Abstract
Being an assembly of protein machines, cells depend on adequate supply of energetic molecules for retaining their homeodynamics. Consequently, mitochondria functionality is ensured by quality control systems and mitochondrial dynamics (fusion/fission). Similarly, proteome stability is maintained by the machineries of the proteostasis network. We report here that reduced mitochondrial fusion rates in Drosophila caused developmental lethality or if induced in the adult accelerated aging. Imbalanced mitochondrial dynamics were tolerable for various periods in young flies, where they caused oxidative stress and proteome instability that mobilized Nrf2 and foxo to upregulate cytoprotective antioxidant/proteostatic modules. Consistently, proteasome inhibition or Nrf2, foxo knock down in young flies exaggerated perturbed mitochondrial dynamics toxicity. Neither Nrf2 overexpression (with concomitant proteasome activation) nor Atg8a upregulation suppressed the deregulated mitochondrial dynamics toxicity, which was mildly mitigated by antioxidants. Thus, despite extensive functional wiring of mitostatic and antioxidant/proteostatic modules, sustained loss-of mitostasis exhausts adaptation responses triggering premature aging.
Keywords: Aging, Drp1, Mitofusins, Mitostasis, Opa1, Proteostasis
Graphical abstract

Highlights
-
•
Reduced mitochondrial fusion rates cause severe organismal toxicity and progeria.
-
•
Perturbed mitostasis activates cytoprotective antioxidant and proteostatic modules.
-
•
Nrf2 or Foxo KD exaggerates the imbalanced mitochondrial dynamics induced toxicity.
-
•
Antioxidants mildly alleviate loss-of mitochondrial dynamics-mediated progeria.
Abbreviations
- ALP
Autophagy Lysosome Pathway
- ARE
Antioxidant Response Element
- C-L
Caspase-Like proteasomal activity
- cncC
cap’-n’-collar isoform-C
- CT-L
Chymotrypsin-Like proteasomal activity
- DOA
autosomal-dominant optic atrophy
- ER
Endoplasmic Reticulum
- foxo
forkhead box, sub-group O
- GLU
Glucose
- GLY
Glycogen
- IMM
Inner Mitochondrial Membrane
- Keap1
Kelch-like ECH-Associated Protein 1
- LHON
Leber hereditary optic neuropathy
- Lon
Lon protease
- mtDNA
mitochondrial DNA
- Mfn1
Mitofusin 1
- Mfn2
Mitofusin 2
- Miro
Mitochondrial Rho GTPase
- NFE2L2/Nrf2
nuclear factor, erythroid 2 like 2
- OMM
Outer Mitochondrial Membrane
- Opa1
Optic atrophy 1
- OXPHOS
Oxidative Phosphorylation
- PN
Proteostasis Network
- ROS
Reactive Oxygen Species
- TREH
trehalose
- Ub
Ubiquitin
- UPP
Ubiquitin Proteasome Pathway
- UPR
Unfolded Protein Response
1. Introduction
Viability of metazoans largely depends on their ability to regulate metabolic processes for producing energetic molecules, as well as on their capacity to mount anti-stress responses for retaining the integrity and functionality of biomolecules (e.g. genome and proteome). At the whole organism level, these pathways and responses require complicated, largely unknown in its details, co-regulation and wiring of cell autonomous and non-autonomous mechanisms [1,2].
Energetic molecules in eukaryotic cells are produced in mitochondria through the integrated action of the oxidative phosphorylation (OXPHOS) protein machines [3,4]. The outer mitochondrial membrane (OMM) prevents the diffusion of small molecules and it also serves as an exchanging platform. The inner mitochondrial membrane (IMM) contains three different areas, namely the inner boundary membrane, the cristae and the cristae junctions, harboring the protein complexes of OXPHOS and the energy producing F1F0 ATP synthase [5]. Mitochondria are highly dynamic structures that undergo rapid remodeling through fusion and fission [6]. Fusion rearranges the matrix content of a damaged mitochondrion with a healthy one, diluting thus unfolded proteome and mutated DNA; whereas, fission is important for either mitochondria division or for the removal of damaged mitochondria by a specific type of autophagy, namely mitophagy [7,8]. In mammals the process of fusion is operated by three GTPases, namely Mitofusin 1 (Mfn1), Mitofusin 2 (Mfn2) and Optic Atrophy 1 (Opa1). Mfns are implicated in OMM fusion, whereas Opa1 is involved in IMM fusion; energetics, apoptosis regulation and cristae remodeling [9]. Pathogenic mutations that disrupt Mfns or Opa1 function cause axonal Charcot-Marie-Tooth disease type 2A or autosomal dominant optic atrophy (ADOA; a mitochondrial disease leading to childhood onset blindness), respectively [10]. The main operator of fission is the dynamin-related protein 1 (Drp1), a cytosolic protein that is recruited to the fission site where by forming a spiral superstructure constricts both OMM and IMM after GTP hydrolysis [11]. Mitochondria have developed their own system of chaperones and proteases to fold or degrade respectively the proteins encoded by the mitochondrial genome or imported into the organelle. This quality system is critical also because mitochondria are the primary source of Reactive Oxygen Species (ROS) which can damage all biomolecules [12]. Lon mitochondrial protease is a conserved AAA + protein that degrades misfolded and/or damaged proteins; it also regulates mitochondrial DNA copies number and transcription [13,14].
Proteome stability in cells is assured by the proteostasis network (PN), a modular integrated system which ensures proteome quality control at both basal conditions and under conditions of proteotoxic stress [15,16]. Key components of the PN are the molecular chaperones and the two main degradation machineries, namely the Autophagy Lysosome- (ALP) and the Ubiquitin Proteasome- (UPP) pathways. ALP is an intracellular self-catabolic process that in mammalian cells comprises the chaperone-mediated autophagy, microautophagy and macroautophagy; during macroautophagy the autophagy related proteins (Atgs) form autophagosomes that capture lipids, proteins or even organelles and transfer them to lysosome for degradation [17]. Mitophagy is a selective to mitochondria autophagic response that regulates their degradation when organelle damage is not repairable [8]. One of the most described pathways is the Pink1/Parkin-mediated mitophagy [18]; yet, mitophagy can also occur in a Parkin-independent way [19]. UPP degrades short-lived poly-ubiquitinated normal proteins and non-functional or misfolded polypeptides and it is composed of the ubiquitin (Ub)-conjugating enzymes and the 26S proteasome [20]. The 26S eukaryotic proteasome comprises the 19S regulatory particles (RP) which are bound to a 20S core particle (CP). The 20S CP consists of four stacked heptameric rings (two of α-type surrounding two of β-type) that form a barrel-like structure; the caspase- (C-L), trypsin- (T-L), and chymotrypsin- (CT-L) like peptidase activities are located at the β1, β2, and β5 proteasomal subunits, respectively. The 19S RP is involved in substrate recognition, deubiquitination, unfolding and translocation into the 20S CP [21]. Proteasomes are also found at the OMM where during activation of the mitochondrial Unfolded Protein Response (UPR) perform the process of OMM-Associated Degradation (OMMAD) [22,23]. Reportedly, UPP also regulates mitochondria dynamics as Mfns are UPP substrates [24].
The PN branches are functionally coordinated by stress sensors, which respond to deregulated proteostasis and/or to increased amounts of stressors. These sensors are mostly short-lived transcription factors, e.g. forkhead box, sub-group O (Foxo), heat shock factor-1 (Hsf1) and nuclear factor, erythroid 2 like 2 (Nrf2); that mobilize cytoprotective genomic responses against various types of stress. The Foxo family members are highly conserved through evolution and regulate oxidative stress levels, energy metabolism, apoptosis, cell cycle, DNA damage repair and mitophagy [25]. Similarly, the Nrf2 signaling pathway plays a crucial role in defenses against oxidative and/or xenobiotic damage by binding to electrophile- (EpREs) or antioxidant- (ARE) response elements of a broad range of phase-II and antioxidant enzymes [26]. Reportedly, in higher metazoans Nrf2 is also involved in the upregulation of UPP and ALP genes during proteotoxic stress [20,27,28], and in the regulation of mitostatic modules involved in energetics and mitochondria biogenesis [29,30].
The functionality of mitochondria, proteostatic modules and anti-stress responses decline during aging and these events seem to fuel the appearance of aging and/or age-related diseases [2,[31], [32], [33]]. Accordingly, age is the major risk factor for several disorders, including diabetes, neurodegeneration and cancer [2,34].
Notably, the mechanistic insights of mitostatic and antioxidant/proteostatic modules functional cross-talk and wiring at the whole organism level remain not well understood. By using Drosophila flies as a model organism, we report here that deregulated mitochondrial dynamics or Lon knock down (KD) caused developmental lethality or if induced in the adult, oxidative stress, proteome instability and aging acceleration. Loss-of mitostasis activated in the young organism cap’-n’-collar isoform-C (cncC; the Nrf2 ortholog in Drosophila) and foxo to trigger the upregulation of cytoprotective antioxidant/proteostatic modules. Yet, neither cncC/Nrf2 overexpression (OE) (with concomitant proteasome activation) nor autophagy induction rescued the damaging effects of imbalanced mitochondrial dynamics; these toxic effects were mildly alleviated by antioxidants.
2. Materials and methods
2.1. Flies stocks
The Drosophila melanogaster wild type and all transgenic lines used in this study are described in Supplemental Materials and Methods. The conditional driver (Gal4GS−Tub) is ubiquitously activated upon dietary administration of RU486 (320 μΜ). In all experiments referring to adult flies, micro-dissected head/thorax somatic tissues were used (equal numbers from mated male/female flies) [35].
2.2. Flies culture, exposure to compounds, mobility and longevity assays
Flies stocks were maintained at 25 °C, 60% relative humidity on a 12-h light:12-h dark cycle and were fed standard medium [36]. All used compounds (RU486, PS-341, Rapamycin, Tiron, 6BIO) were added in flies' culture medium; doses and duration of flies’ exposure to compounds are indicated in figure legends. Tiron and RU486 were obtained from Sigma-Aldrich, PS-341 was from Calbiochem, Rapamycin was from Cayman Chemical Company and 6BIO was a kind gift from Prof. A.L. Skaltsounis (National and Kapodistrian University of Athens, Greece).
Mobility of flies was assayed as described before [27]. For longevity assays female and male flies (equal numbers per sex) were collected and cultured in vials; flies were transferred to vials with fresh food every 3–4 days and deaths were scored daily.
In shown graphs, bars denote mean (n ≥ 2) ± SD; *, indicates P < 0.05 and **, P < 0.01. Gene expression was plotted vs. the respective control set to 1; in all other cases (unless otherwise indicated) control values were set to 100. For survival curves and statistical analyses, the Kaplan-Meier procedure and log-Rank (Mantel-Cox) test were used; significance was accepted at P < 0.05. Statistical analysis of the longevity experiments is reported in Table S1.
Full Methods, description of Statistical analysis and any associated References are available in Supplemental Materials and Methods.
3. Results
3.1. Imbalanced mitochondrial dynamics deregulated organismal physiology and accelerated aging; these effects were severely enhanced after Opa1 KD
To investigate the impact of imbalanced mitochondrial dynamics (Fig. S1A) and to map the cross-talk between mitostatic and proteostatic machineries, we genetically modulated Marf, Drp1 and Opa1 expression levels. Marf (the fly single homologue of Mfns) OE in larvae muscles resulted in hyperfused mitochondria that tended to aggregate perinuclearly, whereas Marf KD induced mitochondria clustering and fragmentation (Figs. S1B and S2). Also, gene expression analysis showed that Marf is likely a negative regulator of Drp1 expression (Fig. S1C). Muscle-targeted perturbation of Marf caused a lipolytic effect in larvae fat body (analogous to the mammalian liver and adipose tissue; Fig. S1D) indicating a cell non-autonomous systemic organ effect. In the adult, inducible ubiquitous Marf OE or KD accelerated age-related loss-of locomotor activity (Fig. S1E) and aging (Figs. S1F and S1G, Table S1). These effects were mostly evident after Marf KD indicating that rebalancing mitochondrial dynamics towards reduced fusion rates were less tolerated compared to fusion upregulation; also evident following Drp1 KD vs. Drp1 OE (Figs. S1H and S1I, Table S1). Consistently, reduced mitochondrial fusion rates were also significantly more toxic in young flies (Figs. S1G and S1I; shaded rectangles in longevity curves), while induced KD of the Marf gene was particularly toxic in aged flies (Fig. S1J).
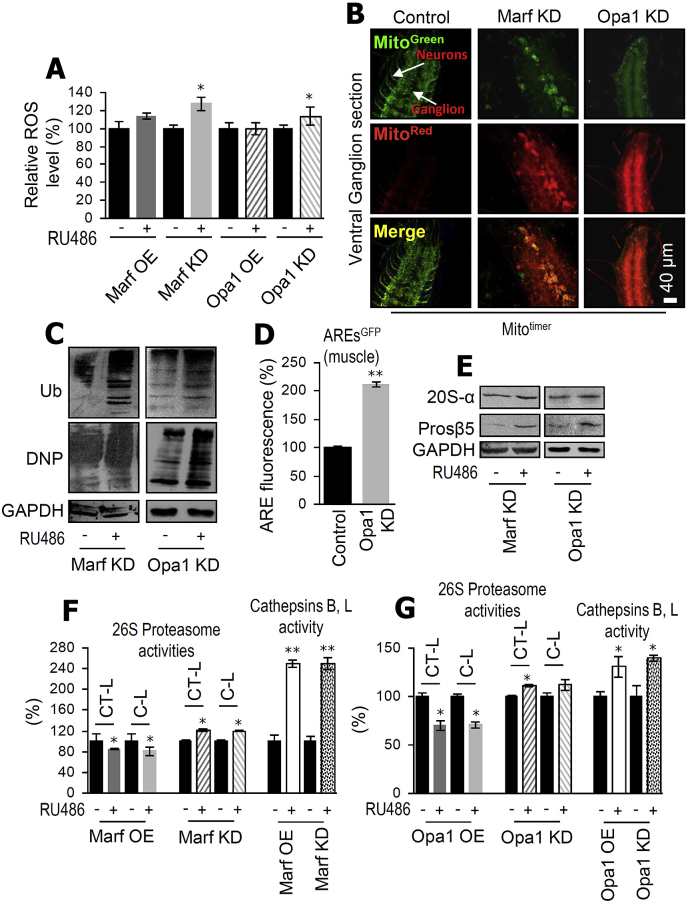
Opa1 OE was not lethal during development, and it did not affect mitochondria distribution or number in either larvae muscle (Fig. 1A) or nervous (Fig. 1B) tissues. Also, although Opa1 OE tended to increase mitochondrial respiratory chain efficiency rates (Fig. 1C and D); it had minor effect on tissue glucose (GLU), trehalose (TREH; a sugar that circulates in flies' hemolymph) or glycogen (GLY) content (Fig. 1E) after targeted expression in muscles and it caused mild lipolysis in the fat body (Fig. 1F). Furthermore, inducible Opa1 OE did not affect flies' locomotion (Fig. 1G) and it mildly reduced flies' longevity (Fig. 1H). On the contrary, Opa1 KD increased mitochondria fragmentation (Figs. 1A and S2), reduced mitochondria number (Figs. 1A, 1B, S2) and caused pupae lethality. Opa1 KD decreased mitochondrial respiration (Fig. 1C and D) and following muscle targeting it reduced tissues' GLU content while it increased TREH levels (Fig. 1E), indicating that flies are becoming hyperglycemic. Also, sustained Opa1 KD in the muscle increased lipolysis in the fat body (Fig. 1F) further supporting cell non-autonomous systemic organ effects. Finally, inducible Opa1 KD in adult flies accelerated age-related loss of locomotion activity (Fig. 1G); was tolerable for a very short period in young flies and sharply reduced longevity (Fig. 1I, Table S1). Consistently to metabolic deregulation, Opa1 KD flies were very sensitive to increased calories even under conditions of leaky [RU486(−)] transgene expression, since exposure to a high sugar diet further reduced pupation (Fig. S3A) and dramatically exacerbated Opa1 KD-mediated loss-of locomotion (Fig. S3B) and progeria (Fig. S3C). Again, Opa1 KD was more toxic in aged (compared to young) flies; yet, this effect was less evident compared to Marf KD due to high toxicity of Opa1 KD under conditions of no inducer addition [RU486(−)] in flies’ culture medium (Fig. 1J).
Fig. 1.
Opa1 KD disrupts mitochondria structure and function; induces metabolic and neuromuscular defects and drastically accelerates aging. (A, B) CLSM visualization of MitoGFP in larval muscle (A; Gal4Mef2) or nervous system (B; Gal4D42) after Opa1 OE or KD. (C) Relative mitochondrial ST3/ST4 respiratory efficiency rates from somatic tissues of flies expressing (or not) the shown transgenes; young mated flies were exposed to RU486 for 7 days. (D) Oxygen consumption values (basal and after Oligomycin addition) during mitochondrial respiration assays. (E) Relative (%) content of GLU, TREH and GLY in larvae tissue after muscle specific (Gal4Mef2) Opa1 OE or KD. (F) CLSM visualization of larvae fat body after BODIPY staining of lipids in shown Opa1 transgenic lines (Gal4Mef2). (G) Climbing activity (%) of middle aged transgenic flies of the indicated genotypes. (H, I) Longevity curves of the indicated transgenic lines; shaded rectangles and shown periods in days indicate the duration of neutral (vs. control) effect of the expressed transgene; viability at 20 days post-Opa1 KD was ∼28%. (J) Longevity curves following induced ubiquitous Opa1 KD in middle aged (30 days old) flies; viability at 20 days post-transgene expression was ∼20%. Statistics of the longevity curves are reported in Table S1. In (C, D), (G–J) the Gal4GS−Tub inducible ubiquitous driver was used. Bars, ± SD; n ≥ 2; *P < 0.05; **P < 0.01.
During our assays we observed that Opa1 KD in male flies was more toxic as compared to females. CLSM analysis showed that Opa1 KD male larvae displayed enhanced mitochondria fragmentation (Fig. 2A), as well as lower mitochondrial respiration and increased leakiness (Fig. 2B) compared to females. Electron microscopy imaging revealed cristae alterations in both genders which, again, were more severe in males as their mitochondria had larger cristae swallowing compared to females, establishing a clear link between inner membrane architecture and functional decline; these effects were accompanied by more severe mitochondria aggregation and myofibrils structure disorganization in males' muscles (Fig. 2C). In support, longevity assays showed that Opa1 KD caused a sharp reduction in males’ lifespan (median longevity, 13 ± 1.14 days) while females survived significantly longer (median longevity, 20 ± 1.51 days) (Fig. 2D, Table S1).
Fig. 2.
KD of Opa1 is increasingly toxic in male flies. (A) CLSM visualization of mitochondria (MitoGFP) in larvae muscles (Gal4Mef2) of male and female flies. (B) Relative mitochondrial ST3/ST4 and FCCP/ST4 values after Opa1 KD in adult male and female flies. (C) EM visualization of mitochondria and myofibril structure after Opa1 KD; arrows indicate disrupted cristae and the rectangle aggregated mitochondria. In (B), (C) the transgene was induced in young mated flies for 7 days. (D) Longevity curves of male and female flies after inducible ubiquitous Opa1 KD. Statistics of the shown longevity curves are reported in Table S1. In (B–D) the Gal4GS−Tub driver was used. Bars, ± SD; n ≥ 2; *P < 0.05; **P < 0.01.
Thus, imbalanced mitochondrial dynamics towards reduced fusion cause developmental lethality, while if induced in adults is intolerable in young organisms and dramatically accelerates aging; these effects were sex-dependent in the case of Opa1 KD. Also, the differential severity (compared to Marf KD) of Opa1 KD-mediated toxicity, highlights the increased organismal sensitivity to alterations in the fusion rates and structural organization of the IMM vs. OMM.
3.2. Mitochondria dysfunction caused oxidative stress and proteome instability inducing the activation of cytoprotective antioxidant and proteostatic modules
To investigate the functional wiring between mitostatic and antioxidant/proteostatic modules we initially focused on perturbed mitochondrial dynamics and found that either Marf or Opa1 KD increased ROS levels (Fig. 3A). In support, by using the MitoTimer reporter (encodes a mitochondria-targeted green fluorescent protein when newly synthesized, which shifts irreversibly to red fluorescence when oxidized) we noted increased mitochondria oxidation after Marf or Opa1 KD (Figs. 3B and S4), which again was more pronounced following Opa1 KD. We then used the redox sensitive GFP (roGFPs) probes to further study the in vivo oxidative load in Marf and Opa1 KD mutants. We used transgenic lines with a cytosolic EGSH probe (UAS-cyto-roGFP2-Grx1) for glutathione redox couple (GSH/GSSG), as well as lines with a cytosolic (UAS-cyto-roGFP2-Orp1) or a mitochondrial (UAS-mito-roGFP2-Orp1) hydrogen peroxide (H2O2) probe [37]. We observed significant changes in the cytosolic GSH redox balance in larvae muscles of both mutants as indicated by the increased 405 nm/488 nm ratio (Figs. S5A and S5B). Following Opa1 KD, H2O2 levels were increased in both the cytosol and mitochondria, while in Marf KD larvae significant H2O2 changes were observed only in mitochondria compartment (Figs. S5C–S5F); consistently, with the MitoTimer reporter analyses, the oxidative load was more pronounced in Opa1 KD larvae compared to Marf KD.
Fig. 3.
Imbalanced mitochondrial dynamics towards reduced fusion causes oxidative stress and proteome instability triggering the activation of antioxidant/proteostatic modules. (A) Relative (%) ROS levels in tissues of young flies after ubiquitous inducible expression of the shown transgenes. (B) CLSM visualization of the MitoTimer reporter in the nervous system of control (+/Gal4Elav) larvae or following Marf or Opa1 KD. (C) Representative immunoblot analysis of tissue protein samples probed for total protein ubiquitination (Ub) and carbonylation (DNP). (D) Relative (%) activation of AREs in larvae following muscle targeted KD of Opa1 (Gal4Mef2). (E) Representative immunoblot analysis of young flies' tissue protein samples probed with antibodies against proteasomal subunits 20S-α and Prosβ5 . (F, G) Relative (%) proteasomal (F) and cathepsins B, L (G) activities in young flies' tissues expressing (or not) ubiquitously the shown transgenes. In (C), (E) GAPDH was used as reference. In (A), (C), (E-G) the transgene was induced in young flies for 7 days by using the Gal4GS−Tub driver. Bars, ± SD; n ≥ 2; *P < 0.05; **P < 0.01.
Mitochondria malfunction increased proteome instability as evidenced by upregulated ubiquitination and carbonylation (Fig. 3C), which correlated with significant induction of the antioxidant response elements (AREs) in Opa1 KD flies (Fig. 3D) suggesting the activation of cellular stress sensors (e.g. cncC/Nrf2). Consistently, Marf KD activated proteasomal and Atg8a genes (Fig. S6A), as well as proteasomal proteins (Fig. 3E), proteasome peptidases and autophagic cathepsins activity (Fig. 3F). As after Marf KD, and despite dissimilar gene expression responses (Fig. S6B) likely due to increased cell death [9,38], Opa1 KD upregulated proteasomal protein subunits (Fig. 3E), proteasome peptidases and autophagic cathepsins activity (Fig. 3G). Based on the observed patterns of proteasomal activities (Fig. 3F and G) we concluded that Marf or Opa1 suppress UPP, while in line with the suggested ALP activation, muscle targeted Opa1 KD in larvae induced the accumulation of ref(2)P (the fly homolog of p62/SQSTM1) (Fig. S6C) and higher mCherry-Atg8a levels (Fig. S6D).
To investigate whether upregulation of antioxidant/proteostatic machineries is a dominant downstream response independently of the intervention that induces loss-of mitostasis we suppressed the expression levels of Lon protease. Similarly, to reduced fusion rates, Lon KD decreased mitochondria number in larvae muscles (Fig. S7A) and nervous tissue (Fig. S7B) and caused developmental lethality, while inducible Lon KD in adults was very toxic in young flies (Fig. S7C, shaded rectangle) and it significantly decreased overall longevity (Fig. S7C). Lon KD in young flies increased ROS (Fig. S7E); massively upregulated proteostatic genes and genes involved in mitochondria biogenesis (Fig. S7F) and it activated proteasome peptidases (Fig. S7G) and cathepsins (Fig. S7H) activity. Moreover, targeted Lon KD in larvae nervous tissues and in muscles caused the upregulation of GFP-Lamp1 (Fig. S7I) and of ref(2)P (Fig. S7J) respectively, further supporting ALP activation. As before, loss-of mitostasis due to Lon KD in aged flies was significantly more toxic as compared to young flies (Fig. S7D).
These findings suggest a tight functional wiring between proteostatic and mitostatic modules.
3.3. Activation of cytoprotective antioxidant/proteostatic modules due to imbalanced mitochondrial dynamics is mediated by cncC/Nrf2 and foxo; consistently, cncC/Nrf2, foxo KD or proteasome inhibition exaggerated loss-of mitostasis toxicity
We then sought to identify the mediators of antioxidant and proteostatic modules activation upon perturbed mitochondrial dynamics. We found that cncC/Nrf2 KD in the Marf KD background abolished proteasomal or mitophagy related genes (Pink1, park) upregulation, while other proteostatic [e.g. Hsp70, ref(2)P, Atg8a] genes remained highly upregulated (Fig. 4A). Downregulation of cncC/Nrf2 in Marf KD flies also resulted in elevated cellular ROS (Fig. 4B), abolished proteasome activation (Fig. 4C), which resulted in proteome over-ubiquitination (Fig. 4D) and had no effect on Marf KD-mediated cathepsins activation (Fig. 4E). It also reduced MitoGFP aggregates in Marf KD larvae (Fig. 4F) likely because of activated foxo enhanced mitophagy [39], and increased mitochondria fragmentation (Fig. 4G). On the contrary, downregulation of foxo in the Marf KD background did not affect proteasomal genes upregulation; yet, it abolished the autophagy/mitophagy genes induction (Fig. 4A). In addition, foxo KD decreased ROS (Fig. 4B), significantly induced proteasome activities (Fig. 4C) resulting in downregulation of proteome ubiquitination (Fig. 4D) and it suppressed cathepsins activation (Fig. 4E); thus, likely the cncC/Nrf2 axis is overactive upon foxo KD. Finally, either cncC/Nrf2 or foxo KD significantly accelerated aging in Marf KD flies (Fig. 4H and I).
Fig. 4.
cncC/Nrf2 and foxo mediate cytoprotective cellular responses after Marf KD in young flies; their inactivation enhances Marf KD-mediated lethality. (A) Relative expression of shown genes in young flies after combined ubiquitous expression (or not) of the indicated transgenes. (B–D) Relative (%) ROS levels (B), proteasome activities (C), proteome ubiquitination (GAPDH was used as loading reference) (D) and cathepsins B, L (E) activity in shown transgenic lines. (F) CLSM viewing of mitochondria (MitoGFP) in larvae muscle (Gal4Mef2) of the indicated genotypes. (G) Relative quantification of mitochondria density (number/area) and length in larvae muscles of the indicated genotypes. (H, I) Longevity curves of the indicated transgenic lines; shaded rectangles highlight the accelerated toxicity (vs. Marf KD flies) in young flies co-expressing the indicated transgenes. Statistics of the shown longevity curves are reported in Table S1. In (A–E) the transgene was induced in young flies for 7 days. In (A–E), (H), (I) the Gal4GS−Tub driver was used. Gene expression was plotted vs. the respective control set to 1; the RpL32/rp49 gene expression was used as input reference. Bars, ± SD; n ≥ 2; *P < 0.05; **P < 0.01.
Gene expression analysis after cncC/Nrf2 or foxo downregulation in Opa1 KD background revealed increased proteasomal genes expression in both cases; whereas autophagy/mitophagy related genes were downregulated after cncC KD and upregulated after foxo KD (Fig. 5A). As in the Marf KD background, downregulation of cncC/Nrf2 parallel to Opa1 KD tended to increase ROS levels (Fig. 5B), with no activated proteasomal peptidases (Fig. 5C), culminating in proteome over-ubiquitination (Fig. 5D) and highly elevated cathepsins activity (Fig. 5E). Moreover, cncC/Nrf2 KD increased mitochondria fragmentation of Opa1 KD larvae (Fig. 5F and G). KD of foxo in the Opa1 KD background decreased ROS (Fig. 5B) and activated C-L proteasomal activity (Fig. 5C); yet, it resulted in increased proteome ubiquitination (Fig. 5D) and it abolished cathepsins activation (Fig. 5E). Downregulation of Opa1 in Drosophila resulted in foxo activation as evidenced by foxo accumulation (Figs. S8A and S8B), as well as by the upregulation of foxo transcriptional target genes in Opa1 KD flies (Figs. S8C and S8D), suggesting that upon loss of Opa1 foxo is being activated to signal the upregulation of catabolic processes. Finally, suppression of either cncC/Nrf2 or foxo was extremely toxic in Opa1 KD flies (Fig. 5H and I).
Fig. 5.
Cytoprotective cell responses after Opa1 KD in young flies are mediated by cncC/Nrf2 and foxo; combined Opa1 and cncC/Nr2 or foxo KD exaggerates the Opa1 KD-mediated toxicity. (A) Relative expression of shown genes in young flies following combined ubiquitous expression (or not) of the shown transgenes. (B–E) Relative (%) ROS levels (B), proteasome activities (C), proteome ubiquitination (GAPDH was used as loading reference) (D) and cathepsins B, L (E) activity in the shown transgenic lines. (F) CLSM viewing of MitoGFP in larvae muscle (Gal4Mef2) of the indicated genotypes. (G) Relative quantification of mitochondria density (number/area) and length in larvae muscles of the indicated genotypes. (H, I) Longevity curves of the shown transgenic lines; shaded rectangles highlight the accelerated toxicity (vs. Opa1 KD) in young flies co-expressing the shown transgenes. Statistics of the longevity curves are reported in Table S1. In (A–E) transgenes were induced for 7 days. In (A–E), (H), (I) the Gal4GS−Tub driver was used. Gene expression was plotted vs. the respective control set to 1; the RpL32/rp49 gene expression was used as input reference. Bars, ± SD; n ≥ 2; *P < 0.05; **P < 0.01.
To further demonstrate the protective role of proteostatic modules upon imbalanced mitochondrial dynamics we treated Opa1 and Marf transgenic lines with the proteasome inhibitor PS-341 (1 μM; [27]). We found that proteasome inhibition enhanced growth retardation and caused lethality in all transgenic animals, with the notable exception of the Opa1 overexpressing larvae (not shown). Gene expression analysis after targeted expression of the transgenes in muscles revealed that PS-341 enhanced the expression of proteostatic genes under conditions of Marf or Opa1 OE but not after Marf or Opa1 KD (Figs. S9A and S9B) and induced proteome over-ubiquitination (Fig. S9C) indicating that stress sensors (e.g. cncC/Nrf2 or foxo) remain largely inactive under severe disruption of cellular homeostasis, i.e. combined loss-of both proteostasis and mitostasis. Exposure of Marf KD flies to PS-341 reduced mitochondria numbers and MitoGFP aggregates likely due to enhanced mitophagy [28] and increased mitochondrial fragmentation in Opa1 KD flies (Figs. S10A and S10B); also, it accelerated the mortality rates of both Marf and Opa1 mutants (Figs. S10C–S10F, Table S1). In support, KD of the rate limiting for protein degradation Prosβ5 proteasomal subunit in the Marf KD background caused activation of proteostatic modules and proteome over-ubiquitination (Figs. S9D and S9E), extreme growth retardation and lethality. Abolishment of proteasome activity in Marf KD larvae also resulted in increased clustering of mitochondria, which co-localized with ref(2)P (Fig. S9F); the latter is recruited to damaged and depolarized mitochondria likely mediating their aggregation for mitophagy [28].
Overall, imbalance of mitochondrial dynamics activates cncC/Nrf2 and foxo to induce cytoprotective genomic responses that in the short term contribute to organismal survival.
3.4. Treatment of flies with antioxidants mildly ameliorated the reduced mitochondrial fusion rates-mediated progeria
Given that mitochondrial malfunction is a hallmark of aging and of several age-related diseases [2] we asked whether the deleterious effects of imbalanced mitochondrial dynamics could be ameliorated by cncC/Nrf2-mediated forced activation of cytoprotective antioxidant/proteostatic (e.g. UPP) modules. We found that cncC/Nrf2 OE in Marf KD or Opa1 KD backgrounds abolished ROS accumulation (Fig. 6A; see also Fig. 3A) and it significantly upregulated proteostatic and mitochondrial biogenesis genes (Fig. S11), proteasome subunits expression (Fig. 6D) and proteasome activities (Fig. 6B); with no effect on lysosomal cathepsins activity (Fig. 6C). Furthermore, CLSM analysis revealed that although cncC/Nrf2 OE increased the number of mitochondria in Marf or Opa1 KD backgrounds their fragmentation persisted (Fig. 6E and F). Yet, despite the activation of cytoprotective modules, cncC/Nrf2 OE shortened the lifespan of both Marf KD (Fig. 6G) and Opa1 KD flies (Fig. 6H). Next, we studied the effects of ALP activation on loss-of mitostasis by combining Marf or Opa1 KD with Atg8a OE. Atg8a OE in the Marf KD background could not overcome (or delay) Marf KD-mediated larval lethality (not shown); elevated ROS levels (Fig. S12A), had no effect on lysosomal cathepsins activity (Fig. S12B) and it induced proteasomal activities (Fig. S12C). Also, it further reduced median longevity of the Marf KD flies (Fig. S12D). CLSM analysis in Opa1 KD larvae showed that the mitochondrial morphology was improved after Atg8a OE (Fig. S13A). Additionally, Atg8a OE in Opa1 KD young flies increased ROS accumulation (Fig. S13B) and it suppressed proteasome (Fig. S13C) and cathepsins activity (Fig. S13D); moreover, Atg8a OE could not improve the lifespan/healthspan of Opa1 KD flies (Fig. S13E). Consistently, treatment of Opa1 KD flies with autophagic inducers, namely rapamycin (Fig. S13F) or caloric restriction (Fig. S13G) was either toxic or had no beneficial effects. These findings indicate that activation of the highly energetically demanding catabolic proteostatic modules likely accelerates the exhaustion of cellular energetics in organisms bearing dysfunctional mitochondria.
Fig. 6.
Combined cncC/Nrf2 OE and Marf or Opa1 KD activates cytoprotective antioxidant/proteostatic modules; yet, it cannot mitigate the imbalanced mitochondrial dynamics-mediated toxicity. (A–C) Relative (%) ROS levels (A), proteasome (B) and cathepsins B, L (C) activities of the shown young transgenic flies. (D) Immunoblot analysis of young flies' tissue protein samples probed with antibodies against proteasomal subunits 20S-α and Prosβ5; GAPDH was used as reference. (E) CLSM imaging of mitochondria (MitoGFP) in larvae muscle (Gal4Mef2) of the indicated genotypes. (F) Relative quantification of mitochondria density (number/area) and length in larvae muscle (Gal4Mef2) of the indicated genotypes. (G, H) Longevity curves of the indicated transgenic lines; the shaded rectangle in (G) highlights the increased toxicity seen in young flies after cncC/Nrf2 OE at the Marf KD background. In (A–D) flies were treated with RU486 for 7 days. Statistics of the longevity curves are reported in Table S1. In (A–D), (G), (H) the Gal4GS−Tub driver was used. Bars, ± SD; n ≥ 2; *P < 0.05; **P < 0.01.
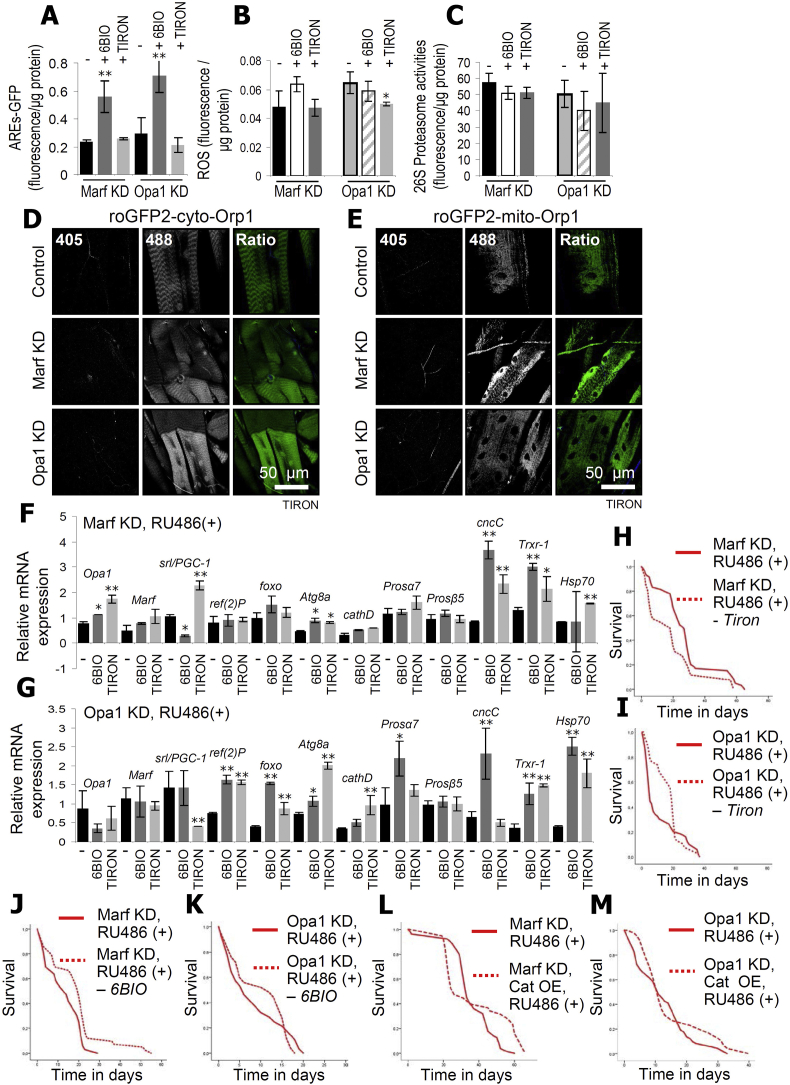
Given the sustained organismal oxidative stress due to mitochondrial dynamics imbalance we then asked whether concomitant treatment with small molecules that act either as ROS scavengers (Tiron; [40]) or activate antioxidant pathways (6BIO; [41,42]) could ameliorate the phenotypes of Marf or Opa1 KD flies. As shown before [42], treatment with 6BIO activated AREs (Fig. 7A), while both 6BIO and Tiron tended (not reaching statistical significance) to suppress ROS levels (Fig. 7B) and proteasome activity (Fig. 7C) in Opa1 KD flies indicating reduced oxidative/proteostatic stress. The effects of Tiron were validated by analyzing the in vivo excitation ratio of the roGFP2 reporters in the cytosol and mitochondria of larvae muscles. CLSM analysis showed that Tiron reduced the excitation at 405 nm and thus the H2O2 levels after Marf or Opa1 KD in flies (Fig. 7D and E). Despite compound-specific genomic responses, 6BIO and Tiron induced the expression of genes involved in antioxidant (Trxr-1) and proteostatic (Atg8a, Hsp70) responses (Fig. 7F and G). Treatment with Tiron mildly improved the median lifespan of Opa1 KD (but not Marf KD) flies (Fig. 6, Fig. 7BIO improved the median lifespan in both Marf and Opa1 KD transgenic lines (Fig. 7J and K). Consistently, coexpression of Catalase [43] in Marf or Opa1 KD genetic backgrounds tended to ameliorate the toxic effects induced at the organismal level by imbalanced mitochondrial dynamics (Fig. 7L, M).
Fig. 7.
Treatment with 6BIO and Tiron reduce oxidative load and activate antioxidant responses at the Marf and Opa1 KD genetic backgrounds. AREs-GFP (A), ROS levels (B) and proteasome activities (C) following ubiquitous Marf or Opa1 KD and treatment (or not) with 6BIO or Tiron. (D, E) CLSM visualization of the cyto-roGFP-Orp1 (D) and mito-roGFP2-Orp1 (E) reporters in larvae muscle, after ubiquitous expression (Gal4Tub) of Marf or Opa1 RNAi transgenes and co-treatment with Tiron. (F, G) Relative expression levels of proteostatic and mitostatic genes in somatic tissues of transgenic flies after ubiquitous expression of Marf (F) or Opa1 (G) RNAi transgenes and co-treatment with 6BIO or Tiron. (H–M) Longevity curves of the indicated transgenic flies after treatment with Tiron (H, I), 6BIO (J, K), or Catalase co-overexpression (L, M); statistics of the shown longevity curves are reported in Table S1. 6BIO was used at a concentration of 200 μM and Tiron of 5 mM. In (A–C), (F–M) the Gal4GS−Tub driver was used. Bars, ± SD; n ≥ 2; *P < 0.05; **P < 0.01.
Thus, perturbation of mitochondrial dynamics is not tolerated in higher metazoans and only treatment with antioxidants can mildly mitigate the induced toxicity.
4. Discussion
We report here that prolonged suppression of mitochondrial fusion or Lon activity in Drosophila caused developmental lethality and when induced in the adult oxidative and proteotoxic stress, neuromuscular defects and accelerated aging. These effects were more intense (especially in high calories diet) after Opa1 KD which induced metabolic deregulation, severe disruption of cristae structure, respiratory deficits and reduced mitochondria number. Consistently, mitochondrial malfunction has been associated with senescence, aging and age-related diseases [2,7,12]. Marf depletion was also found in previous studies to cause lethality in flies [44] and a heterozygous mutation of Opa1 shortened lifespan in Drosophila through increased ROS production [38]. Moreover, depletion of the mitochondrial rhomboid protein (required for mitochondrial fusion during fly spermatogenesis and muscle maturation), led to severe neurological defects, ADOA-like degeneration of photoreceptors and reduced lifespan [45]; while muscle-specific deletion of Opa1 in adult mice induced a precocious senescence phenotype and premature death [46]. Under our experimental setting (inducible transgene expression in identical genotypes) increased fusion rates due to Marf, Opa1 OE or Drp1 KD, although by far less toxic as compared to conditions of increased fission, also decreased longevity suggesting that sustained deregulation of mitochondrial dynamics is toxic. In support, diseases caused by mutations in fission proteins are very rare. A mutation in Drp1 has been associated with a severe type of infantile neurodegeneration in humans [47], while depletion of Drp1 caused embryonic lethality in mice [48]. Notably, reduced mitochondrial fission increased lifespan and resistance to apoptosis in two fungal models [49], whereas Drp1 depletion prolonged longevity but not stress resistance of C. elegans with reduced insulin signaling [50]. Also, enhancing Marf ameliorated neuromuscular dysfunction in Drosophila models of TDP-43 proteinopathies [51], while ablation of Drp1 in mouse liver protected mice from diet-induced obesity [52]; finally, mild Opa1 OE ameliorated the phenotype of two mitochondrial disease mouse models [53]. Thus, the beneficial effects of increased mitochondrial fusion are more evident under stress conditions.
Interestingly, the toxic effects of Opa1 KD were severely enhanced in male flies. Similarly, sex differences have been reported in Opa1+/− flies [38]; yet, these observations were limited to complex II activity. Leber hereditary optic neuropathy (LHON) and ADOA are the two most common inherited optic neuropathies in the population where most affected families harbor mutations in the Opa1 gene. One of the key features of LHON is the gender bias in disease predisposition, with ∼50% of males and only ∼10% of female carriers’ eventually losing vision in their lifetime [54]. Although it is established that mitochondria contribute significantly to sexual differences during aging and this impact likely relates to their maternal inheritance, further studies are needed to clarify the molecular basis of the sex-dependent Opa1 KD phenotypes.
Our observations clearly support a direct cross-talk and functional wiring of mitostatic and proteostatic modules, as loss-of mitostasis (e.g. due to imbalanced mitochondrial dynamics or Lon KD) increased oxidative stress and proteome instability causing the activation of Nrf2 and foxo to trigger in the young organism the upregulation of cytoprotective antioxidant/proteostatic modules. In line with previous findings, these were the antioxidant/UPP pathway for cncC/Nrf2 [20] and autophagy for foxo [39] suggesting the existence of distinct transcriptional targets of the cncC/Nrf2 and foxo axes upon loss-of mitostasis. In support, foxo mediated the UPR and activated autophagy in Opa1−/− mice [46]. cncC/Nrf2 KD at the MarfRNAi background led to significant upregulation of the ref(2)P and Hsp70 genes likely indicating increased mitophagy and also that upon loss-of cncC/Nrf2 activity the PN branch of molecular chaperons takes over. Beyond activation of antioxidant/proteostatic modules, our data also indicated that Nrf2 regulates mitostatic modules and mitochondria number. Consistently, recent reports suggested that Nrf2 controls substrate availability for mitochondrial respiration [55], upregulates electron transport-related genes [56] and affects mitochondria biogenesis [29]; we have fully confirmed these findings in the fly model [27,28,30].
This regulatory circuit and constant monitoring of mitostasis is critical in ensuring organismal survival; yet, our finding suggest that depending on the extent and duration of the damage, loss-of mitostasis can be transiently tolerable in young organisms. Notably, the Nrf2 functionality declines during aging [27,57] and a similar age-related decline in responsiveness was evident in other stress-responsive signaling pathways, such as heat shock factor-1 and hypoxia-inducible factor-1 [58,59]. The noted (yet, not understood in its molecular basis) age-related dysfunction of stress sensors and of most (if not all) proteostatic pathways [16,20,32] likely contributes greatly to the dramatic toxicity of loss-of mitostasis events in aged flies. In support to this notion, we found that proteasome inhibition or cncC/Nrf2, foxo KD in young flies drastically exaggerated the toxicity of imbalanced mitochondrial dynamics. Consistently, to the vital role of UPP in retaining mitostasis, UPP mediates the degradation of IMM space proteins before they arrive at mitochondria [60] and of damaged mitochondrial proteins of the OMM [61]. Also, ubiquitination of Mfns leads to either their proteasomal breakdown or to enhanced mitochondrial fusion [23]. Our recent studies in the fly model have also revealed that proteasome malfunction causes severe loss-of mitostasis, as evidenced by disruption of mitochondrial dynamics, energetics and cristae structure as well as by increased mitophagy [30]. These data suggest that the functionality of antioxidant/proteostatic and mitostatic cellular modules is mutually interdependent providing thus a reasonable explanation for their extended functional wiring in the young organism; our observation that Marf and Opa1 are negative regulators of UPP suggests the existence of direct regulatory links even in the absence of stress. Thus, the stochastic unrepaired dysfunction of even one of these integrated modules will slowly, but inevitably, affect all others resulting in the accelerated collapse of the cellular functionality boundary below a threshold that keeps organism young and disease free.
Given these notions it is not surprising that in our experimental setting loss-of mitostasis is essentially not rescuable; supportively, there are no effective treatments for patients with LHON and DOA [62]. Neither cytoprotective cncC/Nrf2 OE (with concomitant proteasome activation) nor autophagy upregulation could prevent the damaging effects of imbalanced mitochondrial dynamics and in fact even worsened toxicity. We hypothesize that likely this outcome relates to the fact that by being highly energetically demanding, these catabolic processes (UPP, ALP) accelerate the exhaustion of cellular energetics in young organisms bearing dysfunctional mitochondria. In support, we recently found that Nrf2 OE in the fly model increases stress tolerance at the cost of aging acceleration due to metabolic deregulation that leads to Diabetes Type 1 hallmarks [28]. Notably, under conditions of normal mitochondrial functionality mild cncC/Nrf2 or UPP activation can confer protection to stress and even extend the longevity of model organisms [20,28,63,64]; similarly, ALP activation exerted beneficial effects in neurodegeneration models of flies and mice [16,32] and had a renoprotective role in diabetic nephropathy [65]. Also, dietary interventions e.g. mild caloric restriction [66] or treatment with rapamycin [67] that culminate in autophagy activation, extended healthspan of model organisms and delayed accelerated aging; they also improved neuronal function and alleviated genomic stress in DNA repair-deficient mice [68].
Interestingly, genetic or pharmacological suppression of endoplasmic reticulum stress attenuated the lethal defects caused by Marf depletion in Drosophila [44] and prevented muscle atrophy and premature death caused by muscle-specific Opa1 deletion in mice [46]. Consistently, we found that concomitant treatment with small molecules that either act as ROS scavengers (Tiron) or activate antioxidant pathways (6BIO) could mildly mitigate the deleterious effects induced by imbalanced mitochondrial dynamics; these findings were validated by Catalase OE in the genetic backgrounds of Marf or Opa1 KD. In support, a quinone analogue, namely idebenone was shown to partially improve visual function in patients with LHON [69]. Also, vitamin E reversed the glossy eye phenotype caused by eye-targeted expression of mutant Opa1CG8479 in flies [70] and antioxidant treatment partially restored lifespan in Opa1 mutant male flies [38]. Previous studies have shown that 6BIO (an inhibitor of the pleiotropic Gsk-3 enzyme) reduced oxidative stress and exerted anti-aging effects in both the fly model [42] and in normal human cells [71]; worth mentioning is that 6BIO also mildly suppress the insulin/IGF-like axis suggesting pleiotropic effects on cell signaling pathways [42]. Similarly, treatment of flies with another Gsk-3 inhibitor, namely lithium extended flies’ longevity [41]. Nonetheless, whether mild activation of antioxidant/proteostatic pathways can be used to alleviate the adverse effects of mitopathies should be further investigated.
Taken together, our findings indicate that despite immediate cytoprotective responses being triggered in the young organism, due of the extensive wiring and functional integration of mitostatic and antioxidant/proteostatic modules, sustained loss-of mitostasis eventually exhausts the organismal capacity to adapt resulting in accelerated collapse of cellular homeodynamics and reduced cellular/organismal viability. Interestingly, our data suggest that antioxidants likely represent promising modalities for alleviating the severe effects of mitopathies.
Author contributions
IPT designed and supervised the study; SG, ZE, ENT and IPT conducted experiments or interpreted the data; LS contributed reagents, materials, analysis tools; IPT wrote the manuscript.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgements
We acknowledge Maria Eugenia Soriano Garcìa-Cuerva (University of Padua, Italy) for assisting during mitochondria isolation and respiration analysis. We also thank Maria Figueiredo-Pereira (Hunter College, NY, USA) for the Prosβ5 antibody, as well as, Profs Dirk Bohmann (University of Rochester, NY, USA), Jongkyeong Chung (Seoul National University, Korea), Andrea Daga (University of Padua, Italy), Gábor Juhász (Eotvos Lorand University, Hungary) and Alexios-Leandros Skaltsounis (National and Kapodistrian University of Athens, Greece) for fly lines and reagents. Zoi Evangelakou is a recipient of the Ph.D. fellowship from the Hellenic Foundation for Research and Innovation (H.F.R.I. GA 1869). IPT acknowledges funding from the EU project TASCMAR (EU-H2020/634674) and the Hellenic GSRT project BIOIMAGING-GR (MIS 5002755).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101219.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sala A.J., Bott L.C., Morimoto R.I. Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol. 2017;216:1231–1241. doi: 10.1083/jcb.201612111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sala, A.J., Bott, L.C., and Morimoto, R.I. (2017). Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol. 216, 1231-1241. [DOI] [PMC free article] [PubMed]
- 2.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lopez-Otin, C., Blasco, M.A., Partridge, L., Serrano, M., and Kroemer, G. (2013). The hallmarks of aging. Cell 153, 1194-1217. [DOI] [PMC free article] [PubMed]
- 3.Friedman J.R., Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]; Friedman, J.R., and Nunnari, J. (2014). Mitochondrial form and function. Nature 505, 335-343. [DOI] [PMC free article] [PubMed]
- 4.Spinelli J.B., Haigis M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018;20:745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Spinelli, J.B., and Haigis, M.C. (2018). The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 20, 745-754. [DOI] [PMC free article] [PubMed]
- 5.Pernas L., Scorrano L. Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu. Rev. Physiol. 2016;78:505–531. doi: 10.1146/annurev-physiol-021115-105011. [DOI] [PubMed] [Google Scholar]; Pernas, L., and Scorrano, L. (2016). Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annu. Rev. Physiol. 78, 505-531. [DOI] [PubMed]
- 6.Eisner V., Picard M., Hajnóczky G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat. Cell Biol. 2018;20:755–765. doi: 10.1038/s41556-018-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eisner, V., Picard, M., and Hajnoczky, G. (2018). Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat. Cell Biol. 20, 755-765. [DOI] [PMC free article] [PubMed]
- 7.Gumeni S., Trougakos I.P. Cross talk of proteostasis and mitostasis in cellular homeodynamics, ageing, and disease. Oxid. Med. Cell Longev. 2016;4587691 doi: 10.1155/2016/4587691. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gumeni, S., and Trougakos, I.P. (2016). Cross Talk of Proteostasis and Mitostasis in Cellular Homeodynamics, Ageing, and Disease. Oxid. Med. Cell Longev. 4587691. [DOI] [PMC free article] [PubMed]
- 8.Klionsky D.J. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12 doi: 10.4161/auto.36187. 1-222 third ed. [DOI] [PMC free article] [PubMed] [Google Scholar]; Klionsky, D.J., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1-222 [DOI] [PMC free article] [PubMed]
- 9.Cogliati S., Enriquez J.A., Scorrano, Mitochondrial Cristae L. Where beauty meets functionality. Trends Biochem. Sci. 2016;41:261–273. doi: 10.1016/j.tibs.2016.01.001. [DOI] [PubMed] [Google Scholar]; Cogliati, S., Enriquez, J.A., and Scorrano, L. Mitochondrial Cristae. (2016). Where Beauty Meets Functionality. Trends Biochem. Sci. 41, 261-273. [DOI] [PubMed]
- 10.Burté F., Carelli V., Chinnery P.F., Yu-Wai-Man P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 2015;11:11–24. doi: 10.1038/nrneurol.2014.228. [DOI] [PubMed] [Google Scholar]; Burte, F., Carelli, V., Chinnery, P.F., and Yu-Wai-Man, P. (2015). Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 11, 11-24 [DOI] [PubMed]
- 11.van der Bliek A.M., Shen Q., Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a011072. pii: a011072. [DOI] [PMC free article] [PubMed] [Google Scholar]; van der Bliek, A.M., Shen, Q., and Kawajiri, S. (2013). Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5, pii: a011072. [DOI] [PMC free article] [PubMed]
- 12.Suomalainen A., Battersby B.J. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell Biol. 2018;19:77–92. doi: 10.1038/nrm.2017.66. [DOI] [PubMed] [Google Scholar]; Suomalainen, A., and Battersby, B.J. (2018). Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell. Biol. 19, 77-92. [DOI] [PubMed]
- 13.Quirós P.M., Langer T., López-Otín C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]; Quiros, P.M., Langer, T., and Lopez-Otin, C. (2015). New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 16, 345-359. [DOI] [PubMed]
- 14.Friguet B., Bulteau A.L., Petropoulos I. Mitochondrial protein quality control: implications in ageing. Biotechnol. J. 2008;3:757–764. doi: 10.1002/biot.200800041. [DOI] [PubMed] [Google Scholar]; Friguet, B., Bulteau, A.L., and Petropoulos, I. (2008). Mitochondrial protein quality control: implications in ageing. Biotechnol J. 3, 757-764. [DOI] [PubMed]
- 15.Trougakos I.P., Sesti F., Tsakiri E., Gorgoulis V.G. Non-enzymatic post-translational protein modifications and proteostasis network deregulation in carcinogenesis. J. Proteomics. 2013;92:274–298. doi: 10.1016/j.jprot.2013.02.024. [DOI] [PubMed] [Google Scholar]; Trougakos, I.P., Sesti, F., Tsakiri, E., and Gorgoulis, V.G. (2013). Non-enzymatic post-translational protein modifications and proteostasis network deregulation in carcinogenesis. J. Proteomics 92, 274-298. [DOI] [PubMed]
- 16.Klaips C.L., Jayaraj G.G., Hartl F.U. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 2017;217:51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]; Klaips, C.L., Jayaraj, G.G., and Hartl, F.U. (2017). Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 217, 51-63. [DOI] [PMC free article] [PubMed]
- 17.Kaushik S., Cuervo A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018;19:365–381. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kaushik, S., and Cuervo, A.M. (2018). The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365-381. [DOI] [PMC free article] [PubMed]
- 18.Ding W.X., Yin X.M. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012;393:547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ding, W.X., and Yin, X.M. (2012). Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 393, 547-564. [DOI] [PMC free article] [PubMed]
- 19.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., Youle R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lazarou, M., Sliter, D.A., Kane, L.A., Sarraf, S.A., Wang, C., Burman, J.L., Sideris, D.P., Fogel, A.I., and Youle, R.J. (2015). The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309-314. [DOI] [PMC free article] [PubMed]
- 20.Tsakiri E.N., Trougakos I.P. The amazing ubiquitin-proteasome system: structural components and implication in aging. Int. Rev. Cell Mol. Biol. 2015;314:171–237. doi: 10.1016/bs.ircmb.2014.09.002. [DOI] [PubMed] [Google Scholar]; Tsakiri, E.N., and Trougakos, I.P. (2015). The amazing ubiquitin-proteasome system: structural components and implication in aging. Int. Rev. Cell Mol. Biol. 314, 171-237. [DOI] [PubMed]
- 21.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]; Finley, D. (2009). Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477-513. [DOI] [PMC free article] [PubMed]
- 22.Neutzner A., Youle R.J., Karbowski M. Outer mitochondrial membrane protein degradation by the proteasome. Novartis Found. Symp. 2007;287:4–14. discussion 14-20. [PubMed] [Google Scholar]; Neutzner, A., Youle, R.J., and Karbowski, M. (2007). Outer mitochondrial membrane protein degradation by the proteasome. Novartis Found. Symp. 287, 4-14; discussion 14-20. [PubMed]
- 23.Franz A., Kevei É., Hoppe T. Double-edged alliance: mitochondrial surveillance by the UPS and autophagy. Curr. Opin. Cell Biol. 2015;37:18–27. doi: 10.1016/j.ceb.2015.08.004. [DOI] [PubMed] [Google Scholar]; Franz, A., Kevei, E., and Hoppe, T. (2015). Double-edged alliance: mitochondrial surveillance by the UPS and autophagy. Curr Opin Cell Biol. 37, 18-27. [DOI] [PubMed]
- 24.Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., Youle R.J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tanaka, A., Cleland, M.M., Xu, S., Narendra, D.P., Suen, D.F., Karbowski, M., and Youle, R.J. (2010). Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191, 1367-1380. [DOI] [PMC free article] [PubMed]
- 25.Sanchez A.M., Candau R.B., Bernardi H. FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell. Mol. Life Sci. 2014;71:1657–1671. doi: 10.1007/s00018-013-1513-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sanchez, A.M., Candau, R.B., and Bernardi, H. (2014). FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell. Mol. Life Sci. 71, 1657-1671. [DOI] [PMC free article] [PubMed]
- 26.Sykiotis G.P., Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci. Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sykiotis, G.P., and Bohmann, D. (2010). Stress-activated cap'n'collar transcription factors in aging and human disease. Sci. Signal. 3: re3. [DOI] [PMC free article] [PubMed]
- 27.Tsakiri E.N., Sykiotis G.P., Papassideri I.S., Terpos E., Dimopoulos M.A., Gorgoulis V.G., Bohmann D., Trougakos I.P. Proteasome dysfunction in Drosophila signals to an Nrf2-dependent regulatory circuit aiming to restore proteostasis and prevent premature aging. Aging Cell. 2013;12:802–813. doi: 10.1111/acel.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsakiri, E.N., Sykiotis, G.P., Papassideri, I.S., Terpos, E., Dimopoulos, M.A., Gorgoulis, V.G., Bohmann, D., and Trougakos, I.P. (2013). Proteasome dysfunction in Drosophila signals to an Nrf2-dependent regulatory circuit aiming to restore proteostasis and prevent premature aging. Aging Cell 12, 802-813. [DOI] [PMC free article] [PubMed]
- 28.Tsakiri E.N., Gumeni S., Iliaki K.K., Benaki D., Vougas K., Sykiotis G.P., Gorgoulis V.G., Mikros E., Scorrano L., Trougakos I.P. Hyperactivation of Nrf2 increases stress tolerance at the cost of aging acceleration due to metabolic deregulation. Aging Cell. 2019;18 doi: 10.1111/acel.12845. e12845. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsakiri, E.N., Gumeni, S., Iliaki, K.K., Benaki, D., Vougas, K., Sykiotis, G.P., Gorgoulis, V.G., Mikros, E., Scorrano, L., Trougakos, I.P. (2019). Hyperactivation of Nrf2 increases stress tolerance at the cost of aging acceleration due to metabolic deregulation. Aging Cell 18, e12845. [DOI] [PMC free article] [PubMed]
- 29.Palikaras K., Lionaki E., Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521:525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]; Palikaras, K., Lionaki, E., and Tavernarakis, N. (2015). Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525-528. [DOI] [PubMed]
- 30.Tsakiri E.N., Gumeni S., Vougas K., Pendin D., Papassideri I., Daga A., Gorgoulis V., Juhász G., Scorrano L., Trougakos I.P. Proteasome dysfunction induces excessive proteome instability and loss of mitostasis that can be mitigated by enhancing mitochondrial fusion or autophagy. Autophagy. 2019;19:1–17. doi: 10.1080/15548627.2019.1596477. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsakiri, E.N., Gumeni, S., Vougas, K., Pendin, D., Papassideri I., Daga, A., Gorgoulis, V., Juhasz, G., Scorrano, L., and Trougakos, I.P. (2019). Proteasome dysfunction induces excessive proteome instability and loss of mitostasis that can be mitigated by enhancing mitochondrial fusion or autophagy. Autophagy 19, 1-17. [DOI] [PMC free article] [PubMed]
- 31.Grune T. Oxidative stress, aging and the proteasomal system. Biogerontology. 2000;1:31–40. doi: 10.1023/a:1010037908060. [DOI] [PubMed] [Google Scholar]; Grune, T. (2000). Oxidative stress, aging and the proteasomal system. Biogerontology 1, 31-40. [DOI] [PubMed]
- 32.Kaushik S., Cuervo A.M. Proteostasis and aging. Nat. Med. 2015;21:1406–1415. doi: 10.1038/nm.4001. [DOI] [PubMed] [Google Scholar]; Kaushik, S., and Cuervo, A.M. (2015). Proteostasis and aging. Nat. Med. 21, 1406-1415. [DOI] [PubMed]
- 33.Sklirou A., Papanagnou E.D., Fokialakis N., Trougakos I.P. Cancer chemoprevention via activation of proteostatic modules. Cancer Lett. 2018;413:110–121. doi: 10.1016/j.canlet.2017.10.034. [DOI] [PubMed] [Google Scholar]; Sklirou, A., Papanagnou, E.D., Fokialakis, N., and Trougakos, I.P. (2018). Cancer chemoprevention via activation of proteostatic modules. Cancer Lett. 413, 110-121. [DOI] [PubMed]
- 34.Franceschi C., Garagnani P., Morsiani C., Conte M., Santoro A., Grignolio A., Monti D., Capri M., Salvioli S. The continuum of aging and age-related diseases: common mechanisms but different rates. Front. Med. 2019;12(5):61. doi: 10.3389/fmed.2018.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]; Franceschi, C., Garagnani, P., Morsiani, C., Conte, M., Santoro, A., Grignolio, A., Monti, D., Capri, M., Salvioli, S. (2019) The Continuum of Aging and Age-Related Diseases: Common Mechanisms but Different Rates. Front Med (Lausanne). 12, 5:61. [DOI] [PMC free article] [PubMed]
- 35.Tsakiri E.N., Sykiotis G.P., Papassideri I.S., Gorgoulis V.G., Bohmann D., Trougakos I.P. Differential regulation of proteasome functionality in reproductive vs. somatic tissues of Drosophila during aging or oxidative stress. FASEB J. 2013;27:2407–2420. doi: 10.1096/fj.12-221408. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsakiri, E.N., Sykiotis, G.P., Papassideri, I.S., Gorgoulis, V.G., Bohmann, D., and Trougakos, I.P. (2013). Differential regulation of proteasome functionality in reproductive vs. somatic tissues of Drosophila during aging or oxidative stress. FASEB J. 27, 2407-2420. [DOI] [PMC free article] [PubMed]
- 36.Trougakos I.P., Margaritis L.H. Immunolocalization of the temporally "early" secreted major structural chorion proteins, Dvs38 and Dvs36, in the eggshell layers and regions of Drosophila virilis. J. Struct. Biol. 1998;123:111–123. doi: 10.1006/jsbi.1998.4028. [DOI] [PubMed] [Google Scholar]; Trougakos, I.P., and Margaritis, L.H. (1998). Immunolocalization of the temporally "early" secreted major structural chorion proteins, Dvs38 and Dvs36, in the eggshell layers and regions of Drosophila virilis. J. Struct. Biol. 123, 111-123. [DOI] [PubMed]
- 37.Albrecht S.C., Barata A.G., Grosshans J., Teleman A.A., Dick T.P. In vivo mapping of hydrogen peroxide and oxidized glutathione reveals chemical and regional specificity of redox homeostasis. Cell Metab. 2011;14:819–829. doi: 10.1016/j.cmet.2011.10.010. [DOI] [PubMed] [Google Scholar]; Hanson, G.T., Aggeler, R., Oglesbee, D., Cannon, M., Capaldi, R.A., Tsien, R.Y., Remington, S.J. (2004) Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 279(13), 13044-13053. [DOI] [PubMed]
- 38.Tang S., Le P.K., Tse S., Wallace D.C., Huang T. Heterozygous mutation of Opa1 in Drosophila shortens lifespan mediated through increased reactive oxygen species production. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004492. e4492. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tang, S., Le, P.K., Tse, S., Wallace, D.C., and Huang, T. (2009). Heterozygous mutation of Opa1 in Drosophila shortens lifespan mediated through increased reactive oxygen species production. PLoS One 4, e4492. [DOI] [PMC free article] [PubMed]
- 39.Webb A.E., Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem. Sci. 2014;39:159–169. doi: 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Webb, A.E., and Brunet, A. (2014). FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 39, 159-169. [DOI] [PMC free article] [PubMed]
- 40.Han Y.H., Park W.H. Tiron, a ROS scavenger, protects human lung cancer Calu-6 cells against antimycin A-induced cell death. Oncol. Rep. 2009;21:253–261. [PubMed] [Google Scholar]; Han, Y.H., and Park, W.H. (2009). Tiron, a ROS scavenger, protects human lung cancer Calu-6 cells against antimycin A-induced cell death. Oncol. Rep. 21, 253-261. [PubMed]
- 41.Castillo-Quan J.I., Li L., Kinghorn K.J., Ivanov D.K., Tain L.S., Slack C., Kerr F., Nespital T., Thornton J., Hardy J., Bjedov I., Partridge L. Lithium promotes longevity through GSK3/NRF2-dependent hormesis. Cell Rep. 2016;15:638–650. doi: 10.1016/j.celrep.2016.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]; Castillo-Quan, J.I., Li, L., Kinghorn, K.J., Ivanov, D.K., Tain, L.S., Slack, C., Kerr, F., Nespital, T., Thornton, J., Hardy, J., Bjedov, I., and Partridge, L. (2016). Lithium Promotes Longevity through GSK3/NRF2-Dependent Hormesis. Cell Rep. 15, 638-650. [DOI] [PMC free article] [PubMed]
- 42.Tsakiri E.N., Gaboriaud-Kolar N., Iliaki K.K., Tchoumtchoua J., Papanagnou E.D., Chatzigeorgiou S., Tallas K.D., Mikros E., Halabalaki M., Skaltsounis A.L., Trougakos I.P. The indirubin derivative 6-bromoindirubin-3'-oxime activates proteostatic modules, reprograms cellular bioenergetic pathways, and exerts antiaging effects. Antioxidants Redox Signal. 2017;27:1027–1047. doi: 10.1089/ars.2016.6910. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsakiri, E.N., Gaboriaud-Kolar, N., Iliaki, K.K., Tchoumtchoua, J., Papanagnou, E.D., Chatzigeorgiou, S., Tallas, K.D., Mikros, E., Halabalaki, M., Skaltsounis, A.L., and Trougakos, I.P. (2017). The Indirubin Derivative 6-Bromoindirubin-3'-Oxime Activates Proteostatic Modules, Reprograms Cellular Bioenergetic Pathways, and Exerts Antiaging Effects. Antioxid. Redox Signal. 27, 1027-1047. [DOI] [PMC free article] [PubMed]
- 43.Orr W.C., Sohal R.S. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263(5150):1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]; Orr, W.C., Sohal, R.S.(1994) Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 263(5150), 1128-1130. [DOI] [PubMed]
- 44.Debattisti V., Pendin D., Ziviani E., Daga A., Scorrano L. Reduction of endoplasmic reticulum stress attenuates the defects caused by Drosophila mitofusin depletion. J. Cell Biol. 2014;204:303–312. doi: 10.1083/jcb.201306121. [DOI] [PMC free article] [PubMed] [Google Scholar]; Debattisti, V., Pendin, D., Ziviani, E., Daga, A., and Scorrano, L. (2014). Reduction of endoplasmic reticulum stress attenuates the defects caused by Drosophila mitofusin depletion. J. Cell Biol. 204, 303-312. [DOI] [PMC free article] [PubMed]
- 45.McQuibban G.A., Lee J.R., Zheng L., Juusola M., Freeman M. Normal mitochondrial dynamics requires rhomboid-7 and affects Drosophila lifespan and neuronal function. Curr. Biol. 2006;16:982–989. doi: 10.1016/j.cub.2006.03.062. [DOI] [PubMed] [Google Scholar]; McQuibban, G.A., Lee, J.R., Zheng, L., Juusola, M., and Freeman, M. (2006). Normal mitochondrial dynamics requires rhomboid-7 and affects Drosophila lifespan and neuronal function. Curr. Biol. 16, 982-989. [DOI] [PubMed]
- 46.Tezze C., Romanello V., Desbats M.A., Fadini G.P., Albiero M., Favaro G., Ciciliot S., Soriano M.E., Morbidoni V., Cerqua C., Loefler S., Kern H., Franceschi C., Salvioli S., Conte M., Blaauw B., Zampieri S., Salviati L., Scorrano L., Sandri M. Age-associated loss of OPA1 in muscle impacts muscle mass, metabolic homeostasis, systemic inflammation, and epithelial senescence. Cell Metabol. 2017;25:1374–1389. doi: 10.1016/j.cmet.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tezze, C., Romanello, V., Desbats, M.A., Fadini, G.P., Albiero, M., Favaro, G., Ciciliot, S., Soriano, M.E., Morbidoni, V., Cerqua, C., Loefler, S., Kern, H., Franceschi, C., Salvioli, S., Conte, M., Blaauw, B., Zampieri, S., Salviati, L., Scorrano, L., and Sandri, M. (2017). Age-Associated Loss of OPA1 in Muscle Impacts Muscle Mass, Metabolic Homeostasis, Systemic Inflammation, and Epithelial Senescence. Cell Metab. 25, 1374-1389. [DOI] [PMC free article] [PubMed]
- 47.Waterham H.R., Koster J., van Roermund C.W., Mooyer P.A., Wanders R.J., Leonard J.V. A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]; Waterham, H.R., Koster, J., van Roermund, C.W., Mooyer, P.A., Wanders, R.J. and Leonard, J.V. (2007). A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 356, 1736-1741. [DOI] [PubMed]
- 48.Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S.O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y., Taguchi N., Morinaga H., Maeda M., Takayanagi R., Yokota S., Mihara K. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]; Ishihara, N., Nomura, M., Jofuku, A., Kato, H., Suzuki, S.O., Masuda, K., Otera, H., Nakanishi, Y., Nonaka, I., Goto, Y., Taguchi, N., Morinaga, H., Maeda, M., Takayanagi, R., Yokota, S., and Mihara, K. (2009). Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11, 958-966. [DOI] [PubMed]
- 49.Scheckhuber C.Q., Erjavec N., Tinazli A., Hamann A., Nyström T., Osiewacz H.D. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 2007;9:99–105. doi: 10.1038/ncb1524. [DOI] [PubMed] [Google Scholar]; Scheckhuber, C.Q., Erjavec, N., Tinazli, A., Hamann, A., Nystrom, T., and Osiewacz, H.D. (2007). Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 9, 99-105. [DOI] [PubMed]
- 50.Yang C.C., Chen D., Lee S.S., Walter L. The dynamin-related protein DRP-1 and the insulin signaling pathway cooperate to modulate C. elegans longevity. Aging Cell. 2011;10:724–728. doi: 10.1111/j.1474-9726.2011.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang, C.C., Chen, D., Lee, S.S. and Walter, L. (2011). The dynamin-related protein DRP-1 and the insulin signaling pathway cooperate to modulate C. elegans longevity. Aging Cell 10, 724-728. [DOI] [PMC free article] [PubMed]
- 51.Khalil B., Cabirol-Pol M.J., Miguel L., Whitworth A.J., Lecourtois M., Liévens J.C. Enhancing Mitofusin/Marf ameliorates neuromuscular dysfunction in Drosophila models of TDP-43 proteinopathies. Neurobiol. Aging. 2017;54:71–83. doi: 10.1016/j.neurobiolaging.2017.02.016. [DOI] [PubMed] [Google Scholar]; Khalil, B., Cabirol-Pol, M.J., Miguel, L., Whitworth, A.J., Lecourtois, M., and Lievens, J.C. (2017). Enhancing Mitofusin/Marf ameliorates neuromuscular dysfunction in Drosophila models of TDP-43 proteinopathies. Neurobiol. Aging 54, 71-83. [DOI] [PubMed]
- 52.Wang L., Ishihara T., Ibayashi Y., Tatsushima K., Setoyama D., Hanada Y., Takeichi Y., Sakamoto S., Yokota S., Mihara K., Kang D., Ishihara N., Takayanagi R., Nomura M. Disruption of mitochondrial fission in the liver protects mice from diet-induced obesity and metabolic deterioration. Diabetologia. 2015;58:2371–2380. doi: 10.1007/s00125-015-3704-7. [DOI] [PubMed] [Google Scholar]; Wang, L., Ishihara, T., Ibayashi, Y., Tatsushima, K., Setoyama, D., Hanada, Y., Takeichi, Y., Sakamoto, S., Yokota, S., Mihara, K., Kang, D., Ishihara, N., Takayanagi, R., and Nomura, M. (2015). Disruption of mitochondrial fission in the liver protects mice from diet-induced obesity and metabolic deterioration. Diabetologia 58, 2371-2380. [DOI] [PubMed]
- 53.Civiletto G., Varanita T., Cerutti R., Gorletta T., Barbaro S., Marchet S., Lamperti C., Viscomi C., Scorrano L., Zeviani M. Opa1 overexpression ameliorates the phenotype of two mitochondrial disease mouse models. Cell Metabol. 2015;21:845–854. doi: 10.1016/j.cmet.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Civiletto, G., Varanita, T., Cerutti, R., Gorletta, T., Barbaro, S., Marchet, S., Lamperti, C., Viscomi, C., Scorrano, L., and Zeviani, M. (2015). Opa1 overexpression ameliorates the phenotype of two mitochondrial disease mouse models. Cell Metab. 21, 845-854. [DOI] [PMC free article] [PubMed]
- 54.Jurkute N., Yu-Wai-Man P. Leber hereditary optic neuropathy: bridging the translational gap. Curr. Opin. Ophthalmol. 2017;28:403–409. doi: 10.1097/ICU.0000000000000410. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jurkute, N., and Yu-Wai-Man, P. (2017). Leber hereditary optic neuropathy: bridging the translational gap. Curr. Opin. Ophthalmol. 28, 403-409. [DOI] [PMC free article] [PubMed]
- 55.Holmström K.M., Baird L., Zhang Y., Hargreaves I., Chalasani A., Land J.M., Stanyer L., Yamamoto M., Dinkova-Kostova A.T., Abramov A.Y. Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biol. Open. 2013;2:761–770. doi: 10.1242/bio.20134853. [DOI] [PMC free article] [PubMed] [Google Scholar]; Holmstrom, K.M., Baird, L., Zhang, Y., Hargreaves, I., Chalasani, A., Land, J.M., Stanyer, L., Yamamoto, M., Dinkova-Kostova, A.T., and Abramov, A.Y. (2013). Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biol. Open 2, 761-770. [DOI] [PMC free article] [PubMed]
- 56.Misra J.R., Horner M.A., Lam G., Thummel C.S. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 2011;25:1796–1806. doi: 10.1101/gad.17280911. [DOI] [PMC free article] [PubMed] [Google Scholar]; Misra, J.R., Horner, M.A., Lam, G., and Thummel, C.S. (2011). Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 25, 1796-1806. [DOI] [PMC free article] [PubMed]
- 57.Rahman M.M., Sykiotis G.P., Nishimura M., Bodmer R., Bohmann D. Declining signal dependence of Nrf2-MafS-regulated gene expression correlates with aging phenotypes. Aging Cell. 2013;12:554–562. doi: 10.1111/acel.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rahman, M.M., Sykiotis, G.P., Nishimura, M., Bodmer, R., and Bohmann, D. (2013). Declining signal dependence of Nrf2-MafS-regulated gene expression correlates with aging phenotypes. Aging Cell 12, 554-562. [DOI] [PMC free article] [PubMed]
- 58.Frenkel-Denkberg G., Gershon D., Levy A.P. The function of hypoxia-inducible factor 1 (HIF-1) is impaired in senescent mice. FEBS Lett. 1999;462:341–344. doi: 10.1016/s0014-5793(99)01552-5. [DOI] [PubMed] [Google Scholar]; Frenkel-Denkberg, G., Gershon, D., and Levy, A.P. (1999). The function of hypoxia-inducible factor 1 (HIF-1) is impaired in senescent mice. FEBS Lett. 462, 341-344. [DOI] [PubMed]
- 59.Ben-Zvi A., Miller E.A., Morimoto R.I. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. U.S.A. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ben-Zvi, A., Miller, E.A., and Morimoto, R.I. (2009). Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl Acad. Sci. USA 106, 14914-14919. [DOI] [PMC free article] [PubMed]
- 60.Bragoszewski P., Gornicka A., Sztolsztener M.E., Chacinska A. The ubiquitin-proteasome system regulates mitochondrial intermembrane space proteins. Mol. Cell. Biol. 2013;33:2136–2148. doi: 10.1128/MCB.01579-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bragoszewski, P., Gornicka, A., Sztolsztener, M.E., and Chacinska, A. (2013). The ubiquitin-proteasome system regulates mitochondrial intermembrane space proteins. Mol. Cell Biol. 33, 2136-2148. [DOI] [PMC free article] [PubMed]
- 61.Taylor E.B., Rutter J. Mitochondrial quality control by the ubiquitin-proteasome system. Biochem. Soc. Trans. 2011;39:1509–1513. doi: 10.1042/BST0391509. [DOI] [PubMed] [Google Scholar]; Taylor, E.B., and Rutter, J. (2011). Mitochondrial quality control by the ubiquitin-proteasome system. Biochem. Soc. Trans. 39, 1509-1513. [DOI] [PubMed]
- 62.Chun B.Y., Rizzo J.F., 3rd Dominant optic atrophy and leber's hereditary optic neuropathy: update on clinical features and current therapeutic approaches. Semin. Pediatr. Neurol. 2017;24:129–134. doi: 10.1016/j.spen.2017.06.001. [DOI] [PubMed] [Google Scholar]; Chun, B.Y., and Rizzo, J.F. 3rd. (2017). Dominant Optic Atrophy and Leber's Hereditary Optic Neuropathy: Update on Clinical Features and Current Therapeutic Approaches. Semin. Pediatr. Neurol. 24, 129-134. [DOI] [PubMed]
- 63.Gumeni S., Evangelakou Z., Gorgoulis V.G., Trougakos I.P. Proteome stability as a key factor of genome integrity. Int. J. Mol. Sci. 2017;22(10):18. doi: 10.3390/ijms18102036. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gumeni, S., Evangelakou, Z., Gorgoulis, V.G., Trougakos, I.P. (2017) Proteome Stability as a Key Factor of Genome Integrity. Int J Mol Sci. 22, 18(10). [DOI] [PMC free article] [PubMed]
- 64.Vilchez D., Saez I., Dillin A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat. Commun. 2014;5:5659. doi: 10.1038/ncomms6659. [DOI] [PubMed] [Google Scholar]; Vilchez, D., Saez, I., and Dillin, A. (2014). The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat Commun. 5, 5659. [DOI] [PubMed]
- 65.Xu Y., Liu L., Xin W., Zhao X., Chen L., Zhen J., Wan Q. The renoprotective role of autophagy activation in proximal tubular epithelial cells in diabetic nephropathy. J. Diabet. Complicat. 2015;29:976–983. doi: 10.1016/j.jdiacomp.2015.07.021. [DOI] [PubMed] [Google Scholar]; Xu, Y., Liu, L., Xin, W., Zhao, X., Chen, L., Zhen, J., and Wan, Q. (2015). The renoprotective role of autophagy activation in proximal tubular epithelial cells in diabetic nephropathy. J. Diabetes Complications 29, 976-983. [DOI] [PubMed]
- 66.Fontana L., Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell. 2015;161:106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fontana, L., Partridge, L. (2015) Promoting health and longevity through diet: from model organisms to humans. Cell 161, 106-118. [DOI] [PMC free article] [PubMed]
- 67.Li J., Kim S.G., Blenis J. Rapamycin: one drug, many effects. Cell Metabol. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, J., Kim, S.G., and Blenis, J. (2014). Rapamycin: one drug, many effects. Cell Metab. 19, 373-379. [DOI] [PMC free article] [PubMed]
- 68.Vermeij W.P., Dollé M.E., Reiling E., Jaarsma D., Payan-Gomez C., Bombardieri C.R., Wu H., Roks A.J., Botter S.M., van der Eerden B.C., Youssef S.A., Kuiper R.V., Nagarajah B., van Oostrom C.T., Brandt R.M., Barnhoorn S., Imholz S., Pennings J.L., de Bruin A., Gyenis Á., Pothof J., Vijg J., van Steeg H., Hoeijmakers J.H. Restricted diet delays accelerated ageing and genomic stress in DNA-repair-deficient mice. Nature. 2016;537:427–431. doi: 10.1038/nature19329. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vermeij, W.P., Dolle, M.E., Reiling, E., Jaarsma, D., Payan-Gomez, C., Bombardieri, C.R., Wu, H., Roks, A.J., Botter, S.M., van der Eerden, B.C., Youssef, S.A., Kuiper, R.V., Nagarajah, B., van Oostrom, C.T., Brandt, R.M., Barnhoorn, S., Imholz, S., Pennings, J.L., de Bruin, A., Gyenis, A., Pothof, J., Vijg, J., van Steeg, H., and Hoeijmakers, J.H. (2016). Restricted diet delays accelerated ageing and genomic stress in DNA-repair-deficient mice. Nature 537, 427-431. [DOI] [PMC free article] [PubMed]
- 69.Barboni P., Valentino M.L., La Morgia C., Carbonelli M., Savini G., De Negri A., Simonelli F., Sadun F., Caporali L., Maresca A., Liguori R., Baruzzi A., Zeviani M., Carelli V. Idebenone treatment in patients with OPA1-mutant dominant optic atrophy. Brain. 2013;136(Pt 2):e231. doi: 10.1093/brain/aws280. [DOI] [PubMed] [Google Scholar]; Barboni, P., Valentino, M.L., La Morgia, C., Carbonelli, M., Savini, G., De Negri, A., Simonelli, F., Sadun, F., Caporali, L., Maresca, A., Liguori, R., Baruzzi, A., Zeviani, M., and Carelli, V. (2013). Idebenone treatment in patients with OPA1-mutant dominant optic atrophy. Brain 136, (Pt 2):e231. [DOI] [PubMed]
- 70.Yarosh W., Monserrate J., Tong J.J., Tse S., Le P.K., Nguyen K., Brachmann C.B., Wallace D.C., Huang T. The molecular mechanisms of OPA1-mediated optic atrophy in Drosophila model and prospects for antioxidant treatment. PLoS Genet. 2008;4:e6. doi: 10.1371/journal.pgen.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yarosh, W., Monserrate, J., Tong, J.J., Tse, S., Le, P.K., Nguyen, K., Brachmann, C.B., Wallace, D.C., and Huang, T. (2008). The molecular mechanisms of OPA1-mediated optic atrophy in Drosophila model and prospects for antioxidant treatment. PLoS Genet. 4, e6. [DOI] [PMC free article] [PubMed]
- 71.Sklirou A.D., Gaboriaud-Kolar N., Papassideri I., Skaltsounis A.L., Trougakos I.P. 6-bromo-indirubin-3'-oxime (6BIO), a Glycogen synthase kinase-3β inhibitor, activates cytoprotective cellular modules and suppresses cellular senescence-mediated biomolecular damage in human fibroblasts. Sci. Rep. 2017;7:11713. doi: 10.1038/s41598-017-11662-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sklirou, A.D., Gaboriaud-Kolar, N., Papassideri, I., Skaltsounis, A.L., and Trougakos, I.P. (2017). 6-bromo-indirubin-3'-oxime (6BIO), a Glycogen synthase kinase-3β inhibitor, activates cytoprotective cellular modules and suppresses cellular senescence-mediated biomolecular damage in human fibroblasts. Sci. Rep. 7, 11713. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.