Abstract
Background: Oral squamous cell carcinoma (OSCC) is the most frequent oral malignancy in humans. Increasing numbers of studies suggest that circular RNAs (circRNAs) play a major role in tumorigenesis and tumor progression.
Purpose: We aimed to identify differently expressed circRNA in OSCC and adjacent normal tissue. As well as to study the correlation of circRNA expression with clinicopathological feature in patients with OSCC.
Patients and methods: Primary OSCC tissues and adjacent normal tissues from twenty-seven OSCC patients were collected. The expression level of hsa_circ_009755 were analyzed by qRT-PCR.
Results: The measurement results from qRT-PCR revealed that hsa_circ_009755 expression was low in OSCC tissue samples and cell lines, and the low expression of hsa_circ_009755 was closely related to a clinicopathological feature in patients with OSCC. Moreover, the area under the receiver operating characteristic curve was 0.782, which indicates the sensitivity and specificity of the diagnostic value.
Conclusion: Our data indicates that hsa_circ_009755 could serve as a potential biomarker for OSCC diagnosis.
Keywords: oral squamous cell carcinoma, circular RNA, biomarker, hsa_circ_009755
Introduction
Oral cancer is one of the 10 most common cancers worldwide.1 The cancer sites in the oral cavity include the buccal mucosa, floor of the mouth, anterior tongue, alveolar ridge, retromolar trigone, and hard palate.2 The tumors that develop affect basic physiological functions such as chewing, ability to swallow and breathe, sensing, vision, and hearing.2 Therefore, halting disease progression in patients with oral cancer is extremely critical. Oral cancer is a malignant tumor that has traditionally been defined as oral squamous cell carcinoma (OSCC), and ~90% of the cancers are histologically found to originate in squamous cells.1 The prognosis for patients with OSCC has not improved markedly despite therapeutic advances in the case of this cancer and several other malignancies, and the 5-year survival rate from OSCC has increased only modestly in the past 30 years and remains at ~55–60%.3 Therefore, identification of novel therapeutic targets is crucial for OSCC treatment, and to achieve this goal, it is essential to enhance our understanding of the molecular mechanisms underlying OSCC.
The advent of next-generation sequencing has led to an increasingly rapid growth of the catalog of identified DNA and RNA molecules. Circular RNAs (circRNAs), which feature covalently linked 3′ and 5′ ends, constitute a class of RNA recently discovered to be stable and abundant,4,5 and circRNAs not only extensively regulate cellular activities, but also correlate with oncogenesis and/or tumor progression and are implicated in diverse cancers, such as gastric cancer, breast cancer, esophageal squamous cell carcinoma, and hepatocellular carcinoma.6–9 Although circRNAs thus hold the potential to serve as novel biomarkers, little is known regarding the role of circRNAs in OSCC.
By using high-throughput sequencing, microarray of circRNA expression profiles was acquired from patients with OSCCs (n=8) and adjacent normal tissue controls (n=8). We found that hsa_circ_009755 are downregualted in OSCC. Moreover, low-level expression of the circRNA hsa_circ_009755 was associated with I-II T stage (P=0.011) in patients with OSCC. Thus, we used a receiver operating characteristic (ROC) curve to evaluate the diagnostic value of hsa_circ_009755, and our data indicate that hsa_circ_009755 could represent a promising biomarker for OSCC diagnosis.
Materials and methods
Patients and specimens
All primary OSCC tissues and adjacent normal tissues were collected from patients who had undergone surgery at the Department of Oral and Maxillofacial Surgery, Shenzhen Hospital, Peking University (Shenzhen, China), between December 2016 and May 2017. All samples were confirmed by experienced pathologists. The clinical and pathological characteristics of each patient were collected after surgery. Tissue samples were frozen in liquid nitrogen and stored at −80°C. All patients received written informed consent, which was approved by the Clinical Research Ethics Committee of Peking University Health Science Center (IRB00001053-08043). The study was designed and conducted in accordance with the principle of the Declaration of Helsinki.
RNA extraction
Total RNA was isolated from OSCC tissues and adjacent normal tissues by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol, and the concentration of the RNA samples was measured using a NanoDrop ND2000 spectrophotometer (NanoDrop, Wilmington, DE, USA). Quantitative RT-PCR (qRT-PCR) analysis of circRNAs and β-actin gene was performed on an ABI PRISM7500 Sequence Detection System (Applied Biosystems, Life Technologies, Waltham, MA, USA) by using a PrimeScript RT Reagent Kit (Takara Bio, Nojihigashi, Kusatsu, Japan) and SYBR-Green Premix Ex Taq (Takara Bio) as per manufacturer's instructions. The sequences of the primers used were the following: hsa_circ_009755, 5′-TGCAGCTCTGGAAATGCCAAT-3′ (forward) and 5′-TGGTCACGTCACACAGAAATGAA-3′ (reverse); β-actin, 5′-AAACTGGAACGTTGAGAGTG-3′ (forward) and 5′-AGTGGTCTGGCTTTTAGGT-3′ (reverse). The obtained data were analyzed using the ΔCt method.10 Data were plotted as cycle threshold value (Ct value),in which the Ct value obtained from the intensity of the fluorescence. The Ct of target mRNA was normalized by the Ct of the reference gene β-actin and recorded as ΔCt of each sample. ΔCt shows the difference of expression between two genes (Ct of target gene - Ct reference gene). Therefore, higher ΔCt values indicate lower concentrations of the target gene templates and the lower expressions of the corresponding target genes.
Cell culture
Three human OSCC cell lines, SCC15, SCC25, and CAL27, were purchased from the College of Stomatology, Wuhan University (Wuhan, China). Human oral keratinocyte (HOK) cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), where the cells were authenticated based on mycoplasma detection. The cell lines used have been approved by the Clinical Research Ethics Committee of Peking University Health Science Center. SCC15, SCC25, CAL27, and HOK cells were maintained in DMEM (Gibco, Waltham, MA, USA) supplemented with 10% FBS and 50 U/mL penicillin and streptomycin sulfate. All cells were cultured at 37°C in 5% CO2.
Prediction of the circRNA/miRNA/mRNA interaction
The miRNA-binding sites were predicted by Mireap, Miranda (v3.3a), and TargetScan (Version:7.0). The targeted miRNAs and mRNAs were predicted by miRTarBase (v6.1).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5.0 Software for Windows (GraphPad Software, La Jolla, CA, USA) and Service Solutions SPSS software 16.0 (SPSS, Chicago, IL, USA). Differences in the levels of hsa_circ_009755 between OSCC tissues and paired adjacent normal tissues and between OSCC cell lines and HOK cells were assessed using t-tests for paired data. One-way ANOVA was used to examine the correlation between hsa_circ_009755 levels and clinicopathological factors, and the ROC curve was established to evaluate diagnostic values. P<0.05 was considered statistically significant.
Results
Low expression level of hsa_circ_009755 in OSCC tissues
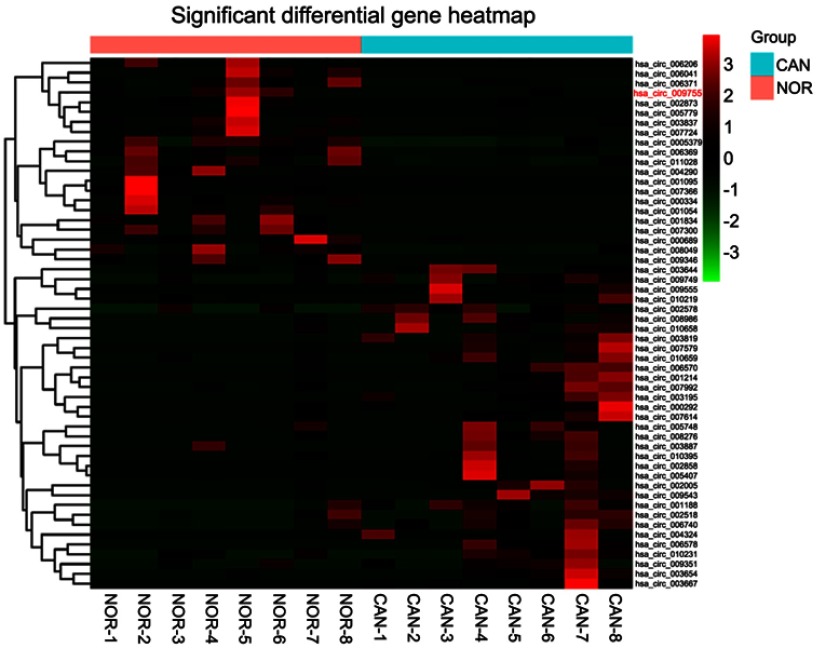
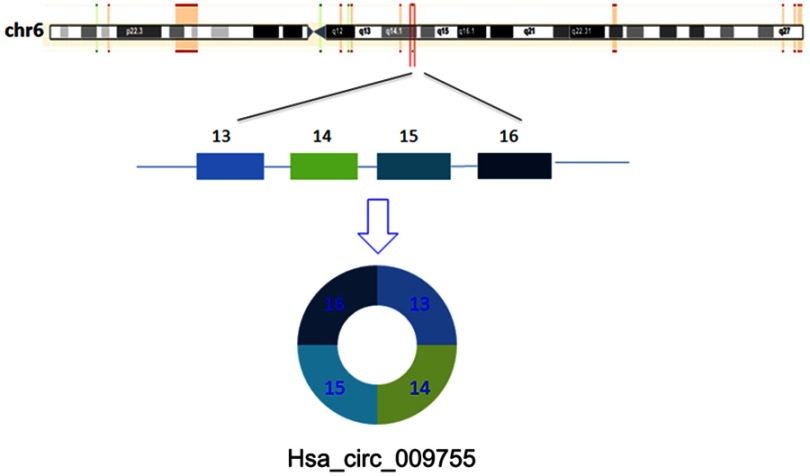
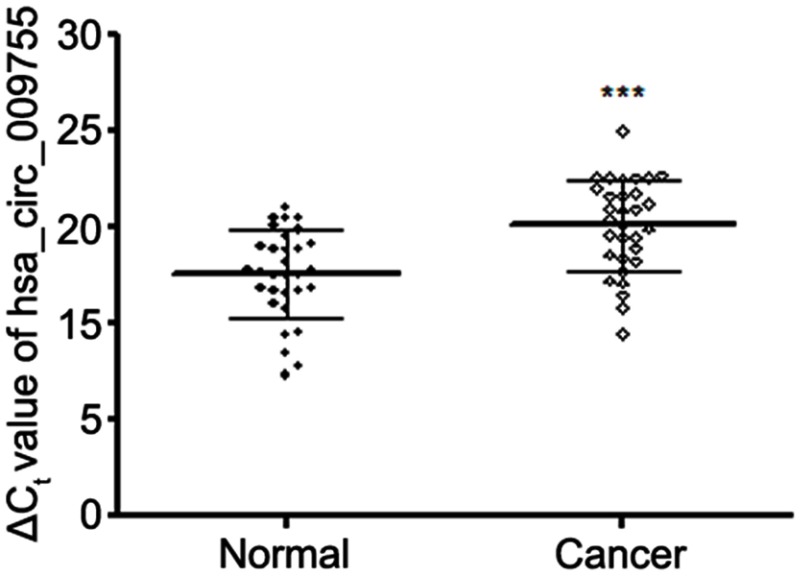
By using high-throughput screening of circRNA microarrays, we found that hsa_circ_009755 was downregulated in OSCC tissues (Figure 1). Furthermore, we confirmed that hsa_circ_009755 is encoded from chr6 (82,169,983–82,247,754). Featuring a spliced sequence length of 305 bp, hsa_circ_009755 (associated gene symbol IBTK) is composed of 4 exons (exons 13–16) (Figure 2). By performing RNA-seq analyses, we determined the expression level of hsa_circ_009755 in each pair of cancer tissue and adjacent nontumorous tissue; hsa_circ_009755 was expressed at significantly lower levels in OSCC tissues than in the corresponding nontumorous tissues (n=27, P<0.001) (Figure 3).
Figure 1.
Differentially expressed circRNAs between OSCC tissues and adjacent nontumorous tissues. The results of hierarchical cluster analysis showed that the differentiable expression profiles of circRNA between the samples (for OSCC, CAN, and NOR). Each column represents the expression of tissue samples. The expression level was indicated from “red” (high expression) to “green” (low expression).
Abbreviations: CAN, cancerous; NOR, normal adjacent nontumorous tissue; OSCC, oral squamous cell carcinoma.
Figure 2.
Characterization of hsa_circ_009755. The circRNA hsa_circ_009755 is encoded from chromosomal region 6q14.1. Four exons in this region (exons 13–16) generate hsa_circ_009755.
Figure 3.
Low-level expression hsa_circ_009755 in OSCC tissues. Expression levels of hsa_circ_009755 in 27 pairs of OSCC tissues are shown relative to the levels in paired adjacent normal tissues. Higher ΔCt values indicate lower expression. Data are expressed as means ± SD; ***P<0.001.
Abbreviation: OSCC, oral squamous cell carcinoma.
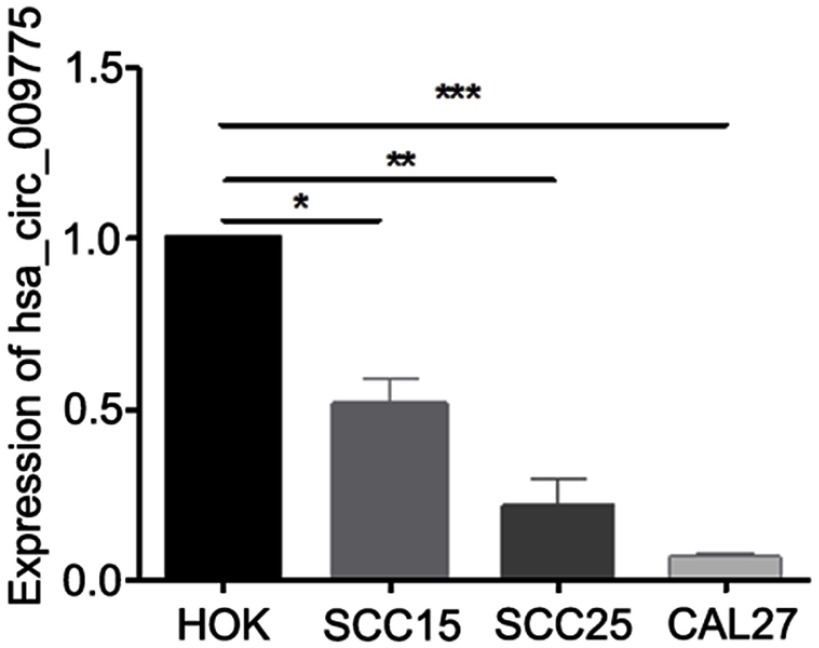
Low-level expression of hsa_circ_009755 in OSCC cell lines
We next measured hsa_circ_009755 expression levels in a normal cell line (HOK) and in 3 human OSCC cell lines, CAL27, SCC15, and SCC25. The expression levels in the tumor cell lines were considerably lower than that in the normal cell line (Figure 4).
Figure 4.
Expression of hsa_circ_009755 in distinct OSCC cell lines. Expression levels of hsa_circ_009755 in 3 OSCC cell lines (SCC15, SCC25, and CAL27) and normal HOKcells were assessed using qRT-PCR. Data are presented as means ± SD; *P<0.01, **P<0.01, ***P<0.001.
Abbreviations: HOK, human oral keratinocyte; OSCC, oral squamous cell carcinoma; qRT-PCR, quantitative RT-PCR.
Correlation between clinicopathological features and hsa_circ_009755 expression levels
The association of hsa_circ_009755 expression with several clinicopathological features of OSCC patients was analyzed (Table 1): We found that hsa_circ_009755 levels were related to T stage (P=0.011), but we did not find association between hsa_circ_009755 levels and other clinicopathological factors (age, gender, tumor differentiation, tumor site, and lymphatic metastasis; P>0.05).
Table 1.
Relationship between hsa_circ_009755 expression levels (ΔCt) in cancer tissue and clinicopathological features of OSCC patients
| Characteristic | Number of patients | Mean ± SD | P-value |
|---|---|---|---|
| Gender | |||
| Male | 21 | 15.15±2.37 | 0.267 |
| Female | 6 | 13.85±2.77 | |
| Age | |||
| <60 | 20 | 14.61±2.65 | 0.376 |
| ≥60 | 7 | 15.59±1.84 | |
| Tumor site | |||
| Tongue | 18 | 14.50±2.74 | 0.565 |
| Buccal | 3 | 15.89±1.50 | |
| Others | 6 | 15.43±1.97 | |
| T stage | |||
| T1–T2 | 13 | 13.64±2.37 | 0.011* |
| T3–T4 | 14 | 15.99±2.04 | |
| Lymph node metastasis | |||
| N0 | 19 | 15.41±2.13 | 0.074 |
| N1–N2 | 8 | 13.56±2.85 | |
| Differentiation | |||
| Well | 22 | 14.69±2.58 | 0.456 |
| Moderate and poor | 5 | 15.62±1.96 |
Note: *Statistically significant.
Abbreviation: OSCC, oral squamous cell carcinoma.
Diagnostic value of hsa_circ_009755 in OSCC
To estimate the diagnostic value of hsa_circ_009755 in OSCC, an ROC curve was constructed for differentiating OSCC tissues from paired adjacent normal tissues. The area under the ROC curve (AUC) was 0.782 (Figure 5A), the cutoff value was 13.22 (Figure 5B), and the corresponding sensitivity and specificity were 70.37% and 77.78%, respectively. The ROC is used to reflect the sensitivity and specificity of continuous variables; the diagnostic value of the variables is high only when the AUC is large. Values below and above the cutoff value are negative and positive values, respectively, and the cutoff value is a schematic combination of sensitivity and specificity. Therefore, the calculated values suggest that hsa_circ_009755 could serve as a potential biomarker for the diagnosis of OSCC.
Figure 5.

Diagnostic value of hsa_circ_009755 in OSCC.
Notes: (A) Area under the ROC curve was 0.78 (95% CI =0.6670–0.8544; P<0.0001), and (B) cutoff of hsa_circ_009755 was 13.22. Sensitivity and specificity were 0.704 and 0.778, respectively. Data are presented as means ± SD; ***P<0.001.
Abbreviations: AUC, area under the ROC curve; OSCC, oral squamous cell carcinoma; ROC, receiver operating characteristic.
Prediction of the circRNA/miRNA/mRNA interaction
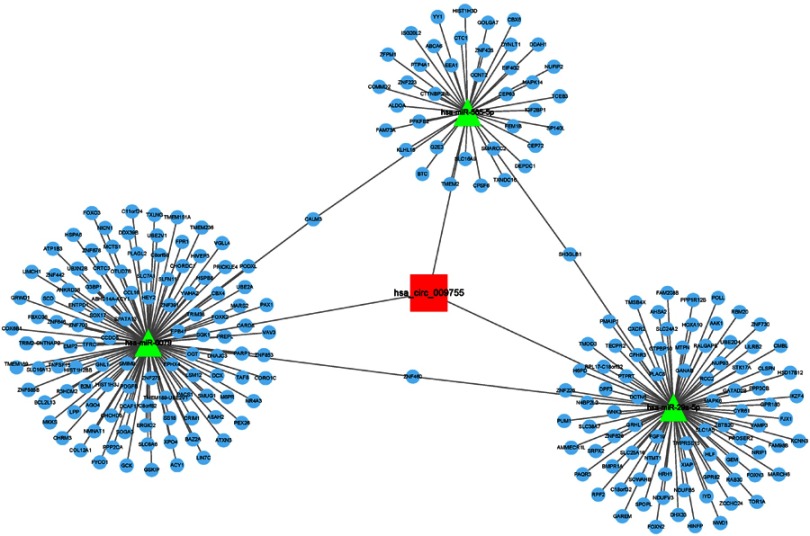
In order to explore the molecular mechanism and function of circRNA hsa_circ_009755, we investigated the potential miRNAs and mRNAs bound to hsa_circ_009755. As shown in Figure 6, hsa_circ_009755 in cancer tissues may interact with has-miR-585-5p, has-miR-29a-5p, and has-miR-6079.
Figure 6.
CircRNA-miRNA-mRNA network of hsa_circ_009755.
Notes: CircRNA: rectangular; miRNA: triangular; mRNA: circular.
Discussion
Detection of circRNAs in RNA viruses was reported as early as in the 1970s,11,12 and isolated reports of circRNA expression from single genes were published in the subsequent decades; however, because of technological and methodological biases, the detected circRNAs were generally regarded as rare splicing “mistakes” until recently.4 Currently, circRNAs are regarded as abundant, stable, conserved, and nonrandom products of RNA splicing that could be involved in the control of gene expression.5 In accordance with previous work, we noted that the abnormal expression of circRNAs might be correlated with disease. For instance, circRNAs can interact with RNA-binding proteins or miRNAs and thereby influence the generation and development of cancer.13,14 Moreover, several types of tumor markers have been investigated, and the expression circRNAs has been reported to differ substantially between tumor cells and normal cells.6,15–20 These studies indicate that circRNAs could serve as potential cancer biomarkers.8,21,22
Li et al found that circITCH was markedly downregulated in esophageal squamous cell carcinoma samples; circITCH harbors sites for concurrently binding several miRNAs, which indicates that this circRNA could function as a cavernous transporter or regulator of miRNAs, and circITCH might inhibit tumor growth by regulating the Wnt/β-catenin signaling pathway.8 Another circRNA reported to be associated with cell proliferation is circFOXO3; intriguingly, as in the case of circITCH, low levels of circFOXO3 appear to be associated with increased cell proliferation.23 Last, circHIPK3 is strongly downregulated in bladder cancer tissues and cell lines, and circHIPK3 overexpression effectively inhibits migration, invasion, and angiogenesis of bladder cancer cells in vitro and suppresses bladder cancer growth and metastasis in vivo; circHIPK3 contains two key binding sites for the miRNA miR-558 and can sponge large amounts of miR-558 and thereby suppress heparanase expression.24
The circRNA hsa_circ_009755 is located at chromosome 6q14.1 on the plus strand, is aligned in a sense orientation to the known protein-coding gene VWA8, and spans exons 13–16; however, the function of this circRNA in tumor progression has remained unknown. Here, we used TaqMan-based RT-PCR analyses to compare hsa_circ_009755 expression levels in OSCC tissues and cell lines with those in normal tissues and cells, and our results showed that hsa_circ_009755 was drastically downregulated in OSCC; thus, hsa_circ_009755 expression might specifically and sensitively indicate the degree of OSCC. Notably, comparison of hsa_circ_009755 expression with clinicopathological characteristics of patients revealed that the circRNA expression level was significantly associated with the T stage. Hsa_circ_009755 might serve as a potential diagnostic biomarker for OSCC.
CircRNA plays an important role in regulating gene expression. It may be used as a gene sponge, an RBP sponge, or a transcription/translation regulator.25–27 We found that hsa_circ_9755 was abnormal low expression in cancer tissues. It is predicted to be related to has-miR-585-5p, has-miR-29a-5p, and has-miR-6079. Our future research plan is to further explain the important role of hsa_circ_009755 in OSCC to unravel the molecular mechanisms of hsa_circ_9755 in OSCC and other cancers.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant no. 81572654), the Basic Research Program of Shenzhen Innovation Council of China (Grant nos. JCYJ20160428173933559, JCYJ20150403091443286 and SZBC2017023), and Sanming Project of Medicine in Shenzhen (SZSM 201512036, Oral and Maxillofacial Surgery Team, Professor Yu Guangyan, Stomatology Hospital Peking University).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lingen MW, Kalmar JR, Karrison T, Speight PM. Critical evaluation of diagnostic aids for the detection of oral cancer. Oral Oncol. 2008;44(1):10–22. doi: 10.1016/j.oraloncology.2007.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colevas AD, Yom SS, Pfister DG, et al. NCCN guidelines insights: head and neck cancers, version 1.2018. J Natl Compr Canc Netw. 2018;16(5):479–490. doi: 10.6004/jnccn.2018.0026 [DOI] [PubMed] [Google Scholar]
- 3.Rhodus NL, Kerr AR, Patel K. Oral cancer: leukoplakia, premalignancy, and squamous cell carcinoma. Dent Clin North Am. 2014;58(2):315–340. doi: 10.1016/j.cden.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 4.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. Preiss T, ed PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2012;19:141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang H-F, Zhang X-Z, Liu B-G, Jia G-T, Li W-L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7(7):1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu M, Xu Y, Chen Y, Yan F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–144. doi: 10.1016/j.biopha.2016.12.097 [DOI] [PubMed] [Google Scholar]
- 8.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo W, Zhang J, Zhang D, et al. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8(29):48169–48177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song H, Sun W, Ye G, et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. doi: 10.1186/1479-5876-11-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340(6131):440–441. doi: 10.1126/science.1238522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Q, Wang Y, Cao M, et al. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc Natl Acad Sci. 2012;109(10):3938–3943. doi: 10.1073/pnas.1117815109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY). 2017;9(6):1585–1594. doi: 10.18632/aging.101254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Wang G, Ding C, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8(37):61687–61697. doi: 10.18632/oncotarget.18671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6(1). doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang P, Qiu Z, Jiang Y, et al. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/β-catenin signaling pathway. Oncotarget. 2016;7(39):63449–63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi: 10.1111/febs.14132 [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–4561. doi: 10.1038/onc.2017.89 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Kong Z, Wan X, Zhang Y, et al. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;493:1217–1223. doi: 10.1016/j.bbrc.2017.07.162 [DOI] [PubMed] [Google Scholar]
- 20.Chen -L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Li C, Tan C, Liu X. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53(6):359–365. doi: 10.1136/jmedgenet-2016-103758 [DOI] [PubMed] [Google Scholar]
- 22.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR‐558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi: 10.15252/embr.201643581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 26.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 27.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. Embo J. 2013;32(7):923–925. doi: 10.1038/emboj.2013.53 [DOI] [PMC free article] [PubMed] [Google Scholar]