Abstract
Dystrophinopathies are multi-system disorders that affect the skeletal musculature, the cardio-respiratory system and the central nervous system. The systematic screening of suitable biofluids for released or altered proteins promises new insights into the highly complex pathophysiology of X-linked muscular dystrophy. However, standard detection approaches using antibody-based assays often fail to reproducibly detect low-abundance protein isoforms in dilute biological fluids. In contrast, mass spectrometric screening approaches enable the proteome-wide identification of minor protein changes in biofluids. This report describes the findings from the comparative proteomic analysis of whole saliva samples from wild type versus the established mdx-4cv mouse model of highly progressive muscular dystrophy, focusing on the kallikrein protein family. Kallikrein-1 (Klk1) and 13 Klk1-related peptidases were identified in saliva and serum from normal mice. Comparative proteomics revealed elevated saliva levels of the Klk1-related peptidases Klk1-b1, Klk1-b5 and Klk-b22, as well as an increased Klk-1 concentration, which agrees with higher Klk-1 levels in serum from mdx-4cv mice. This indicates altered cellular signaling, extracellular matrix remodeling and an altered immune response in the mdx-4cv mouse, and establishes liquid biopsy procedures as suitable bioanalytical tools for the systematic survey of complex pathobiochemical changes in animal models of muscular dystrophy.
Keywords: Dystrophinopathy, Kallikrein-1, Kallikrein-related peptidase, Klk, Saliva proteomics, X-linked muscular dystrophy
Highlights
-
•
Dystrophin deficiency is associated with proteome-wide changes in saliva.
-
•
Proteomics of the mdx-4cv mouse revealed increased levels of saliva kallikrein-1.
-
•
Saliva Klk1-related peptidases b1, b5 and b22 are increased in the mdx-4cv mouse.
-
•
Saliva changes indicate altered cellular signaling in the mdx-4cv mouse.
-
•
Proteome-wide alterations in saliva agree with an altered immune response.
1. Introduction
The systematic identification of pathobiochemical changes in proteins is the focus of quantitative body fluid proteomics, aiming at both the improved understanding of alterations in local versus multi-system disease processes and the discovery of proteome-wide changes [1]. If distinct alterations in the protein composition of biological fluids, such as blood, cerebrospinal fluid, bronchoalveolar lavage fluid, saliva, tears, sweat or urine, mirror disease-related alterations, then these body fluids present an excellent starting point for the in-depth analysis of complex pathophysiological processes. Although the plasma/serum and urine proteomes represent well established sources for the discovery of new disease markers [2], [3], [4], the existence of a small number of highly abundant protein species and the extensive dynamic range of proteins in these biofluids present challenging analytical obstacles [5]. The 15 most abundant proteins in urine and plasma account for approximately 60% and 90% of the total biofluid proteome, respectively [6], [7]. In contrast, this class of highly abundant proteins makes up only approximately 30% of the total saliva protein complement, as recently confirmed by the advanced mass spectrometric cataloguing of the human saliva proteome [8].
The saliva proteome is primarily composed of protein species derived from the oral cavity, but also infiltrating molecules from plasma. This includes many protein species with a considerable diagnostic, prognostic or disease/therapy-monitoring potential, such as enzymes, cytokines, hormones and circulating protein factors [9]. Hence, analyzing the protein constituents of saliva offers potentially a comprehensive insight into both local and systemic changes due to physiological adaptations or pathological insults [10]. Saliva diagnostics is already established for the evaluation of oral diseases, including the detection of dental caries, gingivitis and periodontal diseases, and the autoimmune Sjogren's syndrome that is characterized by salivary and lacrimal gland dysfunction, as well as oral cancer and certain types of head and neck cancer [11]. In addition, circulating marker proteins found in saliva have the potential to be also useful for the clinical evaluation of common systemic diseases, such as diabetes mellitus, cardiovascular disorders, cancers of the pancreas, breast, lungs or prostate, infectious diseases, renal abnormalities and neurological disorders [12].
In this report, the proteomic characterization of saliva has been extended to studying the mdx-4cv mouse model of the most frequently inherited neuromuscular disorder of early childhood, Duchenne muscular dystrophy [13]. Primary abnormalities in the Dmd gene cause the almost complete absence of the membrane cytoskeletal protein dystrophin in skeletal muscle tissues and result in the collapse of the sarcolemmal dystrophin-associated glycoprotein complex [14]. Loss of membrane integrity in dystrophin-deficient muscles triggers a complex pathogenesis that is characterized by abnormal excitation-contraction coupling, impaired ion homeostasis, fibre degeneration, reactive myofibrosis and sterile inflammation [15], [16], [17]. Subtypes of skeletal muscles are differentially affected in dystrophinopathies. Muscle wasting causes initially proximal lower limb and truncal weakness, which is followed by the progressive degeneration of upper limb and distal muscles [18]. In addition to the affected skeletal musculature, secondary complications occur in the respiratory system, the heart and the central nervous system of Duchenne patients [19], [20].
Saliva proteomics was carried out with samples from the established mdx-4cv mouse model of dystrophinopathy [21], which exhibits characteristic proteome-wide abnormalities in the skeletal musculature, heart and brain [22], [23], [24]. Proteomic findings were compared to results from the recent mass spectrometric profiling of mdx-4cv serum [25] and revealed significantly elevated levels of kallikrein-1 and kallikrein-related peptidases [26]. The kallikrein/kinin and bradykinin signaling axis of vasoactive peptides is involved in blood pressure regulation, tissue homeostasis and renal function. In addition, the endogenous kallikrein/kinin system was shown to be involved in various neurological diseases [27] and appears to play a protective role in dystrophic mdx muscle [28]. Building on these findings and the recent proteomic identification of greatly reduced levels of the kallikrein-1 related peptidase Klk1-b9 in mdx muscle [29], it was of interest to study the status of kallikreins in mdx-4cv saliva specimens. The establishment of elevated saliva kallikrein-1 and kallikrein-related peptidases, shown in this report using comparative proteomics, may be related to altered cellular signaling, remodeling of the extracellular matrix and an altered immune response in the mdx-4cv mouse model of dystrophinopathy.
2. Materials and methods
2.1. Materials
The general analytical grade reagents and materials used in this study were sourced from GE Healthcare (Little Chalfont, Buckinghamshire, UK), BioRad Laboratories (Hemel-Hempstead, Hertfordshire, UK) and Sigma Chemical Company (Dorset, UK). Ultrapure acrylamide stock solutions were purchased from National Diagnostics (Atlanta, GA, USA), sequencing grade modified trypsin was obtained from Promega (Madison, WI, USA) and protease inhibitors were purchased from Roche Diagnostics (Mannheim, Germany). For the filter-aided sample preparation method FASP, Vivacon 500 with 30 kDa molecular mass cut-off spin filters were acquired from Sartorius (Göttingen, Germany).
2.2. Saliva samples from the mdx-4cv mouse model of Duchenne muscular dystrophy
The mdx-4cv mouse is a mutant variant of the spontaneous mdx model of Duchenne muscular dystrophy that has been generated by chemical mutagenesis [30]. For the proteomic profiling of biofluids, saliva samples were obtained from 6-month-old control C57BL/6 mice and age-matched mdx-4cv mice through the Bioresource Unit of the University of Bonn [22], where mice were kept under standard conditions according to German legislation on the use of animals in experimental research. A small crystal of Vitamin C was carefully placed in the mouth of mice immediately post mortem, thereby inducing a salivating reflex. The produced saliva was suctioned off with a micro pipette and the oral cavity irrigated with 10μl of phosphate-buffered saline. The collected saliva was then immediately placed in liquid nitrogen. Samples were transported to Maynooth University on dry ice in accordance with the Department of Agriculture (animal by-product register number 2016/16 to the Department of Biology, National University of Ireland, Maynooth) and stored at − 80 °C prior to analysis. To minimize degradation of salivary proteins, protease inhibitor cocktails [24] were added to individual samples from wild type and mdx-4 cv mice.
2.3. Proteolytic digestion of salivary proteins
The protein concentration of saliva samples was equalized with label-free solubilisation buffer (6 M urea, 2 M thiourea, 10 mM Tris, pH 8.0 in LC-MS grade water), and 30 µg of protein was processed by the filter-aided sample preparation (FASP) method, as described in detail by Wiśniewski et al. [31], using a trypsin to protein ratio of 1:25 (protease:protein). Following overnight digestion and elution of peptides from the spin filter, 2% trifluoroacetic acid (TFA) in 20% acetonitrile (ACN) was added to the filtrates (3:1 (v/v) dilution). The peptides were then purified using Pierce C18 spin columns from Thermo Fisher Scientific (Dublin, Ireland), dried through vacuum centrifugation and re-suspended in mass spectrometry loading buffer (2% ACN, 0.05% TFA in LC-MS grade water). Peptides were vortexed, sonicated and briefly centrifuged at 14,000×g and the supernatant transferred to mass spectrometry vials for label-free liquid chromatography mass spectrometry (LC-MS/MS).
2.4. Label-free liquid chromatography mass spectrometry
The LC-MS/MS analysis of salivary peptides was performed using an Ultimate 3000 NanoLC system (Dionex Corporation, Sunnyvale, CA, USA) coupled to a Q-Exactive mass spectrometer (Thermo Fisher Scientific). 800 ng of each digested sample was loaded by an auto-sampler onto a C18 trap column (C18 PepMap, 300 µm id × 5 mm, 5 µm particle size, 100 Å pore size; Thermo Fisher Scientific). The trap column was switched on-line with an analytical Biobasic C18 Picofrit column (C18 PepMap, 75 µm id × 50 cm, 2 µm particle size, 100 Å pore size; Dionex). Peptides were eluted over a 65-min binary gradient [solvent A: 2% (v/v) ACN and 0.1% (v/v) formic acid in LC-MS grade water and solvent B: 80% (v/v) ACN and 0.1% (v/v) formic acid in LC-MS grade water]: 3% solvent B for 5 min, 3–10% solvent B for 5 min, 10–40% solvent B for 30 min, 40–90% solvent B for 5 min, 90% solvent B for 5 min and 3% solvent B for 10 min [32]. The column flow rate was set to 0.3 µL/min. Data were acquired with Xcalibur software (Thermo Fisher Scientific). The mass spectrometer was externally calibrated and operated in positive, data-dependent mode. A full survey MS scan was performed in the 300–1700 m/z range with a resolution of 140,000 (m/z 200) and a lock mass of 445.12003. Collision-induced dissociation (CID) fragmentation was carried out with the fifteen most intense ions per scan and at 17,500 resolution. Within 30 s a dynamic exclusion window was applied. An isolation window of 2 m/z and one microscan were used to collect suitable tandem mass spectra.
2.5. Protein identification and quantification
Proteins present in the wild type and the mdx-4cv salivary proteomes were initially identified using Proteome Discoverer 1.4 against Sequest HT (SEQUEST HT algorithm, licence Thermo Scientific, registered trademark University of Washington, USA) using the UniProtKB/Swiss-Prot database, with 25,041 sequences for Mus musculus. The following search parameters were used for protein identification: (i) peptide mass tolerance set to 10 ppm, (ii) MS/MS mass tolerance set to 0.02 Da, (iii) an allowance of up to two missed cleavages, (iv) carbamidomethylation set as a fixed modification and (v) methionine oxidation set as a variable modification [32]. Peptides were filtered using a minimum XCorr score of 1.5 for 1, 2.0 for 2, 2.25 for 3 and 2.5 for 4 charge states, with peptide probability set to high confidence. XCorr is a search-dependent score employed by the SEQUEST HT search engine in Proteome Discoverer, and reflects the number of fragment ions that are common to two different peptides with the same precursor mass. Since the XCorr value is dependent upon the number of identified fragment ions, its value is usually higher for larger peptides. XCorr scores are filtered based on charge state, whereby larger XCorr thresholds are used for higher charge states. For quantitative analysis, samples were evaluated with MaxQuant software (version 1.6.1.0) and the Andromeda search engine used to explore the detected features against the UniProtKB/SwissProt database for Mus musculus. The following search parameters were used: i) first search peptide tolerance of 20 ppm, ii) main search peptide tolerance of 4.5 ppm, iii) cysteine carbamidomethylation set as a fixed modification, iv) methionine oxidation set as a variable modification, v) a maximum of two missed cleavage sites and vi) a minimum peptide length of seven amino acids. The false discovery rate (FDR) was set to 1% for both peptides and proteins using a target-decoy approach [8]. Relative quantification was performed using the MaxLFQ algorithm [33]. The “proteinGroups.txt” file produced by MaxQuant was further analyzed in Perseus (version 1.5.1.6). Proteins that matched to the reverse database or a contaminants database or that were only identified by site were removed. The LFQ intensities were log2 transformed, and only proteins found in all eight replicates in at least one group were used for further analysis. Data imputation was performed to replace missing values with values that simulate signals from peptides with low abundance chosen from a normal distribution specified by a downshift of 1.8 times the mean standard deviation of all measured values and a width of 0.3 times this standard deviation [34]. A two-sample t-test was performed using p < 0.05 on the post imputated data to identify statistically significant differentially abundant proteins.
3. Results and discussion
3.1. Proteomic identification of kallikrein isoform Klk1 and Klk1-related peptidases in saliva and serum
The recent proteomic survey of the mdx mouse model of dystrophinopathy confirmed that the dystrophin isoform Dp427-M represents the most reduced protein species in the dystrophic quadriceps femoris muscle and established the kallikrein-1 (Klk1) related peptidase Klk1-b9 as the second most reduced muscle-associated protein [29]. In addition, proteomic profiling of the dystrophic mdx-4cv mouse revealed increased Klk1 levels in serum [25]. Based on these findings, it was of interest to evaluate the presence of kallikreins and related peptidases in normal saliva, as compared to more widely researched serum samples [4], and investigate potential changes in saliva from dystrophic rodents. As listed in Table 1, Klk1 and 13 Klk1-related peptidases, i.e. isoforms b1, b3, b4, b5, b8, b9, b11, b16, b21, b22, b24, b26 and b27 [26], were identified by mass spectrometry in both saliva and serum from normal mice. The mass spectrometrically identified total saliva protein population and the differential presence of protein species in saliva versus serum samples are listed in the accompanying Data-in-Brief publication [35].
Table 1.
Mass spectrometric identification of kallikrein-1 and kallikrein-related peptidases in mouse saliva and serum.
| Accession Number | Protein Name | Number of unique peptides in saliva | % Sequence coverage in saliva | Number of unique peptides in serum | % Sequence coverage in serum |
|---|---|---|---|---|---|
| P15947 | Kallikrein-1 (Klk1) | 2 | 27.59 | 3 | 38.7 |
| P00755 | Klk1-related peptidase b1 | 4 | 41.76 | 10 | 72.8 |
| P00756 | Klk1-related peptidase b3 | 4 | 39.46 | 8 | 77.01 |
| P00757 | Klk1-related peptidase b4 | 6 | 33.98 | 10 | 43.75 |
| P15945 | Klk1-related peptidase b5 | 4 | 30.27 | 6 | 47.51 |
| P07628 | Klk1-related peptidase b8 | 7 | 49.43 | 8 | 70.5 |
| P15949 | Klk1-related peptidase b9 | 5 | 45.98 | 10 | 68.97 |
| P15946 | Klk1-related peptidase b11 | 5 | 44.83 | 13 | 73.56 |
| P04071 | Klk1-related peptidase b16 | 7 | 44.83 | 9 | 60.92 |
| Q61759 | Klk1-related peptidase b21 | 2 | 37.55 | 2 | 49.43 |
| P15948 | Klk1-related peptidase b22 | 5 | 27.03 | 13 | 71.81 |
| Q61754 | Klk1-related peptidase b24 | 3 | 38.4 | 4 | 42.59 |
| P36369 | Klk1-related peptidase b26 | 3 | 42.53 | 5 | 57.47 |
| Q9JM71 | Klk1-related peptidase b27 | 3 | 41.83 | 6 | 58.17 |
3.2. Comparative proteomic profiling of kallikrein-1 and Klk1-related peptidases in saliva from the mdx-4cv mouse model of dystrophinopathy
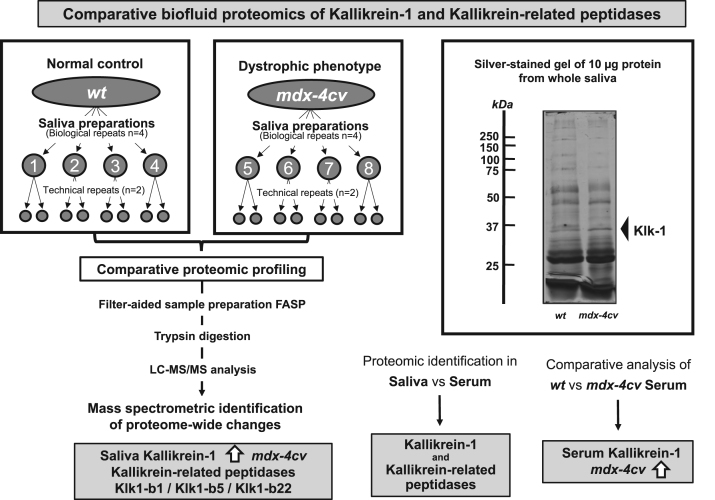
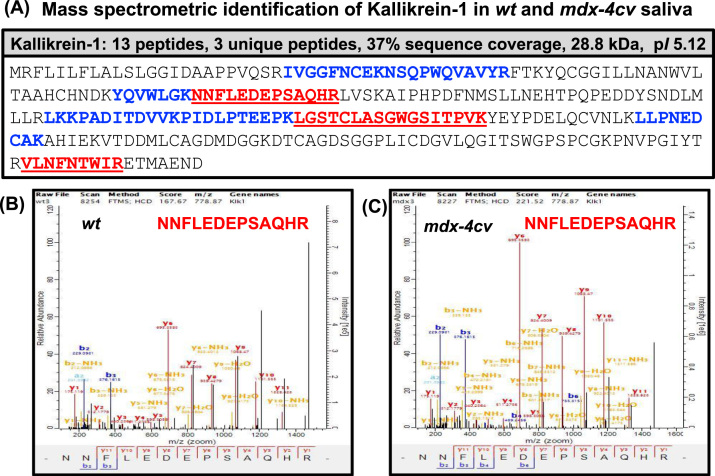
Following the proteomic cataloguing of Klk1 and related peptidases in wild type saliva, whole saliva samples from normal versus the established mdx-4cv mouse model of highly progressive X-linked muscular dystrophy were analyzed by mass spectrometry. Fig. 1 gives an overview of the proteomic workflow and the number of biological and technical repeats used in this comparative study, as well as a summary of the main findings with a focus on altered kallikreins. The representative silver-stained gel of whole saliva specimens shows a relatively comparable expression pattern of proteins in wild type versus mdx-4cv samples with only a few differences in the intensity of individual protein bands (Fig. 1). The mass spectrometric identification of Klk1 of apparent 28.8 kDa, which apparent protein band is marked in the SDS-PAGE gel at the approximate 30 kDa region (Fig. 1), established its presence in mdx-4cv saliva with a 37% sequence coverage with 13 peptides and 3 unique peptides (Fig. 2A). Representative MS/MS scans are shown for the unique peptide NNFLEDEPSAQHR that was used to identify the Klk-1 isoform in both wild type and mdx-4cv samples, which illustrates higher intensities of peaks from mdx-4cv peptides compared to wild type peptides (Fig. 2B, C). Additional MS/MS scans of unique peptides used to identify kallikrein are provided in the accompanying Data in Brief article [35].
Fig. 1.
Comparative proteomic profiling of kallikrein and related peptidases in saliva from the mdx-4cv model of Duchenne muscular dystrophy. Shown is the bioanalytical workflow and major findings of the mass spectrometric characterization of changes in kallikrein-1 and related peptidases. A representative silver-stained protein gel of whole saliva samples from wild type versus dystrophic mice is shown. The relative position of the approximate 30 kDa Klk-1 band is marked by an arrowhead. Molecular mass standards are indicated on the left of the gel.
Fig. 2.
Proteomic identification of kallikrein isoform Klk1 in saliva from the mdx-4cv mouse model of Duchenne muscular dystrophy. (A) Shown is the sequence coverage of kallikrein as determined by LC-MS/MS analysis. Unique peptide sequences are underlined and all identified peptide regions are shown in bold. Panels (B) and (C) show representative MS/MS scans of the unique Klk-1 peptide NNFLEDEPSAQHR, which was identified and compared in wild type versus mdx-4cv saliva, respectively.
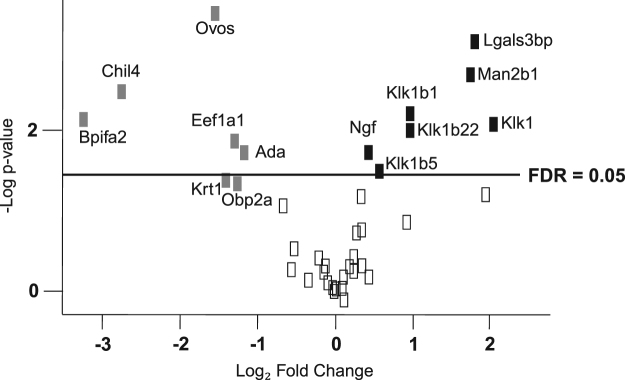
As listed in Table 2, comparative proteomics revealed elevated saliva levels of the Klk1-related peptidases Klk1-b1, Klk1-b5 and Klk-b22, as well as an increased Klk-1 concentration, which agrees with higher Klk-1 levels in serum from mdx-4cv mice [25]. In addition, galectin-3-binding protein, lysosomal alpha-mannosidase and beta-nerve growth factor were found to be increased in mdx-4cv saliva. Mass spectrometric analysis identified a reduced concentration for adenosine deaminase, odorant-binding protein 2a, elongation factor-1 (alpha 1 and 2), type II keratin (cytoskeletal 1), ovostatin, chitinase-like protein 4 and the BPI fold-containing family A member 2 in mdx-4cv saliva (Table 2). Fig. 3 shows a volcano plot of all identified proteins based on relative abundance differences between wild type and mdx-4cv saliva, illustrating the distribution of quantified proteins according to p-value and fold change.
Table 2.
List of proteins with an altered abundance in whole saliva samples from the mdx-4cv mouse model of Duchenne muscular dystrophy as revealed by comparative mass spectrometry-based proteomics.
| Protein ID | Protein Name | Gene Name | Number of peptides | Number of unique peptides | p-value | Fold change | Score |
|---|---|---|---|---|---|---|---|
| P15947 | Kallikrein-1 | Klk1 | 13 | 3 | 0.008229 | 4.16 | 189.7 |
| Q07797 | Galectin-3-binding protein | Lgals3bp | 4 | 4 | 0.000788 | 3.54 | 33.4 |
| O09159 | Lysosomal alpha-mannosidase | Man2b1 | 21 | 21 | 0.001934 | 3.36 | 323.3 |
| P15948 | Kallikrein 1-related peptidase b22 | Klk1b22 | 14 | 9 | 0.010188 | 1.97 | 323.3 |
| P00755 | Kallikrein 1-related peptidase b1 | Klk1b1 | 14 | 6 | 0.006031 | 1.96 | 215.4 |
| P15945 | Kallikrein 1-related peptidase b5 | Klk1b5 | 13 | 5 | 0.031962 | 1.49 | 323.3 |
| P01139 | Beta-nerve growth factor | Ngf | 4 | 4 | 0.018827 | 1.34 | 323.3 |
| P03958 | Adenosine deaminase | Ada | 4 | 4 | 0.017819 | 0.44 | 56.6 |
| Q8K1H9 | Odorant-binding protein 2a | Obp2a | 3 | 3 | 0.045699 | 0.42 | 75.9 |
| P10126;P62631 | Elongation factor-1, | Eef1a1 | 7 | 7 | 0.013068 | 0.41 | 100.4 |
| alpha 1 and 2 | Eef1a2 | ||||||
| P04104 | Keratin, type II, cytoskeletal 1 | Krt1 | 10 | 5 | 0.039762 | 0.37 | 323.3 |
| Q3UU35 | Ovostatin homolog | Ovos | 6 | 6 | 0.000347 | 0.34 | 323.3 |
| Q91Z98 | Chitinase-like protein 4 | Chil4 | 7 | 7 | 0.003153 | 0.15 | 95.2 |
| P07743 | BPI fold-containing family A member 2 | Bpifa2 | 6 | 6 | 0.007214 | 0.11 | 289.8 |
Fig. 3.
Volcano plot of all identified proteins based on relative abundance differences between wild-type and mdx-4cv saliva. The volcano plot illustrates the distribution of quantified saliva proteins according to p-value (-log10 p-value) and fold change (log2 mean LFQ intensity difference). Grey and black boxes represent individual protein species that were identified to exhibit a statistically significant (p-value < 0.05) decrease or increase in their abundance in mdx-4cv saliva, respectively. All protein-presenting boxes above the line are those which also passed an FDR criteria of 0.05.
Kallikreins have a great potential as general markers of degenerative processes [36] and have previously been identified in saliva by various proteomic approaches [37]. Kallikreins of approximately 30 kDa appear to be relatively abundant protein species within the saliva proteome and are stable constituents of this body fluid. Thus, although proteins are only minor components of watery saliva [10], the kallikreins can be readily identified by sensitive biochemical methodology in dilute saliva samples. This establishes kallikreins as potential biomarker candidates of secondary changes due to dystrophin deficiency in the mdx-4cv mouse model of Duchenne muscular dystrophy. However, an important modulating factor to take into account is the potential effect of prednisolone treatment on kallikrein levels. Gene expression profiles of prednisolone-treated mdx mice showed a delayed over-expression of genes encoding kallikreins 13, 16 and 26 [38]. Based on the initial proteomic identification of saliva kallikreins as new biomarker candidates, as reported here, it will be crucial to carry out a more detailed characterization of potential confounding factors in future analyses. This would include the evaluation of possible influences of saliva kallikrein levels due to general life style issues, physical activity, emotional stress, disease-unrelated neuromuscular trauma, inflammation, metabolic abnormalities, co-morbidities such as cardiomyopathic complications in dystrophinopathy, diet, pharmacological side effects and/or surgical procedures, as discussed below.
At the level of pathophysiological mechanisms, elevated levels of circulating kallikrein isoform Klk-1 and Klk1-related peptidases suggest altered cellular signaling at the level of the kallikrein-kinin system and the regulation of tissue homeostasis [28], as well as remodeling of the extracellular matrix and a modification of the immune response in X-linked muscular dystrophy [39]. Thus, comparative saliva proteomics is capable of establishing distinct changes in the biofluid protein complement and this study confirmed that liquid biopsy procedures are suitable bioanalytical tools for the systematic survey of complex pathobiochemical changes in muscular dystrophy [4].
3.3. Saliva as a suitable biofluid for non-invasive and systemic sampling approaches in neuromuscular pathology
Routine clinical biofluid analyses in the field of muscle pathology are usually restricted to simple blood tests with a focus on muscle-derived serum proteins that are indicative of skeletal muscle damage, including creatine kinase, myoglobin, alanine aminotransferase, aspartate aminotransferase and a variety of glycolytic enzymes [40]. However, in the case of childhood disorders of the neuromuscular system, these serum markers are relatively non-specific and potentially susceptible to (i) modifying effects due to co-morbidities, (ii) considerable age-related changes during childhood development, (iii) fluctuations during disease progression, (iv) obscuring influences of patient gender and ethnicity, (v) prone to changes due to general life style, nutritional status, metabolic imbalances and physical activity levels, (vi) modifying effects of intramuscular injections, surgery or drug treatments, and (vii) seasonal variations in concentration levels [4]. Taken into account the above listed issues with existing diagnostic markers of muscle damage in a disease of early childhood, the findings reported here are of considerable bioanalytical interest and establish whole saliva samples as a suitable source for the establishment of superior biomarker signatures [41].
Importantly, the more evenly distributed concentration range of saliva-associated protein species, as compared to serum/plasma and urine, allows the routine usage of whole saliva specimens without the need for elaborative pre-fractionation steps in large-scale proteomic surveys [8], [9], [10]. This establishes the saliva proteome as an ideal non-invasive and systemic sampling measure for the discovery of proteome-wide changes. Whole saliva is a highly complex mixture of fluids, which are crucial for the physiological functioning, preservation and maintenance of tissues in the oral cavity. Saliva derives mostly from the secretory activities of salivary glands, including the major submandibular, paratoid and sublingual glands and the minor labial, palatine, lingual and buccal glands [42]. Kallikrein, which acts as an enzyme that converts kininogen to the vasodilator bradykinin, is secreted by the acinar cells of major salivary glands. However, salivary glands exhibit a high permeability for their surrounding capillaries, which allows the infiltration of circulating peptides and proteins from blood. The proteomic comparison of wild type saliva and wild type serum revealed a 57% commonality in protein species. Thus, saliva kallikreins might originate to a large extent from blood and this might explain the similar changes of Klk1 in blood and saliva of the mdx-4cv mouse model of dystrophinopathy, as established here by proteomic analysis.
3.4. Biofluid markers of the molecular pathogenesis of muscular dystrophy
The newly established increase in kallikreins in both saliva and serum from the dystrophic mdx-4cv mouse is an appropriate addition to the list of already established biofluid marker candidates of X-linked muscular dystrophy. Biofluid marker research has mostly focused on the effects of dystrophin deficiency on plasma/serum specimens and urine [4]. A change of abundance in a great variety of muscle-derived proteins, mostly related to fibre degeneration, cellular stress and fibrosis, and non-muscle markers of secondary pathological processes, such as sterile inflammation, has been identified by mass spectrometric analysis [43]. This includes proteins or protein fragments of muscle components derived from the sarcomere, the membrane cytoskeleton, the non-sarcomeric cytoskeletal network, the extracellular matrix and major classes of organelles, such as the sarcolemma, the sarcoplasmic reticulum and mitochondria. These changes in the biofluid proteome reflect complex pathophysiological alterations due to progressive cellular degeneration and enhanced surface membrane rupturing. Abnormal Ca2+-handling at the level of the plasma membrane, cytosol and sarcoplasmic reticulum results in increased Ca2+-fluxes through the sarcolemma, elevated Ca2+-levels in the sarcosol and impaired luminal Ca2+-buffering, which triggers enhanced fibre proteolysis and skeletal muscle necrosis [17]. These major pathophysiological changes appear to be associated with considerable fluctuations in the protein constituents of biofluids.
Since the signaling axes of kallikreins and members of the matrix metalloproteinase family were shown to intersect and this may influence metalloproteinase function in the extracellular matrix under conditions of pathological dysregulation [44], kallikreins might be involved in the modification of the extracellular matrix [39]. In dystrophin-deficient muscle, significant increases in various collagens and the matricellular protein periostin [45], as well as the extracellular matrix proteins asporin, decorin, dermatopontin and prolargin [46], are typical features of reactive myofibrosis and contractile tissue damage [16]. Thus, kallikreins might act as modulating factors in the deposition of these myofibrotic protein aggregates in the mdx-4cv mouse. The recent proteomic profiling of highly purified sarcolemma vesicles from mdx-4cv muscle indicates the up-regulation of proteins involved in immune cell infiltration [47], which agrees with a robust inflammatory response by the innate immune system due to chronic skeletal muscle damage [48]. This process might also involve kallikreins that were previously shown to assist in immune invasion processes [39].
4. Conclusions
Using comparative proteomic profiling, elevated Klk1 levels were identified in both saliva and serum, suggesting that these biofluids are suitable for testing new disease marker candidates in animal models of progressive muscular dystrophy. Changes in kallikreins appear to be appropriate diagnostic targets, since these proteins are easily assessable by non-invasive or minimally invasive sampling procedures. Saliva Klk1 should therefore be considered a potential biomarker candidate for studying the mdx-4cv mouse model of dystrophinopathy, which might be suitable to establish a highly sensitive and robust assay system that is applicable for repeated diagnostic procedures. Changes in kallikrein concentration should be convenient for the development of a cost-effective assay system that is relatively specific for disease status and progression, and should not be prone to sampling errors. Solid biomarkers are especially needed for the improved monitoring of experimental therapeutic approaches during pre-clinical and clinical testing [49]. Considerable advances have been made over the last few years in the development of standardized saliva collection devices [50], therefore it should be relatively easy to apply saliva-based biomarker tests to the field of muscular dystrophy diagnostics.
Novel biomarker candidates, such as kallikreins, might be useful to evaluate new pharmacological substances that target ion homeostasis, muscle growth, fibre regeneration, blood flow, the cellular stress response, energy metabolism and/or myofibrosis, as well as the inflammatory response and oxidative stress in muscular dystrophy. A superior biomarker signature of dystrophic changes, as mirrored by proteome-wide changes in easily assessable biofluids, would also be beneficial for the routine evaluation of advanced therapeutic approaches, such as stem cell treatment, myoblast transfer, gene transfer, exon skipping, stop codon read-through, utrophin replacement and CRISPR/Cas9 genome editing. In the long-term, based on this type of applied muscular dystrophy research, newly characterized biomarkers can then hopefully be tested for their effectiveness to improve the diagnosis, prognosis and therapy monitoring of patients suffering from dystrophinopathy.
Acknowledgements
Research was supported by a Hume scholarship from Maynooth University, and project grants from Muscular Dystrophy Ireland and the Irish Health Research Board (HRB/MRCG-2016-20). The Q-Exactive quantitative mass spectrometer was funded under the Research Infrastructure Call 2012 by Science Foundation Ireland (SFI-12/RI/2346/3).
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.05.006.
Contributor Information
Sandra Murphy, Email: sandra.murphy@mu.ie.
Margit Zweyer, Email: margit.zweyer@ukbonn.de.
Rustam R. Mundegar, Email: mundegar@uni-bonn.de.
Dieter Swandulla, Email: dieter.swandulla@ukbonn.de.
Kay Ohlendieck, Email: kay.ohlendieck@mu.ie.
Appendix A. Transparency document
Supplementary material
References
- 1.Csősz É., Kalló G., Márkus B., Deák E., Csutak A., Tőzsér J. Quantitative body fluid proteomics in medicine – a focus on minimal invasiveness. J. Proteom. 2017;153:30–43. doi: 10.1016/j.jprot.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Geyer P.E., Kulak N.A., Pichler G., Holdt L.M., Teupser D., Mann M. Plasma proteome profiling to assess human health and disease. Cell Syst. 2016;2:185–195. doi: 10.1016/j.cels.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Beasley-Green A. Urine proteomics in the era of mass spectrometry. Int Neurourol. J. 2016;20:S70–S75. doi: 10.5213/inj.1612720.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy S., Zweyer M., Mundegar R.R., Swandulla D., Ohlendieck K. Proteomic serum biomarkers for neuromuscular diseases. Expert Rev. Proteom. 2018;15:277–291. doi: 10.1080/14789450.2018.1429923. [DOI] [PubMed] [Google Scholar]
- 5.Hortin G.L., Sviridov D. The dynamic range problem in the analysis of the plasma proteome. J. Proteom. 2010;73:629–636. doi: 10.1016/j.jprot.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Gianazza E., Miller I., Palazzolo L., Parravicini C., Eberini I. With or without you – proteomics with or without major plasma/serum proteins. J. Proteom. 2016;140:62–80. doi: 10.1016/j.jprot.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Nagaraj N., Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J. Proteome Res. 2011;10:637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 8.Grassl N., Kulak N.A., Pichler G., Geyer P.E., Jung J., Schubert S., Sinitcyn P., Cox J., Mann M. Ultra-deep and quantitative saliva proteome reveals dynamics of the oral microbiome. Genome Med. 2016;8:44. doi: 10.1186/s13073-016-0293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castagnola M., Cabras T., Iavarone F., Fanali C., Nemolato S., Peluso G., Bosello S.L., Faa G., Ferraccioli G., Messana I. The human salivary proteome: a critical overview of the results obtained by different proteomic platforms. Expert Rev. Proteom. 2012;9:33–46. doi: 10.1586/epr.11.77. [DOI] [PubMed] [Google Scholar]
- 10.Kaczor-Urbanowicz K.E., Martin Carreras-Presas C., Aro K., Tu M., Garcia-Godoy F., Wong D.T. Saliva diagnostics – current views and directions. Exp. Biol. Med. (Maywood) 2017;242:459–472. doi: 10.1177/1535370216681550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Podzimek S., Vondrackova L., Duskova J., Janatova T., Broukal Z. Salivary markers for periodontal and general diseases. Dis. Markers. 2016;2016:9179632. doi: 10.1155/2016/9179632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C.Z., Cheng X.Q., Li J.Y., Zhang P., Yi P., Xu X., Zhou X.D. Saliva in the diagnosis of diseases. Int. J. Oral Sci. 2016;8:133–137. doi: 10.1038/ijos.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guiraud S., Aartsma-Rus A., Vieira N.M., Davies K.E., van Ommen G.J., Kunkel L.M. The pathogenesis and therapy of muscular dystrophies. Annu Rev. Genom. Hum. Genet. 2015;16:281–308. doi: 10.1146/annurev-genom-090314-025003. [DOI] [PubMed] [Google Scholar]
- 14.Murphy S., Ohlendieck K. The biochemical and mass spectrometric profiling of the dystrophin complexome from skeletal muscle. Comput. Struct. Biotechnol. J. 2015;14:20–27. doi: 10.1016/j.csbj.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin J., Tajrishi M.M., Ogura Y., Kumar A. Wasting mechanisms in muscular dystrophy. Int. J. Biochem. Cell Biol. 2013;45:2266–2279. doi: 10.1016/j.biocel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holland A., Murphy S., Dowling P., Ohlendieck K. Pathoproteomic profiling of the skeletal muscle matrisome in dystrophinopathy associated myofibrosis. Proteomics. 2016;16:345–366. doi: 10.1002/pmic.201500158. [DOI] [PubMed] [Google Scholar]
- 17.Allen D.G., Whitehead N.P., Froehner S.C. Absence of dystrophin disrupts skeletal muscle signaling: roles of Ca2+, reactive oxygen species, and nitric oxide in the development of muscular dystrophy. Physiol. Rev. 2016;96:253–305. doi: 10.1152/physrev.00007.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yiu E.M., Kornberg A.J. Duchenne muscular dystrophy. J. Paediatr. Child Health. 2015;51:759–764. doi: 10.1111/jpc.12868. [DOI] [PubMed] [Google Scholar]
- 19.Anderson J.L., Head S.I., Rae C., Morley J.W. Brain function in Duchenne muscular dystrophy. Brain. 2002;125:4–13. doi: 10.1093/brain/awf012. [DOI] [PubMed] [Google Scholar]
- 20.Birnkrant D.J., Ararat E., Mhanna M.J. Cardiac phenotype determines survival in Duchenne muscular dystrophy. Pediatr. Pulmonol. 2016;51:70–76. doi: 10.1002/ppul.23215. [DOI] [PubMed] [Google Scholar]
- 21.Tichy E.D., Mourkioti F. A new method of genotyping MDX4CV mice by PCR-RFLP analysis. Muscle Nerve. 2017;56:522–524. doi: 10.1002/mus.25566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy S., Zweyer M., Mundegar R.R., Henry M., Meleady P., Swandulla D., Ohlendieck K. Concurrent label-free mass spectrometric analysis of dystrophin isoform Dp427 and the myofibrosis marker collagen in crude extracts from mdx-4cv skeletal muscles. Proteomes. 2015;3:298–327. doi: 10.3390/proteomes3030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy S., Zweyer M., Henry M., Meleady P., Mundegar R.R., Swandulla D., Ohlendieck K. Label-free mass spectrometric analysis reveals complex changes in the brain proteome from the mdx-4cv mouse model of Duchenne muscular dystrophy. Clin. Proteom. 2015;12:27. doi: 10.1186/s12014-015-9099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy S., Dowling P., Zweyer M., Mundegar R.R., Henry M., Meleady P., Swandulla D., Ohlendieck K. Proteomic analysis of dystrophin deficiency and associated changes in the aged mdx-4cv heart model of dystrophinopathy-related cardiomyopathy. J. Proteom. 2016;145:24–36. doi: 10.1016/j.jprot.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Murphy S., Dowling P., Zweyer M., Henry M., Meleady P., Mundegar R.R., Swandulla D., Ohlendieck K. Proteomic profiling of mdx-4cv serum reveals highly elevated levels of the inflammation-induced plasma marker haptoglobin in muscular dystrophy. Int. J. Mol. Med. 2017;39:1357–1370. doi: 10.3892/ijmm.2017.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pathak M., Wong S.S., Dreveny I., Emsley J. Structure of plasma and tissue kallikreins. Thromb. Haemost. 2013;110:423–433. doi: 10.1160/TH12-11-0840. [DOI] [PubMed] [Google Scholar]
- 27.Nokkari A., Abou-El-Hassan H., Mechref Y., Mondello S., Kindy M.S., Jaffa A.A., Kobeissy F. Implication of the Kallikrein-Kinin system in neurological disorders: quest for potential biomarkers and mechanisms. Prog. Neurobiol. 2018 doi: 10.1016/j.pneurobio.2018.01.003. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acuña M.J., Salas D., Córdova-Casanova A., Cruz-Soca M., Céspedes C., Vio C.P., Brandan E. Blockade of Bradykinin receptors worsens the dystrophic phenotype of mdx mice: differential effects for B1 and B2 receptors. J. Cell Commun. Signal. 2017 doi: 10.1007/s12079-017-0439-x. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy S., Brinkmeier H., Krautwald M., Henry M., Meleady P., Ohlendieck K. Proteomic profiling of the dystrophin complex and membrane fraction from dystrophic mdx muscle reveals decreases in the cytolinker desmoglein and increases in the extracellular matrix stabilizers biglycan and fibronectin. J. Muscle Res. Cell Motil. 2017;38:251–268. doi: 10.1007/s10974-017-9478-4. [DOI] [PubMed] [Google Scholar]
- 30.Chapman V.M., Miller D.R., Armstrong D., Caskey C.T. Recovery of induced mutations for X chromosome-linked muscular dystrophy in mice. Proc. Natl. Acad. Sci. USA. 1989;86:1292–1296. doi: 10.1073/pnas.86.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 32.Murphy S., Ohlendieck K. Mass spectrometric identification of dystrophin, the protein product of the Duchenne muscular dystrophy gene, in distinct muscle surface membranes. Int. J. Mol. Med. 2017;40:1078–1088. doi: 10.3892/ijmm.2017.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deslyper G., Colgan T.J., Cooper A.J., Holland C.V., Carolan J.C. A proteomic investigation of hepatic resistance to ascaris in a murine model. PLoS Negl. Trop. Dis. 2016;10:e0004837. doi: 10.1371/journal.pntd.0004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.S. Murphy, P. Dowling, M. Zweyer, M. Henry, P. Meleady, R.R. Mundegar, D. Swandulla, K. Ohlendieck. Comparative proteomic profiling of mouse saliva and serum from wild type versus the dystrophic mdx-4cv mouse model of dystrophinopathy. Data in Brief (Submitted for publication). [DOI] [PMC free article] [PubMed]
- 36.Scorilas K. Mavridis. Predictions for the future of kallikrein-related peptidases in molecular diagnostics. Expert Rev. Mol. Diagn. 2014;14:713–722. doi: 10.1586/14737159.2014.928207. [DOI] [PubMed] [Google Scholar]
- 37.Robinson S., Niles R.K., Witkowska H.E., Rittenbach K.J., Nichols R.J., Sargent J.A., Dixon S.E., Prakobphol A., Hall S.C., Fisher S.J., Hardt M. A mass spectrometry-based strategy for detecting and characterizing endogenous proteinase activities in complex biological samples. Proteomics. 2008;8:435–445. doi: 10.1002/pmic.200700680. [DOI] [PubMed] [Google Scholar]
- 38.Fisher I., Abraham D., Bouri K., Hoffman E.P., Muntoni F., Morgan J. Prednisolone-induced changes in dystrophic skeletal muscle. FASEB J. 2005;19:834–836. doi: 10.1096/fj.04-2511fje. [DOI] [PubMed] [Google Scholar]
- 39.Sotiropoulou G., Pampalakis G. Kallikrein-related peptidases: bridges between immune functions and extracellular matrix degradation. Biol. Chem. 2010;391:321–331. doi: 10.1515/BC.2010.036. [DOI] [PubMed] [Google Scholar]
- 40.Rebalka I.A., Hawke T.J. Potential biomarkers of skeletal muscle damage. Biomark. Med. 2014;8:375–378. doi: 10.2217/bmm.13.163. [DOI] [PubMed] [Google Scholar]
- 41.Ohlendieck K. Novel proteomic biomarkers for skeletal muscle diseases. Biomark. Med. 2017;11:409–412. doi: 10.2217/bmm-2017-0069. [DOI] [PubMed] [Google Scholar]
- 42.Carpenter G.H. The secretion, components, and properties of saliva. Annu. Rev. Food Sci. Technol. 2013;4:267–276. doi: 10.1146/annurev-food-030212-182700. [DOI] [PubMed] [Google Scholar]
- 43.Dowling P., Holland A., Ohlendieck K. Mass spectrometry-based identification of muscle-associated and muscle-derived proteomic biomarkers of dystrophinopathies. J. Neuromuscul. Dis. 2014;1:15–40. [PubMed] [Google Scholar]
- 44.Yoon H., Blaber S.I., Li W., Scarisbrick I.A., Blaber M. Activation profiles of human kallikrein-related peptidases by matrix metalloproteinases. Biol. Chem. 2013;394:137–147. doi: 10.1515/hsz-2012-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holland A., Dowling P., Meleady P., Henry M., Zweyer M., Mundegar R.R., Swandulla D., Ohlendieck K. Label-free mass spectrometric analysis of the mdx-4cv diaphragm identifies the matricellular protein periostin as a potential factor involved in dystrophinopathy-related fibrosis. Proteomics. 2015;15:2318–2331. doi: 10.1002/pmic.201400471. [DOI] [PubMed] [Google Scholar]
- 46.Ohlendieck K., Swandulla D. Molecular pathogenesis of Duchenne muscular dystrophy-related fibrosis. Pathologe. 2017;38:21–29. doi: 10.1007/s00292-017-0265-1. [DOI] [PubMed] [Google Scholar]
- 47.Murphy S., Zweyer M., Henry M., Meleady P., Mundegar R.R., Swandulla D., Ohlendieck K. Proteomic analysis of the sarcolemma-enriched fraction from dystrophic mdx-4cv skeletal muscle. J. Proteom. 2018 doi: 10.1016/j.jprot.2018.01.015. (In press) [DOI] [PubMed] [Google Scholar]
- 48.Rosenberg A.S., Puig M., Nagaraju K., Hoffman E.P., Villalta S.A., Rao V.A., Wakefield L.M., Woodcock J. Immune-mediated pathology in Duchenne muscular dystrophy. Sci. Transl. Med. 2015;7:299rv4. doi: 10.1126/scitranslmed.aaa7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guiraud S., Davies K.E. Pharmacological advances for treatment in Duchenne muscular dystrophy. Curr. Opin. Pharmacol. 2017;34:36–48. doi: 10.1016/j.coph.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Khurshid Z., Zohaib S., Najeeb S., Zafar M.S., Slowey P.D., Almas K. Human saliva collection devices for proteomics: an update. Int. J. Mol. Sci. 2016;17:E846. doi: 10.3390/ijms17060846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material