Abstract
Dichlorodiphenyltrichloroethane (DDT) has previously been shown to promote the epigenetic transgenerational inheritance of adult onset disease in rats. The current study investigated the potential that sperm epimutation biomarkers can be used to identify ancestral induced transgenerational obesity and associated pathologies. Gestating F0 generational female rats were transiently exposed to DDT during fetal gonadal sex determination, and the incidence of adult-onset pathologies was assessed in the subsequent F1, F2, and F3 generations. In addition, sperm differential DNA methylation regions (DMRs) that were associated with specific pathologies in the transgenerational F3 generation males were investigated. There was an increase of testis disease and early-onset puberty in the F2 generation DDT lineage males. The F3 generation males and females had significant increases in the incidence of obesity and multiple disease. The F3 generation DDT males also had significant increases in testis disease, prostate disease, and late onset puberty. The F3 generation DDT females had increases in ovarian and kidney disease. Epigenetic alterations of the germline are required for the transgenerational inheritance of pathology. Therefore, the F3 generation sperm was collected to examine DMRs for the ancestrally exposed DDT male population. Unique sets of DMRs were associated with late onset puberty, prostate disease, kidney disease, testis disease, obesity, and multiple disease pathologies. Gene associations with the DMR were also identified. The epigenetic DMR signatures identified for these pathologies provide potential biomarkers for transgenerationally inherited disease susceptibility.
Keywords: DDT, epigenetics, transgenerational, obesity, DNA methylation, diagnostics
Introduction
Epigenetic transgenerational inheritance is a non-genetic form of inheritance defined as ‘the germline transmission of epigenetic information between generations that promotes phenotypic variation in the absence of continued direct environmental exposures’ [1]. An example of an environmental class of compounds that can promote the epigenetic transgenerational inheritance of disease is endocrine disruptors that interfere with the action of endocrine hormones [2]. A number of previous studies have revealed that environmental toxicants such as the fungicide vinclozolin [1, 3], the herbicide atrazine [4, 5], plasticizers such as bisphenol A [6] and phthalates [6], the insect repellant diethyltoluamide with the insecticide permethrin [7], the pesticide methoxychlor [8], a hydrocarbon mixture jet fuel [9], and industrial pollutants benzo[a]pyrene [10], biocide tributyltin [11], mercury [12], and dioxins [13, 14] promote the epigenetic transgenerational inheritance of disease susceptibility and sperm epimutations. The transgenerational disease pathologies observed include testis [6, 15–17], prostate [14, 15], ovarian [6–9, 14, 17], uterine [13, 18], kidney disease [8, 14, 15, 17], immune abnormalities [15], behavioral alterations [4, 19], tumor development [15], and obesity [17].
Environmental exposures in early development can alter cellular differentiation that increases the risk of chronic disease later in life [20–22]. Epigenetic developmental plasticity allows an organism to respond to the surrounding environment during cell differentiation, changing the phenotype and gene expression without modifying the DNA sequence [23]. Methylation of CpG dinucleotide residues is one of the most investigated forms of epigenetic regulation [24]. DNA methylation is generally stable in adult differentiated somatic cells. In contrast, the epigenome goes through developmental cascades of methylation changes during critical windows of development in order to promote the cell-type-specific gene expression patterns needed [15, 25, 26]. Changes in the environmental conditions during these critical windows, such as nutritional imbalances and environmental toxicants, can disrupt these processes and permanently alter the methylation patterns of the cell [25, 27]. During the development of primordial germ cells, there is an erasure of DNA methylation during migration in the genital ridge [28–32]. Aberrant methylation of the germline due to environmental insults would then cause germline epimutations that have the potential to be inherited [33]. Exposure of an F0 generation gestating female to an environmental toxicant also exposes the F1 generation fetus and the germline within the F1 generation fetus that will develop into the F2 generation. A germline epimutation can impact the stem cells in the embryo, which can potentially alter all somatic cells in the subsequent generation. When the altered germline methylation patterns are heritable to the subsequent F3 generations, similar to imprinted-like genes, the transmission of these epimutations is called epigenetic transgenerational inheritance [34, 35] and impacts many different somatic cells and tissues.
Dichlorodiphenyltrichloroethane (DDT) has been previously shown to promote the epigenetic transgenerational inheritance of disease [17]. Historically, DDT was one of the most commonly used pesticides used to combat insect vectors of contagious diseases [36]. DDT was banned in the USA in 1973, but it is still recommended by the World Health Organization for indoor residual spray [37]. A number of epidemiological studies have suggested that exposure to DDT and its metabolites are associated with an increase in obesity and insulin resistance [38–40]. Obesity is rapidly increasing in frequency and is associated with severe health risks [41, 42]. The latest data from 2015–16 show a 39.8% overall prevalence of obesity in the USA [43]. The onset of obesity is often associated with over-nutrition and a sedentary lifestyle, but other factors increase the susceptibility to obesity and metabolic disease [44]. India has experienced a 5-fold increase of type II diabetes over the last three decades with a predisposition to obesity already present at birth in much of the population [45, 46]. In addition, India is by far the largest consumer of DDT worldwide [36]. Although a large number of factors may contribute to this increased incidence of obesity, the potential contribution of ancestral toxicant exposures in the induction of obesity susceptibility requires further investigation.
Previous studies on DDT have focused primarily on F0 or F1 generation exposures [47]. Actions on the F0 and F1 generations are direct exposure and have been previously shown to increase the susceptibility to obesity [40, 48], neurological disease [49], and impaired semen quality [50, 51]. Developmental exposure to DDT and its metabolites can transgenerationally increase the incidence of obesity, testicular, ovarian, renal, and pancreatic pathologies in the F3 generation through epigenetic changes in the germline [17, 52, 53]. Previous studies have shown that epimutations in sperm are exposure-specific and may be used as biomarkers in order to assess susceptibility to disease, and should not be considered causally linked to the pathology [4, 16]. Therefore, epigenetic marks such as differential DNA methylation regions (DMRs) termed epimutations may be used to identify a DDT-specific epigenetic signature to serve as biomarkers for DDT-induced epigenetic transgenerational inheritance of disease susceptibility.
The current study used a new colony of experimental rats and further examined the observation that the exposure of a gestating female during fetal gonadal sex determination to DDT can promote the epigenetic transgenerational inheritance of obesity and disease. The hypothesis investigated is that epigenetic biomarkers for disease can be identified and used for identification of transgenerational disease susceptibility due to ancestral exposure to DDT. The study examines the potential actions of DDT to influence the epigenetic transgenerational inheritance, but was not designed for risk assessment. Transgenerational disease pathologies examined include testis, prostate, kidney, ovary disease, and obesity in 1-year-old Sprague Dawley rats in the F1, F2, and F3 generation control and DDT lineages. Although we compared pathologies in the different generations and between sexes, the epigenetic analysis was restricted to only the transgenerational F3 generation males. This was due to the distinct molecular mechanism of direct exposure toxicity in F1 and F2 generations compared to the transgenerational germline-mediated transgenerational mechanism in the F3 generation [54]. Therefore, we investigated the potential for pathology-specific epigenetic alterations to be used as biomarkers to detect transgenerational adult-onset disease.
Results
Pathology Analysis
The actions of control vehicle [dimethylsulfoxide (DMSO)] exposed and DDT (25 mg/kg body weight) treatments administered to female rats (F0 generation) during Days 8–14 of gestation were investigated. The control vehicle DMSO was not found to influence pathology incidence compared to control wildtype animals [15, 16]. The dose of DDT used is an anticipated environmental exposure [49, 55–58]. The F1 generation (direct fetal exposure), F2 generation (direct germline exposure), and F3 generation (transgenerational) rats of control and DDT lineages were aged to 1 year then euthanized for analysis. No inbreeding (sibling or cousin crosses) was performed to eliminate inbreeding artifacts [1]. The testis, prostate, kidney, and ovary were collected and examined for the presence of specific histological abnormalities as described in the Methods. No effect was observed on litter size, sex ratio, or weaning weights (P > 0.05) for any generation (Supplementary Fig. S1). Therefore, negligible overt toxicity to DDT was observed in the direct in utero exposed F1 generation lineage.
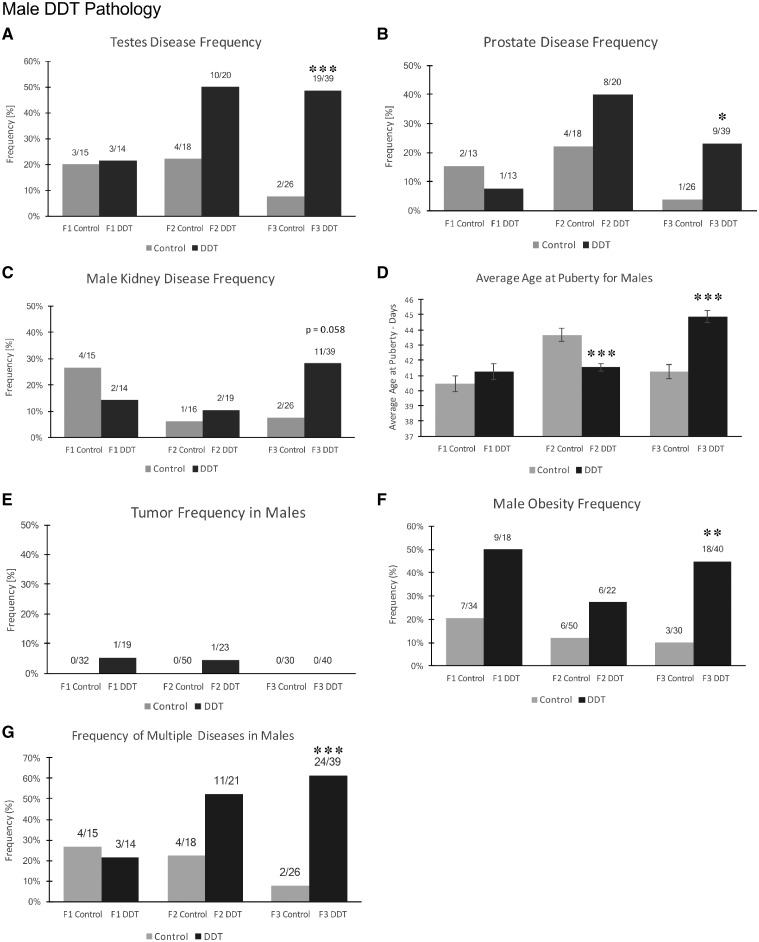
For the purposes of this study, an animal was considered to have a diseased tissue if the number of histological abnormalities was markedly increased (i.e. >2 SD) compared to that of the controls for that tissue, as described for each tissue in Methods. The incidences of testis, prostate, and male kidney disease were not significantly different between the control and DDT lineages for either the F1 or F2 generation animals at one year of age (Fig. 1A and B). There was a trend toward an increased rate of testis and prostate disease in the F2 generation DDT rats; however, it was not statistically significant. The F1 and F2 generations had reduced numbers of animals so higher variability is anticipated. The incidence of testis disease was significantly increased in the F3 generation (P < 0.001) DDT lineage compared to control (Fig. 1A). The incidence of prostate disease was also found to be increased in F3 generation males (P < 0.05) (Fig. 1B). Although there appeared to be a small change in the incidence of prostate disease in the control lineage F2 and F3 generation males, there was no statistical significance within the two control generations (P < 0.15). The F2 generation DDT rats had an increase in prostate disease that was not significantly increased. The disease ratio is actually higher in the F2 generation animals than the F3 generation DDT rats. There was an increase of kidney disease with a P = 0.058 in the F3 generation males (Fig. 1C). The pubertal analysis identified early pubertal onset in males in the F2 generation DDT lineage (F2 generation DDT n = 35 vs. control n = 32), and a late pubertal onset in males in the F3 generation DDT lineage (F3 generation DDT n = 35 vs. control n = 33) (Fig. 1D).
Figure 1:
pathology analysis in F1, F2, and F3 generation control and DDT lineage 1-year-old male rats. (A) Testis disease frequency, (B) prostate disease frequency, (C) male kidney disease frequency, (D) average age at puberty for males, (E) tumor frequency in males, (F) male obesity frequency, and (G) multiple disease. The pathology number ratio with total animal number is listed for each bar graph (A–G), statistical significance is represented with the P-value indicated (*P < 0.05; **P < 0.01; ***P < 0.001)
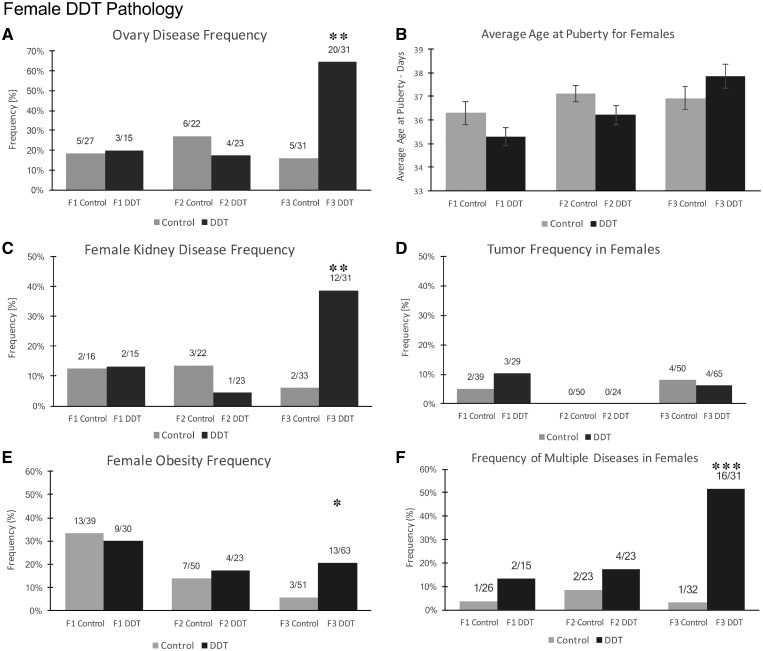
The incidence of female ovarian and kidney disease was not found to be significantly different between the control versus DDT lineages in the F1 or F2 generations. However, they were both found to be significantly increased in DDT lineage F3 generation females (P < 0.001) (Fig. 2A and C). There were no female pubertal abnormalities observed in any generation (Fig. 2B). Tumor development was also monitored in males and females, and there was no change in incidence observed between control and DDT lineages in any generation (Fig. 2D). The F3 generation 1-year males and females were also assessed for behavioral alterations using an elevated plus maze and open field analysis, as described in the Methods. No significant effects on male or female behaviors were observed (Supplementary Fig. S2).
Figure 2:
pathology analysis in F1, F2, and F3 generation control and DDT lineage 1-year-old female rats. (A) Ovary disease frequency, (B) average age at puberty for females, (C) female kidney disease frequency, (D) tumor disease frequency, (E) female obesity frequency, and (F) multiple disease. The pathology number ratio with total animal number is listed for each bar graph (A–F), statistical significance is represented with the P-value indicated (*P < 0.05; **P < 0.01; ***P < 0.001)
The body weight, body-mass index (BMI), abdominal adiposity, and adipocyte cell size were analyzed in order to assess the incidence of obesity in DDT lineage males and females as described in the Methods. There was no significant difference in average weight at euthanization in the F1, F2, and F3 generation control and DDT lineage males and females (Supplementary Fig. S1A and B). The incidence of obesity was not found to be different within the control versus DDT lineage F1 or F2 generation males and females. However, it was found to be significant in F3 generation males (P < 0.01) and females (P < 0.05) (Fig. 1F and 2E). Although the female obesity is statistically significant in the F3 generation DDT females, it is important to note that the ratios of disease have not significantly changed and the incidence of obesity between the F1 generation female controls and the F3 generation female controls decreased.
The specific disease associated with each individual animal is shown in Table 1 and Supplementary Tables S1–S3. This information was used for the analysis of multiple (≥2) disease incidence. The incidence of multiple disease in the F1 generation males or females and F2 generation males or females was not significantly different between control and DDT lineages. The incidence of multiple diseases in F3 generation DDT lineage males and females was significantly increased in comparison to the control lineage (Fig. 1G and 2F). Therefore, transgenerational pathologies (F3 generation) of late puberty, obesity, testis, prostate, and multiple disease were observed in the DDT lineage males. Obesity, ovarian, kidney, and multiple disease transgenerational pathologies (F3 generation) were observed in the DDT lineage females.
Table 1:
F3 generation DDT lineage male individuals pathology
| Puberty | Testis | Prostate | Kidney | Tumor | Lean | Obese | Multiple disease | Total disease | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rat ID | Early | Late | |||||||||
| DM1 | 14D5-3-3-8 | − | + | + | − | − | − | − | + | + | 3 |
| DM2 | 14D5-3-3-9 | − | − | + | + | + | − | + | − | + | 4 |
| DM3 | 14D5-3-3-10 | − | − | + | + | − | − | − | − | + | 2 |
| 14D5-3-3-11 | − | − | |||||||||
| 14D5-3-3-12 | − | + | |||||||||
| 14D5-3-3-13 | − | − | |||||||||
| DM4 | 14D5-3-4-4 | − | − | − | − | − | − | − | + | − | 1 |
| DM5 | 14D5-3-4-5 | − | − | − | + | + | − | − | − | + | 2 |
| DM6 | 14D5-3-21-6 | + | + | − | − | − | − | + | 2 | ||
| DM7 | 14D5-3-21-7 | + | − | + | − | − | − | + | 2 | ||
| DM8 | 14D5-3-21-8 | − | − | − | − | − | − | − | 0 | ||
| DM9 | 14D5-3-21-9 | + | − | − | − | − | + | + | 2 | ||
| DM10 | 14D5-3-21-10 | + | − | − | − | − | + | + | 2 | ||
| DM11 | 14D8-3-10-6 | − | − | − | − | − | + | − | 1 | ||
| DM12 | 14D8-3-10-7 | + | − | − | − | − | − | − | 1 | ||
| 14D8-3-10-8 | − | + | |||||||||
| 14D8-3-10-10 | − | + | |||||||||
| DM13 | 14D8-3-10-11 | − | + | − | − | − | − | − | − | − | 1 |
| DM14 | 14D8-3-10-12 | − | − | − | − | + | − | − | 1 | ||
| DM15 | 14D8-3-10-13 | − | + | − | + | − | − | − | − | + | 2 |
| DM16 | 14D10-3-5-6 | − | + | + | − | − | − | + | − | + | 3 |
| DM17 | 14D10-3-5-7 | − | + | + | − | + | − | + | − | + | 4 |
| DM18 | 14D10-3-5-8 | − | + | + | − | + | − | − | − | + | 3 |
| DM19 | 14D10-3-5-9 | − | + | − | + | + | − | − | − | + | 3 |
| DM20 | 14D10-3-5-10 | − | + | − | − | + | − | − | − | + | 2 |
| DM21 | 14D10-3-5-11 | − | + | − | − | + | − | + | − | + | 3 |
| DM22 | 14D11-3-6-10 | − | + | + | − | + | − | − | + | + | 4 |
| DM23 | 14D11-3-6-11 | − | − | − | − | − | − | − | − | − | 0 |
| DM24 | 14D11-3-6-12 | − | − | − | − | + | − | − | + | + | 2 |
| DM25 | 14D11-3-6-13 | − | − | + | − | − | − | − | + | + | 2 |
| DM26 | 14D12-3-7-7 | − | − | + | − | − | − | − | + | + | 2 |
| DM27 | 14D12-3-7-8 | − | − | − | − | − | − | − | − | − | 0 |
| 14D12-3-7-9 | − | − | |||||||||
| 14D12-3-7-10 | − | − | |||||||||
| 14D12-3-7-11 | − | − | |||||||||
| 14D12-3-7-12 | − | − | |||||||||
| DM28 | 14D13-3-8-9 | − | − | − | − | − | − | − | − | − | 0 |
| DM29 | 14D13-3-8-10 | − | − | − | − | − | − | − | + | − | 1 |
| 14D13-3-8-11 | − | − | |||||||||
| 14D13-3-8-12 | − | − | |||||||||
| DM30 | 14D15-3-23-2 | − | − | − | − | − | |||||
| DM31 | 14D15-3-23-3 | − | + | - | − | − | + | + | 2 | ||
| DM32 | 14D15-3-23-4 | − | + | + | − | − | + | + | 3 | ||
| DM33 | 14D15-3-23-5 | + | − | − | − | − | + | + | 2 | ||
| DM34 | 14D15-3-23-6 | − | − | + | − | − | − | − | + | + | 2 |
| 14D15-3-23-10 | − | − | |||||||||
| DM35 | 14D15-3-25-3 | − | − | − | − | − | + | − | 1 | ||
| DM36 | 14D15-3-25-4 | − | − | − | − | − | − | − | 0 | ||
| DM37 | 14D15-3-25-5 | + | − | − | − | − | − | − | 1 | ||
| DM38 | 14D15-3-25-6 | − | + | − | − | − | + | + | 2 | ||
| DM39 | 14D15-3-25-7 | − | − | − | − | − | + | − | 1 | ||
| DM40 | 14D15-3-25-8 | − | − | − | − | − | + | − | 1 | ||
| Affected | 0 | 13 | 18 | 9 | 11 | 0 | 5 | 19 | 24 | 34 | |
| Population | 35 | 35 | 39 | 39 | 39 | 40 | 40 | 40 | 39 | 39 | |
| Multiple ≥2 | Total ≥1 | ||||||||||
Disease Associated Sperm Epimutations
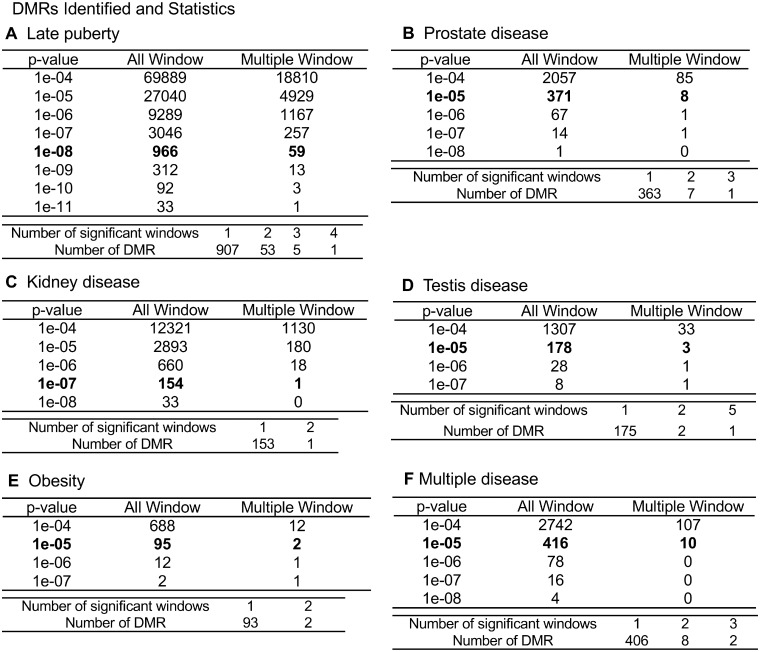
Previous studies have shown several environmental toxicants can promote DMRs in the F3 generation sperm [1, 16, 59]. A recent study found ancestral exposure to DDT induces epigenetic alterations in sperm DNA methylation, non-coding RNA, and histone retention sites with distinct differences between the F1, F2, and F3 generations [60]. The current study investigated the sperm disease-specific epimutations in F3 generation DDT lineage males. DMRs from the F3 generation DDT lineage sperm samples were identified that associated with the specific pathologies identified. The majority of the DMRs were single 100-bp windows, but some of them were multiple-window DMRs. The genome was bioinformatically fragmented into 100-bp segments (i.e. windows) to identify statistically significant differences in DNA methylation (i.e. read depth changes) to identify the DMRs [60]. Both single- and multiple-window data sets are presented with different P-values in Fig. 3 for all disease-specific biomarkers. Although DMRs at different P-values for the different pathologies are potentially important, the selected DMR data sets at P < 1e-08 for late puberty, P < 1e-07 for kidney disease, and P < 1e-05 for prostate disease, testis disease, obesity, and multiple disease were further investigated. The differing P-values were used to allow a more balanced comparative analysis of the disease-specific DMRs. The lists of the specific disease DMRs (chromosomal locations, size, CpG density, increased/decreased methylation ratios, and associated genes) are presented in Supplementary Tables S4–S9.
Figure 3:
disease-specific DMR analysis. The number of DMRs found using different P-value cutoff thresholds are presented. The All Window column shows all DMRs. The Multiple Window column shows the number of DMRs containing at least two significant windows. The number of DMRs with each specific number of significant windows is shown: (A) late puberty, (B) prostate disease, (C) kidney disease, (D) testis disease, (E) obesity, and (F) multiple disease
Animals were separated into groups based on whether they had a specific pathology or not. Comparison of these groups using the methylated DNA immunoprecipitation (MeDIP)-Seq data identified a DMR data set or signature that was associated with each pathology. The analyses identified epimutation signatures comprised 966 DMRs at P < 1e-08 for late puberty, 371 DMRs at P < 1e-05 for prostate disease, 178 DMRs at P < 1e-05 for testis disease, 154 DMRs at P < 1e-07 for kidney disease, 95 DMRs at P < 1e-05 for obesity, and 416 DMRs at P < 1e-05 as an epimutation signature for multiple disease (Fig. 3A–F). The list of DMRs for late puberty, prostate disease, kidney disease, testis disease, obesity, and multiple disease is presented in the Supplementary Tables S4–S9.
Compared to those of late puberty and kidney disease, the obesity and other disease DMR signatures were more variable. However, the presence of multiple diseases within rats may be confounding factors when identifying specific signatures for a particular disease. Supplementary Table S3 details specific pathologies for the F3 generation DDT lineage males used in the study. For example, there were only 5 out of 17 males with obesity that were not paired with other pathologies. Eight of the animals with obesity also had testis disease while the others also had prostate or kidney disease. Therefore, the association of obesity with other diseases may act as confounding factors in the identification of specific epigenetic disease signatures.
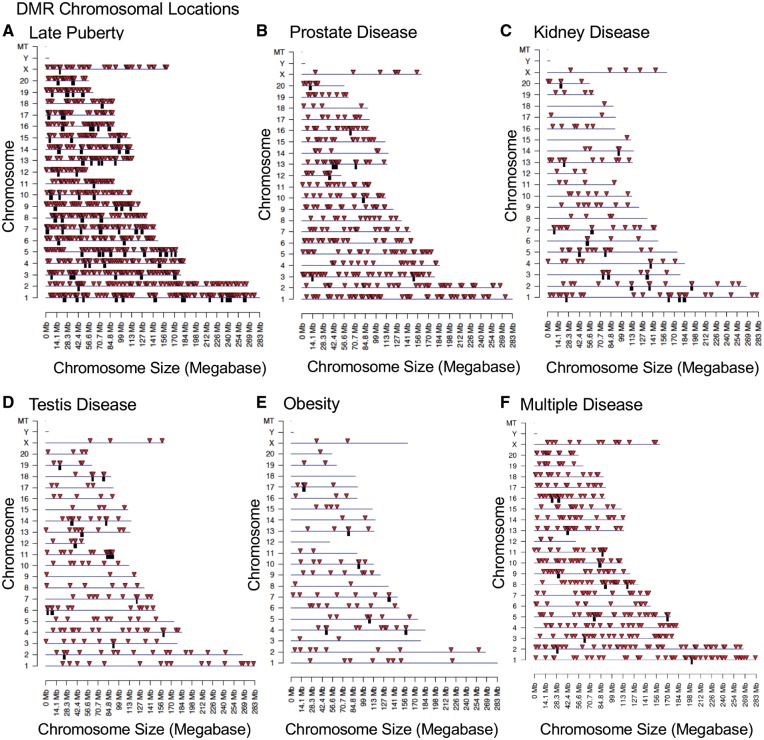
The chromosomal locations of the DDT lineage DMRs associated with the late puberty phenotype, prostate disease, kidney disease, testis disease, and multiple disease are presented (Fig. 4A–F). DMRs or epimutation signatures show that all chromosomes but Y are involved. The DMR chromosomal locations of the DDT obesity phenotype show that all chromosomes but 12, 18, and Y are involved (Fig. 4E). All epimutation signatures investigated contained DMR clusters represented as black boxes. Each disease epimutation signature was characterized by examining the DMR genomic features. All potential biomarkers (i.e. DMRs) had a CpG density of primarily 1–2 CpG per 100 bp (Supplementary Fig. S3). These low-density CpG regions are referred to as CpG deserts [61]. All potential disease biomarkers had a DMR length of ∼1 kb (Supplementary Fig. S4). Therefore, the potential disease biomarkers had an average of 10–20 CpGs per DMR.
Figure 4:
chromosomal locations of disease-specific DMRs. DMR locations on the individual chromosomes are presented. (A) Late puberty, all DMRs at a P-value threshold of 1e-08 are shown. (B) Prostate disease, all DMRs at a P-value threshold of 1e-05 are shown. (C) Kidney disease, all DMRs at a P-value threshold of 1e-07 are shown. (D) Testis disease, all DMRs at a P-value threshold of 1e-05 are shown. (E) Obesity, all DMRs at a P-value threshold of 1e-05 are shown. (F) Multiple disease, all DMRs at a P-value threshold of 1e-05 are shown. The red arrowhead indicates the DMR location and black box a cluster of DMRs
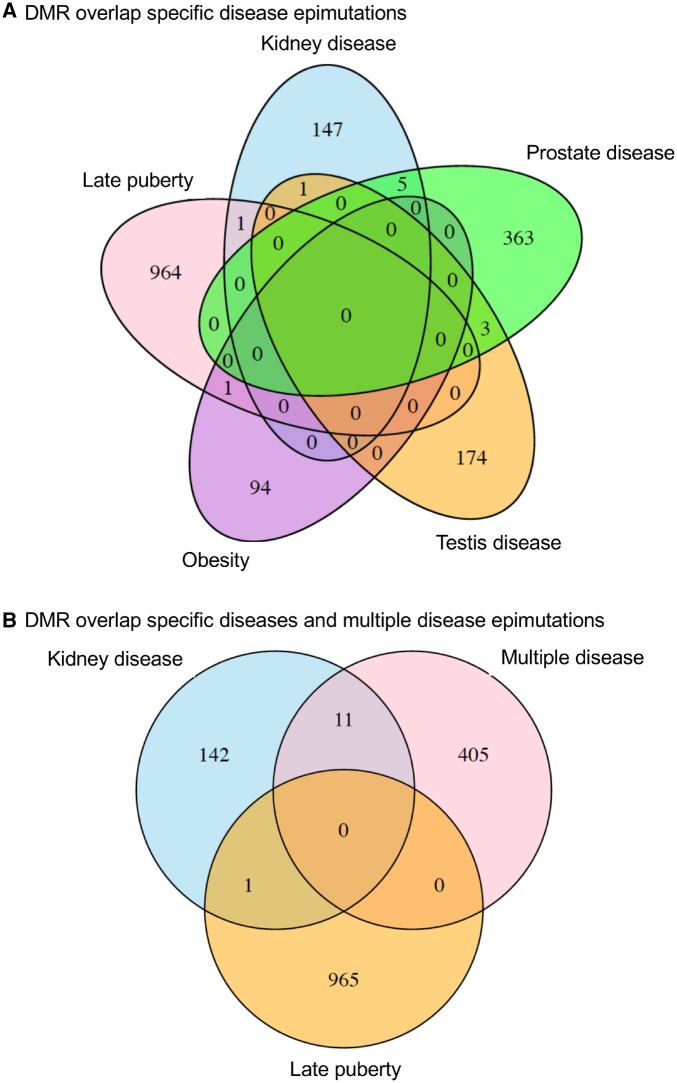
The specific disease DMRs identified appear to be present in the majority of the animals analyzed with that disease in comparison to the non-disease population. Analysis of the individual DMRs for each specific disease signature determined that the significant read depth alterations were present in nearly all the animals used to generate the DMRs and that specific signature. Therefore, the majority of animals do appear to have the DMR signature identified at the P-value threshold selected. The specific disease DMRs (epimutations) were found to have minimal overlap with those of the other diseases (Fig. 5A). There is minimal DMR overlap between multiple disease, kidney disease, and late puberty (Fig. 5B). Therefore, the disease-specific DMRs appear to be potential biomarkers unique for each disease.
Figure 5:
DMR overlap Venn diagram for specific disease DMRs. (A) The overlaps for specific diseases and (B) overlaps with multiple disease are presented
A principle component analysis (PCA) of the DMRs in each of the specific pathology epimutation signatures was performed. Generally, when using the DMRs associated with pathologies the DMRs cluster separately from those associated with individuals without those pathologies (Supplementary Fig. S5). Distinct clusterings for late puberty, prostate disease, kidney disease, testis disease, obesity, and multiple disease were observed using the DMR PCA (Supplementary Fig. S5A–F). Interestingly, the obesity analysis resulted in two separate clusters of the obese individuals (Supplementary Fig. S5E). The DMR PCA analyses suggest that the control versus DDT lineage DMRs are distinct. The genome-wide PCA analysis was also performed and shown to provide separation for the puberty, kidney, obesity, and testis diseases, and less separation for the prostate and multiple diseases.
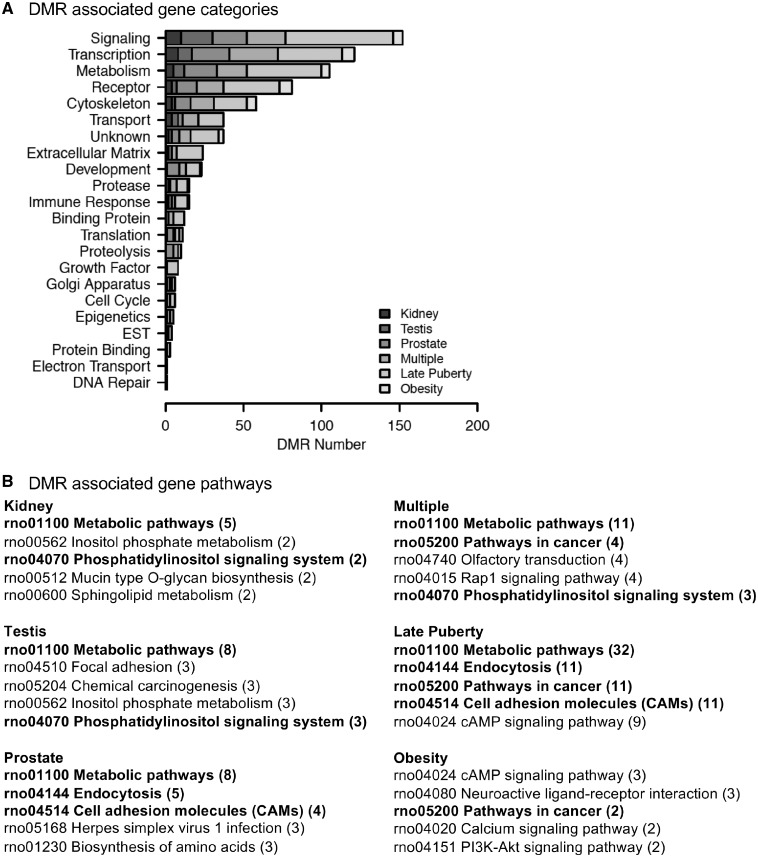
The DMR-associated genes were identified and listed in Supplementary Tables S4–S9. These genes were identified within 10 kb of the DMRs to include a promoter of the gene. The genes were categorized for functional relationships, and the DMR-associated gene categories are presented in Fig. 6A. The predominant categories of each disease DMR set were signaling, transcription, metabolism, and receptors. Potential gene pathways associated with the DMR-associated genes for each disease group of DMRs are listed in Fig. 6B. Signaling pathways were the more common pathways observed.
Figure 6:
DMR gene associations. (A) DMR-associated gene functional categories (genes within 10-kb DMR). (B) KEGG pathways containing DMR-associated genes. Number of DMR-associated genes in pathway is provided in brackets. Pathways in bold are common to at least two pathology DMR sets
Discussion
The current study was designed to further investigate the transgenerational actions of DDT, and investigate potential biomarkers for specific transgenerational pathologies. Classic toxicology studies only evaluate direct exposure of an individual while future generations are not assessed. Previously, transgenerational pathologies such as obesity, testis disease, ovarian disease, and kidney disease were observed in the F3 generation of DDT lineage animals [17]. The results of the current study suggest that future risk assessment studies should consider multigenerational or transgenerational impacts. Pathologies can differ between generations as the F1 generation involves direct exposure, the F2 generation involves both direct exposure to the germline and generational actions, while the F3 generation is mediated through the germline transgenerationally [35]. The F1 generation that is directly exposed would have somatic effects, and potential changes in development while the F3 generation would involve epigenomic alterations of the germline that could subsequently affect all somatic epigenomes and transcriptomes. This potentially increases the susceptibility to disease in the F3 generation compared to the F1 and F2 generations. Therefore, the molecular mechanisms are distinct for each generation between direct exposure and transgenerational germline-mediated ancestral exposure [3]. It should be noted that the higher numbers of F3 generation control and DDT lineage animals were evaluated compared to the lower numbers evaluated for the F1 and F2 generations. This increases the statistical sensitivity to detect abnormalities in the F3 generation, and should be taken into consideration when interpreting results. Therefore, the trend toward an increase in testis abnormalities in F2 generation DDT males may become significant with a larger colony size, but more studies with an increased n-value are needed in order to make that determination. The differences between generations in the pathologies observed in the study appear to be due in part to the direct exposure of the F1 and F2 generations versus the transgenerational F3 generation. The emphasis of the current study is to compare control and DDT lineage animals within the same generations. The current study focused on the transgenerational F3 generation for epigenetic analysis of potential transgenerational disease biomarkers.
Pathologies assessed included testis disease, ovarian disease, prostate disease, kidney disease, and obesity. The F1 generation DDT lineage rats did not have any significant difference in disease incidence compared to controls. Therefore, the in utero direct exposure did not promote somatic cell effects that increase adult-onset disease susceptibility. However, it is proposed that DDT altered the germline to induce heritable epimutations that increase the incidence of disease in later life of offspring. The F3 generation DDT rats had increased rates of testis disease, prostate disease, female kidney disease, ovarian disease, obesity, delayed onset male puberty, and multiple disease. These results are similar to previous observations in a transgenerational model of DDT exposure [17].
Pathologies and abnormalities were evaluated in DDT lineage rats in comparison with DMSO control lineage rats, as DMSO was the vehicle in which DDT was dissolved when preparing treatment solutions for injection into the F0 generation females. A recent study has compared the effects of ancestral exposure to DMSO with exposure to phosphate-buffered saline, and found no differences in disease rates in subsequent generations between these two control populations [62]. This suggests that DMSO exposure itself has little effect on transgenerational disease incidence.
Increased abdominal adiposity and weight were used to assess obesity in a previous study of transgenerational inheritance of disease in ancestral exposure to DDT [17]. To further investigate the obese phenotype previously observed in the F3 generation, a BMI was established and gonadal fat pad tissue was analyzed for cell size. There was no significant increase in obesity between the F1 and F2 generation DDT lineage animals. Both the male and female F3 generation DDT lineage animals had a significant increase in the obese phenotype compared to controls. Future studies will be needed to investigate the contribution of ancestral exposures and the epigenetic transgenerational inheritance of obesity.
The total level of pathology per rat was assessed. There was a significant increase of multiple diseases (≥2 diseases) in the F3 generation DDT lineage males and females. In the F3 generation DDT lineage, 51.6% of females and 61.5% of males had at least two or more pathologies. The results correlate to previous observations in a transgenerational model of DDT exposure [17]. Other epigenetic transgenerational inheritance models of environmental toxicant exposure have also observed similar trends including atrazine, dioxin, and methoxychlor exposure [4, 8, 14]. The incidence of multiple chronic diseases has risen in the last two decades [63]. Greater than 85% of the human population has at least one chronic disease and approximately one in four people in the world have multiple chronic diseases [64, 65]. Environmentally induced epigenetic transgenerational inheritance of disease susceptibility should be considered in the etiology of this multiple chronic disease condition.
The individual sperm epigenomes were analyzed in the F3 generation DDT lineage in order to potentially identify epimutations that may correlate with specific pathologies seen within the F3 generation. The individuals with the specific pathology were grouped and compared to those without the specific pathology within the F3 generation DDT lineage males. This analysis was performed within the same treatment group and generation in order to avoid the potential confounding factor of differences in methylation specific to the ancestral exposure. Therefore, the analysis identified sperm epimutation signatures for each specific transgenerational disease (Fig. 5). The chromosomal locations of the late puberty, obesity, kidney, testis, prostate, and multiple disease DMRs and DMR clusters for potential epimutation signatures are shown in Fig. 4. However, the level of disease within the F3 generation DDT lineage males was very high and there were only five total individuals without any disease (Table 1). Future analysis of these and other animals with no pathology may be useful to identify disease preventative epigenetics. Therefore, all comparisons had individuals in the disease group with additional other disease, and non-disease-specific groups with individuals with other disease. The presence of other diseases is a confounder in the analysis that is expected to increase variation in the data, and reduce the significance of the epimutation disease signatures identified. Although we identified disease-specific signatures in the current analysis, future studies will need to include larger groups of individuals without multiple disease. The current study clearly demonstrates the feasibility to identify disease-specific epigenetic biomarkers and potential diagnostics for disease. The presence of co-morbidities and multiple disease needs to be considered a confounder in this type of analysis.
Interestingly, the obesity epimutation signature demonstrated separate clusters of disease populations versus the single cluster in the non-disease population (Supplementary Fig. S5). This suggests multiple disease parameters, as expected with obesity, may be contributing to the pathology. This may limit the ability of a specific obesity diagnostic to be developed. The multiple comorbidities of obesity include cardiovascular disease, type II diabetes, non-alcoholic fatty liver disease, certain cancers, stroke, and kidney disease [66–69]. Therefore, a certain mixture of these pathologies may lead to the etiology of an individual’s obesity, but not be the same for other obese individuals. Although, it may be difficult to obtain a specific obesity epimutation biomarker, it is possible that expanded populations that have negligible other diseases may allow a more significant obesity signature to be identified.
This is one of the first demonstrations that a transgenerational disease- or pathology-specific epigenetic biomarker could be potentially identified and correlated with the majority of the animals with the pathology. A biomarker involves the association of the molecular component with the disease. This does not suggest a causal relationship to the disease, but simply a biomarker for the associated disease. A diagnostic requires the validation of the biomarkers with blinded test sets of individuals and data, as well as more extensive statistical analysis with increased numbers of individuals. Although the current study demonstrates the potential existence of epigenetic biomarkers for disease, future studies are needed to assess their diagnostic capabilities. The observation of distinct epimutation signatures for each disease or pathology suggests that epigenetic diagnostics or biomarkers for transgenerational disease may be disease specific and unique. The non-disease comparisons suggest a significance for the epimutation signatures identified; however, future analysis will need to evaluate larger populations of individuals without disease to reduce the confounding effects of multiple disease. Therefore, epigenetic biomarkers or diagnostics provide preliminary evidence for preconception diagnosis of increased susceptibility to transgenerational disease in offspring. This concept could have a significant impact on preventative medicine and public health outcomes in regards to the early identification and management of disease.
Conclusions
The current study demonstrates that DDT exposure of a gestating female during gonadal sex determination of the fetus promotes the epigenetic transgenerational inheritance of disease and sperm epimutations. No significant pathology was detected in the F1 generation, but a significant increase in disease and pathology was observed in the F3 generation DDT lineage male and female rats. Therefore, future assessment of exposure toxicity needs to consider transgenerational impacts. A transgenerational increase in testis disease, prostate disease, female kidney disease, ovarian disease, obesity in both males and females, and increase in multiple disease susceptibility was observed in both males and females. The F3 generation males were used to identify potential unique signatures (groups) of DMRs associated with late puberty, prostate disease, kidney disease, testis disease, obesity, and multiple diseases. This provides a preliminary proof of concept that epigenetic biomarkers for disease can be identified, and potentially used in the future to diagnose disease and disease susceptibility.
Methods
Animal Studies and Breeding
Sprague Dawley®™SD®™ male and female rats were fed a standard diet and given water ad lib then pair-mated at 70–100 days of age [70]. On gestational days 8 through 14 [71], the pregnant females were administered daily intraperitoneal injections of DDT (25 mg/kg BW/day) or DMSO (vehicle). The dose of DDT used is an anticipated environmental exposure [49, 55–58]. The p,p′-DDT was obtained from Chem Services (West Chester, PA, USA), and was injected as a 25 mg/ml solution in DMSO as previously described [17]. Treatment lineages are designated ‘control’ and ‘DDT’ lineages. The treated gestating female rats were considered to be the F0 generation. These were new rat colonies and treatment lineages started in 2014, rather than a re-analysis or rats from previous studies [17]. The F1 generation was the offspring of the F0 generation rats. The F2 generation was obtained after breeding the F1 generation control or DDT lineage males and females at 70–90 days of age. The F2 generation rats were bred to obtain F3 generation offspring. No sibling or cousin breedings were performed. The F1–F3 generation offspring were not themselves treated directly with DDT. The control and DDT lineages were housed in the same room and racks with lighting, food, and water as previously described [15, 16, 54]. All experimental protocols for the procedures with rats were pre-approved by the Washington State University Animal Care and Use Committee (protocol IACUC # 6252), and all methods were performed in accordance with the relevant guidelines and regulations.
Tissue Harvest and Histology Processing
Twelve-month-old rats were euthanized via CO2 inhalation and cervical dislocation prior to tissue harvest. Body weight and length were measured at dissection time. Testis, prostate, ovary, kidney, and gonadal fat were fixed in Bouin’s solution (Sigma) followed by 70% ethanol, then processed for paraffin embedding, sectioning and hematoxylin and eosin (H & E) staining by standard procedures for histopathological examination by Nationwide Histology, Spokane, WA, USA [70].
Histopathology Examination and Disease Classification
Testis, prostate, kidney, and ovary histopathological evaluations were performed to determine the incidence of disease in each tissue [70]. The tissues evaluated histologically were selected from previous literature showing them to have pathology in transgenerational models [1, 4, 6–9, 14, 15, 17, 59] with an emphasis on reproductive organs. The Washington Animal Disease Diagnostic Laboratory (WADDL) at the Washington State University College of Veterinary Medicine has board certified veterinary pathologists and assisted in initially establishing the criteria for the pathology analyses and identifying parameters to assess [15]. Histopathology readers were trained to recognize the specific abnormalities evaluated for this study in rat testis, ventral prostate, ovary, and kidney (see below). Three different pathology readers were used for each tissue and blinded to the treatment groups. A set of quality control (QC) slides was generated for each tissue and was read by each reader prior to evaluating any set of experimental slides. These QC slide results are monitored for reader accuracy and concordance. WADDL was consulted when any questions developed. WADDL performed full necropsies as required on animals that died prior to the time of scheduled sacrifice at 1 year, and performed tumor evaluations in the current study.
As previously described [4, 54], testis histopathology criteria included the presence of vacuoles in the seminiferous tubules, azoospermic atretic seminiferous tubules, the presence of sloughed spermatogenic cells in center of the tubule, a lack of a tubule lumen, and increased apoptosis of germ cells as determined by TUNEL assay. As previously described [72, 73], prostate histopathology criteria included the presence of vacuoles in the glandular epithelium, atrophic glandular epithelium encompassing more than one-third of a gland, and hyperplasia of prostatic gland epithelium. Kidney histopathology criteria included markedly reduced size of glomerulus, thickened Bowman’s capsule, and the presence of proteinaceous fluid-filled cysts >50 μm in diameter. Ovary sections were assessed for the two pathologies of primordial follicle loss and ovarian cysts as previously described [74]. Ovarian cysts have little or no granulosa cell layer, a smooth border, and are 50–250 μm (small cysts) or >250 μm (large cysts) in diameter. A cut-off was established to declare a tissue ‘diseased’ based on the mean number of histopathological abnormalities plus 2 SDs from the mean of control group tissues, as assessed by each of the three individual observers blinded to the treatment groups. This number was used to classify rats into those with and without testis, ovary, prostate, or kidney disease in each lineage. A rat tissue section was finally declared ‘diseased’ only when at least two of the three observers marked the same tissue section ‘diseased’.
Age of puberty was defined in females as the age of vaginal opening, and in males as the age of balano-preputial separation.
Obesity Phenotype Analysis
Obesity was assessed with an increase in adipocyte size (area), BMI, and abdominal adiposity as previously described [70]. Previous studies have used these parameters to assess toxicant impacts on transgenerational obesity [4, 6, 9, 17]. The parameters for the adipocyte area in females are <2618 μm for lean, between 2618 and 4643 μm for non-obese, and >4643 μm for obese. The parameters for the adipocyte area in males are <2526 μm for lean, between 2526 and 3979 μm for non-obese, and >3979 μm for obese. The parameters for BMI in females are <0.6081 g/cm2 for lean, between 0.6081 and 0.7971 g/cm2 for non-obese, and >0.7971 g/cm2 for obese. The parameters for BMI in males are <0.8196 g/cm2 for lean, between 0.8196 and 1.0354 g/cm2 for non-obese, and >1.0354 g/cm2 for obese.
Behavior Analysis
An elevated plus maze and open-field test were performed for behavioral analysis as previously described [19, 70, 75]. Elevated plus maze data were obtained from 19 DDT lineage males, 29 DDT lineage females, 17 control males, and 27 control females. Open-field data were obtained from 24 DDT lineage males, 41 DDT females, 18 control males, and 34 control females.
Statistical Analyses for Histopathological Analysis
Pubertal age and behavioral parameters were analyzed using a Student’s t-test. For results expressed as the proportion of affected animals that exceeded a pre-determined threshold (testis, prostate, kidney or ovary disease frequency, tumor frequency, obese frequency, multiple disease frequency), groups were analyzed using Fisher’s exact test. The incidence of disease in rats from each lineage was assessed and the proportion of individual disease and multiple disease incidences was computed. For the individual diseases, only those rats that had histopathology assessed are included in the computation. For the multiple diseases, the total number of diseases for each rat was assessed, and the number added up for each of the rats. The single- or multiple-disease proportions are listed in Supplementary Tables S1–S3.
MeDIP-Seq
DNA was isolated from caudal epididymal sperm as previously described [70]. Briefly, the epididymis from each rat was dissected free of fat and connective tissue, a small cut made to the cauda and the tissue placed in 6 ml of phosphate-buffered saline for 20 min at room temperature. The epididymal tissue was coarsely minced and the released sperm centrifuged at 4000 × g for 5 min and pellet resuspended in NIM buffer and stored at −20°C until processed further. Fifty to 100 μl of rat sperm suspension were used for DNA extraction. The suspension was sonicated for 5 s (Fisher Sonic Dismembrator, model 300, power 25) to lyse any somatic cells as sperm heads are resistant to sonication [76, 77], then centrifuged 5 min at 6000 × g, supernatant discarded and sperm washed again to remove somatic cells and debris.
Each F3 generation individual’s sperm DNA sample was analyzed individually. MeDIP and MeDIP-Seq were performed as previously described [70]. Sequencing libraries were created from MeDIP (single stranded) DNA using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (San Diego, CA) starting at step 1.4 (second strand synthesis) of the manufacturer’s protocol to generate double-stranded DNA from the single-stranded DNA received through the MeDIP procedure. Afterwards, the manufacturer’s protocol was followed. Next-generation sequencing was performed at the WSU Spokane Genomics core using Illumina HiSeq 2500 with a PE50 application, with a read size of ∼50 bp and ∼25 million reads per sample.
Statistics and Bioinformatics
The basic read quality was verified using summaries produced by the FastQC program. The new data were cleaned and filtered to remove adapters and low-quality bases using Trimmomatic [78]. The reads for each MeDIP sample were mapped to the Rnor 6.0 rat genome using Bowtie2 [79] with default parameter options. The mapped read files were then converted to sorted BAM files using SAMtools [80]. The DMRs were identified through a non-biased analysis such that all DMRs at a specific P-value threshold were used for the disease biomarker. The reference window was broken into 100-bp windows in order to identify DMRs. The duplicate reads likely to be PCR artifacts were removed prior to the DMR analysis. The MEDIPS R package [81] was used to calculate differential coverage between control and exposure sample groups. The edgeR P-value [82] was used to determine the relative difference between the two groups for each genomic window. DMRs were considered as having windows with an edgeR P-value that was less than an arbitrarily selected threshold. The DMR edges were extended until no genomic window with an edgeR P-value <0.1 remained within 1000 bp of the DMR. CpG density and other information were then calculated for the DMR based on the reference genome. A PCA on all DMR sites for the control and DDT lineage comparison was performed [83] with the use of the prcomp function in R (www.R-project.org). In addition, the false-discovery rate (FDR) adjusted P-values were calculated. At an FDR P-value threshold of <0.05 the DMRs at for late puberty and kidney disease were validated. The FDR P-value threshold of 0.3 or 0.2 for DMRs at P < 1e-05 for prostate disease, testis disease, obesity, and multiple disease was obtained. This FDR is less stringent than the traditional standard of 0.1. Therefore, the data sets for prostate, testis, obesity, and multiple disease have higher variability.
DMRs were annotated using the biomaRt R package [84] to access the Ensembl database [85]. The genes that overlapped with DMR were then input into the KEGG pathway search [86, 87] to identify associated pathways. The DMR-associated genes were then sorted into functional groups using information provided by the DAVID [88] and Panther [89] databases incorporated into an internal curated database (www.skinner.wsu.edu under genomic data). All molecular data have been deposited into the public database at NCBI (GEO # GSE114032) and R code computational tools available at GitHub (https://github.com/skinnerlab/MeDIP-seq) and www.skinner.wsu.edu.
Availability of Data and Material
All molecular data have been deposited into the public database at NCBI (GEO # GSE114032) and R code computational tools available at GitHub (https://github.com/skinnerlab/MeDIP-seq) and www.skinner.wsu.edu.
Supplementary Material
Acknowledgements
We acknowledge Dr Millissia Ben Maamar, Ms Michelle Pappalardo, Ms Hannah Kimbel, Ms Deepika Kubsad, Ms Marlee Lawley, Ms Heidi Skinner, Mr Ryan Thompson, Ms Jayleana Barton, and Dr Jennifer L.M. Thorson for technical assistance. We also acknowledge Ms Amanda Quilty for assistance in editing the manuscript and Ms Heather Johnson for assistance in preparation of the manuscript. We thank Dr Gerlinde Metz, University of Lethbridge, Canada, for behavioral data analysis advice. This study was supported by a John Templeton Foundation grant (61174) to M.K.S. and NIH grant (ES012974) to M.K.S.
Conflict of interest statement. None declared.
References
- 1. Anway MD, Cupp AS, Uzumcu M, Skinner MK.. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005;308:1466–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vandenberg LN, Colborn T, Hayes TB, Heindel JJ, Jacobs DR Jr, Lee DH, Shioda T, Soto AM, vom Saal FS, Welshons WV.. Hormones and endocrine-disrupting chemicals: low-dose effects and nonmonotonic dose responses. Endocr Rev 2012;33:378–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beck D, Sadler-Riggleman I, Skinner MK.. Generational comparisons (F1 versus F3) of vinclozolin induced epigenetic transgenerational inheritance of sperm differential DNA methylation regions (epimutations) using MeDIP-Seq. Environ Epigenet 2017;3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McBirney M, King SE, Pappalardo M, Houser E, Unkefer M, Nilsson E, Sadler-Riggleman I, Beck D, Winchester P, Skinner MK.. Atrazine induced epigenetic transgenerational inheritance of disease, lean phenotype and sperm epimutation pathology biomarkers. PLoS One 2017;12:e0184306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hao C, Gely-Pernot A, Kervarrec C, Boudjema M, Becker E, Khil P, Tevosian S, Jegou B, Smagulova F.. Exposure to the widely used herbicide atrazine results in deregulation of global tissue-specific RNA transcription in the third generation and is associated with a global decrease of histone trimethylation in mice. Nucleic Acids Res 2016;44:9784–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner M.. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One 2013;8:e55387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner M.. Pesticide and insect repellent mixture (permethrin and DEET) induces epigenetic transgenerational inheritance of disease and sperm epimutations. Reprod Toxicol 2012;34:708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Manikkam M, Haque MM, Guerrero-Bosagna C, Nilsson E, Skinner MK.. Pesticide methoxychlor promotes the epigenetic transgenerational inheritance of adult onset disease through the female germline. PLoS One 2014;9:e102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tracey R, Manikkam M, Guerrero-Bosagna C, Skinner MK.. Hydrocarbons (jet fuel JP-8) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. Reprod Toxicol 2013;36:104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mohamed el SA, Song WH, Oh SA, Park YJ, You YA, Lee S, Choi JY, Kim YJ, Jo I, Pang MG.. The transgenerational impact of benzo(a)pyrene on murine male fertility. Hum Reprod 2010;25:2427–33. [DOI] [PubMed] [Google Scholar]
- 11. Chamorro-Garcia R, Diaz-Castillo C, Shoucri BM, Kaech H, Leavitt R, Shioda T, Blumberg B.. Ancestral perinatal obesogen exposure results in a transgenerational thrifty phenotype in mice. Nat Commun 2017;8:2012.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carvan M Jr, Kalluvila TA, Klingler RH, Larson JK, Pickens M, Mora-Zamorano FX, Connaughton VP, Sadler-Riggleman I, Beck D, Skinner MK.. Mercury-induced epigenetic transgenerational inheritance of abnormal neurobehavior is correlated with sperm epimutations in zebrafish. PLoS One 2017;12:e0176155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bruner-Tran Kl, Ding T, Yeoman KB, Archibong A, Arosh JA, Osteen KG.. Developmental exposure of mice to dioxin promotes transgenerational testicular inflammation and an increased risk of preterm birth in unexposed mating partners. PLoS One 2014;9:e105084.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK.. Dioxin (TCDD) induces epigenetic transgenerational inheritance of adult onset disease and sperm epimutations. PLoS One 2012;7:e46249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anway MD, Leathers C, Skinner MK.. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology 2006;147:5515–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK.. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS One 2012;7:e31901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Skinner MK, Manikkam M, Tracey R, Guerrero-Bosagna C, Haque MM, Nilsson E.. Ancestral dichlorodiphenyltrichloroethane (DDT) exposure promotes epigenetic transgenerational inheritance of obesity. BMC Med 2013;11:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bruner-Tran KL, Osteen KG.. Developmental exposure to TCDD reduces fertility and negatively affects pregnancy outcomes across multiple generations. Reprod Toxicol 2011;31:344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crews D, Gillette R, Scarpino SV, Manikkam M, Savenkova MI, Skinner MK.. Epigenetic transgenerational inheritance of altered stress responses. Proc Natl Acad Sci USA 2012;109:9143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin-Gronert MS, Ozanne SE.. Mechanisms underlying the developmental origins of disease. Rev Endocr Metab Disord 2012;13:85–92. [DOI] [PubMed] [Google Scholar]
- 21. Barker DJ. The developmental origins of chronic adult disease. Acta Paediatr Suppl 2004;93:26–33. [DOI] [PubMed] [Google Scholar]
- 22. Hanson MA, Gluckman PD.. Early developmental conditioning of later health and disease: physiology or pathophysiology? Physiol Rev 2014;94:1027–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bernal AJ, Jirtle RL.. Epigenomic disruption: the effects of early developmental exposures. Birth Defects Res A Clin Mol Teratol 2010;88:938–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klose RJ, Bird AP.. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 2006;31:89–97. [DOI] [PubMed] [Google Scholar]
- 25. Murphy SK, Jirtle RL.. Imprinting evolution and the price of silence. Bioessays 2003;25:577–88. [DOI] [PubMed] [Google Scholar]
- 26. Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics 2011;6:838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Das R, Hampton DD, Jirtle RL.. Imprinting evolution and human health. Mamm Genome 2009;20:563–72. [DOI] [PubMed] [Google Scholar]
- 28. Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA.. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 2002;117:15–23. [DOI] [PubMed] [Google Scholar]
- 29. Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F.. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 2002;129:1807–17. [DOI] [PubMed] [Google Scholar]
- 30. Sato S, Yoshimizu T, Sato E, Matsui Y.. Erasure of methylation imprinting of Igf2r during mouse primordial germ-cell development. Mol Reprod Dev 2003;65:41–50. [DOI] [PubMed] [Google Scholar]
- 31. Shovlin TC, Durcova-Hills G, Surani A, McLaren A.. Heterogeneity in imprinted methylation patterns of pluripotent embryonic germ cells derived from pre-migratory mouse germ cells. Dev Biol 2008;313:674–81. [DOI] [PubMed] [Google Scholar]
- 32. Hackett JA, Reddington JP, Nestor CE, Dunican DS, Branco MR, Reichmann J, Reik W, Surani MA, Adams IR, Meehan RR.. Promoter DNA methylation couples genome-defence mechanisms to epigenetic reprogramming in the mouse germline. Development 2012;139:3623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Skinner M, Guerrero-Bosagna C, Haque MM, Nilsson E, Bhandari R, McCarrey J.. Environmentally induced transgenerational epigenetic reprogramming of primordial germ cells and subsequent germline. PLoS One 2013;8:e66318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jirtle RL, Skinner MK.. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007;8:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Skinner MK. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod Toxicol 2008;25:2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van den Berg H. Global status of DDT and its alternatives for use in vector control to prevent disease. Environ Health Perspect 2009;117:1656–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.WHO. The Use of DDT in Malaria Vector Control – WHO Position Statement. 2011. http://apps.who.int/iris/bitstream/10665/69945/1/WHO_HTM_GMP_2011_eng.pdf (26 April 2019, date last accessed).
- 38. Rignell-Hydbom A, Lidfeldt J, Kiviranta H, Rantakokko P, Samsioe G, Agardh CD, Rylander L.. Exposure to p,p'-DDE: a risk factor for type 2 diabetes. PLoS One 2009;4:e7503.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee DH, Porta M, Jacobs DR Jr, Vandenberg LN.. Chlorinated persistent organic pollutants, obesity, and type 2 diabetes. Endocrine Reviews 2014;35:557–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cano-Sancho G, Salmon AG, La Merrill MA.. Association between exposure to p,p'-DDT and its metabolite p,p'-DDE with obesity: integrated systematic review and meta-analysis. Environ Health Perspect 2017;125:096002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.NHLBI. Obesity Education Initiative Expert Panel on the Identification, Evaluation, and Treatment of Obesity in Adults (US). Clinical Guidelines on the Identification, Evaluation, and Treatment of Overweight and Obesity in Adults: The Evidence Report 1998. https://www.ncbi.nlm.nih.gov/books/NBK2003/ (26 April 2019, date last accessed).
- 42. Caballero B. The global epidemic of obesity: an overview. Epidemiol Rev 2007;29:1–5. [DOI] [PubMed] [Google Scholar]
- 43. Hales CM, Carroll MD, Fryar CD, Ogden CL.. Prevalence of Obesity Among Adults and Youth: United States, 2015–2016. Hyattsville, MD: National Center for Health Statistics, 2017. (NCHS Data Brief). [PubMed] [Google Scholar]
- 44. McAllister EJ, Dhurandhar NV, Keith SW, Aronne LJ, Barger J, Baskin M, Benca RM, Biggio J, Boggiano MM, Eisenmann JC. et al. Ten putative contributors to the obesity epidemic. Crit Rev Food Sci Nutr 2009;49:868–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kurpad AV, Varadharajan KS, Aeberli I.. The thin-fat phenotype and global metabolic disease risk. Curr Opin Clin Nutr Metab Care 2011;14:542–7. [DOI] [PubMed] [Google Scholar]
- 46. Yajnik CS, Fall CH, Coyaji KJ, Hirve SS, Rao S, Barker DJ, Joglekar C, Kellingray S.. Neonatal anthropometry: the thin-fat Indian baby. The Pune Maternal Nutrition Study. Int J Obes 2003;27:173–80. [DOI] [PubMed] [Google Scholar]
- 47. Guimaraes RM, Asmus CI, Meyer A.. DDT reintroduction for malaria control: the cost-benefit debate for public health. Cad Saude Publica 2007;23:2835–44. [DOI] [PubMed] [Google Scholar]
- 48. La Merrill M, Karey E, Moshier E, Lindtner C, La Frano MR, Newman JW, Buettner C.. Perinatal exposure of mice to the pesticide DDT impairs energy expenditure and metabolism in adult female offspring. PLoS One 2014;9:e103337.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ATSDR, Agency for Toxic Substances and Diseases Registry (ATSDR)/US Public Health Service. Toxicological Profile for 4,4'-DDT, 4,4'-DDE, 4,4'-DDD (Update). Atlanta, GA: ATSDR, 1994. [Google Scholar]
- 50. Aneck-Hahn NH, Schulenburg GW, Bornman MS, Farias P, de Jager C.. Impaired semen quality associated with environmental DDT exposure in young men living in a malaria area in the Limpopo Province, South Africa. J Androl 2006;28:423–34. [DOI] [PubMed] [Google Scholar]
- 51. Messaros BM, Rossano MG, Liu G, Diamond MP, Friderici K, Nummy-Jernigan K, Daly D, Puscheck E, Paneth N, Wirth JJ.. Negative effects of serum p,p'-DDE on sperm parameters and modification by genetic polymorphisms. Environ Res 2009;109:457–64. [DOI] [PubMed] [Google Scholar]
- 52. Song Y, Wu N, Wang S, Gao M, Song P, Lou J, Tan Y, Liu K.. Transgenerational impaired male fertility with an Igf2 epigenetic defect in the rat are induced by the endocrine disruptor p,p'-DDE. Hum Reprod 2014;29:2512–21. [DOI] [PubMed] [Google Scholar]
- 53. Song Y, Yang L.. Transgenerational pancreatic impairment with Igf2/H19 epigenetic alteration induced by p,p'-DDE exposure in early life. Toxicol Lett 2017;280:222–31. [DOI] [PubMed] [Google Scholar]
- 54. Nilsson E, Sadler-Riggleman I, Skinner MK.. Environmentally induced epigenetic transgenerational inheritance of disease. Environ Epigenet 2018;4:1–13. dvy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guillette LJ Jr, Gross TS, Masson GR, Matter JM, Percival HF, Woodward AR.. Developmental abnormalities of the gonad and abnormal sex hormone concentrations in juvenile alligators from contaminated and control lakes in Florida. Environ Health Perspect 1994;102:680–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kirman CR, Aylward LL, Hays SM, Krishnan K, Nong A.. Biomonitoring equivalents for DDT/DDE. Regul Toxicol Pharmacol 2011;60:172–80. [DOI] [PubMed] [Google Scholar]
- 57. de Jager C, Aneck-Hahn NH, Bornman MS, Farias P, Leter G, Eleuteri P, Rescia M, Spano M.. Sperm chromatin integrity in DDT-exposed young men living in a malaria area in the Limpopo Province, South Africa. Hum Reprod 2009;24:2429–38. [DOI] [PubMed] [Google Scholar]
- 58. Wassie F, Spanoghe P, Tessema DA, Steurbaut W.. Exposure and health risk assessment of applicators to DDT during indoor residual spraying in malaria vector control program. J Expo Sci Environ Epidemiol 2012;22:549–58. [DOI] [PubMed] [Google Scholar]
- 59. Guerrero-Bosagna C, Settles M, Lucker B, Skinner M.. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One 2010;5:e13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Skinner MK, Ben Maamar M, Sadler-Riggleman I, Beck D, Nilsson E, McBirney M, Klukovich R, Xie Y, Tang C, Yan W.. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenetics Chromatin 2018;11:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Skinner MK, Guerrero-Bosagna C.. Role of CpG deserts in the epigenetic transgenerational inheritance of differential DNA methylation regions. BMC Genomics 2014;15:692.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kubsad D, Nilsson EE, King SE, Sadler-Riggleman I, Beck D, Skinner MK.. Assessment of glyphosate induced epigenetic transgenerational inheritance of pathologies and sperm epimutations: generational toxicology. Sci Rep 2019;9:6372.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ward BW, Schiller JS.. Prevalence of multiple chronic conditions among US adults: estimates from the National Health Interview Survey, 2010. Prev Chronic Dis 2013;10:E65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Murray CJL, Barber RM, Foreman KJ, Abbasoglu Ozgoren A, Abd-Allah F, Abera SF, Aboyans V, Abraham JP, Abubakar I, Abu-Raddad LJ. et al. Global, regional, and national disability-adjusted life years (DALYs) for 306 diseases and injuries and healthy life expectancy (HALE) for 188 countries, 1990-2013: quantifying the epidemiological transition. Lancet 2015;386:2145–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ward BW, Black LI.. State and regional prevalence of diagnosed multiple chronic conditions among adults aged >/=18 years - United States, 2014. MMWR Morb Mortal Wkly Rep 2016;65:735–8. [DOI] [PubMed] [Google Scholar]
- 66. Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, Fruchart JC, James WP, Loria CM, Smith SC Jr. et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 2009;120:1640–5. [DOI] [PubMed] [Google Scholar]
- 67. Marchesini G, Brizi M, Bianchi G, Tomassetti S, Bugianesi E, Lenzi M, McCullough AJ, Natale S, Forlani G, Melchionda N.. Nonalcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes 2001;50:1844–50. [DOI] [PubMed] [Google Scholar]
- 68. Ninomiya JK, L’Italien G, Criqui MH, Whyte JL, Gamst A, Chen RS.. Association of the metabolic syndrome with history of myocardial infarction and stroke in the Third National Health and Nutrition Examination Survey. Circulation 2004;109:42–6. [DOI] [PubMed] [Google Scholar]
- 69. Thomas G, Sehgal AR, Kashyap SR, Srinivas TR, Kirwan JP, Navaneethan SD.. Metabolic syndrome and kidney disease: a systematic review and meta-analysis. CJASN 2011;6:2364–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nilsson E, King SE, McBirney M, Kubsad D, Pappalardo M, Beck D, Sadler-Riggleman I, Skinner MK.. Vinclozolin induced epigenetic transgenerational inheritance of pathologies and sperm epimutation biomarkers for specific diseases. PLoS One 2018;13:e0202662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nilsson EE, Anway MD, Stanfield J, Skinner MK.. Transgenerational epigenetic effects of the endocrine disruptor vinclozolin on pregnancies and female adult onset disease. Reproduction 2008;135:713–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Anway MD, Skinner MK.. Transgenerational effects of the endocrine disruptor vinclozolin on the prostate transcriptome and adult onset disease. Prostate 2008;68:517–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Taylor JA, Richter CA, Ruhlen RL, vom Saal FS.. Estrogenic environmental chemicals and drugs: mechanisms for effects on the developing male urogenital system. J Steroid Biochem Mol Biol 2011;127:83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nilsson E, Larsen G, Manikkam M, Guerrero-Bosagna C, Savenkova M, Skinner M.. Environmentally induced epigenetic transgenerational inheritance of ovarian disease. PLoS One 2012;7:e36129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS One 2008;3:e3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Calvin HI. Isolation of subfractionation of mammalian sperm heads and tails. Methods Cell Biol 1976;13:85–104. [DOI] [PubMed] [Google Scholar]
- 77. Huang TT Jr, Yanagimachi R.. Inner acrosomal membrane of mammalian spermatozoa: its properties and possible functions in fertilization. Am J Anat 1985;174:249–68. [DOI] [PubMed] [Google Scholar]
- 78. Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Langmead B, Salzberg SL.. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L.. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 2014;30:284–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Robinson MD, McCarthy DJ, Smyth GK.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pearson K. LIII. On lines and planes of closest fit to systems of points in space. The London, Edinburgh, and Dublin Philosophical Magazine and Journal of Science 1901;2:559–72. [Google Scholar]
- 84. Durinck S, Spellman PT, Birney E, Huber W.. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009;4:1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S. et al. Ensembl 2015. Nucleic Acids Res 2015;43:D662–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kanehisa M, Goto S.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M.. Data, information, knowledge and principle: back to metabolism in KEGG. Nucl Acids Res 2014;42:D199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang da W, Sherman BT, Lempicki RA.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 89. Mi H, Muruganujan A, Casagrande JT, Thomas PD.. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013;8:1551–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All molecular data have been deposited into the public database at NCBI (GEO # GSE114032) and R code computational tools available at GitHub (https://github.com/skinnerlab/MeDIP-seq) and www.skinner.wsu.edu.