli | 103 | F30-Ch2-116 | 1.53 ± 0.01ª | 1.69 ± 0.00a | −0.46 ± 0.13b | 0.73 ± 0.00e | – | – | – | – | – | – | – |
| 104 | F-H1-384 | 1.56 ± 0.26ª | 1.67 ± 0.08ª | −0.40 ± 0.09b | 0.90 ± 0.00d | – | – | – | – | – | – | – | |
| 105 | H2-200 | 1.76 ± 0.07ª | 1.82 ± 0.11ª | −0.42 ± 0.03b | 0.66 ± 0.01f | – | – | – | – | – | – | – | |
| 106 | H1F-130 | 1.77 ± 0.11ª | 1.78 ± 0.08ª | −0.43 ± 0.09b | 0.64 ± 0.00f | – | – | – | – | – | – | – | |
| 107 | Cq1F-246 | 1.74 ± 0.32ª | 1.80 ± 0.20ª | −0.40 ± 0.10b | 0.82 ± 0.02d | – | + | – | – | – | – | – | |
| 108 | F-H3–450 | 1.69 ± 0.04ª | 1.73 ± 0.08ª | −0.39 ± 0.18b | 0.53 ± 0.02g | – | – | – | – | – | – | – | |
| Ec. casseliflavus /gallinarum | 82 | FMy3-26 | n.d. | n.d. | n.d. | n.d. | – | + | + | n.d. | n.d. | n.d. | n.d. |
| 83 | F-My4-243 | n.d. | n.d. | n.d. | n.d. | + | + | + | n.d. | n.d. | n.d. | n.d. | |
| 84 | P1-1 | n.d. | n.d. | n.d. | n.d. | + | – | + | n.d. | n.d. | n.d. | n.d. | |
| Ec. casseliflavus | 85 | FN3-310 | n.d. | n.d. | n.d. | n.d. | + | + | – | n.d. | n.d. | n.d. | n.d. |
| 86 | F-P1-150 | n.d. | n.d. | n.d. | n.d. | + | + | – | n.d. | n.d. | n.d. | n.d. | |
| 87 | F30-P1-182 | n.d. | n.d. | n.d. | n.d. | + | + | – | n.d. | n.d. | n.d. | n.d. | |
| 88 | FCh3-31 | n.d. | n.d. | n.d. | n.d. | – | - | + | n.d. | n.d. | n.d. | n.d. | |
| Ec. faecalis | 89 | FMy1-8 | n.d. | n.d. | n.d. | n.d. | ++ | + | – | + | – | – | – |
| Ec. hirae | 90 | F-Ch2-102 | n.d. | n.d. | n.d. | n.d. | + | + | – | n.d. | n.d. | n.d. | n.d. |
| 91 | F-N1-266 | n.d. | n.d. | n.d. | n.d. | – | + | – | n.d. | n.d. | n.d. | n.d. | |
| 92 | F30-P1-160 | n.d. | n.d. | n.d. | n.d. | – | + | – | n.d. | n.d. | n.d. | n.d. | |
| 93 | F-N1-272 | n.d. | n.d. | n.d. | n.d. | – | + | – | n.d. | n.d. | n.d. | n.d. | |
| Ec. mundtii | 94 | F30-Ch2-119 | n.d. | n.d. | n.d. | n.d. | – | – | – | n.d. | n.d. | n.d. | n.d. |
| Ec. faecium | 95 | F30-P1-154 | n.d. | n.d. | n.d. | n.d. | + | + | – | n.d. | n.d. | n.d. | n.d. |
| 96 | F-N1-291 | n.d. | n.d. | n.d. | n.d. | + | + | – | n.d. | n.d. | n.d. | n.d. | |
| 97 | F-Mr2-358 | n.d. | n.d. | n.d. | n.d. | – | – | – | n.d. | n.d. | n.d. | n.d. | |
| Ec. durans | 98 | F-H2-428 | n.d. | n.d. | n.d. | n.d. | + | – | – | n.d. | n.d. | n.d. | n.d. |
T: tyramine, H: histidine, P: putrescine, C: cadaverine.
Mean values (± standard deviation) within the same column followed by different superscript letters are significantly different (p < 0.05) by DGC test.
Production of Fruity Esters
The specific esterase activity of hydrolysis (EAh) of α-naphthyl derivatives (α-NA) with carbon chains of C2, C3, C4, C8, C10, and C12 as carbon substrates using CFE was studied. Then, the REA for fruity ethyl ester biosynthesis was determined for selected strains.
From the total LAB isolates, 45 LAB strains belonging to 5 genera (Lactococcus, Weissella, Leuconostoc, Lactobacillus, and Enterococcus) showed EAh with at least two α-NA derivatives (Figure 3); all strains were active against α-NA-C2 and α-NA-C3 and none against α-NA-C12. The EAh on α-NA-C2, α-NA-C3, α-NA-C4 and α-NA-C8 was widely distributed among the LAB strains studied, whereas only Lc. lactis subsp. lactis F-Cq1-484-2, Leuc. mesenteroides FMy1-2 and Lc. lactis subsp. lactis FN3-308 hydrolyzed α-NA-C10. Lc. lactis FN3-317 showed the highest EAh value (32.38 ± 8.45 U/mg) on α-NA-C2, whereas Lb. rhamnosus H3F-210 showed the highest values on α-NA-C3 and α-NA-C4 (20.89 ± 6.49 and 19.76 ± 2.26 U/mg, respectively). On the other hand, all strains of Lb. brevis showed low EAh values, while the behavior of all enterococci was similar with respect to hydrolysis of α-NA-C3 and α-NA-C4 substrates.
Figure 3.
Esterase activity of hydrolysis of α-naphthyl derivatives (carbon chains of C2, C3, C4, C8, and C10) by selected LAB strains. Specific esterase activity values are indicated for each substrate. The strain numbers correspond to the codes given in Table 5.
Based on the results obtained, 8 strains were selected to further study the production of ethyl fruity esters. In general, varied activity values (0.97–22.43 U/mg) of ethyl ester biosynthesis were detected, being both strain- and substrate-dependent (Table 6). All strains produced ethyl acetate in diverse concentrations (1.55–28.75 nmol/mg protein) when using butanoic acid as substrate. Moreover, most strains produced at least one additional ester (ethyl propionate and/or ethyl butanoate) from the above mentioned substrate, with the exception for W. minor G1-E-19. Also, 4 strains could produce esters from hexanoic acid (the 2 strains of Lc. lactis, Leuc. pseudomesenteroides F30-G1-38, and Lb. rhamnosus H3F-210). High levels of short-chain ethyl esters were found, namely EtC2, EtC3, and EtC4 from butanoic acid and EtC2 and EtC3 from hexanoic acid, whereas no formation of esters of higher carbon atoms (EtC5, EtC6, EtC8, and EtC10) were detected with any of the substrates used. The highest REA was found for the strains Lb. rhamnosus H3F-210 and W. minor G1-E-19, with values of 22.43 ± 1.70 U/mg protein for ethyl butanoate and 17.97 ± 1.20 U/mg protein for ethyl acetate, respectively. The latter compound was the only ester formed by the W. minor strain that was also the only one producing a single type of ester. The highest ethyl propionate activity corresponded to Lc. lactis FMy2-21-2 (4.95 ± 0.83 U/mg protein), this value being 4 to 5 times lower than the maximum values detected for the biosynthesis of ethyl acetate and ethyl butanoate. Lc. lactis FN3-317 was the only strain that produced the 3 types of ethyl esters detected while Lb. brevis F-G2-31 presented remarkable values of ethyl acetate and ethyl butanoate activity (11.33 ± 1.05 and 4.86 ± 0.8 U/mg protein, respectively), despite being the strain with the lowest EAh among the selected strains (1.35 ± 0.72 and 1.23 ± 0.06 U/mg protein on α-NA-C2 and α-NA-C4, respectively).
Table 6.
Specific activity for the production of fruity ethyl esters by selected LAB strains.
| Strain | Specific ethyl ester-producing activity (U/mg protein) | |||||
|---|---|---|---|---|---|---|
| Butanoic acid + Ethanol | Hexanoic acid + Ethanol | |||||
| EtC2 | EtC3 | EtC4 | EtC2 | EtC3 | EtC4 | |
| 2 Lc. lactis FMy2-21-2 | 1.89 ± 0.76e | 4.95 ± 0.83a | n.d. | n.d. | 2.92 ± 0.64a | n.d. |
| 4 Lc. lactis FN3-317 | 3.42 ± 0.72 | 4.07 ± 0.75a | 7.85 ± 0.95b | n.d. | 3.71 ± 0.70a | n.d. |
| 21 W. minor G1-E-19 | 17.97 ± 1.20a | n.d. | n.d. | n.d. | n.d. | n.d. |
| 24 Leuc. mesenteroides FMy-1-2 | 4.38 ± 0.83d | n.d. | 1.94 ± 0.60d | n.d. | n.d. | n.d. |
| 29 Leuc. mesenteroides F-G1-7 | 0.97 ± 0.35e | 2.61 ± 0.60b | n.d. | n.d. | n.d. | n.d. |
| 62 Leuc. pseudomesenteroides F30-G1-38 | 6.41 ± 0.90c | n.d. | 2.35 ± 0.75d | 8.29 ± 01.10a | n.d. | n.d. |
| 76 Lb. brevis F-G2-31 | 11.33 ± 1.05b | n.d. | 4.86 ± 0.80c | n.d. | n.d. | n.d. |
| 81 Lb. rhamnosus H3F-210 | 2.88 ± 0.85e | n.d. | 22.43 ± 1.70a | 8.77 ± 1.20a | n.d. | n.d. |
Only the results for the esters of fatty acids with 2 to 4 C atoms are shown (no esters of fatty acids with 5 to 10 C were detected). EtC2, ethyl acetate; EtC3, ethyl propionate; EtC4, ethyl butanoate; n.d., not detected. Mean values (± standard deviation) within the same column followed by different superscript letters are significantly different (p < 0.05) by Tukey test.
Remarkably, when comparing the EAh and REA values of the strains capable of producing ethyl esters, the ability to hydrolyze esters of a certain chain length did not always correspond with the ability to produce ethyl esters of the same length (Figure 4).
Figure 4.

Esterase activity of biosynthesis and hydrolysis of ethyl esters by 8 LAB strains. 1, Lc. lactis FMy2-21-2; 2, Lc. lactis FN3-317; 3, W. minor G1-E-19; 4, Leuc. mesenteroides FMy-1-2; 5, Leuc. mesenteroides F-G1-7; 6, Leuc. pseudomesenteroides F30-G1-38; 7, Lb. brevis F-G2-31; and 8, Lb. rhamnosus H3F-210.
Properties of Technological Interest
Acidifying Capacity and Growth Rate
LAB are characterized by rapid acidification during growth due to the production of organic acids, such as lactic acid, acetic acid, and formic acid, depending on their ability to ferment carbohydrates. The acidifying capacity of 67 LAB strains from tropical fruits and flowers was evaluated by determining their pH curves during a 24 h-period, considering that the established time of fruit and vegetable fermentation is between 15 and 24 h (Di Cagno et al., 2013). From the pH values obtained, ΔpH8, ΔpH24, and maximum acidification rates (Vmax) were calculated. Likewise, microbial growth (OD600 and maximum specific growth rate, μmax) were determined (Table 5).
The maximum ΔpH8 and ΔpH24 values in FSM corresponded to Leuc. pseudomesenteroides FN2-284 and Lb. brevis G2-E-50, respectively while the maximum value of Vmax corresponded to Lc. lactis subsp. lactis FN3-317. The highest μmax value corresponded to Leuc. pseudomesenteroides F-G1-13. It is interesting to highlight that in some cases strains belonging to the same species showed a similar behavior; for example, all strains of Lc. lactis subsp. lactis showed high acidification and growth rate values. Most strains of W. minor and Lactobacillus showed low acidifying capacity and low μmax values. For the remaining species, the acidifying behavior was strain-dependent. It is important to emphasize that out of 67 strains assayed, 66 (except for W. fabalis Cq1-277) reached a pH value ≤4.5 within 24 h of fermentation, which is an essential requirement for fruit fermentation. Additionally, 10 strains decreased the pH to 4.5 in only 6 h of incubation, whereas 26 did so after 8 h (data not shown).
As expected, a positive correlation between the acidifying capacity and the growth rate of the strains was found.
As maximum Vmax and μmax values were strain-dependent, the existence of statistically significant differences between the values of each parameter of all strains was studied. In general, limited statistically significant variability between the ΔpH and Vmax values was found, unlike the μmax values, which were more diverse, allowing grouping of the strains into 8 categories significantly different among each other (a-h, Table 5).
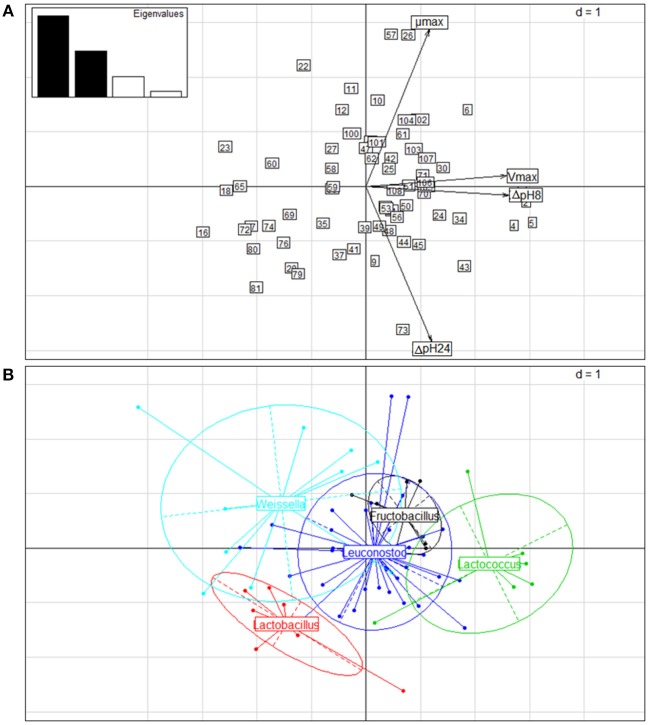
The results of the acidifying capacity and growth rates of the strains were also subjected to PCA. Two principal components (PC) accounted for 82.79% of the total variance. Figure 5 shows the biplot of the PCA for the first (PC1) and the second (PC2) PC, which explained 52.86 and 29.93% of the total variance, respectively. PC1 (vertical axis) separated strains displaying the highest values of ΔpH8 and Vmax to the right, whereas to the left samples appeared with low variable values. PC2 (horizontal axis) separated upwards those strains with higher μmax and downwards those with higher ΔpH24 values (Figure 5A). Considering the genus as classification factor (Figure 5B), the Lactococcus strains presented ΔpH8 and Vmax values higher than the rest of the genera, while the genus Lactobacillus differed from the rest throughout horizontal axis, showing higher ΔpH24 values but lower μmax values than the rest. The genera Leuconostoc, Weissella, and Fructobacillus could not be clearly differentiated from each other using these 2 PC analyses, probably due to the close phylogenetic relationship among these three genera belonging to the Leuconostocaceae family.
Figure 5.
Principal component analysis (PCA) of the kinetic parameters of 67 LAB strains isolated from wild fruits and flowers in Northern Argentina. (A) Biplot of PCA obtained considering ΔpH8, ΔpH24, Vmax and μmax of the strains grown in FSM medium at 30°C for 24 h. Arrows correspond to eigenvectors for the kinetic parameters. The numbers of the strains correspond to the codes presented in Table 5. (B) Biplot of the strain distribution obtained, considering the genus as classification factor.
Pectinolytic Activity
Pectinases are enzymes that break down pectin substances that are used in the industry to increase the yield and clarity of fruit juices. Pectic polysaccharides are normal components of plant tissues, thus LAB present on fruits and vegetable surfaces could harbor pectinolytic activity as a niche adaptation feature. From the total evaluated strains (43), 51.2% showed a clearance zone of pectin depolymerization around the assayed spots. The largest hydrolysis halos of citrus pectin were found around growing colonies of all lactococci, particularly for the strain Lc. lactis subsp. lactis FN3-317. Among the enterococci, 12 strains hydrolyzed citrus pectin, whereas almost all lactobacilli, except for Lb. brevis G2-E-50 and Lb. plantarum G2-E-39, and 14 strains of Leuc. pseudomesenteroides exhibited pectinase activity. On the other hand, this property was generally absent in the Fructobacillus species (except for F. tropaeoli Cq1F-246), all strains of Leuc. mesenteroides (with the exception of F30-P1-181) and almost all Weissella isolates, among which W. minor G1-E-21 and 3 strains of W. cibaria (FMy1-3, FMy2-21-1 and FMy2- 18) were pectinolytic strains (Table 5).
Cinnamoyl Esterase Activity
Some LAB show esterase activity on plant phenolic compounds (esters of hydroxycinnamic acids), releasing the corresponding acid whose beneficial properties for human health have been widely studied. The capacity of the strains assayed to hydrolyze ethyl ferulate was qualitatively evaluated. From the total LAB strains studied, only 12 strains showed hydrolysis of ethyl ferulate, although weak activity was found as compared to the control strain Lb. fermentum ATCC 14932. Four Lc. lactis strains, 1 W. minor, 1 Leuc. mesenteroides, 2 Leuc. pseudomesenteroides, 1 Lb. plantarum, and 4 Ec. casseliflavus showed small halos of positive cinnamoyl esterase activity, among which Lc. lactis F-Cq1-484-2 presented a larger and sharper halo (Table 5).
Production of Biogenic Amines
The intake of foods with high levels of BA may cause toxicological problems. In some fermented foods, either spontaneously or through the use of starter cultures, BA may be present as result of microbial metabolism. Thus, the inclusion of BA-producing strains in starter cultures should be avoided. The ability of 67 LAB strains to produce BA, particularly histamine, tyramine, putrescine, and cadaverine, was studied (Table 5). All Enterococcus strains were excluded because they frequently produce BA and carry virulence factors, and their intentional use is highly questioned in food processing. However, a tyramine-producing Ec. faecalis strain was used as positive control (Bover-Cid and Holzapfel, 1999).
Among the 67 strains evaluated, only 3 produced tyramine, which was visualized by purple coloration around colonies on tyrosine-containing agar media. The capacity to produce BA was rarely widespread among the flower- and fruit-origin strains studied.
As the number of strains belonging to each studied genus was substantially different, the percentage of strains of the same genus capable of displaying a specific property should be considered. Thus, it is important to highlight that would be necessary to increase the number of strains of a particular genus (i.e., Lactococcus in this study) to be able to conclude about how frequent a particular characteristic is detected.
Discussion
Microbiological studies of 12 types of wild fruits and flowers disseminated in the province of Tucumán, Argentina, revealed that the estimated total microbial population fluctuated between 105 and 109 CFU/g and 104 and 105 CFU/g on fruits and flowers, respectively, in coincidence with other studies indicating values between 105 and 107 CFU/g on fruits and vegetables (Di Cagno et al., 2010, 2015). Regarding LAB populations present in this type of niches, the present study showed that colony counts obtained by direct isolation were variable and dependent on the samples, being in general between <102-104 CFU/g, or not detectable, such as in fruits as passion fruit, custard apple, medlar, and mulberries. On the contrary, high LAB counts were recorded in the order of 107 and 105 CFU/g in fig and papaya samples, respectively, these values being similar to those reported by other authors for different fruits, whereas Fessard et al. (2016) reported the same counts for papaya. Di Cagno et al. (2010, 2011a) found that mesophilic LAB populations on blackberries, prunes, kiwis, and papayas ranged between 102 and 104 CFU/g, whereas on pineapple the counts were higher (104-105 CFU/g). Bae et al. (2006) found that LAB were present in 102 and 104 CFU/mL in grape juice homogenates, whereas counts in masau tropical fruit were 103 CFU/g (Nyanga et al., 2007).
From the 12 wild fruits and flowers studied, 673 isolates were confirmed as LAB; the isolated LAB/sample average ratio (673/12) being higher than for most other works available in the literature for this types of matrices, although the isolation protocols were similar; some differences occurred in the sample type and treatments applied (commercial or wild, washed or unwashed, direct isolation or by culture enrichment) in each case (Bae et al., 2006; Trias Mansilla et al., 2008; Chen et al., 2010; Naeem et al., 2012). Bae et al. (2006) only found LAB by direct plating in 4 out of 43 batches of grape homogenates studied and could isolate 160 LAB mainly by culture enrichment. Chen et al. (2010) isolated 88 LAB from blackberry samples from 5 different farms in Taiwan, whereas Di Cagno et al. (2010) isolated 104 LAB from pineapple. While Emerenini et al. (2013) obtained 105 LAB isolates from different fruits (citrus, bananas, tomatoes) and vegetables (pumpkin and green vegetables), Garcia et al. (2016) isolated 50 LAB from samples of Barbados cherry, mango, soursop, and strawberry.
Strain biodiversity in the fruit and flower samples assayed was estimated according to the band profiles obtained by using the (GTG)5-PCR genotyping technique. The rep-PCR technique has been recognized as a simple technique, with high discriminatory power, low cost, suitable for handling large numbers of strains, and for classifying a wide range of Gram (+) and Gram (–) bacteria at species, subspecies, and/or strain level (Gevers et al., 2001). Rep-PCR has been used for typing LAB from different origins by several authors (Gevers et al., 2001; Ouadghiri et al., 2005; Vasilopoulos et al., 2008; Sengun et al., 2009; Papalexandratou et al., 2011b; Perin and Nero, 2014; Šalomskiene et al., 2015; Sáez et al., 2017). In this work, 673 isolates were grouped into 95 different clusters according to their band profiles, indicating higher species and strain diversity of LAB in the samples examined than that found in other fruits and vegetables (Bae et al., 2006; Chen et al., 2010; Di Cagno et al., 2010; Askari et al., 2012; Emerenini et al., 2013; Wu et al., 2014); (Verón et al., 2017).
The isolated LAB strains belonged to 21 different species of the genera Enterococcus, Fructobacillus, Lactobacillus, Lactococcus, Leuconostoc, and Weissella. The genera Fructobacillus, Lactobacillus, and Weissella were less spread among the samples studied, Enterococcus and Leuconostoc being the most widely distributed genera. Many of the LAB species isolated in this work are typically associated with the plant environment. Ec. faecalis, Ec. faecium, Ec.mundtii, and Ec. casseliflavus are commonly plant-associated species; while Ec. durans and Ec. hirae have been found in veterinary materials among other habits (Holzapfel and Wood, 2014). Fructobacillus species have originally been found in fructose-rich habitats such as flowers, fruit surfaces, fermented fruits, and the guts of insects (Holzapfel and Wood, 2014; Endo et al., 2018). Lactobacillus species are usually found in nutrient-rich habitats such as foods, feeds, surface of plants, animals and humans. The ecological role in nature of plant-associated lactobacilli is still poorly understood, as their occurrence is only sporadic they are not considered plant symbionts but rather epiphytic by some researchers (Duar et al., 2017). Duar et al. (2017) classified Lb. plantarum and Lb. rhamnosus as nomadic lifestyle species while Lb. brevis as free-living species. Leuconostocs are associated with different habitats; Leuc. citreum has been isolated from clinical sources and from food such as kimchi and wheat sourdoughs; in general, the typical habitats of Leuc. mesenteroides subsp. dextranicum and Leuc. mesenteroides are fermentable plant raw materials, fruit/vegetable mashes (e.g., sauerkraut) and fruit juices; Leuc. pseudomesenteroides has been normally isolated from dairy, food, and clinical sources (Holzapfel and Wood, 2014). Weissella species inhabit diverse niches such as vegetable-, fish-, and meat-derived fermented foods; W. cibaria has been detected in fermented foods of vegetable origin as well as W. fabalis, which has been isolated from cocoa bean fermentation; W. minor has been found from sludge of milking machine, fermented fruits and vegetables, and fermented dry salami (Fessard and Remize, 2017). Lactococcus species have been found on the surface of plants and animals and products of these origins, but the best recognized habitats are raw milk, cheese, and other dairy products (Holzapfel and Wood, 2014).
In the present study, the most abundant species according to the number of isolates were Leuc. pseudomesenteroides (155) and F. tropaeoli (140), the latter being restricted to samples of custard apple, fig, and khaki. These three fruits possess the highest total sugar content values (12.87, 16.26, and 12.53 g per 100 g of fruit, respectively) from the eight fruits studied here (USDA Food Composition Databases, United States Department of Agriculture, Agricultural Research Service, www.ndb.nal.usda.gov/ndb/). In addition, fig and khaki were the samples from which the majority of LAB were isolated (Table 3). In contrast to these results, the species most frequently reported in these matrices were W. cibaria/confusa and especially Lb. plantarum. Although the latter is considered ubiquitous and is frequently the major part of the lactic population of fruits and vegetables due to its metabolic versatility (Di Cagno et al., 2015), only 3 Lactobacillus species were isolated from the samples of our study: Lb. plantarum (1 strain, guava), Lb. brevis (4 strains, guava; 1 strain, custard apple flower), and Lb. rhamnosus (2 strains, fig). Lb. rhamnosus is often a dairy-associated species but it has been isolated once from fruit or vegetable by other authors (Ceapa et al., 2015). Similarly, Di Cagno et al. (2010) reported the isolation of Lb. rossiae from pineapple although it is a species frequently isolated from sourdough; in another study the meat-associated species Lb. curvatus was isolated from pepper by the same authors (Di Cagno et al., 2010). On the other hand, Verón et al. (2017) only isolated strains of Lb. plantarum (2) and F. fructosus (2) from samples of prickly pear fruit. Askari et al. (2012) reported that Lactobacillus and Leuconostoc were the most abundant genera in raisins and figs, followed by Streptococcus and to a lesser extent Pediococcus, whereas in fresh grape juice Bae et al. (2006) found that Lb. lindneri was the most frequent species and in a smaller ratio Lb. kunkeei and Ec. durans. In the fig samples of the present study, Fructobacillus was the most abundant genus (4 different species), whereas Enterococcus, Lactobacillus, Lactococcus, and Leuconostoc were only represented by one strain. Some FLAB have been naturally found in figs (Antunes et al., 2002; Endo et al., 2009). Lb. plantarum was also the dominant species in pineapple while Lb. rossiae and W. cibaria were also present (Di Cagno et al., 2010); Emerenini et al. (2013) identified as Lb. plantarum 8 out of 22 LAB from fruits and vegetables while this species was also the most abundant in fruit pulps (Garcia et al. (2016) and fresh fruits (Naeem et al. (2012). Nyanga et al. (2007) found that the lactic microbiota present in samples of mulberries was composed of Ec. durans, Ec. faecium, Leuc. mesenteroides, and Leuc. pseudomesenteroides, whereas Chen et al. (2010) found that W. cibaria was the dominant species in mulberries from Taiwan together with strains of Lb. plantarum, Lc. lactis and, similarly to this work, Leuc. pseudomesenteroides. In the papaya samples, Enterococcus was the predominant genus (Ec. casseliflavus, Ec. faecium, and Ec. hirae), whereas strains of Lc. lactis and Leuc. mesenteroides were also isolated; other authors isolated strains of W. cibaria, Lb. plantarum, Lb. pentosus, Leuc. holzapfelii, and Leuc. mesenteroides/pseudomesenteroides (Fessard et al., 2016). Custard apple fruits showed a broad LAB species diversity, such as Ec. hirae, Ec. mundtii, F. durionis, F. tropaeoli, Leuc. pseudomesenteroides, and W. cibaria, although a low number of isolates were obtained. The only authors who reported LAB isolation from custard apple were Trias Mansilla et al. (2008), who found one strain of Leuc. mesenteroides. Regarding the presence of LAB on flowers, Ec. casseliflavus was isolated from the 3 types of flowers analyzed in our study, whereas Lc. lactis was present on flowers of passion fruit and medlar. On medlar flowers, as well as on the fruits, Leuc. pseudomesenteroides was present, whereas on the passion fruit flowers a Leuc. mesenteroides strain was found and W. cibaria was the most abundant species. Other Lactobacillus species, such as Lb. ozensis (Kawasaki et al., 2011a), Lb. florum (Endo et al., 2010), Lb. floricola (Kawasaki et al., 2011b), Lb. sakei and Lb. kunkeei (Linjordet, 2016) in addition to Ec. faecium (Di Cagno et al., 2011a) and Enterococcus spp. (Linjordet, 2016) were isolated from wild flowers. Linjordet (2016) conducted a detailed study, during which W. viridescens and W. ceti, Lc. lactis, Lc. garvieae, and F. fructosus were isolated. Some of these LAB species, such as F. fructosus, Lb. kunkeei, and Ec. faecium, has also been isolated from the pollinator Apis mellifera L. (Carina Audisio et al., 2011; Pachla et al., 2018). The lactic microbiota of guava, passion fruit, medlar, khaki, and flowers of passion fruit, custard apple, and medlar was reported for the first time during the present study. No new LAB species could be isolated from the samples studied.
The carbohydrates present in the isolation media as well as the isolation methods employed strongly influence the success for the isolation of LAB from specific niches. Direct isolation is generally used if cells are present in high numbers, as in feces and fermented foods; however, if microbial cells are present in low numbers and specific species are sought, culture enrichment before bacterial isolation should be applied. This methodology presents the disadvantage that only some species, showing fast cell growth, will grow at the cost of others, eliminating competition from those with slower growth. One of the main factors responsible for this selection is the growth substrate, since the use of specific carbon sources is generally different at species level (Endo et al., 2011a). In this work, culture enrichment using fructose as a carbon source for the isolation of FLAB was conducted. This approach enabled not only the isolation of FLAB but also other LAB species which were present in very low numbers. Several authors isolated LAB from flowers and fruits using this methodology (Antunes et al., 2002; Bae et al., 2006; Endo et al., 2009, 2010, 2011a; Naeem et al., 2012).
In general, glucose is the most easily metabolizable substrate for the majority of microorganisms, including LAB, and therefore the most used carbohydrate for bacterial isolation and culturing (Antunes et al., 2002). However, several studies have suggested that some species have evolved by adapting to their niches to survive and preferring to metabolize other specific carbohydrates; this may be the case for FLAB that, when inhabiting fructose-rich niches such as fruits, may have lost their ability to mainly metabolize glucose during adaptation and hence preferring fructose as a carbon source to grow (Endo et al., 2011a; Endo, 2012; Filannino et al., 2019). In our work, the use of fructose allowed isolating the fructophilic species F. fructosus and F. pseudoficulneus, and to increase the number of fructophilic isolates of F. durionis and F. tropaeoli. Endo et al. (2009) described the isolation protocol for FLAB, by which they could isolate F. pseudoficulneus from banana and fig samples and Lb. kunkeei and F. fructosus from different flowers.
It has been claimed that the microbial population present on plants and their parts, including flowers and fruits, may be subjected to nutritional fluctuations, and physicochemical and environmental conditions, as well as to dispersal events (Samuni-Blank et al., 2014), reasons by which each fruit, flower or plant might carry a particular microbiota in a specific geographical region and on a specific time-point. Plant-associated habitats (roots, leaves, flowers, fruits or decaying tissues) differ in their local availability of nutrients and physicochemical conditions, conditioning the range of potential microbiota. For instance, floral nectar has been regarded merely as a sweet aqueous secretion offered by flowering plants to attract pollinators. Nevertheless, pollinators act not only as pollen vectors, but at the same time they can transport microorganisms from flower to flower (Alvarez-Perez et al., 2012). Also, it has been demonstrated that nectar microbial community may vary among different plant species (Fridman et al., 2012). All this background, might explain the variations regarding LAB diversity present on flowers and fruits reported in the literature so far.
Recently, there has been particular interest in the use of autochthonous fruit LAB as starter cultures to be used in the manufacture of differentiated and/or functional fruit-based foods (Trias et al., 2008; Di Cagno et al., 2011a,b, 2015, 2016; Mousavi et al., 2011; Filannino et al., 2013). Fruits and flowers are fructose-rich plant parts that heterofermentative LAB inhabit; many of these bacteria being able to reduce this sugar and to produce mannitol (Filannino et al., 2018). From 24 positive mannitol-producing strains, 6 strains of the genera Leuconostoc and Fructobacillus synthesized mannitol in concentrations higher than 5 g/L from 10 g/L of fructose present in the culture medium. According to the literature, several strains of Lactobacillus, Leuconostoc, Fructobacillus, and Oenococcus are capable of producing mannitol from fructose (Saha and Racine, 2011). Although to date Lb. intermedius NRRL B-3693 is the LAB strain with the highest capacity to produce mannitol reported so far (Saha and Nakamura, 2003; Saha, 2006a,b,c; Saha and Racine, 2010), strains of Leuconostoc and Fructobacillus have been described as very good mannitol producers (von Weymarn et al., 2002; Fontes et al., 2009; Carvalheiro et al., 2011; Ruiz Rodríguez et al., 2017b). Likewise, all described species of the genus Fructobacillus, normally isolated from fructose-rich niches such as flowers, fruits and insect intestines, can convert fructose into mannitol as a result of their peculiar fructophilic metabolism (Endo et al., 2009, 2014; Endo, 2012). Filannino et al. (2016) reported that all FLAB strains isolated from bee intestines, including 5 strains of F. fructosus, produced mannitol from fructose. Other plant-associated species such as Lb. florum were capable of producing mannitol in culture media containing glucose and fructose (Tyler et al., 2016), and as observed for F. tropaeoli F-H3-450 and F. fructosus F-H2-401 during the present study.
Diacetyl is undoubtedly another industrially interesting compound that contributes to the flavor of many fermented foods and can be naturally synthesized by LAB. The ability to form diacetyl was present in all genera, except for Weissella; strains of Lb. rhamnosus showing the highest production. The most important diacetyl-producing LAB species are Lc. lactis, Lactobacillus spp., S. thermophilus, and Leuc. mesenteroides, besides the subspecies Lc. lactis biovar. diacetylactis (Ruiz Rodríguez et al., 2017a). Filannino et al. (2014) studied the behavior of strains of Lb. plantarum isolated from fermented fruits and vegetables and found that diacetyl was the ketone present in the highest concentration.
Some LAB possess esterase activity (Liu et al., 2014), capable of hydrolyzing and producing esters, which play a fundamental role in the flavor of fermented foods. In the present work, the EAh was strain-specific; in addition, substrate specificity was different among different LAB genera and species. Although not many studies on LAB strains isolated from fruit sources exist, this observation coincides with that reported for strains using other matrices (Oliszewski et al., 2007; Pérez-Martín et al., 2013). The EAh on α-NA-C2, α-NA-C3, α-NA-C4, and α-NA-C8 derivatives was distributed among the LAB strains studied; however, a tendency of the strains to be more active against short-chain substrates such as α-NA-C2, α-NA-C3, and α-NA-C4 was found. Similarly, Oliszewski et al. (2007) reported that all strains of indigenous goat milk and cheese LAB showed EAh on α-NA-C2 to α-NA-C6. Matthews et al. (2007) also found that 9 strains of Oenococcus, Lactobacillus, and Pediococcus species, isolated from commercial starter cultures for vinification and from olive products, showed greater activity on short-chain esters (C2-C8) compared to long ones (C10-C18). Likewise, Taboada et al. (2014) found that 22 LAB strains from goat dairy products showed activity on α-NA-C2, α-NA-C3, α-NA-C4 and α-NA-C8. The preferential or exclusive hydrolysis of α-NA esters derived from C2-C6 fatty acids by LAB was detected by other authors too (Gobbetti et al., 1996; Katz et al., 2002). On the contrary, Pérez-Martín et al. (2013) observed that most of the 243 wine strains of Oenococcus, Lactobacillus, Pediococcus, and Enterococcus species from different grape varieties could more easily hydrolyse esters of 8 and more carbon atoms. The EAh values determined in the present work were varied, the Lc. lactis and Lb. rhamnosus strains showing the maximum activity values, whereas the lowest corresponding to strains of Lactobacillus and Enterococcus species. Oliszewski et al. (2007) reported that enterococci showed the highest activities on α-NA-C4 and α-NA-C6, whereas Lb. rhamnosus ETC14 showed the highest specific EAh on α-NA butyrate and caproate. Taboada et al. (2014) also reported that a Lb. rhamnosus strain showed the highest activity on α-NA acetate. Similarly, the strain Lb. rhamnosus H3F-210 of our study showed the highest activity values on α-NA-C3 and α-NA-C4. Nardi et al. (2002) found that the esterase activity EstA was responsible for most of the ester-producing activity in Lc. lactis. Most of the strains of LAB species present in wine, namely Oenococcus, Pediococcus and Lactobacillus, possess esterase activity (Sumby et al., 2010). Further, several studies highlight the biosynthesis of fruity esters by O. oeni strains (Costello et al., 2013; Sumby et al., 2013).
As mentioned elsewhere, in addition to hydrolysis, esterases also have the ability to synthesize esters by esterification of fatty acids and ethanol. These ethyl esters, even in very low amounts, play an important role in the development of the fruity organoleptic characteristics of some foods (Taboada et al., 2014). In this work, the ability to produce ethyl esters through esterification by 8 selected LAB strains with EAh was evaluated. REA values were variable and dependent on both the strain and substrate used, in coincidence with the findings of Abeijón Mukdsi et al. (2009) and Costello et al. (2013), working with LAB strains from milk and goat and sheep cheese origin and wines, respectively. When studying particularly the biosynthesis of ethyl butanoate by dairy LAB, (Liu et al., 1998) found that this ability was variable and species- and strain-dependent. All the strains studied in the present work could synthesize at least one ethyl ester from the two substrates assayed as well as the strains studied by (Costello et al., 2013). The fruit- and flower-origin strains of the present study produced ethyl acetate, ethyl propionate, and ethyl butanoate from butanoic acid and hexanoic acid, unlike the vinification LAB strains that produced mainly ethyl butanoate, ethyl hexanoate, and ethyl octanoate from the corresponding fatty acid precursors (Costello et al., 2013), and of other dairy-origin strains that produced mainly ethyl butanoate and ethyl hexanoate (Abeijón Mukdsi et al., 2009). All the strains tested produced ethyl acetate, although to a different extent, in contrast to the observations of Abeijón Mukdsi et al. (2009), who did not detect the biosynthesis of this ethyl ester by LAB. In the present work, the studied strains produced 3 types of ethyl esters when butanoic acid was used as substrate, unlike when using hexanoic acid, which led to the biosynthesis of ethyl acetate (2 strains) and ethyl propionate (2 strains). In contrast, Abeijón Mukdsi et al. (2009) found that some strains could synthesize mainly EtC6 from butanoic acid, whereas Costello et al. (2013), in agreement to the present findings, did not detect biosynthesis of EtC4 from the corresponding fatty acid. Regarding the biosynthesis of ethyl butanoate, strains of Lb. rhamnosus, Lc. lactis, and Lb. brevis showed the highest biosynthesis activity, whereas strains of Leuc. mesenteroides and Leuc. pseudomesenteroides showed the lowest capacity. Also, Liu et al. (1998) observed that strains of Lc. lactis showed moderate ethyl butanoate biosynthesis activity and strains of Lb. rhamnosus and Lb. paracasei subsp. paracasei showed greater potential than those of lactococci, whereas the capacity of Leuconostoc strains was variable.
As mentioned above, all strains studied produced ethyl acetate from butanoic acid, which is not a direct precursor of this ester. The mechanism involved in this phenomenon is still unknown. Some authors (Liu et al., 2003; Abeijón Mukdsi et al., 2009) suggested that other mechanisms of ester biosynthesis in addition to esterification could be involved; Liu et al. (2004) hypothesized that ethyl esters could be synthesized non-enzymatically.
Noticeably, when comparing the EAh and REA activities of the strains of this study, their ability to hydrolyze esters of a certain length was not always correlated with the capacity to produce ethyl esters of the same length (Figure 4); these results may be explained by the presence of more than one esterase enzyme with different specificities (Oliszewski et al., 2007).
Native cultures are preferred to allochthonous starters for food fermentation since indigenous strains display shorter latency phases and better acidification capacity. For the selection of autochthonous strains as starter cultures for fruit and vegetable fermentations, the bacterial capacity to lower the matrix pH to values below 4.5 should be considered essential to achieve the inhibition of unwanted microorganisms from the early stages of fermentation (Di Cagno et al., 2013). Since rapid growth and acidification rates are conventional criteria for the selection of starter cultures, these parameters were studied. In general, acidification kinetics and growth parameters were variable among the LAB strains examined, in coincidence with findings of LAB from plant matrices (Filannino et al., 2014; Fessard et al., 2016, 2017). Although the absolute values of the parameters studied were strain-dependent, a slight tendency of lactococci and fructobacilli to grow and acidify more rapidly than the other bacteria was noticed, the lowest values being observed for Weissella and Lactobacillus strains. Fessard et al. (2016) studied 10 LAB strains isolated from papaya, tomato, and cabbage, belonging to Leuconostoc, Weissella and Lactobacillus, and found Vmax values of 0.10- 0.15 U pH/h when growing the strains in MRS. These values were lower than those found in the present work and much lower than for other LAB (0.2-1.2 U pH/h) (Latrille et al., 1992; Xanthopoulos et al., 2001; Cachon et al., 2002). The same researchers found that strains of W. cibaria showed the highest acidifying potential. More recently, Fessard et al. (2017) observed that half of 28 strains of Leuconostoc, Lactococcus, Lactobacillus, and Fructobacillus examined exhibited Vmax values in the range of 0.155–0.22 U pH/h. Analyzing some particular cases in our study, the strain of Lb. plantarum showed values of Vmax = 0.21 U pH/h, similar to those found by Filannino et al. (2014) for Lb. plantarum strains isolated from cherry and pineapple in MRS (0.23- 0.21 U pH/h, respectively). Although acidification kinetics has been widely used as a tool to monitor fermentation performance, this parameter it is not frequently used in vegetable or fruit fermentations and the LAB starters related to them (Fessard et al., 2016). Different microbial acidifying capacities are needed depending on the type of fermented product; on one side, rapid acidification (i.e., L. plantarum) till pH ≤ 4.5 is desired in fruit juice fermentation (Di Cagno et al., 2010) while on the other side, for a few vegetables mild acidification is preferred (Lee et al., 2011) as in the case of kimchi, where over souring is one of the most serious defect. Then, autochthonous strains of Lb. sakei are selected because of their mild acid-producing properties (Lee et al., 2011). Thus, the diversity on the growth and acidification parameters shows the potential of the strains of our study to be used in starter culture formulations for different fermentation processes.
In nature, several microorganisms are capable of synthesizing pectinases, a complex set of hydrolytic enzymes that cleave pectic substances that constitute a large part of the vegetable raw materials (Sakellaris et al., 1989; Karam and Belarbi, 1995; Pedrolli et al., 2009; Prathyusha and Suneetha, 2011). Although this microbial enzymatic activity may cause unwanted softening of fermented vegetables, as in pickles (Buckenhüskes, 1993), this pectinolytic activity would be desirable in starters in the fruit juice industry, since it would help to control the viscosity of these products (Di Cagno et al., 2013). Currently, little information about the pectinolytic activity of LAB is available although several pectinase and pectinase-like coding genes have been recently annotated (http://www.ncbi.nlm.nih.gov/GenBank/). In our work, 43 strains representative of the 6 genera assayed capable of hydrolyzing citrus pectin were found. Sakellaris et al. (1988, 1989) described the extracellular polygalacturonase (PG) activity of a Lb. plantarum strain and purified and characterized this enzyme. Karam and Belarbi (1995) studied the presence of pectinolytic activity in 80 LAB strains isolated from milk in Algeria, of which only 4 strains (2 Lb. casei, 1 Lb. plantarum, and 1 Lc. lactis) produced PG and/or pectin esterase. Vidhyasagar et al. (2013) studied pectin degradation by LAB isolated from fermented foods and found strains of P. pentosaceus, Leuc. lactis and Lb. plantarum subsp. argentoratensis able to hydrolyze pectin. Chatterjee et al. (2016) found that the presence of pectin significantly improved bacterial growth and titratable acidity in LAB and Bifidobacterium cultures, concluding that it could be used as a potential prebiotic. The pectinolytic activity of strains of Enterococcus and Fructobacillus species was qualitatively revealed for the first time in the present work. Further studies are needed to characterize these enzymes.
Considering that human tissues and biological fluids do not possess esterases capable of hydrolyzing esters of phenolic acids (for example, chlorogenic acid), bacterial cinnamoyl esterases present in starter cultures could enrich plant matrices in free phenolic acids with high bioavailability for man (Filannino et al., 2018). For this reason, the presence of cinnamoyl esterases in LAB from flowers and fruits was evaluated. Only 14% of the examined strains showed weak cinnamoyl esterase activity compared to the control. The species capable of hydrolyzing ethyl ferulate were Lc. lactis, W. minor, Lb. plantarum, Ec. casseliflavus, Leuc. mesenteroides, and Leuc. pseudomesenteroides, being the strain Lc. lactis F-Cq1-484-2 the most remarkable whose halo, although larger and more defined than the others, was less evident than that of the positive control. Many of the strains with enzymatic activity on phenolic compounds were isolated from raw materials or fermented foods with a high content of these compounds or from human samples (Sánchez-Maldonado et al., 2011; Di Cagno et al., 2015). Abeijón Mukdsi (2009) isolated the strain Lb. fermentum CRL 1446 with high cinnamoyl ester activity, mainly on methyl ferulate, from an Argentinean goat cheese, whereas Xu et al. (2017) isolated 4 strains of Lactobacillus with feruloyl esterase activity from ensiled corn stover. Esteban-Torres et al. (2013) studied the ability to hydrolyze hydroxycinnamic esters by the human saliva strain Lb. plantarum WCFS1 and found that it could partially hydrolyze methyl ferulate and methyl p-coumarate. Later, the same authors (Esteban-Torres et al., 2015) studied the cinnamoyl ester activity of 28 Lb. plantarum strains; only 7 could hydrolyze methyl ferulate or methyl caffeate. To date, studies on cinnamoyl esterase activity of LAB strains isolated from flowers and fruits remain scarce. Interestingly, data on Lc. lactis, W. minor, Ec. casseliflavus, Leuc. mesenteroids, and Leuc. pseudomesenteroides strains with esterase activity on hydroxycinnamic acids was reported in the present study for the first time.
Histamine is recognized as the causative agent of scombroid poisoning, whereas tyramine consumption has been linked to food-induced migraines and hypertensive crisis in patients who consume monoamine oxidase inhibitor drugs. In turn, putrescine and cadaverine can potentiate the toxicity of the previous amines and, in addition, be precursors of carcinogenic nitrosamines. Apart from these toxicological aspects, the appearance of relatively high levels of certain BA indicates deterioration and/or defective food elaboration (Bover-Cid and Holzapfel, 1999). From all strains evaluated, only three (Lb. lactis, Leuc. mesenteroides and Leuc pseudomesenteroides) produced only tyramine, indicating that the formation of BA was not widely spread among the fruit- and flower-origin LAB assayed. Similarly, tyramine was the main BA formed by Enterococcus, Carnobacterium, and some Lactobacillus strains in the studies conducted by Bover-Cid and Holzapfel (1999) and by Leuconostoc strains in the work of Moreno-Arribas et al. (2003). To date, very few strains of Lb. curvatus, Lb. buchneri, and Lb. brevis as producers of putrescine and cadaverine were detected (Bover-Cid and Holzapfel, 1999; Moreno-Arribas et al., 2003) and only significant levels of histamine biosynthesis were reported in strains of O. oeni (Landete et al., 2005). On the other hand, Tomita et al. (2017) found that heterofermentative LAB species were characterized as producers of high levels of tyramine.
Conclusion
The results obtained during the present study supported the hypothesis that LAB strains isolated from fruits and flowers from Northern Argentina could be exploited from a biotechnological point of view. Strains capable of producing mannitol, organic acids, and aroma compounds were found; in addition, strains harboring cinnamoyl esterase, pectinase, and esterase activities, interesting properties to be used in fruit food matrices, were detected. Differences between the results obtained for the fruit- and flower-origin LAB strains of the present study and those available in the literature could be explained by the diversity of substrates, fermentation protocols, and analyses used; but more importantly, they could be inherent to the microbial diversity existing in wild niches belonging to different regions of the world. In this sense, this work provided a deeper insight into the lactic microbiota present on tropical fruits and flowers. In addition, the LAB strains isolated harbored interesting functional properties to be used in starter culture formulations for fruit-based fermented food products.
Author Contributions
LRR carried out the majority of the experiments and wrote the manuscript. FM and JB carried out some of the experiments. RM directed the esterases experiments. LDV and EH corrected the manuscript. FeM directed the work and corrected the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by grants Préstamo BID PICT 2014-312 from FONCyT and PIP 003 from CONICET from Argentina and the BAFF (Belgian Argentinean Fermented Foods) project from VUB, Brussels, Belgium. We thank Dr. Sabrina Volentini for her assistance in DNA sequencing and Dr. Claudia Abeijón Mukdsi for her assistance and advise during the esterase activity experiments. LRR, FM, and JB are the recipients of fellowships from CONICET, Argentina.
Footnotes
Funding. The experimental work of this study was supported by grants Préstamo BID PICT 2014–312 from FONCyT and PIP 003 from CONICET from Argentina and the BAFF (Belgian Argentinean Fermented Foods) project from VUB, Brussels, Belgium.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01091/full#supplementary-material
References
- Abeijón Mukdsi M.C. (2009). Esterases of Lactic Acid Bacteria in Fermented Foods. Ph. D. Thesis, Universidad Nacional de Tucumán, Argentina. [Google Scholar]
- Abeijón Mukdsi M.C., Medina R.B., Alvarez M.d,.F, González S.N. (2009). Ester synthesis by lactic acid bacteria isolated from goat's and ewe's milk and cheeses. Food Chemistry 117, 241–247. 10.1016/j.foodchem.2009.03.105 [DOI] [Google Scholar]
- Alvarez-Perez S., Herrera C.M., de Vega C. (2012). Zooming-in on floral nectar: a first exploration of nectar-associated bacteria in wild plant communities. FEMS Microbiol. Ecol. 80, 591–602. 10.1111/j.1574-6941.2012.01329.x [DOI] [PubMed] [Google Scholar]
- Anderson K.E., Sheehan T.H., Mott B.M., Maes P., Snyder L., Schwan M.R., et al. (2013). Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS ONE 8:e83125. 10.1371/journal.pone.0083125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes A., Rainey F.A., Nobre M.F., Schumann P., Ferreira A.M., Ramos A., et al. (2002). Leuconostoc ficulneum sp. nov., a novel lactic acid bacterium isolated from a ripe fig, and reclassification of Lactobacillus fructosus as Leuconostoc fructosum comb. nov. Int. J.Syst. Evol. Microbiol. 52, 647–655. 10.1099/00207713-52-2-647 [DOI] [PubMed] [Google Scholar]
- Askari G., Kahouadji A., Khedid K., Charof R., Mennane Z. (2012). Screenings of lactic acid bacteria isolated from dried fruits and study of their antibacterial activity. Middle East J. Sci. Res. 11, 209–215. [Google Scholar]
- Bae S., Fleet G., Heard G. (2006). Lactic acid bacteria associated with wine grapes from several Australian vineyards. J. Appl. Microbiol. 100, 712–727. 10.1111/j.1365-2672.2006.02890.x [DOI] [PubMed] [Google Scholar]
- Bover-Cid S., Holzapfel W.H. (1999). Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 53, 33–41. 10.1016/S0168-1605(99)00152-X [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Brown T.A. (1995). Purification of DNA from living cells, in Gene Cloning: An Introduction, eds Chapman & Hall (Manchester: Stanley Thornes; ), 27–51. [Google Scholar]
- Buckenhüskes H.J. (1993). Selection criteria for lactic acid bacteria to be used as starter cultures for various food commodities. FEMS Microbiol. Rev. 12, 253–271. 10.1016/0168-6445(93)90067-J [DOI] [Google Scholar]
- Cachon R., Jeanson S., Aldarf M., Divies C. (2002). Characterisation of lactic starters based on acidification and reduction activities. Le Lait 82, 281–288. 10.1051/lait:2002010 [DOI] [Google Scholar]
- Camu N., De Winter T., Verbrugghe K., Cleenwerck I., Vandamme P., Takrama J.S., et al. (2007). Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl. Environ. Microbiol. 73, 1809–1824. 10.1128/AEM.02189-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carina Audisio M., Torres M.J., Sabat,é D.C., Ibarguren C., Apella M.C. (2011). Properties of different lactic acid bacteria isolated from Apis mellifera L. bee-gut. Microbiol. Res. 166, 1–13. 10.1016/j.micres.2010.01.003 [DOI] [PubMed] [Google Scholar]
- Carvalheiro F., Moniz P., Duarte L.C., Esteves M.P., Gírio F.M. (2011). Mannitol production by lactic acid bacteria grown in supplemented carob syrup. J. Indust. Microbiol. Biotechnol. 38, 221–227. 10.1007/s10295-010-0823-5 [DOI] [PubMed] [Google Scholar]
- Ceapa C., Lambert J., van Limpt K., Wels M., Smokvina T., Knol J., et al. (2015). Correlation of Lactobacillus rhamnosus genotypes and carbohydrate utilization signatures determined by phenotype profiling. Appl. Environ. Microbiol. 81, 5458–5470. 10.1128/AEM.00851-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambel L., Chelo I.M., Zé-,Zé L., Pedro L.G., Santos M.A., Tenreiro R. (2006). Leuconostoc pseudoficulneum sp. nov., isolated from a ripe fig. Int. J. Syst. Evol. Microbiol. 56, 1375–1381. 10.1099/ijs.0.64054-0 [DOI] [PubMed] [Google Scholar]
- Chatterjee M., Manuel G.A.S., Hassan S.S. (2016). Effect of fruit pectin on growth of lactic acid bacteria. J. Probiot. Health. 2, 147–150. 10.4172/2329-8901.1000147 [DOI] [Google Scholar]
- Chen Y.-S., Wu H.-C., Yanagida F. (2010). Isolation and characteristics of lactic acid bacteria isolated from ripe mulberries in Taiwan. Braz. J. Microbiol. 41, 916–921. 10.1590/S1517-83822010000400010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello P., Siebert T., Solomon M., Bartowsky E. (2013). Synthesis of fruity ethyl esters by acyl coenzyme A: alcohol acyltransferase and reverse esterase activities in Oenococcus oeni and Lactobacillus plantarum. J. Appl. Microbiol. 114, 797–806. 10.1111/jam.12098 [DOI] [PubMed] [Google Scholar]
- De Man J., Rogosa d., Sharpe M.E. (1960). A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23, 130–135. 10.1111/j.1365-2672.1960.tb00188.x [DOI] [Google Scholar]
- Dhakal R., Bajpai V.K., Baek K.-H. (2012). Production of GABA (γ-aminobutyric acid) by microorganisms: a review. Braz. J. Microbiol. 43, 1230–1241. 10.1590/S1517-83822012000400001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cagno R., Cardinali G., Minervini G., Antonielli L., Rizzello C.G., Ricciuti P., et al. (2010). Taxonomic structure of the yeasts and lactic acid bacteria microbiota of pineapple (Ananas comosus L. Merr.) and use of autochthonous starters for minimally processing. Food Microbiol. 27, 381–389. 10.1016/j.fm.2009.11.012 [DOI] [PubMed] [Google Scholar]
- Di Cagno R., Coda R., De Angelis M., Gobbetti M. (2013). Exploitation of vegetables and fruits through lactic acid fermentation. Food Microbiol. 33, 1–10. 10.1016/j.fm.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Di Cagno R., Filannino P., Gobbetti M. (2015). Vegetable and fruit fermentation by lactic acid bacteria, in Biotechnology of Lactic Acid Bacteria: Novel Applications, eds Mozzi F., Raya R. R., Vignolo G. M. (Chichester: John Wiley & Sons; ), 216–230. 10.1002/9781118868386.ch14 [DOI] [Google Scholar]
- Di Cagno R., Filannino P., Vincentini O., Lanera A., Cavoski I., Gobbetti M. (2016). Exploitation of Leuconostoc mesenteroides strains to improve shelf life, rheological, sensory and functional features of prickly pear (Opuntia ficus-indica L.) fruit puree. Food Microbiol. 59, 176–189. 10.1016/j.fm.2016.06.009 [DOI] [PubMed] [Google Scholar]
- Di Cagno R., Minervini G., Rizzello C.G., De Angelis M., Gobbetti M. (2011a). Effect of lactic acid fermentation on antioxidant, texture, color and sensory properties of red and green smoothies. Food Microbiol. 28, 1062–1071. 10.1016/j.fm.2011.02.011 [DOI] [PubMed] [Google Scholar]
- Di Cagno R., Surico R.F., Minervini G., Rizzello C.G., Lovino R., Servili M., et al. (2011b). Exploitation of sweet cherry (Prunus avium L.) puree added of stem infusion through fermentation by selected autochthonous lactic acid bacteria. Food Microbiol. 28, 900–909. 10.1016/j.fm.2010.12.008 [DOI] [PubMed] [Google Scholar]
- Di Cagno R., Surico R.F., Paradiso A., De Angelis M., Salmon J.C., Buchin S., et al. (2009). Effect of autochthonous lactic acid bacteria starters on health-promoting and sensory properties of tomato juices. Int. J. Food Microbiol. 128, 473–483. 10.1016/j.ijfoodmicro.2008.10.017 [DOI] [PubMed] [Google Scholar]
- Di Rienzo J.A., Guzmán A.W., Casanoves F. (2002). A multiple-comparisons method based on the distribution of the root node distance of a binary tree. J. Agric. Biol. Environ. Stat. 7, 129–142. 10.1198/10857110260141193 [DOI] [Google Scholar]
- Donaghy J., Kelly P., McKay A. (1998). Detection of ferulic acid esterase production by Bacillus spp. and lactobacilli. Appl. Microbiol. Biotechnol. 50, 257–260. 10.1007/s002530051286 [DOI] [PubMed] [Google Scholar]
- Duar R.M., Lin X.B., Zheng J., Martino M.E., Grenier T., Pérez-Muñoz M.E., et al. (2017). Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 41, S27–S48. 10.1093/femsre/fux030 [DOI] [PubMed] [Google Scholar]
- Edwards C., Haag K., Collins M., Hutson R., Huang Y. (1998). Lactobacillus kunkeei sp. nov.: a spoilage organism associated with grape juice fermentations. J. Appl. Microbiol. 84, 698–702. 10.1046/j.1365-2672.1998.00399.x [DOI] [PubMed] [Google Scholar]
- Emerenini E., Afolabi O., Okolie P., Akintokun A. (2013). Isolation and molecular characterization of lactic acid bacteria isolated from fresh fruits and vegetables using nested PCR analysis. Br. Microbiol. Res. J. 3, 368–377. 10.9734/BMRJ/2013/2520 [DOI] [Google Scholar]
- Endo A. (2012). Fructophilic lactic acid bacteria inhabit fructose-rich niches in nature. Microb. Ecol. Health Dis. 23:18563. 10.3402/mehd.v23i0.18563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A., Futagawa-Endo Y., Dicks L. (2011a). Influence ofcarbohydrates on the isolation of lactic acid bacteria. J. Appl. Microbiol. 110, 1085–1092. 10.1111/j.1365-2672.2011.04966.x [DOI] [PubMed] [Google Scholar]
- Endo A., Futagawa-Endo Y., Dicks L.M. (2009). Isolation and characterization of fructophilic lactic acid bacteria from fructose-rich niches. Syst. Appl. Microbiol. 32, 593–600. 10.1016/j.syapm.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Endo A., Futagawa-Endo Y., Sakamoto M., Kitahara M., Dicks L.M. (2010). Lactobacillus florum sp. nov., a fructophilic species isolated from flowers. Int. J. Syst. Evol. Microbiol. 60, 2478–2482. 10.1099/ijs.0.019067-0 [DOI] [PubMed] [Google Scholar]
- Endo A., Irisawa T., Futagawa-Endo Y., Sonomoto K., Itoh K., Takano K., et al. (2011b). Fructobacillus tropaeoli sp. nov., a fructophilic lactic acid bacterium isolated from a flower. Int. J. Syst. Evol. Microbiol. 61, 898–902. 10.1099/ijs.0.023838-0 [DOI] [PubMed] [Google Scholar]
- Endo A., Maeno S., Tanizawa Y., Kneifel W., Arita M., Dicks L., et al. (2018). Fructophilic lactic acid bacteria, a unique group of fructose-fermenting microbes. Appl. Environ. Microbiol. 84, e01290–e01218. 10.1128/AEM.01290-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A., Okada S. (2008). Reclassification of the genus Leuconostoc and proposals of Fructobacillus fructosus gen. nov., comb. nov., Fructobacillus durionis comb. nov., Fructobacillus ficulneus comb. nov. and Fructobacillus pseudoficulneus comb. nov. Int. J. Syst. Evol. Microbiol. 58, 2195–2205. 10.1099/ijs.0.65609-0 [DOI] [PubMed] [Google Scholar]
- Endo A., Salminen S. (2013). Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst. Appl. Microbiol. 36, 444–448. 10.1016/j.syapm.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Endo A., Tanaka N., Oikawa Y., Okada S., Dicks L. (2014). Fructophilic characteristics of Fructobacillus spp. may be due to the absence of an alcohol/acetaldehyde dehydrogenase gene (adhE). Curr. Microbiol. 68, 531–535. 10.1007/s00284-013-0506-3 [DOI] [PubMed] [Google Scholar]
- Esteban-Torres M., Landete J. M., Reverón I., Santamaría L., de las Rivas B., Muñoz R. (2015). A Lactobacillus plantarum esterase active on a broad range of phenolic esters. App. Environ. Microb. 81, 3235–3242. 10.1128/AEM.00323-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban-Torres M., Reverón I., Mancheño J.M., de las Rivas B., Muñoz R. (2013). Characterization of a feruloyl esterase from Lactobacillus plantarum. Appl. Environ. Microbiol. 79, 5130–5136. 10.1128/AEM.01523-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessard A., Bourdon E., Payet B., Remize F. (2016). Identification, stress tolerance, and antioxidant activity of lactic acid bacteria isolated from tropically grown fruits and leaves. Can. J. Microbiol. 62, 550–561. 10.1139/cjm-2015-0624 [DOI] [PubMed] [Google Scholar]
- Fessard A., Kapoor A., Patche J., Assemat S., Hoarau M., Bourdon E., et al. (2017). Lactic fermentation as an efficient tool to enhance the antioxidant activity of tropical fruit juices and teas. Microorganisms 5:23. 10.3390/microorganisms5020023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessard A., Remize F. (2017). Why are Weissella spp. not used as commercial starter cultures for food fermentation? Fermentation 3:38 10.3390/fermentation3030038 [DOI] [Google Scholar]
- Filannino P., Azzi L., Cavoski I., Vincentini O., Rizzello C.G., Gobbetti M., et al. (2013). Exploitation of the health-promoting and sensory properties of organic pomegranate (Punica granatum L.) juice through lactic acid fermentation. Int. J. Food Microbiol. 163, 184–192. 10.1016/j.ijfoodmicro.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Filannino P., Cardinali G., Rizzello C.G., Buchin S., De Angelis M., Gobbetti M., et al. (2014). Metabolic responses of Lactobacillus plantarum strains during fermentation and storage of vegetable and fruit juices. Appl. Environ. Microbiol. 80, 2206–2215. 10.1128/AEM.03885-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filannino P., Di Cagno R., Addante R., Pontonio E., Gobbetti M. (2016). Metabolism of fructophilic lactic acid bacteria isolated from the Apis mellifera L. bee gut: phenolic acids as external electron acceptors. Appl. Environ. Microbiol. 82, 6899–6911. 10.1128/AEM.02194-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filannino P., Di Cagno R., Gobbetti M. (2018). Metabolic and functional paths of lactic acid bacteria in plant foods: get out of the labyrinth. Curr. Opin. Biotechnol. 49, 64–72. 10.1016/j.copbio.2017.07.016 [DOI] [PubMed] [Google Scholar]
- Filannino P., Di Cagno R., Tlais A.Z.A., Cantatore V., Gobbetti M. (2019). Fructose-rich niches traced the evolution of lactic acid bacteria toward fructophilic species. Crit. Rev. Microbiol. 45, 65–81. 10.1080/1040841X.2018.1543649 [DOI] [PubMed] [Google Scholar]
- Fontes C.P., Honorato T.L., Rabelo M.C., Rodrigues S. (2009). Kinetic study of mannitol production using cashew apple juice as substrate. Bioproc. Biosyst. Eng. 32, 493–499. 10.1007/s00449-008-0269-6 [DOI] [PubMed] [Google Scholar]
- Fridman S., Izhaki I., Gerchman Y., Halpern M. (2012). Bacterial communities in floral nectar. Environ. Microbiol. Rep. 4, 97–104. 10.1111/j.1758-2229.2011.00309.x [DOI] [PubMed] [Google Scholar]
- Garcia E.F., Luciano W.A., Xavier D.E., da Costa W.C., de Sousa Oliveira K., Franco O.L., et al. (2016). Identification of lactic acid bacteria in fruit pulp processing byproducts and potential probiotic properties of selected Lactobacillus strains. Front. Microbiol. 7:1371. 10.3389/fmicb.2016.01371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D., Huys G., Swings J. (2001). Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 205, 31–36. 10.1111/j.1574-6968.2001.tb10921.x [DOI] [PubMed] [Google Scholar]
- Gobbetti M., Fox P., Stepaniak L. (1996). Esterolytic and lipolytic activities of mesophilic and thermophilic lactobacilli. Ital. J. Food Sci. 8, 127–135. [Google Scholar]
- Hebert E.M., Mamone G., Picariello G., Raya R.R., Savoy G., Ferranti P., et al. (2008). Characterization of the pattern of α-and β-casein breakdown and release of a bioactive peptide by a cell envelope proteinase from Lactobacillus delbrueckii subsp. lactis CRL 581. Appl. Environ. Microbiol. 74, 3682–3689. 10.1128/AEM.00247-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert E.M., Raya R.R., Tailliez P., de Giori G.S. (2000). Characterization of natural isolates of Lactobacillus strains to be used as starter cultures in dairy fermentation. Int. J. Microbiol. 59, 19–27. 10.1016/S0168-1605(00)00282-8 [DOI] [PubMed] [Google Scholar]
- Holzapfel W.H., Wood B.J.B. (2014). Lactic Acid Bacteria: Biodiversity and Taxonomy. West Sussex: John Wiley & Sons, Ltd; 10.1002/9781118655252 [DOI] [Google Scholar]
- Janani K., Kumar G., Rao B. (2011). Screening of pectinase producing microorganisms from agricultural waste dump soil. Asian J. Biochem. Pharmaceut. Res. 2, 329–337. [Google Scholar]
- Karam N., Belarbi A. (1995). Detection of polygalacturonases and pectin esterases in lactic acid bacteria. World J. Microbiol. Biotechnol. 11, 559–563. 10.1007/BF00286373 [DOI] [PubMed] [Google Scholar]
- Katz M., Medina R., Gonzalez S., Oliver G. (2002). Esterolytic and lipolytic activities of lactic acid bacteria isolated from ewe's milk and cheese. J. Food Protect. 65, 1997–2001. 10.4315/0362-028X-65.12.1997 [DOI] [PubMed] [Google Scholar]
- Kawasaki S., Kurosawa K., Miyazaki M., Sakamoto M., Ohkuma M., Niimura Y. (2011a). Lactobacillus ozensis sp. nov., isolated from mountain flowers. Int. J. Syst. Evol. Microbiol. 61, 2435–2438. 10.1099/ijs.0.027847-0 [DOI] [PubMed] [Google Scholar]
- Kawasaki S., Kurosawa K., Miyazaki M., Yagi C., Kitajima Y., Tanaka S., et al. (2011b). Lactobacillus floricola sp. nov., lactic acid bacteria isolated from mountain flowers. Int. J. Syst. Evol. Microbiol. 61, 1356–1359. 10.1099/ijs.0.022988-0 [DOI] [PubMed] [Google Scholar]
- King N. (1948). Modification of Vogues–Proskauer test for rapid colorimetric determination of acetyl methyl carbinol plus diacetyl in butter. Dairy Industries 13, 860–866. [Google Scholar]
- Landete J.M., Ferrer S., Pardo I. (2005). Which lactic acid bacteria are responsible for histamine production in wine? J. Appl. Microbiol. 99, 580–586. 10.1111/j.1365-2672.2005.02633.x [DOI] [PubMed] [Google Scholar]
- Latrille E., Picque D., Perret B., Corrieu G. (1992). Characterizing acidification kinetics by measuring pH and electrical conductivity in batch thermophilic lactic fermentations. J. Ferment. Bioeng. 74, 32–38. 10.1016/0922-338X(92)90264-U [DOI] [Google Scholar]
- Lee H., Yoon H., Ji Y., Kim H., Park H., Lee J., et al. (2011). Functional properties of Lactobacillus strains isolated from kimchi. Int. J. Food Microbiol. 145, 155–161. 10.1016/j.ijfoodmicro.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Lefeber T., Janssens M., Camu N., De Vuyst L. (2010). Kinetic analysis of strains of lactic acid bacteria and acetic acid bacteria in cocoa pulp simulation media toward development of a starter culture for cocoa bean fermentation. Appl. Environ. Microbiol. 76, 7708–7716. 10.1128/AEM.01206-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong K.-,h., Chen Y.-S., Pan S.-F., Chen J.-J., Wu H.-C., Chang Y.-C., et al. (2014). Diversity of lactic acid bacteria associated with fresh coffee cherries in Taiwan. Curr. Microbiol. 68, 440–447. 10.1007/s00284-013-0495-2 [DOI] [PubMed] [Google Scholar]
- Linjordet M.S. (2016). A Comparative Analysis of Lactic Acid Bacteria Isolated From Honeybee Gut and Flowers, With Focus on Phylogeny and Plasmid Profiling. Master Thesis, Norwegian University of Life Sciences, Ås. [Google Scholar]
- Liu S.-Q., Holland R., Crow V. (1998). Ethyl butanoate formation by dairy lactic acid bacteria. Int. Dairy J. 8, 651–657. 10.1016/S0958-6946(98)00100-9 [DOI] [Google Scholar]
- Liu S.-Q., Holland R., Crow V. (2003). Ester synthesis in an aqueous environment by Streptococcus thermophilus and other dairy lactic acid bacteria. Appl. Microbiol. Biotechnol. 63, 81–88. 10.1007/s00253-003-1355-y [DOI] [PubMed] [Google Scholar]
- Liu S.Q., Holland R., Crow V.L. (2004). Esters and their biosynthesis in fermented dairy products: a review. Int. Dairy J. 14, 923–945. 10.1016/j.idairyj.2004.02.010 [DOI] [Google Scholar]
- Liu W., Pang H., Zhang H., Cai Y. (2014). Biodiversity of lactic acid bacteria, in Lactic Acid Bacteria, eds Zhang H., Cai Y. (Dordrecht: Springer; ), 103–203. 10.1007/978-94-017-8841-0_2 [DOI] [Google Scholar]
- Matthews A., Grbin P.R., Jiranek V. (2007). Biochemical characterisation of the esterase activities of wine lactic acid bacteria. Appl. Microbiol. Biotechnol. 77, 329–337. 10.1007/s00253-007-1173-8 [DOI] [PubMed] [Google Scholar]
- Moraes P.M., Perin L.M., Silva Júnior A., Nero L.A. (2013). Comparison of phenotypic and molecular tests to identify lactic acid bacteria. Braz. J. Microbiol. 44, 109–112. 10.1590/S1517-83822013000100015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Arribas M.V., Polo M.C., Jorganes F., Muñoz R. (2003). Screening of biogenic amine production by lactic acid bacteria isolated from grape must and wine. Int. J. Food Microbiol. 84, 117–123. 10.1016/s0168-1605(02)00391-4 [DOI] [PubMed] [Google Scholar]
- Mousavi Z., Mousavi S., Razavi S., Emam-Djomeh Z., Kiani H. (2011). Fermentation of pomegranate juice by probiotic lactic acid bacteria. World J. Microbiol. Biotechnol. 27, 123–128. 10.1007/s11274-010-0436-1 [DOI] [Google Scholar]
- Mozzi F., Vaningelgem F., Hebert E.M., Van der Meulen R., Foulquie Moreno M.R., Font de Valdez G., et al. (2006). Diversity of heteropolysaccharide-producing lactic acid bacterium strains and their biopolymers. Appl. Environ. Microbiol. 72, 4431–4435. 10.1128/AEM.02780-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem M., Ilyas M., Haider S., Baig S., Saleem M. (2012). Isolation characterization and identification of lactic acid bacteria from fruit juices and their efficacy against antibiotics. Pakistan J. Bot. 44, 323–328. [Google Scholar]
- Nardi M., Fiez-Vandal C., Tailliez P., Monnet V. (2002). The EstA esterase is responsible for the main capacity of Lactococcus lactis to synthesize short chain fatty acid esters in vitro. J. Appl. Microbiol. 93, 994–1002. 10.1046/j.1365-2672.2002.01793.x [DOI] [PubMed] [Google Scholar]
- Neveling D.P., Endo A., Dicks L.M. (2012). Fructophilic Lactobacillus kunkeei and Lactobacillus brevis isolated from fresh flowers, bees and bee-hives. Curr. Microbiol. 65, 507–515. 10.1007/s00284-012-0186-4 [DOI] [PubMed] [Google Scholar]
- Nyanga L.K., Nout M.J., Gadaga T.H., Theelen B., Boekhout T., Zwietering M.H. (2007). Yeasts and lactic acid bacteria microbiota from masau (Ziziphus mauritiana) fruits and their fermented fruit pulp in Zimbabwe. Int. J. Food Microbiol. 120, 159–166. 10.1016/j.ijfoodmicro.2007.06.021 [DOI] [PubMed] [Google Scholar]
- Oliszewski R., Medina R.B., Gonzalez S.N., Chaia A.B.P. (2007). Esterase activities of indigenous lactic acid bacteria from Argentinean goats' milk and cheeses. Food Chem. 101, 1446–1450. 10.1016/j.foodchem.2006.03.053 [DOI] [Google Scholar]
- Olofsson T.C., Butler È., Markowicz P., Lindholm C., Larsson L., Vásquez A. (2014). Lactic acid bacterial symbionts in honeybees–an unknown key to honey's antimicrobial and therapeutic activities. Int. Wound J. 13, 668–679. 10.1111/iwj.12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong Y. Y., Siang Tan W., Rosfarizan M., Chan E.S., Ti Tey B. (2012). Isolation and identification of lactic acid bacteria from fermented red dragon fruit juices. J. Food Sci. 77, M560–M564. 10.1111/j.1750-3841.2012.02894.x [DOI] [PubMed] [Google Scholar]
- Ortiz M.E., Bleckwedel J., Raya R.R., Mozzi F. (2013). Biotechnological and in situ food production of polyols by lactic acid bacteria. Appl. Microbiol. Biotechnol. 97, 4713–4726. 10.1007/s00253-013-4884-z [DOI] [PubMed] [Google Scholar]
- Ouadghiri M., Amar M., Vancanneyt M., Swings J. (2005). Biodiversity of lactic acid bacteria in Moroccan soft white cheese (Jben). FEMS Microbiol. Lett. 251, 267–271. 10.1016/j.femsle.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Pachla A., Wicha M., Ptaszynska A.A., Borsuk G., Łaniewska–Trokenheim Ł., Małek W. (2018). The molecular and phenotypic characterization of fructophilic lactic acid bacteria isolated from the guts of Apis mellifera L. derived from a Polish apiary. J. Appl. Genet. 59, 503–514. 10.1007/s13353-018-0467-0 [DOI] [PubMed] [Google Scholar]
- Papalexandratou Z., Falony G., Romanens E., Jimenez J.C., Amores F., Daniel H.-M., et al. (2011a). Species diversity, community dynamics, and metabolite kinetics of the microbiota associated with traditional Ecuadorian spontaneous cocoa bean fermentations. Appl. Environ. Microbiol. 77, 7698–7714. 10.1128/AEM.05523-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalexandratou Z., Vrancken G., De Bruyne K., Vandamme P., De Vuyst L. (2011b). Spontaneous organic cocoa bean box fermentations in Brazil are characterized by a restricted species diversity of lactic acid bacteria and acetic acid bacteria. Food Microbiol. 28, 1326–1338. 10.1016/j.fm.2011.06.003 [DOI] [PubMed] [Google Scholar]
- Patra F., Tomar S.K., Arora S. (2009). Technological and functional applications of low-calorie sweeteners from lactic acid bacteria. J. Food Sci. 74, R16–R23. 10.1111/j.1750-3841.2008.01005.x [DOI] [PubMed] [Google Scholar]
- Patra F., Tomar S.K., Rajput Y.S., Singh R. (2011). Characterization of mannitol producing strains of Leuconostoc species. World J. Microbiol. Biotechnol. 27, 933–939. 10.1007/s11274-010-0536-y [DOI] [Google Scholar]
- Pedrolli D.B., Monteiro A.C., Gomes E., Carmona E.C. (2009). Pectin and pectinases: production, characterization and industrial application of microbial pectinolytic enzymes. Open Biotechnol. J. 3, 9–18. 10.2174/1874070700903010009 [DOI] [Google Scholar]
- Pérez-Martín F., Seseña S., Izquierdo P.M., Palop M.L. (2013). Esterase activity of lactic acid bacteria isolated from malolactic fermentation of red wines. Int. J. Food Microbiol. 163, 153–158. 10.1016/j.ijfoodmicro.2013.02.024 [DOI] [PubMed] [Google Scholar]
- Perin L.M., Nero L.A. (2014). Antagonistic lactic acid bacteria isolated from goat milk and identification of a novel nisin variant Lactococcus lactis. BMC Microbiol. 14, 36–45. 10.1186/1471-2180-14-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontonio E., Di Cagno R., Tarraf W., Filannino P., De Mastro G., Gobbetti M. (2018). Dynamic and assembly of epiphyte and endophyte lactic acid bacteria during the life cycle of origanum vulgare L. Front. Microbiol. 9:1372. 10.3389/fmicb.2018.01372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospiech A., Neumann B. (1995). A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 11, 217–218. 10.1016/S0168-9525(00)89052-6 [DOI] [PubMed] [Google Scholar]
- Prathyusha K., Suneetha V. (2011). Bacterial pectinases and their potent biotechnological application in fruit processing juice production industry: a review. J. Phytol. 3, 16–19. [Google Scholar]
- Quinto E.J., Jiménez P., Caro I., Tejero J., Mateo J., Girbés T. (2014). Probiotic lactic acid bacteria: a review. Food Nutrit. Sci. 5, 1765–1775. 10.4236/fns.2014.518190 [DOI] [Google Scholar]
- RStudio-Team (2016). RStudio: Integrated Development for R. RStudio, Inc. Boston, MA. Available online at: http://www.rstudio.com/
- Ruiz Rodríguez L., Bleckwedel J., Eugenia Ortiz M., Pescuma M., Mozzi F. (2017a). Lactic acid bacteria, in Industrial Biotechnology: Microorganisms, eds Wittmann C., Liao J. C. (Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; ), 395–451. 10.1002/9783527807796.ch11 [DOI] [Google Scholar]
- Ruiz Rodríguez L.G., Aller K., Bru E., De Vuyst L., Hebert E.M., Mozzi F. (2017b). Enhanced mannitol biosynthesis by the fruit origin strain Fructobacillus tropaeoli CRL 2034. Appl. Microbiol. Biotechnol. 101, 6165–6177. 10.1007/s00253-017-8395-1 [DOI] [PubMed] [Google Scholar]
- Russell A.L., Ashman T.-L. (2019). Associative learning of flowers by generalist bumble bees can be mediated by microbes on the petals. Behav. Ecol. arz011. 10.1093/beheco/arz011 [DOI] [Google Scholar]
- Sáez G.D., Hébert E.M., Saavedra L., Zárate G. (2017). Molecular identification and technological characterization of lactic acid bacteria isolated from fermented kidney beans flours (Phaseolus vulgaris L. and P. coccineus) in northwestern Argentina. Food Res. Int. 102, 605–615. 10.1016/j.foodres.2017.09.042 [DOI] [PubMed] [Google Scholar]
- Saha B.C. (2006a). Effect of salt nutrients on mannitol production by Lactobacillus intermedius NRRL B-3693. J. Indust. Microbiol. Biotechnol. 33, 887–890. 10.1007/s10295-006-0140-1 [DOI] [PubMed] [Google Scholar]
- Saha B.C. (2006b). A low-cost medium for mannitol production by Lactobacillus intermedius NRRL B-3693. Appl. Microbiol. Biotechnol. 72, 676–680. 10.1007/s00253-006-0364-z [DOI] [PubMed] [Google Scholar]
- Saha B.C. (2006c). Production of mannitol from inulin by simultaneous enzymatic saccharification and fermentation with Lactobacillus intermedius NRRL B-3693. Enzyme Microb. Technol. 39, 991–995. 10.1016/j.enzmictec.2006.02.001 [DOI] [Google Scholar]
- Saha B.C., Nakamura L.K. (2003). Production of mannitol and lactic acid by fermentation with Lactobacillus intermedius NRRL B-3693. Biotechnol. Bioeng. 82, 864–871. 10.1002/bit.10638 [DOI] [PubMed] [Google Scholar]
- Saha B.C., Racine F.M. (2010). Effects of pH and corn steep liquor variability on mannitol production by Lactobacillus intermedius NRRL B-3693. Appl. Microbiol. Biotechnol. 87, 553–560. 10.1007/s00253-010-2552-0 [DOI] [PubMed] [Google Scholar]
- Saha B.C., Racine F.M. (2011). Biotechnological production of mannitol and its applications. Appl. Microbiol. Biotechnol. 89, 879–891. 10.1007/s00253-010-2979-3 [DOI] [PubMed] [Google Scholar]
- Sakellaris G., Nikolaropoulos S., Evangelopoulos A. (1988). Polygalacturonase biosynthesis by Lactobacillus plantarum: effect of cultural conditions on enzyme production. J. Appl. Microbiol. 65, 397–404. 10.1111/j.1365-2672.1988.tb01908.x [DOI] [Google Scholar]
- Sakellaris G., Nikolaropoulos S., Evangelopoulos A. (1989). Purification and characterization of an extracellular polygalacturonase from Lactobacillus plantarum strain BA 11. J. Appl. Microbiol. 67, 77–85. 10.1111/j.1365-2672.1989.tb04957.x [DOI] [Google Scholar]
- Šalomskiene J., Abraitiene A., Jonkuviene D., Mačioniene I., Repečkiene J. (2015). Selection of enhanced antimicrobial activity posing lactic acid bacteria characterised by (GTG)(5)-PCR fingerprinting. J. Food Sci. Technol. 52, 4124–4134. 10.1007/s13197-014-1512-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuni-Blank M., Izhaki I., Laviad S., Bar-Massada A., Gerchman Y., Halpern M. (2014). The role of abiotic environmental conditions and herbivory in shaping bacterial community composition in floral nectar. PLoS ONE 9:e99107. 10.1371/journal.pone.0099107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Maldonado A., Schieber A., Gänzle M. (2011). Structure–function relationships of the antibacterial activity of phenolic acids and their metabolism by lactic acid bacteria. J. Appl. Microbiol. 111, 1176–1184. 10.1111/j.1365-2672.2011.05141.x [DOI] [PubMed] [Google Scholar]
- Sengun I.Y., Nielsen D.S., Karapinar M., Jakobsen M. (2009). Identification of lactic acid bacteria isolated from Tarhana, a traditional Turkish fermented food. Int. J. Food Microbiol. 135, 105–111. 10.1016/j.ijfoodmicro.2009.07.033 [DOI] [PubMed] [Google Scholar]
- Siezen R.J., Bachmann H. (2008). Genomics of dairy fermentations. Microb. Biotechnol. 1, 435–442. 10.1111/j.1751-7915.2008.00067.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smid E., Kleerebezem M. (2014). Production of aroma compounds in lactic fermentations. Ann. Rev. Food Sci. Technol. 5, 313–326. 10.1146/annurev-food-030713-092339 [DOI] [PubMed] [Google Scholar]
- Soares M.M., Silva R.d, Gomes E. (1999). Screening of bacterial strains for pectinolytic activity: characterization of the polygalacturonase produced by Bacillus sp. Revista de Microbiologia 30, 299–303. 10.1590/S0001-37141999000400002 [DOI] [Google Scholar]
- Sumby K.M., Grbin P.R., Jiranek V. (2010). Microbial modulation of aromatic esters in wine: current knowledge and future prospects. Food Chem. 121, 1–16. 10.1016/j.foodchem.2009.12.004 [DOI] [Google Scholar]
- Sumby K.M., Jiranek V., Grbin P.R. (2013). Ester synthesis and hydrolysis in an aqueous environment, and strain specific changes during malolactic fermentation in wine with Oenococcus oeni. Food Chem. 141, 1673–1680. 10.1016/j.foodchem.2013.03.087 [DOI] [PubMed] [Google Scholar]
- Taboada N.V., Lopez Alzogaray M.S., Abeijon Mukdsi M.C., Medina R.B. (2014). Esterase activities and biochemical properties of lactic acid bacteria isolated from goat' s milk cheese in Argentina. J. Agric. Sci. Technol. B 4 752–760. 10.17265/2161-6264/2014.09.008 [DOI] [Google Scholar]
- Tomita S., Saito K., Nakamura T., Sekiyama Y., Kikuchi J. (2017). Rapid discrimination of strain-dependent fermentation characteristics among Lactobacillus strains by NMR-based metabolomics of fermented vegetable juice. PLoS ONE 12:e0182229. 10.1371/journal.pone.0182229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trias Mansilla R., Bañeras Vives L., Montesinos Segu,í E., Badosa Roma,ñó E. (2008). Lactic acid bacteria from fresh fruit and vegetables as biocontrol agents of phytopathogenic bacteria and fungi. Int. Microbiol. 11, 231–236. 10.2436/20.1501.01.66 [DOI] [PubMed] [Google Scholar]
- Trias R., Bañeras L., Badosa E., Montesinos E. (2008). Bioprotection of Golden Delicious apples and Iceberg lettuce against foodborne bacterial pathogens by lactic acid bacteria. Int. J. Food Microbiol. 123, 50–60. 10.1016/j.ijfoodmicro.2007.11.065 [DOI] [PubMed] [Google Scholar]
- Tyler C., Kopit L., Doyle C., Yu A., Hugenholtz J., Marco M. (2016). Polyol production during heterofermentative growth of the plant isolate Lactobacillus florum 2F. J. Appl. Microbiol. 120, 1336–1345. 10.1111/jam.13108 [DOI] [PubMed] [Google Scholar]
- Varghese L., Rizvi A., Gupta A. (2013). Isolation, screening and biochemical characterization of pectinolytic microorganism from soil sample of Raipur city. J. Biol. Chem. Res. 30, 636–643. [Google Scholar]
- Vasilopoulos C., Ravyts F., De Maere H., De Mey E., Paelinck H., De Vuyst L., et al. (2008). Evaluation of the spoilage lactic acid bacteria in modified-atmosphere-packaged artisan-type cooked ham using culture-dependent and culture-independent approaches. J. Appl. Microbiol. 104, 1341–1353. 10.1111/j.1365-2672.2007.03664.x [DOI] [PubMed] [Google Scholar]
- Vásquez A., Forsgren E., Fries I., Paxton R.J., Flaberg E., Szekely L., et al. (2012). Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS ONE 7:e33188. 10.1371/annotation/3ac2b867-c013-4504-9e06-bebf3fa039d1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verón H.E., Di Risio H.D., Isla M.I., Torres S. (2017). Isolation and selection of potential probiotic lactic acid bacteria from Opuntia ficus-indica fruits that grow in Northwest Argentina. LWT-Food Sci. Technol. 84, 231–240. 10.1016/j.lwt.2017.05.058 [DOI] [Google Scholar]
- Versalovic J., Schneider M., De Bruijn F.J., Lupski J.R. (1994). Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5, 25–40. [Google Scholar]
- Vidhyasagar V., Saraniya A., Jeevaratnam K. (2013). Identification of pectin degrading lactic acid bacteria from fermented food sources. Int. J. Adv. Life Sci. 6, 8–12. [Google Scholar]
- von Weymarn N., Kiviharju K., Leisola M. (2002). High-level production of D-mannitol with membrane cell-recycle bioreactor. J. Indust. Microbiol. Biotechnol. 29, 44–49. 10.1038/sj.jim.7000262 [DOI] [PubMed] [Google Scholar]
- Wu J.-,j., Du R.-,p., Gao M., Sui Y.-,q., Wang X. (2014). Identification and characterization of lactic acid bacteria isolated from tomato pomace. Ann. Microbiol. 64, 1849–1855. 10.1007/s13213-013-0798-3 [DOI] [Google Scholar]
- Xanthopoulos V., Petridis D., Tzanetakis N. (2001). Characterization and classification of Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus strains isolated from traditional Greek yogurts. J. Food Sci. 66, 747–752. 10.1111/j.1365-2621.2001.tb04632.x [DOI] [Google Scholar]
- Xu Z., He H., Zhang S., Guo T., Kong J. (2017). Characterization of feruloyl esterases produced by the four Lactobacillus species: L. amylovorus, L. acidophilus, L. farciminis and L. fermentum, isolated from ensiled corn stover. Front. Microbiol. 8, 941. 10.3389/fmicb.2017.00941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida F., Srionnual S., Chen Y.S. (2008). Isolation and characteristics of lactic acid bacteria from koshu vineyards in Japan. Lett. Appl. Microbiol. 47, 134–139. 10.1111/j.1472-765X.2008.02398.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.