Abstract
Background
Gene expression profiling (GEP) has been integrated into cancer treatment decision‐making in multiple neoplasms. We prospectively evaluated the prognostic utility of the 31‐GEP test (DecisionDx‐Melanoma, Castle Biosciences, Inc) in cutaneous melanoma (CM) patients undergoing sentinel node biopsy (SNB).
Methods
One hundred fifty‐nine patients (age 26‐88) diagnosed with melanoma between 01/2013 and 8/2015 underwent SNB and concurrent GEP testing. GEP results were reported as low‐risk Class 1 (subclasses 1A and 1B) or high‐risk Class 2 (subclasses 2A and 2B). Statistical analyses were performed with chi‐square analysis, t tests, log‐rank tests, and Cox proportional hazard models. Recurrence‐free survival (RFS) and distant metastasis‐free survival (DMFS) were estimated using Kaplan‐Meier method.
Results
Median follow‐up was 44.9 months for event‐free cases. Median Breslow thickness was 1.4 mm (0.2‐15.0 mm). There were 117 Class 1 and 42 Class 2 patients. Gender, age, Breslow thickness, ulceration, SNB positivity, and AJCC stage were significantly associated with GEP classification (P < 0.05 for all). Recurrence and distant metastasis rates were 5% and 1% for Class 1 patients compared with 55% and 36% for Class 2 patients. Sensitivities of Class 2 and SNB for recurrence were 79% and 34%, respectively. Of 10 SNB‐positive/Class 2 patients, 9 recurred. By multivariate analysis, only SNB result and GEP class were statistically associated with both RFS (P = 0.008 and 0.0001) and DMFS (P = 0.019 and 0.001).
Conclusions
Gene expression profiling Class 2 result and SNB positivity were independently associated with recurrence and distant metastasis in primary CM patients. GEP testing may have additive prognostic utility in initial staging work‐up of these patients.
Keywords: cutaneous melanoma, gene expression profile, prognosis
1. INTRODUCTION
The incidence of melanoma is increasing. For 2018, the number of new cases of melanoma is estimated to reach more than 87,000 and the number of melanoma‐related deaths will be more than 9,000.1 The standard for prognostication of melanoma patients is the American Joint Committee on Cancer's (AJCC) staging system. It is based on clinical and histopathologic variables such as Breslow thickness, ulceration, nodal involvement, in transit or satellite deposit, and presence of distant metastases.2, 3, 4 While each successive revision of the staging system has resulted in a more accurate reflection of patient prognosis, significant variance in survival still exists within each stage under the current AJCC classification.
Gerami et al first reported a prognostic gene expression profiling (GEP) test utilizing the 31‐gene panel for use in patients with cutaneous melanoma (CM).5 This GEP test is comprised of 28 discriminating genes and 3 control genes, which are evaluated using formalin‐fixed paraffin‐embedded tissue from the primary melanoma tumor. The test reports results as Class 1 (low risk, with subclasses 1A and 1B) and Class 2 (high risk, with subclasses 2A and 2B), which are derived from comparison of the gene expression from the tested tumor to a training set of 164 melanoma cases with known long‐term clinical outcomes.5 Since the initial development and validation study, several retrospective and prospective studies have demonstrated that GEP classification is significantly correlated with recurrence‐free survival (RFS), distant metastasis‐free survival (DMFS), overall, and melanoma‐specific survival.6, 7, 8, 9, 10
To further evaluate the prognostic utility of the GEP test, we conducted a prospective observational study in patients undergoing wide excision and sentinel node biopsy (SNB) for treatment of their primary CM at our institution. The GEP test was performed at the time of SNB. Patients were followed at regular intervals. We observed that GEP classification and SNB result were associated with both RFS and DMFS by multivariate analysis. The use of GEP in combination with current AJCC classification may further improve the prognostication schema for patients with primary CM.
2. MATERIALS AND METHODS
2.1. Patient population
One hundred seventy‐four clinically node‐negative patients diagnosed with CM between January 2013 and August 2015 opted for GEP testing and underwent SNB and wide excision of their primary tumor at the Department of Surgery, Saint Louis University, St. Louis, Missouri. Informed consent was obtained under an Institutional Review Board‐approved protocol. SNB was performed with same day pre‐operative lymphoscintigram with Tc‐99 tagged radioactive tracer followed by intraoperative lymphatic mapping with Lymphazurin blue dye. GEP testing was prospectively performed by Castle Biosciences, Inc, Friendswood, Texas, as previously described.5, 6, 10 Fifteen patients had insufficient tumor for GEP testing. The final study cohort was comprised of 159 patients with both GEP and SNB results.
2.2. Follow‐up
Patient follow‐up was performed as described below. AJCC stage I and II patients underwent physical exam (PE), chest X‐ray (AP and lateral views), and laboratory evaluation with complete blood count and complete metabolic panel every 6 months for the first 2 years then yearly for the subsequent 3 years. Stage III patients underwent baseline brain MRI and PET/CT or CT of chest/abdomen/pelvis (CT/CAP) followed by completion node dissection for SNB‐positive patients and subsequently followed every 3 months with PE, MRI of brain, CT C/A/P, or PET/CT and laboratory evaluation for the first year, then every 4 months for the second year, and every 6 months for the subsequent 3 years. All stage III patients were referred to Medical Oncology for discussion regarding adjuvant therapy. Patient information was prospectively entered into a secured database.
2.3. Statistical analysis
Comparisons between GEP class and covariates were made using chi‐square tests for categorical variables and t tests for continuous variables. Survival outcomes were defined as the time between date of diagnosis and date of disease recurrence (RFS) or distant metastasis (DMFS) or last follow‐up. Survival curves were estimated by the nonparametric Kaplan‐Meier method. Log‐rank tests were used for univariate analysis of categorical variables to determine differences between curves. Univariate analysis of continuous variables was performed using the Cox proportional hazard regression method and all statistically significant variables are reported. Multivariate Cox proportional hazards models including all variables significant in univariate analyses were used to examine the association of GEP class with RFS or DMFS. P value < 0.05 was considered significant. All statistical analyses were two‐tailed.
3. RESULTS
3.1. Patient demographics
For the cohort with SNB and GEP results (n = 159; Table 1), the median Breslow thickness was 1.4 mm (range: 0.2‐15.0 mm). The median age was 59 (range: 26‐88) and the majority (61.6%) of patients were male. Thirty‐eight patients (24%) had ulceration of their primary tumors. Distribution of primary site was 16.4% head and neck (n = 26), 40.9% extremity (n = 65), and 42.8% truncal location (n = 68). SNB was positive for metastatic melanoma in 20 patients (12.6%). Three patients had satellite deposits in wide excision specimen with negative SNB. Thus, 23 patients had pathologic AJCC stage III melanoma following surgery. There were 117 Class 1 (91 subclass 1A and 26 subclass 1B) and 42 Class 2 patients (12 subclass 2A and 30 subclass 2B). GEP classification was significantly associated with gender, age, ulceration, Breslow thickness, SNB positivity, and AJCC stage (P = 0.009, 0.0001, <0.0001, <0.0001, 0.011, and <0.0001, respectively; Table 1).
Table 1.
Patient demographics
|
GEPa Class 1 (n = 117) |
GEP Class 2 (n = 42) |
P value | |
|---|---|---|---|
| Gender | 0.009 | ||
| Male | 65 (55%) | 33 (79%) | |
| Female | 52 (45%) | 9 (21%) | |
| Age | 55.8 (SDb =14.5) | 66.0 (SD = 13.9) | 0.0001 |
| Site | 0.5507 | ||
| Extremity | 48 (41%) | 17 (40%) | |
| Head & Neck | 17 (15%) | 9 (21%) | |
| Trunk | 52 (44%) | 16 (38%) | |
| Ulceration | < 0.0001 | ||
| No | 107 (91%) | 14 (33%) | |
| Yes | 10 (9%) | 28 (67%) | |
| Breslow thickness | 1.4 (SD = 1.1) | 3.7 (SD = 2.7) | < 0.0001 |
| T stage | < 0.0001 | ||
| T1 | 50 (43%) | 1(2%) | |
| T2 | 51 (44%) | 11 (26%) | |
| T3 | 13 (10%) | 14 (33%) | |
| T4 | 3 (3%) | 16 (38%) | |
| SNBc result | 0.0105 | ||
| Positive | 10 (9%) | 10 (24%) | |
| Negative | 107 (91%) | 32 (76%) | |
| AJCCd stage | < 0.0001 | ||
| Stage I | 91 (78%) | 5 (12%) | |
| Stage II | 16 (13%) | 24 (57%) | |
| Stage III | 10 (9%) | 13 (31%) |
gene expression profile
standard deviation
sentinel node biopsy
American Joint Committee on Cancer
Within the stage III cohort, 11 patients received adjuvant high‐dose interferon‐alfa 2b or pegylated interferon‐alfa 2b. Two stage III patients enrolled in clinical trials. Ten stage III patients opted for expectant observation. With a median follow‐up time of 44.9 months for event‐free cases, 29 patients experienced recurrence with a median time to recurrence of 13.3 months (range: 1.6‐51.4 months). Of the 29 patients with recurrence, 10 (34%) were SNB‐positive and 19 (66%) were SNB‐negative (Table 2). For GEP, 23 (79%) patients who had recurrence were GEP Class 2, while 6 (21%) were Class 1. Of the 19 node‐negative patients who experienced recurrence, 14 (74%) were Class 2. Nine of 10 patients with a positive sentinel node and GEP Class 2 melanoma recurred. Eleven of 13 patients with AJCC stage III and GEP Class 2 melanoma recurred, including 7 patients who received adjuvant therapy (Table 3). The vast majority (9 of 11, 82%) of first recurrences in the AJCC stage III/GEP Class 2 cohort were to distant visceral sites. At the time of last follow‐up, 9 patients in the GEP Class 2 group had expired due to any cause compared to one in the GEP Class 1 group.
Table 2.
|
Recurrence‐free n (% of row) |
With recurrence n (% of row) |
|
|---|---|---|
| Class 1 (n = 117) | 111 (95%) | 6 (5%) |
| Class 2 (n = 42) | 19 (45%) | 23 (55%) |
| SNB‐negative (n = 139) | 122 (88%) | 19 (14%) |
| SNB‐positive (n = 20) | 10 (50%) | 10 (50%) |
| Class 1/SNB‐negative (n = 107) | 102 (95%) | 5 (5%) |
| Class 1/SNB‐positive (n = 10) | 9 (90%) | 1 (10%) |
| Class 2/SNB‐negative (n = 32) | 18 (56%) | 14 (44%) |
| Class 2/SNB‐positive (n = 10) | 1 (10%) | 9 (90%) |
gene expression profile
sentinel node biopsy
Table 3.
| AJCC stage | GEP class (n) | Recurrence‐free |
With recurrence n, site of first recurrence; second recurrence |
|---|---|---|---|
| Stage I | Class 1 (91) | 88 |
1 in transit 2 nodal |
| Class 2 (5) | 5 | 0 | |
| Stage II | Class 1 (16) | 14 |
1 subcutaneous 1 in transit |
| Class 2 (24) | 12 |
1 local subcutaneous 1 subcutaneous 1 local; lung 3 in transit 1 nodal; bone 1 nodal; brain 1 lung/adrenal/subcutaneous 3 lung |
|
| Stage III | Class 1 (10) | 9 | 1 in transit/liver |
| Class 2 (13) | 2 |
1 subcutaneous; brain 2 in transit 1 in transit; small bowel/liver 1 in transit; liver 2 brain 1 lung 1 lung/liver 1 lung/subcutaneous 1 liver |
American Joint Committee on Cancer
gene expression profile
3.2. Recurrence‐free and distant‐metastasis‐free survival analysis
On univariate analysis, Breslow thickness, ulceration, SNB result, and GEP class were significantly associated with RFS and DMFS (P < 0.001 for all variables; Tables 4 and 5), while age was only significant for RFS (P = 0.0113). Tumor location and gender were not statistically significant for either outcome in univariate analysis (P > 0.05), and thus, along with age for RFS, were not included in subsequent multivariate analysis. In multivariate analysis, the hazard ratios (HR) for GEP Class 2 were 9.2 (P < 0.001, 95% confidence interval (CI) = 3.0‐28.5) and 19 (P < 0.01, 95% CI = 2.12‐170.5) for RFS and DMFS. SNB result was also associated with RFS and DMFS (P < 0.02, HR = 3.5, 3.7, 95% CI = 1.4‐9.1, 1.2‐11.3, respectively). Breslow thickness was not nominally significant for DMFS in multivariate analyses (P = 0.06, HR = 1.2, 95% CI = 1.0‐1.4) but was statistically significant for RFS (P = 0.015, HR = 1.15, 95% CI = 1.01‐1.31). Ulceration was not found to be significant for either endpoint in multivariate analyses. For GEP Class 1 patients, the observed 3‐year RFS (Figure 1A) and DMFS (Figure 1B) rates were 96.6% and 99.1%, respectively, compared to 47.4% and 64.1%, respectively, for GEP Class 2 patients (P < 0.0001 for Class 1 vs Class 2 for both endpoints). GEP subclass provided additional stratification with 3‐year RFS and DMFS rates for subclass 2B cases of 39.5% and 59.6% (Figure S1).
Table 4.
Recurrence‐free survival analysis
| Univariate analysis | Multivariate analysis | |
|---|---|---|
| Age |
P = 0.011 HRa 1.0; 95% CIb 1.0‐1.1 |
P = 0.14 HR 1.0; 95% CI 0.99‐1.1 |
| Breslow thickness |
P < 0.0001 HR 1.4; 95% CI 1.3‐1.5 |
P = 0.015 HR 1.2; 95% CI 1.0‐1.3 |
| Ulceration |
P < 0.0001 HR 5.4; 95% CI 2.6‐11.4 |
P = 0.69 HR 0.8; 95% CI 0.3‐2.1 |
| SNBc results |
P < 0.0001 HR 5.1; 95% CI 2.4‐11.1 |
P = 0.008 HR 3.5; 95% CI 1.4‐9.1 |
| GEPd class |
P < 0.0001; HR 15.0; 95% CI 6.1‐37.0 |
P = 0.0001 HR 9.2; 95% CI 3.0‐28.5 |
hazard ratio
confidence interval
sentinel node biopsy
gene expression profile
Table 5.
Distant metastasis‐free survival
| Univariate analysis | Multivariate analysis | |
|---|---|---|
| Breslow thickness |
P < 0.0001 HRa 1.5; 95% CIb 1.3‐1.7 |
P = 0. 06 HR 1.2; 95% CI 1.0‐1.4 |
| Ulceration |
P < 0.0001 HR 12.6; 95% CI 4.0‐38.9 |
P = 0.14 HR 2.5; 95% CI 0.7‐8.5 |
| SNBc results |
P < 0.0001 HR 7.8; 95% CI 3.0‐20.3 |
P = 0. 019 HR 3.75; 95% CI 1.2‐11.3 |
| GEPd class |
P = 0.0001; HR 55.1; 95% CI 7.3‐415.9 |
P = 0.009 HR 19.0; 95% CI 2.1‐170.5 |
hazard ratio
confidence interval
sentinel node biopsy
gene expression profile
Figure 1.
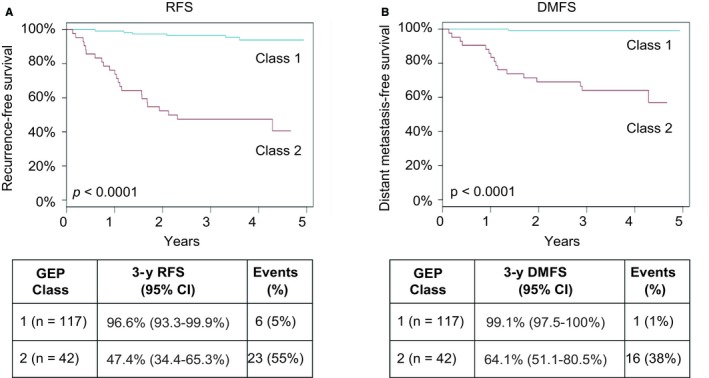
Kaplan‐Meier analysis of recurrence‐free survival (RFS; A) and distant metastasis‐free survival (DMFS; B) by gene expression profiling (GEP) class in the prospective cohort (n = 159). P‐values were calculated by log rank test. Tables report number of patients for each GEP class, 3‐yr survival rates with 95% confidence intervals, the number of overall events for a given outcome and class, and percentages of the class experiencing the event
4. DISCUSSION
GEP molecular class was an independent prognostic variable for disease recurrence in this single‐institution prospective study. SNB result was also an independent prognostic variable for disease recurrence. GEP class was significantly associated with but independent from other clinical and histopathological prognostic variables such as gender, age, Breslow thickness, and SNB status. Of the 10 deaths from all causes observed in the current study, 90% occurred in patients with a GEP Class 2 tumor, which is in agreement with prior publications.5, 8
The current study has a median follow‐up duration of 3.5 years overall and 3.7 years for patients without a recurrence. The median time to recurrence in this cohort was 13.3 months (1.1 years) and 90% of recurrences occurred by 30.1 months (2.5 years), indicating follow‐up duration was adequate to confirm the accuracy of the GEP test and that surveillance imaging for early identification of metastatic disease can be performed in that time period. Although melanoma recurrence can occur decades after initial diagnosis,11 the time duration between initial diagnosis and disease recurrence, i.e. disease‐free interval, has been shown to be prognostic of melanoma patient outcome.12, 13, 14, 15 Patients with early recurrence likely harbor tumors with more aggressive phenotype than those with delayed recurrence. In addition, low tumor burden has been demonstrated to be a robust predictor of outcome for surgical management of metastasis and for novel immune checkpoint therapies.16 Thus, identification of patients with early recurrence and development of effective adjuvant therapy for these patients would likely have significant impact on overall melanoma patient outcomes.
Both GEP class and SNB positivity were independently prognostic of disease recurrence in this prospective study. The combination of GEP test and SNB may improve our ability to determine prognosis in patients with primary CM, as has been previously demonstrated.6, 10 Gerami et al reported that the combination of SNB and GEP testing indeed allowed for separation of primary melanoma patients into distinct subgroups with low, moderate, and high risk for recurrence.6 Zager et al reported similar results in an independent cohort of 523 patients.10 Thus, use of both SLNB and GEP testing in the staging work‐up of primary melanoma patients may allow separation of groups at very low risk, moderate risk, and very high risk for recurrence.
Two recent large retrospective analyses have shown the additive prognostic value of GEP testing in early stage melanoma patients especially in stage II melanoma patients.9, 10 While Zager et al did not observe a statistical significance in DMFS for stage I subgroup (n = 264), Gastman et al did show a statistical significance in DMFS with a larger stage I cohort (n = 333). Due to the small sample size in this prospective cohort, further analysis specific to stage subgroups or age subgroups would not likely yield meaningful results. With the coming maturation of the large multicenter prospective study of GEP testing in primary melanoma patients,8 further subgroup analysis of this type may yield more information regarding the subgroup‐specific utility of GEP testing.
Recently, other molecular testing approaches for prognostication of primary melanoma have been proposed. Nsengimana et al performed an independent validation of a whole‐genome mRNA profiling classification.17, 18 Archival tumor tissues from 300 patients (224 primary and 76 metastatic) were evaluated. Gene signature classification was significantly correlated with Breslow thickness, ulceration, mitotic rate, and melanoma‐specific survival. Other gene expression profiles for melanoma have been described and reported additive prognostic values in addition to clinical and histological factors.19, 20 MicroRNA‐21, ‐137, and ‐203 have been evaluated in primary melanoma tissues and found to be independently associated with survival.21, 22, 23 Down‐regulation of miRNA‐150‐5p and miRNA142‐3p/142‐5p duplex correlated with poor survival in metastatic melanoma patients.24 However, no prospective validations of these approaches have been performed.
Following the validation of several efficacious agents against metastatic melanoma,25, 26, 27, 28, 29 therapies are now been applied in the adjuvant setting. Over the last 2 decades, four agents have been approved for adjuvant use in stage III melanoma: interferon, ipilimumab, nivolumab, and the combination of dabrafenib and trateminib.30, 31, 32, 33 Pembrolizumab has also shown efficacy in stage III patients.34 While only high‐dose interferon is approved for use in T4 Stage II melanoma patients, clinical trials testing PD‐1 inhibitors in stage IIB‐IIC patients are currently ongoing. In light of these trials, identification of a subset of high risk Stage II melanoma would allow for rational clinical trial design and expedient determination of the clinical efficacy of adjuvant targeted and immunotherapies. Furthermore, the observation that 11 of 13 Stage III GEP Class 2 patients recurred within 2 years suggests these patients could have harbored occult disease at the time of primary melanoma presentation and should be treated aggressively. Of interest, 28 patients in this study had both GEP testing and BRAF mutation status analyzed. An association between the two molecular markers was not observed for these patients, as BRAF was mutated in only three of the patients—one Class 1 and two Class 2 (Fisher's Exact P = 0.59). GEP testing can identify melanoma patients at high risk for recurrence, however clinical trials are needed for these high‐risk patients to determine the optimal treatment strategy, in particular for adjuvant therapy decision making.
The results from this single‐center prospective study confirm prior retrospective and prospective studies in support of the use of GEP testing in combination with current staging procedures for prognostication of CM. While prior clinical utility studies have been published to indicate current clinical applications of this test for patient management,35, 36, 37, 38, 39 the ongoing expansion of adjuvant therapy choices for CM patients supports the need for accurate risk assessment in patients eligible for adjuvant therapy and suggests additional potential utility for this prognostic tool.
DISCLOSURES
ECH is on the speaker bureaus of Amgen, Inc and Castle Biosciences, Inc
AUTHOR CONTRIBUTIONS
Study conception and design: Hsueh, Schwartz, Hurley, Patel. Acquisition of data: Keller, Chang, Patel, Lizalek, Hsueh. Analysis and interpretation of data: Hsueh, Keller, Schwartz. Drafting of manuscript: Keller, Schwartz, Chang, Hurley, Hsueh. Critical revision: Keller, Schwartz, Hsueh.
Supporting information
ACKNOWLEDGMENTS
The authors appreciate the editorial support from Brooke Middlebrook, Kristen Meldi Plasseraud, and Sarah J. Kurley with statistical assistance from Kyle R. Covington, all from Castle Biosciences, Inc.
Keller J, Schwartz TL, Lizalek JM, et al. Prospective validation of the prognostic 31‐gene expression profiling test in primary cutaneous melanoma. Cancer Med. 2019;8:2205–2212. 10.1002/cam4.2128
Funding information
This study was supported by the Saint Louis University Cancer Center.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7‐30. 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2. Balch CM, Gershenwald JE, Soong S‐J, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27(36):6199‐6206. 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. 10.1245/s10434-010-0985-4 [DOI] [PubMed] [Google Scholar]
- 4. Gershenwald JE, Scolyer RA, Hess KR, et al. Melanoma staging: evidence‐based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017. 67(6):472–492. 10.3322/caac.21409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gerami P, Cook RW, Wilkinson J, et al. Development of a prognostic genetic signature to predict the metastatic risk associated with cutaneous melanoma. Clin Cancer Res. 2015;21(1):175–183. 10.1158/1078-0432.CCR-13-3316. [DOI] [PubMed] [Google Scholar]
- 6. Gerami P, Cook RW, Russell MC, et al. Gene expression profiling for molecular staging of cutaneous melanoma in patients undergoing sentinel lymph node biopsy. J Am Acad Dermatol. 2015;72(5): 780–785.e3. 10.1016/j.jaad.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 7. Greenhaw BN, Zitelli JA, Brodland DG. Estimation of prognosis in Invasive Cutaneous Melanoma: an independent study of the accuracy of a gene expression profile test. Dermatol Surg. 2018;44(12):1494–1500. 10.1097/DSS.0000000000001588 [DOI] [PubMed] [Google Scholar]
- 8. Hsueh EC, DeBloom JR, Lee J, et al. Interim analysis of survival in a prospective, multi‐center registry cohort of cutaneous melanoma tested with a prognostic 31‐gene expression profile test. J Hematol Oncol. 2017;10(1):152 10.1186/s13045-017-0520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gastman BR, Gerami P, Kurley SJ, Cook RW, Leachman S, Vetto JT. Identification of patients at risk for metastasis using a prognostic 31‐gene expression profile in subpopulations of melanoma patients with favorable outcomes by standard criteria. J Am Acad Dermatol. 2018; 80(1): 149–157.e4. 10.1016/j.jaad.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 10. Zager JS, Gastman BR, Leachman S, et al. Performance of a prognostic 31‐gene expression profile in an independent cohort of 523 cutaneous melanoma patients. BMC Cancer. 2018;18(1):130 10.1186/s12885-018-4016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Faries MB, Steen S, Ye X, Sim M, Morton DL. Late recurrence in melanoma: clinical implications of lost dormancy. J Am Coll Surg. 2013;217(1): 27–34. 10.1016/j.jamcollsurg.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Beek E, Balm A, Nieweg OE, Hamming‐Vrieze O, Lohuis P, Klop W. Treatment of regional metastatic melanoma of unknown primary origin. Cancers. 2015;7(3):1543–1553. 10.3390/cancers7030849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murali R, Brown PT, Kefford RF, et al. Number of primary melanomas is an independent predictor of survival in patients with metastatic melanoma. Cancer. 2012;118(18):4519–4529. 10.1002/cncr.27693. [DOI] [PubMed] [Google Scholar]
- 14. Petersen RP, Hanish SI, Haney JC, et al. Improved survival with pulmonary metastasectomy: an analysis of 1720 patients with pulmonary metastatic melanoma. J Thorac Cardiovasc Surg. 2007;133(1):104–110. 10.1016/j.jtcvs.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 15. Essner R, Lee JH, Wanek LA, Itakura H, Morton DL. Contemporary surgical treatment of advanced‐stage melanoma. Arch Surg. 2004;139(9): 961–966. 10.1001/archsurg.139.9.961 [DOI] [PubMed] [Google Scholar]
- 16. Poklepovic AS, Carvajal RD. Prognostic value of low tumor burden in patients with melanoma. Oncology (Williston Park). 2018;32(9):e90–e96. [PubMed] [Google Scholar]
- 17. Nsengimana J, Laye J, Filia A, et al. Independent replication of a melanoma subtype gene signature and evaluation of its prognostic value and biological correlates in a population cohort. Oncotarget. 2015;6(13):11683–11693. 10.18632/oncotarget.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harbst K, Staaf J, Lauss M, et al. Molecular profiling reveals low‐ and high‐grade forms of primary melanoma. Clin Cancer Res. 2012;18(15):4026–4036. 10.1158/1078-0432.CCR-12-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brunner G, Heinecke A, Falk TM, et al. A prognostic gene signature expressed in primary cutaneous melanoma: synergism with conventional staging. JNCI Cancer Spectrum, 2018;2(3): pky032 10.1093/jncics/pky032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gartrell RD, Marks DK, Rizk EM, et al. Validation of Melanoma Immune Profile (MIP), a prognostic immune gene prediction score for stage II‐III Melanoma. Clin Cancer Res. 2019. 10.1158/1078-0432.CCR-18-2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang Li, Lv X, Li J, et al. The status of microRNA‐21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. 2012;114(6):582–588. 10.1016/j.acthis.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 22. Li N. Low expression of Mir‐137 predicts poor prognosis in cutaneous melanoma patients. Med Sci Monit. 2016;22:140–144. 10.12659/MSM.895207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang K, Zhang Z‐W. Expression of miR‐203 is decreased and associated with the prognosis of melanoma patients. Int J Clin Exp Pathol. 2015;8(10):13249–13254. [PMC free article] [PubMed] [Google Scholar]
- 24. Tembe V, Schramm S‐J, Stark MS, et al. MicroRNA and mRNA expression profiling in metastatic melanoma reveal associations with BRAF mutation and patient prognosis. Pigment Cell Melanoma Res. 2015;28(3):254–266. 10.1111/pcmr.12343. [DOI] [PubMed] [Google Scholar]
- 25. Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367(18):1694–1703. 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ascierto PA, McArthur GA, Dréno B, et al. Cobimetinib combined with vemurafenib in advanced BRAF(V600)‐mutant melanoma (coBRIM): updated efficacy results from a randomised, double‐blind, phase 3 trial. Lancet Oncol. 2016;17(9):1248–1260. 10.1016/S1470-2045(16)30122-X. [DOI] [PubMed] [Google Scholar]
- 27. Robert C, Schachter J, Long GV, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372(26):2521–2532. 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 28. Topalian SL, Sznol M, McDermott DF, et al. Survival, durable tumor remission, and long‐term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol. 2014;32(10):1020–1030. 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andtbacka R, Kaufman HL, Collichio F, et al. Talimogene Laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33(25):2780–2788. 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 30. Eggermont A, Chiarion‐Sileni V, Grob J‐J, et al. Prolonged survival in stage III Melanoma with Ipilimumab adjuvant therapy. N Engl J Med. 2016;375(19):1845–1855. 10.1056/NEJMoa1611299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Long GV, Hauschild A, Santinami M, et al. Adjuvant Dabrafenib plus Trametinib in stage III BRAF‐Mutated Melanoma. N Engl J Med. 2017;377(19):1813–1823. 10.1056/NEJMoa1708539. [DOI] [PubMed] [Google Scholar]
- 32. Weber J, Mandala M, Del Vecchio M, et al. Adjuvant Nivolumab versus Ipilimumab in resected stage III or IV Melanoma. N Engl J Med. 2017;377(19):1824–1835. 10.1056/NEJMoa1709030. [DOI] [PubMed] [Google Scholar]
- 33. Kirkwood JM, Strawderman MH, Ernstoff MS, Smith TJ, Borden EC, Blum RH. Interferon alfa‐2b adjuvant therapy of high‐risk resected cutaneous melanoma: the Eastern Cooperative Oncology Group Trial EST 1684. J Clin Oncol. 1996;14(1):7–17. 10.1200/JCO.1996.14.1.7. [DOI] [PubMed] [Google Scholar]
- 34. Eggermont A, Blank CU, Mandala M, et al. Adjuvant Pembrolizumab versus placebo in resected stage III melanoma. N Engl J Med. 2018;378(19):1789–1801. 10.1056/NEJMoa1802357. [DOI] [PubMed] [Google Scholar]
- 35. Berger AC, Davidson RS, Poitras JK, et al. Clinical impact of a 31‐gene expression profile test for cutaneous melanoma in 156 prospectively and consecutively tested patients. Curr Med Res Opin. 2016;32(9):1599–1604. 10.1080/03007995.2016.1192997. [DOI] [PubMed] [Google Scholar]
- 36. Svoboda RM, Glazer AM, Farberg AS, Rigel DS. Factors affecting dermatologists' use of a 31‐Gene expression profiling test as an adjunct for predicting metastatic risk in cutaneous melanoma. J Drugs Dermatol. 2018;17(5):544–547. [PubMed] [Google Scholar]
- 37. Schuitevoerder D, Heath M, Cook RW, et al. Impact of gene expression profiling on decision‐making in clinically node negative melanoma patients after surgical staging. J Drugs Dermatol. 2018;17(2):196–199. [PubMed] [Google Scholar]
- 38. Farberg AS, Glazer AM, White R, Rigel DS. Impact of a 31‐gene expression profiling test for cutaneous melanoma on dermatologists' clinical management decisions. J Drugs Dermatol. 2017;16(5):428–431. [PubMed] [Google Scholar]
- 39. Dillon LD, Gadzia JE, Davidson RS, et al. Multicenter clinical impact evaluation of a 31‐gene expression profile test for management of melanoma patients. Skin J Cutan Med. 2018;2(2). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


