Abstract
Introduction
The measurement of specific volatile organic compounds in breath has been proposed as a potential diagnostic for a variety of diseases. The most well-studied bacterial lung infection in the breath field is that caused by Pseudomonas aeruginosa.
Objectives
To determine a discriminatory core of molecules in the “breath-print” of mice during a lung infection with four strains of P. aeruginosa (PAO1, PA14, PAK, PA7). Furthermore, we attempted to extrapolate a strain-specific “breath-print” signature to investigate the possibility of recapitulating the genetic phylogenetic groups (Stewart et al. Pathog Dis 71(1), 20–25, 2014. https://doi.org/10.1111/2049–632X.12107).
Methods
Breath was collected into a Tedlar bag and shortly after drawn into a thermal desorption tube. The latter was then analyzed into a comprehensive multidimensional gas chromatography coupled with a time-of-flight mass spectrometer. Random forest algorithm was used for selecting the most discriminatory features and creating a prediction model.
Results
Three hundred and one molecules were significantly different between animals infected with P. aeruginosa, and those given a sham infection (PBS) or inoculated with UV-killed P. aeruginosa. Of those, nine metabolites could be used to discriminate between the three groups with an accuracy of 81%. Hierarchical clustering showed that the signature from breath was due to a specific response to live bacteria instead of a generic infection response. Furthermore, we identified ten additional volatile metabolites that could differentiate mice infected with different strains of P. aeruginosa. A phylogram generated from the ten metabolites showed that PAO1 and PA7 were the most distinct group, while PAK and PA14 were interspersed between the former two groups.
Conclusions
To the best of our knowledge, this is the first study to report on a ‘core’ murine breath print, as well as, strain level differences between the compounds in breath. We provide identifications (by running commercially available analytical standards) to five breath compounds that are predictive of P. aeruginosa infection.
Keywords: Breath, Volatile organic compounds (VOCs), Pseudomonas aeruginosa, Comprehensive gas chromatography-time-of-flight mass spectrometer (GC×GC ToF MS)
1. Introduction
The measurement of specific volatile organic compounds (VOCs) in breath has been proposed as a potential diagnostic approach for a variety of diseases. Already, the Food and Drug Administration (FDA) has approved several breath tests for use in the clinic. For example, breath is used as a diagnostic for Helicobacter pylori-induced gastritis (Gisbert and Pajares 2004), response to asthma therapy (Kharitonov et al. 1994; Silkoff et al. 2004), and more recently for third-grade heart transplant rejection (Phillips et al. 2004). The latter of these tests utilizes a suite of volatile metabolites (or VOCs) in breath, identified via gas chromatography-mass spectrometry (GC-MS). Among the different products generated as part of normal cellular processes (i.e., DNA, RNA, proteins, metabolites), metabolites are reflective of biochemical activity, and can be more appropriately correlated to the phenotype, and subsequently to the actual clinical status (Patti et al. 2012). For this reason, the use of VOCs for the diagnosis of both acute and chronic respiratory infections is an area of intense research. These studies are extensively reviewed in (Sethi et al. 2013).
An important question in the field of breath diagnostics is whether bacterial infections by different strains of a single organism will have a universal breath print. Acute and chronic lung infections caused by Pseudomonas aeruginosa are the most well-studied bacterial lung infections in the field of breath research (Lyczak et al. 2000). For example, work from our group as well as others has expanded the volatile metabolome of P. aeruginosa compounds using in vitro (Bean et al. 2016), in vivo (breath) (Robroeks et al. 2010), and ex vivo (BAL fluid, sputum) samples (Nasir et al. 2018; Shestivska et al. 2012). In vitro studies of various clinical strains of P. aeruginosa suggest the presence of a core volatile metabolome (Bean et al. 2016; Frimmersdorf et al. 2010) that can be useful for pathogen identification. However, no studies to date have shown the presence of such a signature in breath.
The mouse model of P. aeruginosa lung infection has been employed previously in the field of breath analysis (Bean et al. 2015; Zhu et al. 2013). These studies utilized secondary electrospray ionization-mass spectrometry (SESI-MS) to demonstrate that distinctive volatile patterns can be used to differentiate between mice infected with different bacterial species. However, the identification of the specific discriminatory VOCs was not possible using this technique (Bean et al. 2015; Zhu et al. 2013). Identifications of the VOCs is important for understanding the metabolic pathways involved during response to infection and to guide antibiotic development. To be able to provide this information, we used comprehensive two-dimensional gas chromatography (GC × GC) coupled with time-of-flight mass spectrometry (ToF MS), one of the most powerful tools available for the analysis of volatile metabolites (Tranchida et al. 2015).
The aim of this study was to identify a discriminatory core breath-print derived from P. aeruginosa infection by different strains (i.e., PAO1, PA14, PAK, and PA7) and to evaluate the volatile metabolic relationship between these strains. To investigate the possible origins of the volatile compounds detected, the immune response of the host was activated by inoculating the mice with UV-killed P. aeruginosa, thereby excluding metabolites produced by live P. aeruginosa from the exhaled breath profile.
2. Materials and methods
2.1. Bacterial strains and inoculant preparation
Four P. aeruginosa strains were used in the study: PAO1 (Jacobs et al. 2003), PA14 (Rahme et al. 1995), PAK (Minamishima et al. 1968), and PA7 (Roy et al. 2010). All strains were cultured aerobically (24 h; 37 °C under agitation) in 20 mL of LB-Lennox (BD Diagnostics, Franklin Lakes, NJ, USA). Prior to inoculation in the murine lung, the cultures were washed three times and re-suspended in phosphate-buffered saline (PBS, pH 7.4) to the desired final concentration. The actual bacterial burden was verified by culturing the inoculant at 37 °C overnight on LB agar plates, after appropriate dilution, to calculate the bacterial cell density (CFU/mL). Additional mice were exposed to 50 mL of PBS by oropharyngeal aspiration as a negative control (Sham group). An aliquot of the PA14 and PAK inoculant was deactivated by exposing the inoculant to UV radiation for 20 min at 10 cm from the UV source, an empirically-determined radiative dose regime (UV-killed group).
2.2. Murine airway exposure and breath collection
All mice were housed for two weeks before starting the experiment in the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited animal facility at the University of Vermont (Burlington, VT, USA). The protocol for animal infection, respiratory physiology measurements, and breath collection was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Vermont, in accordance with AAALAC guidelines.
Female C57BL/6J mice (10–12 weeks, weight range 18–22 g) were purchased from The Jackson Laboratories (Bar Harbor, ME, USA). The animals were housed in the AAALAC approved vivarium in plastic cages with cedar bedding with maximum 5 mice per cage with a dark/light cycle of 12 h. The bedding was changed once weekly. Mice were provided with standard mouse chow, LabDiets RMH 3000, and tap water from bottles ad lib. Mice were inoculated with 50 μL of a PBS solution containing live or UV-killed P. aeruginosa at ~ 107 CFU via oropharyngeal aspiration, as previously reported (Bean et al. 2015). The sham group was inoculated with 50 μL of sterile PBS but otherwise treated identically to the infected mice. The original study design called for eight animals per group, plus three animals to optimize the breath collection protocol (PA14), but the final numbers were affected by the loss of three animals during collection. Thus a total of 48 mice were used: seven in the PBS group, eight in the UV-killed group (4 PA14 and 4 PAK), and 33 in the Live PA group (8 PAO1, 11 PA14, 6 PA7, 8 PAK).
Twenty-four hours after either P. aeruginosa or PBS inoculation, mice were anesthetized with intraperitoneal sodium pentobarbital (90 mg/kg) and their tracheas cannulated with an 18 gauge cannula. The tracheal cannula was connected to a flexiVent small animal ventilator (flexiVent, SCIREQ, Montreal, QC, Canada), and ventilated at 200 breaths/minute at a positive end-expiratory pressure (PEEP) of 3 cmH2O. Depth of anesthesia was monitored by regular toe pinch and if necessary additional aesthesia was provided. The exhaled breath was collected in a 1L Tedlar bag (SKC, Eighty Four, PA, USA) housed within a plexiglass box where the pressure was maintained at 3 cmH2O to guarantee that a constant PEEP was maintained throughout the procedure. Breath from the ventilator was collected at 200 breaths/min for 30–40 min (Stewart et al. 2014). At the end of the breath collection, mice were euthanized with an intraperitoneal overdose of sodium pentobarbital (150 mg/kg) followed by blood and lung harvest (see below). The VOCs in breath were concentrated by drawing the entire contents of the Tedlar bag (1 L) onto custom-made 60 mm long fritted glass thermal desorption packed with 5 mm of Carbopack-Y 60/80 mesh, 5 mm of Carbopack-X 60/80 mesh, and 5 mm of Carboxen-1000 60/80 mesh (Supelco, Bellefonte, PA, USA) via a vacuum pump, at an average flow rate of 170 mL/min. The loading onto thermal desorption tubes was completed within 5 min of breath collection. Tubes were capped and stored at 4 °C and analyzed using the GC×GC-TOFMS system within 2 weeks of collection. The full protocol for breath collection can be found in previous publications (Franchina et al. 2018; Zhu et al. 2013a, 2014). Room air samples (n = 28) were collected by attaching the Tedlar bag to the ventilator in the absence of a mouse.
2.3. Collection and analysis of bronchoalveolar lavage fluid, lung tissue, and blood
The bronchoalveolar lavage (BAL) fluid was collected by instilling 1 mL of PBS (at room temperature) via tracheal cannula just after cessation of breath collection. The BAL fluid was split in two aliquots. One aliquot was centrifuged and the pellet containing the cellular material was stained using a HEMA 3 kit (Fisher Scientific); first fixed in a methanol solution for 2 min, then stained with Eosin Y for 2 min, and finally stained in Azure A and methylene blue for 30 s. The cells were differentiated by counting 300 cells/slide, identifying eosinophils, neutrophils, macrophages, and lymphocytes (measurements performed in triplicate). The second aliquot was cultured neat (no dilutions) on Pseudomonas Isolation Agar (PIA) and LB agar plates as an additional confirmatory test for the presence of bacterial cells in the lungs at the moment of breath collection, and the presence of visible colonies was assessed after incubation at 37 °C. After collection of BAL fluid, a blood sample was drawn via heart puncture into 1.3 mL microtubes with EDTA and then cultured neat on PIA and LB as described for BAL. Then the lungs were harvested and homogenized. The excised lungs were trimmed free from adjacent tissue and the lung lobes (approximately 0.3 g) placed in sterile gentleMACS M tubes (Miltenyi Biotec, Auburn, CA, USA) with 3 mL of PBS. The lungs were homogenized by running the RNA_01 program (a default method) on a gentleMACS dissociator. Lung bacterial cell counts were obtained by plating an aliquot of the lung homogenate on PIA and LB agar plates and incubating them at 37 °C.
2.4. Analytical instrumentation and statistical analysis
Thermal desorption tubes were analyzed using a Pegasus 4D (LECO Corporation, St. Joseph, MI, USA) GC×GC-ToFMS instrument with an Agilent 7890 GC equipped with a Thermal Desorption Unit (TDU), Cooled Inlet System (CIS), and a Multi-Purpose Sampler (MPS) auto-sampler (Gerstel, Linthicum Heights, MD, USA). Details of the sample desorption, chromatography, and mass spectrometry experimental conditions, as well as the alignment procedure and pre-cleaning steps, are summarized in Supplementary Material (Table S1).
All statistical analyses were performed using R (version 3.3.0). Data were log-transformed and normalized using Probabilistic Quotient Normalization (PQN) (Zhu et al. 2013b). The ‘limma’ package was used to assess and address batch effect in our dataset caused by the temporal spacing between different experiments. Additional information is reported in the Supplementary Material.
To test for statistical significance, the Kruskal–Wallis test (Kruskal and Wallis 1952) with Benjamini-Hochberg (BH) correction (Benjamini and Hochberg 1995) was used, and a significance level of p < 0.05 was selected. The Random Forest (RF) machine learning algorithm was used to train a model and select putative discriminatory VOCs to distinguish between breath samples belonging to different inoculation groups (Breiman 2001) (details on algorithm reported in Supplementary Material). A 10-fold cross validation scheme was used to calculate accuracy and identify discriminatory compounds. RF uses an internal validation scheme (2/3rd training and 1/3rd testing) to compute accuracy (out-of-bag error) and compound importance. The latter is determined by permuting the values of each feature in the test samples and predicting the class labels of these samples. The association with the class labels is lost by randomly permuting the values of the feature. The difference in prediction accuracy before and after permuting the values of features averaged over all trees, is a measure of feature importance.
Hierarchical clustering analysis was used to assess the expression profile of discriminatory VOCs. The multivariate expression profile of strain specific VOCs selected by RF analysis was visualized as an unrooted phylogenetic tree (distance metric = Euclidean distance). The flowchart of data processing highlighting the number of volatile metabolites retained after each step is described in Fig. 1.
Fig. 1.
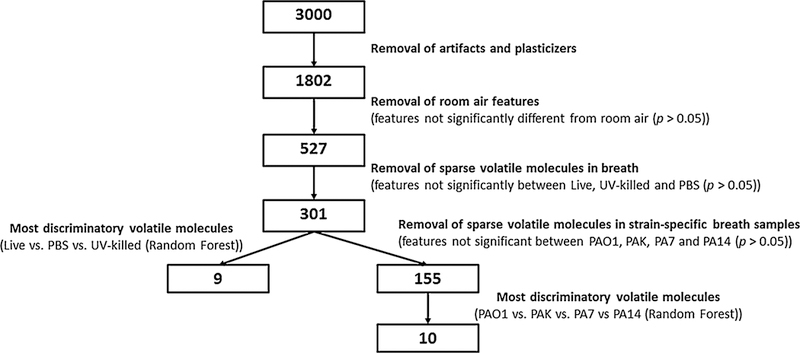
Data reduction methodology for volatile molecules reported in the study. Numbers in boxes represent the number of VOCs remaining at each step of data processing
3. Results
3.1. Murine infection confirmation
The microorganism content of the inoculants was verified by bacterial counts, and all ranged between 2 and 3 × 107 CFU/mL. The establishment of lung infection was verified by bacterial counts (CFU/lung) in lung homogenates and by evaluation of leukocyte differentiation in BAL fluid. Bacterial counts from homogenized lungs of mice infected with live P. aeruginosa (this group is designated as “Live-group” regardless of the specific strain) at the time of breath sampling (24 h after inoculation) ranged between 1 × 103 and 1 × 107 CFU/lung (Supplementary Fig. 1a). The absence of viable bacteria in the mice inoculated with PBS (group designation: “PBS”), or UV-killed PAK or PA14 (group designation: “UV-killed”) was also confirmed. The range of neutrophil percentage from BAL fluid of mice infected with P. aeruginosa was between 49.7% and 99.0%, confirming acute infection (Supplementary Fig. 1b). We did not observe a significant difference in the percentage of neutrophils recovered from BAL fluid between mice infected with live P. aeruginosa and those infected with UV-killed PAK (Supplementary Fig. 1b). There was, however, a significant difference (p < 0.001) between neutrophils recovered in BAL fluid of mice infected with UV-killed PA14 and all other groups (excluding PBS). This may be reflective of a wide spectrum with respect to immune response robustness within this group of mice 24 h post-inoculation. No neutrophils were recovered from mice inoculated with PBS.
3.2. P. aeruginosa-specific volatile molecules in murine breath
The data matrix obtained from the analysis of the exhaled breath was cleaned (eliminating known contaminants and artifacts) and batch-corrected as described in the Supplementary Material and depicted in Supplementary Figures S2a and S2b. The number of peaks was then further reduced by the removal of features that were not statistically different between exhaled breath and room air samples (p ≥ 0.05), resulting in a total of 527 peaks. Hierarchical clustering and a heatmap of these 527 peaks did not show an obvious visual discrimination of the Live-group (PA14, PA7, PAK, and PAO1) from the PBS and UV-killed groups (Supplementary Figure S3). Hence, the data matrix was reduced to 301 features via the removal of breath features that were not significantly different (using the Kruskal–Wallis test) between Live-group, PBS, and UV-killed. Heatmap and hierarchical clustering analysis using these volatile molecules demonstrated clearer structure with respect to separation between groups (Fig. 2a).
Fig. 2.
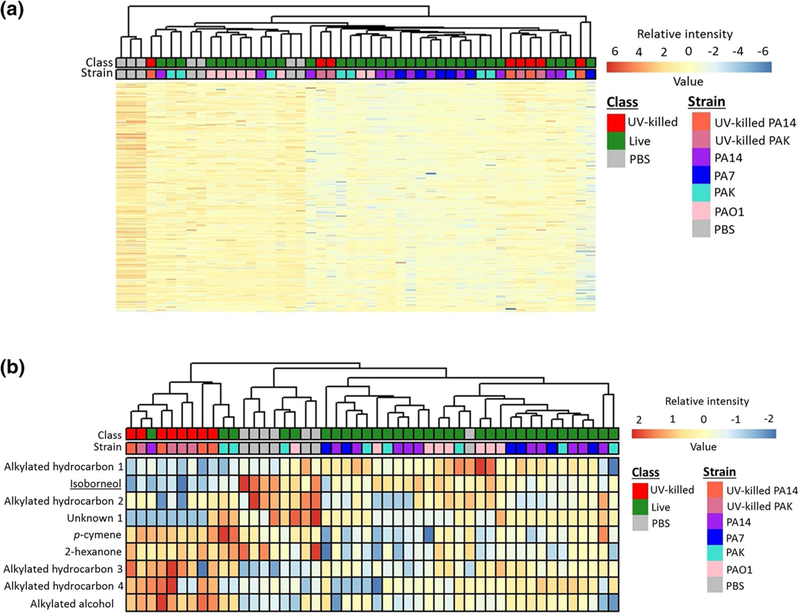
Heatmap and hierarchical clustering analysis of breath profiles: a using 301 volatile molecules that were significantly different between Live-group, PBS, and UV-killed groups (p< 0.05, Kruskal–Wallis with Benjamini–Hochberg correction), and b using nine discriminatory volatile molecules. The identity of underlined volatile metabolites was confirmed using analytical standards
To identify a predictive suite of volatiles, a machine learning algorithm was applied to build a three-class model that could differentiate between the Live-group, PBS, and UV-killed. The Random Forest algorithm was used because of its ability to deal with highly collinear data and its robustness to the influence of different types of outliers (Smolinska et al. 2014). A panel of nine volatile metabolites were selected (Table 1) which were able to classify mice breath samples based on their group identity with a cross-validated accuracy of 81%. Heatmap and hierarchical cluster analysis of the nine discriminatory volatile molecules shows three main clusters, corresponding to the three classes tested, namely Live-group, PBS, and UV-killed (Fig. 2b), with five animals from the Live-group being misclassified. We hypothesize that this misclassification may be due to the substantial variability of the murine immune response to P. aeruginosa infection, as demonstrated by (Bean et al. 2015).
Table 1.
Putative identifications of volatile molecules selected in both models, along with Chemical Abstracts Service (CAS), Mass spectra similarity match (MS), Linear retention index (LRI) calculated experimentally, first and second dimension retention time (1D RT and 2D RT), and references where the specific compounds was previously identified
| Volatile molecule | IUPAC name | Chemical class | CAS | MS | LRI Exp | 1D RT (min:sec) | 2D RT (sec) | Reference |
|---|---|---|---|---|---|---|---|---|
| Live, UV-killed, PBS-groups model | ||||||||
| Alkylated hydro-carbon 1 | Hyd | 991 | 26:27.7 | 0.7 | ||||
| Alkylated hydro-carbon 2 | Hyd | 861 | 18:43.7 | 0.7 | ||||
| Alkylated hydro-carbon 3 | Hyd | 1507 | 48:41.1 | 0.7 | ||||
| Alkylated hydro-carbon 4 | Hyd | 1272 | 26:27.7 | 0.8 | ||||
| Isoborneol | (1R,3R,4R)-4,7,7- trimethylbi- cyclo[2.2.1] heptan-3-ol |
Alc | 124–76-5 | 891 | 1231 | 37:42.8 | 1.3 | |
| Alkylated alcohol | Alc | 108–11-2 | 800 | 1050 | 28:32.6 | 1.0 | ||
| p-cymene | 1-methyl-4-propan- 2-ylbenzene |
Aro | 527–84-4 | 800 | 1157 | 33:40.1 | 1.0 | |
| 2-Hexanone | hexan-2-one | Ket | 591–78-6 | 859 | 854 | 18:20.8 | 0.9 | Haick et al. (2014) |
| Unknown 1 | Unk | 1142 | 32:42.7 | 1.3 | ||||
| Strain versus strain model | ||||||||
| 2-Hydroxyethyl acetate | 2-hydroxyethyl acetate | Ace | 542–59-6 | 750 | 964 | 24:46.9 | 0.7 | |
| 1,3-Dimethylcy-clopentanol | 1,3-dimethylcyclo-pentan-1-ol | Alc | 19550–46-0 | 840 | 909 | 21:25.2 | 1.2 | |
| 2-Methyl-2-bute-nal, (E)- | (E)-2-methylbut-2-enal | Ald | 497–03-0 | 850 | 606 | 11:59.8 | 0.9 | |
| 4-Cyclopentene-1,3-dione | cyclopent-4-ene-1,3-dione | Ket | 930–60-9 | 810 | 975 | 25:29.3 | 0.2 | |
| Aldehyde 1 | Ald | 627 | 08:52.0 | 0.8 | ||||
| Alkylated hydro-carbon 5 | Hyd | 1057 | 28:45.2 | 0.7 | ||||
| Alkylated hydro-carbon 6 | Hyd | 946 | 21:25.2 | 0.6 | ||||
| Cyclohexanol | Cyclohexanol | Alc | 108–93-0 | 912 | 951 | 24:00.9 | 1.6 | Goeminne et al. (2012) |
| Methylated fatty acid 1 |
FAME | 654 | 09:56.7 | 0.9 | ||||
| Methylated fatty acid 2 |
FAME | 847 | 17:59.0 | 0.8 | ||||
The identity of underlined compounds was confirmed using analytical standards IUPAC International Union of Pure and Applied Chemistry, CAS Chemical abstract service, MS Mass spectral similarity, LRIExp Experimental Linear Retention Index, Alc alcohol, Ald aldehyde, Aro aromatic, Ket ketone, FAME fatty acid methyl ester, Hyd hydrocarbon, Unk unknown
The expression profile of the nine selected features for each group (Live-group, PBS, and UV-killed) was examined (Fig. 3) using boxplots. Five features were significantly different between the Live-group and PBS, but only one (alkylated hydrocarbon 1) was more abundant in the former group. Eight features were significantly different between the Live-group and the UV-killed, with three features (alkylated hydrocarbon 1, isoborneol, and unknown 1) significantly more abundant in the Live-group than in the UV-killed group, which may be related to the contribution of live bacterial metabolism to the VOC profile in breath. Five features were highly abundant in the UV-killed group compared to the Live-group, namely alkylated hydrocarbons 3 and 4, alkylated alcohol, p-cymene, and 2-hexanone. Four features: alkylated hydrocarbon 1, isoborneol, alkylated hydrocarbon 2, and unknown 1 were significantly more abundant in the PBS group compared to the UV-killed and Live-group. One feature (alkylated hydrocarbon 3) was significantly less abundant in PBS compared to UV-killed and Live-group. Four features, namely alkylated hydrocarbons 1 and 4, alkylated alcohol, and unknown 1, were not significantly different between PBS and the Live-group.
Fig. 3.
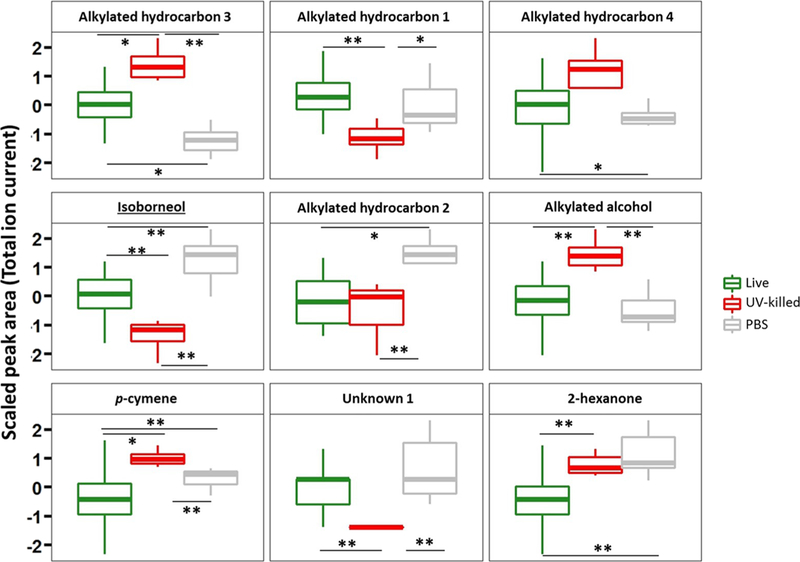
Boxplot of nine volatile metabolites used to discriminate Live-group versus PBS versus UV-killed. * p < 0.05; ** p < 0.01. The lower, middle and upper lines of the box correspond to the first, second and third quartiles (the 25th, 50th, and 75th percentiles, respectively). The upper whisker extends from the upper line to the largest value no further than 1.5 × IQR from the line (where IQR is the inter-quartile range). The lower whisker extends from the bottom line to the smallest value at most 1.5 × IQR of the range. Data beyond the end of the whiskers are called “outlying” points and are plotted individually
3.3. Strain-level identification in mice with P. aeruginosa lung infection
To assess whether volatile molecules in exhaled breath during infection can be used to identify specific bacterial strains, we first performed a multiclass statistical test and removed the features that were not statistically different between two or more strains (p ≥ 0.05). The data matrix was thus reduced to 155 features (Fig. 1). Two main clusters were visualized using hierarchical clustering analysis (Supplementary Figure S4). The breath samples of mice inoculated with PAO1 and PA7 clustered well and separately from each other, while PA14 and PAK were interspersed almost equally between the two clusters (Supplementary Figure S3). Following the same procedure used for creating the previous model, features were reduced to the most discriminatory set using Random Forest to predict strain-level differences. A panel of ten volatile molecules (reported in Table 1) was selected and used to build a phylogenetic tree to visualize the groupings of the breath metabolomes (Fig. 4), confirming that PA7 and PAO1 formed two distinct clusters, while PAK and PA14 were interspersed across these clusters. The expression profile of the ten selected features is reported in Fig. 5. PAO1 and PA7 presented the most distinct profiles compared to the other strains, and generally PAO1 had the highest expression levels of the ten volatile molecules, while PA7 had the lowest.
Fig. 4.
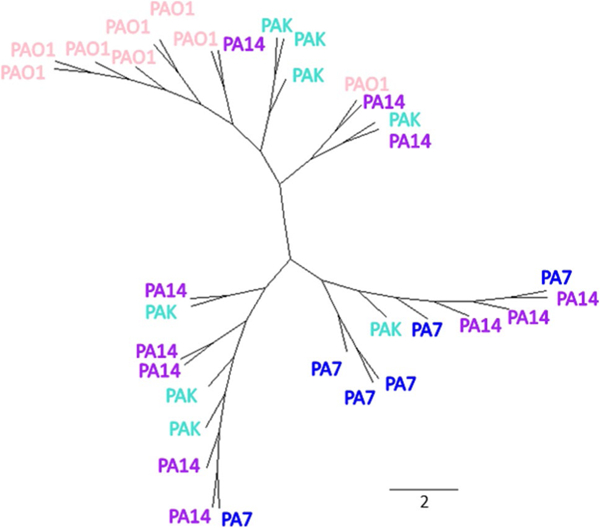
An unrooted phylogenetic tree using ten discriminatory volatile molecules from the breath of mice infected with PA7, PAO1, PA14, and PAK. The length of the scale bar represents the Euclidean distance calculated based on the multivariate volatile molecule profile using ten compounds
Fig. 5.
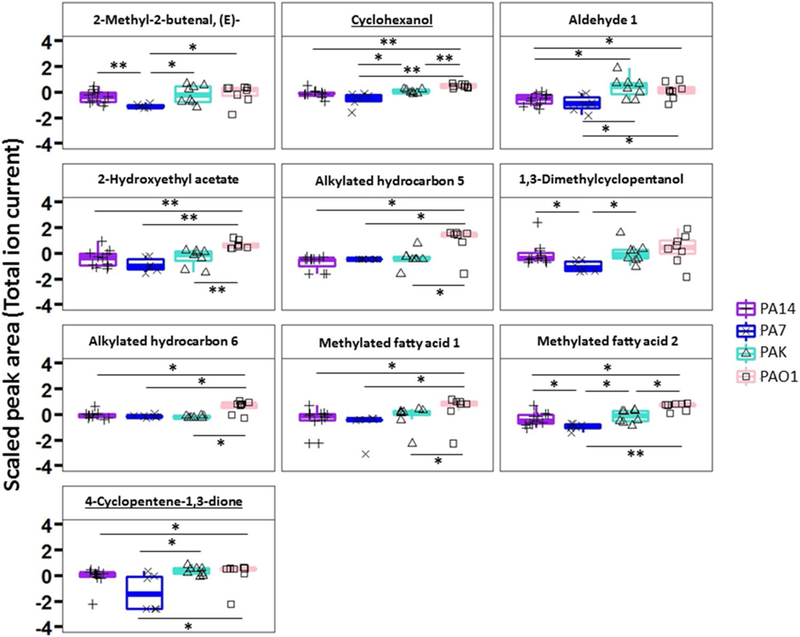
Boxplot of ten volatile metabolites used to differentiate PA14 versus PA7 versus PAK versus PAO1 breath samples (* p< 0.05; ** p< 0.01). The lower, middle and upper lines of the box correspond to the first, second and third quartiles (the 25th, 50th, and 75th percentiles). The upper whisker extends from the upper line to the largest value no further than 1.5 × IQR from the line (where IQR is the inter-quartile range). The lower whisker extends from the bottom line to the smallest value at most 1.5 × IQR of the range. Data beyond the end of the whiskers are called “outlying” points and are plotted individually
3.4. Identification of the most discriminatory compounds
Combining the features selected from the two models discussed above, a list of 19 metabolites was generated (Table 1). The identity of five compounds (p-cymene, 2-hexanone, isoborneol, cyclopent-4-ene-1,3-dione, and cyclohexanol) were confirmed by running commercially-available analytical standards. Three compounds were putatively identified based only on their mass spectral similarity (commercial standards were not available). For these three, the MS library search identified a single candidate with a similarity match score substantially higher than the subsequent match score. However, when this was not the case, only a compound class was reported. Six compounds were defined as “alkylated hydrocarbons” (since the exact identification of compounds in this class is nearly impossible based only on mass spectral similarity, due to their intense fragmentation into the MS ion source), two as fatty acid methyl esters (FAMEs), and one as an aldehyde. Only one compound remained completely unknown (similarity match < 600), though it was reproducibly present. Cyclohexanol has been previously reported to be produced by Pseudomonas strains (Giorgio et al. 2015), and 2-hexanone was previously reported in the headspace of sputum from patients infected with P. aeruginosa (Goeminne et al. 2012).
4. Discussion
To the best of our knowledge, this is the first study to report on a “core” volatile metabolome (nine compounds) for P. aeruginosa detection in an in vivo murine model. Evidence of an in vitro core metabolome for P. aeruginosa, irrespective of bacterial genetic background, carbon source, or growth phase was demonstrated by Frimmersdorf et al. (Frimmersdorf et al. 2010). Bean et al., showed that this observation could be translated broadly to the VOC metabolome by analyzing the headspace of 24 isolates of P. aeruginosa grown in culture and putatively identifying 51 VOCs consistently produced by all strains (Bean et al. 2016). We have demonstrated that using nine core compounds from breath, we were able to identify P. aeruginosa-infected mice with an accuracy of 81%. Animals that were misclassified had similar CFU/lung and cell differentiation data as the rest of the samples in that group. The high variability of the host-response to P. aeruginosa infection may contribute to breath variability and has been described previously (Bean et al. 2015). We note the clear discrimination of the UV-killed group from both PBS and the Live-group, which suggests that the breath signature is not likely to be related to a generic response to lung irritation, but may instead be associated with the specific presence of P. aeruginosa. We acknowledge that this study design does not include the measurement of breath from mice infected with other bacterial species, an important future direction of this work.
Although a conclusive discussion cannot be performed, we can speculate on the possible host-origin of a subset of compounds, such as alkylated hydrocarbon 3, alkylated hydrocarbon 4, alkylated alcohol, p-cymene, and 2-hexanone, which were significantly more abundant in the breath of mice inoculated with the UV-killed bacterium. On the other hand, alkylated hydrocarbon 1, isoborneol, and unknown 1 are produced predominantly by the Live-group infections, and can therefore be hypothesized to be produced by P. aeruginosa (or the interaction between P. aeruginosa and the murine host).
We also identified a P. aeruginosa strain-related signature from the breath of infected animals. Genomic studies have characterized the relationship of core strains of P. aeruginosa (Stewart et al. 2014), revealing three distinct phylogenetic groups to which the strains here belonged: Group 1 (to which PAO1 and PAK belonged), Group 2 (to which PA14 belonged) and Group 3 (to which PA7 belonged). Breath metabolite signatures are much more complicated since breath is composed of volatile molecules generated not only by the bacterium, but also by the host, and the specific host-pathogen interaction. We used the multivariate volatile molecule profile of the ten discriminatory compounds to investigate whether a ‘suite’ of VOCs was more representative of the inter-strain variation rather than a single compound, as previously tested by Shestivska et al. (Shestivska et al. 2012). The phylogenic relations that we were able to visualize in Fig. 4, although not fully recapitulating the same genomic relationship, allows a certain degree of strain-specificity.
Among the most discriminatory metabolites, hydrocarbons were the most enriched compound class (32% of discriminatory compounds), in agreement with the finding that correlates their presence to the oxidative stress occurring during inflammation (Haick et al. 2014). Only two compounds that we identified were previously reported in relation to Pseudomonas, namely cyclohexanol and 2-hexanone (Giorgio et al. 2015; Goeminne et al. 2012). The latter was significantly more abundant in the breath of UV-killed and PBS infected mice compared to the Live-group, suggesting that the molecule may be metabolized by the pathogen. In addition, KEGG database identified two microbial pathways associated with cyclohexanol; degradation of aromatic compounds and microbial metabolism in diverse environments.
5. Conclusion
To the best of our knowledge, this is the first study to report on a core breath metabolome for murine P. aeruginosa infection. Although additional studies will be necessary to define a P. aeruginosa-specific breath signature, our work has shown that a suite of nine volatile molecules in breath can be used to discriminate between mice inoculated with Live bacteria, UV-killed bacteria, and PBS. In addition, for the first time, we defined a P. aeruginosa strain-specific breath signature using ten additional volatile metabolites. Our results demonstrate that the volatile metabolomes of PAO1-and PA7-infected mice were most distinct relative to one another, whereas mice infected with PAK and PA14 were interspersed between the former strains. This is the first time that breath volatile compounds discriminatory for P. aeruginosa infection in mice were reliably identified. Altogether, we confirmed the identity of five breath compounds (by running commercially available analytical standards) that are predictive of P. aeruginosa infection. In addition, three compounds were putatively identified based only on their mass spectra similarity (commercial standards were not available).
Supplementary Material
Acknowledgements
Financial support for this work was provided by Hitchcock Foundation and the National Institutes of Health (NIH, Project # R21AI12107601). MN and CAR were supported by the Burroughs Wellcome Fund institutional program grant unifying population and laboratory based sciences to Dartmouth College (Grant#1014106). CAR was additionally supported by a T32 training grant (T32LM012204, PI: Tor D Tosteson).
Footnotes
Data availability The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest All authors report no potential conflicts of interest.
Ethical approval This article does not contain any studies with human participants performed by any of the authors.
Research involving animal rights All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Electronic Supplementary Material The online version of this article (https://doi.org/10.1007/s11306–018-1461–6) contains supplementary material, which is available to authorized users.
References
- Bean HD, Jiménez-Díaz J, Zhu J, & Hill JE (2015). Breathprints of model murine bacterial lung infections are linked with immune response. European Respiratory Journal, 45(1), 181–190. 10.1183/09031936.00015814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean HD, Rees CA, & Hill JE (2016). Comparative analysis of the volatile metabolomes of Pseudomonas aeruginosa clinical isolates. Journal of Breath Research, 10, 047102. 10.1088/1752-7155/10/4/047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, & Hochberg Y (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological), 57(1), 289–300. [Google Scholar]
- Breiman L (2001). Random forests. Machine Learning, 45(1), 5–32. 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Franchina FA, Mellors TR, Aliyeva M, Wagner J, Daphtary N, Lundblad LKA, et al. (2018). Towards the use of breath for detecting mycobacterial infection: A case study in a murine model. Journal of Breath Research, 12(2), 26008 10.1088/1752-7163/aaa016. [DOI] [PubMed] [Google Scholar]
- Frimmersdorf E, Horatzek S, Pelnikevich A, Wiehlmann L, & Schomburg D (2010). How Pseudomonas aeruginosa adapts to various environments: A metabolomic approach. Environmental Microbiology, 12(6), 1734–1747. 10.1111/j.1462-2920.2010.02253.x. [DOI] [PubMed] [Google Scholar]
- Giorgio A, De Stradis A, Lo Cantore P, & Iacobellis NS (2015). Biocide effects of volatile organic compounds produced by potential biocontrol rhizobacteria on Sclerotinia sclerotiorum. Frontiers in Microbiology, 6, 1056 10.3389/fmicb.2015.01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisbert JP, & Pajares JM (2004). Review article: 13C-urea breath test in the diagnosis of Helicobacter pylori infection—A critical review. Alimentary Pharmacology and Therapeutics, 20(10), 1001–1017. 10.1111/j.1365-2036.2004.02203.x. [DOI] [PubMed] [Google Scholar]
- Goeminne PC, Vandendriessche T, Van Eldere J, Nicolai BM, Hertog ML, A., T., M., & Dupont LJ (2012). Detection of Pseudomonas aeruginosa in sputum headspace through volatile organic compound analysis. Respiratory Research, 13, 87 10.1186/1465-9921-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haick H, Broza YY, Mochalski P, Ruzsanyi V, & Amann A (2014). Assessment, origin, and implementation of breath volatile cancer markers. Chemical Society Reviews, 43, 1423–1449. 10.1039/c3cs60329f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, et al. (2003). Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America, 100(24), 14339–14344. 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonov SA, Yates D, Robbins RA, Logan-Sinclair R, Shine-bourne EA, & Barnes PJ (1994). Increased nitric oxide in exhaled air of asthmatic patients. Lancet, 343(8890), 133–135. [DOI] [PubMed] [Google Scholar]
- Kruskal WH, & Wallis WA (1952). Use of ranks in one-criterion variance analysis. Journal of the American Statistical Association, 47(260), 583–621. [Google Scholar]
- Lyczak JB, Cannon CL, & Pier GB (2000). Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes and Infection, 2(9), 1051–1060. 10.1016/S1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- Minamishima Y, Takeya K, Ohnishi Y, & Amako K (1968). Physicochemical and biological properties of fibrous Pseudomonas bacteriophages. Journal of Virology, 2(3), 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir M, Bean HD, Smolinska A, Rees CA, Zemanick ET, & Hill JE (2018). Volatile molecules from bronchoalveolar lavage fluid can ‘rule-in’ Pseudomonas aeruginosa and ‘rule-out’ Staphylococcus aureus infections in cystic fibrosis patients. Scientific Reports, 8(1), 826 10.1038/S41598-017-18491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, & Siuzdak G (2012). Metabolomics: The apogee of the omics trilogy. Nature Reviews Molecular Cell Biology, 13(4), 263–269. 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M, Boehmer JP, Cataneo RN, Cheema T, Greenberg J, Kobashigawa J, et al. (2004). Heart allograft rejection: Detection with breath alkanes in low levels (the HARDBALL Study) methods: Results. The Journal of heart and lung Transplantation, 23(6), 701–708. 10.1016/j.healun.2003.07.017. [DOI] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, & Ausubel FM (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science, 268(5219), 1899–1902. [DOI] [PubMed] [Google Scholar]
- Robroeks CMHHT, van Berkel JJBN, Dallinga JW, Jöbsis Q, Zimmermann LJI, Hendriks HJE, et al. (2010). Metabolomics of volatile organic compounds in cystic fibrosis patients and controls. Pediatric Research, 68(1), 75–80. 10.1203/PDR.0b013e3181df4ea0. [DOI] [PubMed] [Google Scholar]
- Roy PH, Tetu SG, Larouche A, Elbourne L, Tremblay S, Ren Q, et al. (2010). Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PloS ONE, 5(1), e8842 10.1371/journal.pone.0008842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi S, Nanda R, & Chakraborty T (2013). Clinical application of volatile organic compound analysis for detecting infectious diseases. Clinical Microbiology Reviews, 26(3), 462–475. 10.1128/CMR.00020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestivska V, Spanel P, Dryahina K, Sovova K, Smith D, Musilek M, & Nemec A (2012). Variability in the concentrations of volatile metabolites emitted by genotypically different strains of Pseudomonas aeruginosa. Journal of Applied Microbiology, 113, 701–713. 10.1111/j.1365-2672.2012.05370.x. [DOI] [PubMed] [Google Scholar]
- Silkoff PE, Carlson M, Bourke T, Katial R, Ögren E, & Szefler SJ (2004). The Aerocrine exhaled nitric oxide monitoring system NIOX is cleared by the US Food and Drug Administration for monitoring therapy in asthma. The Journal of Allergy and Clinical Immunology, 114(5), 1241–1256. 10.1016/j.jaci.2004.08.042. [DOI] [PubMed] [Google Scholar]
- Smolinska A, Hauschild A-C, Fijten RRR, Dallinga JW, Baumbach J, & van Schooten FJ (2014). Current breath-omics—A review on data pre-processing techniques and machine learning in metabolomics breath analysis. Journal of Breath Research, 8(2), 027105. 10.1088/1752-7155/8/2/027105. [DOI] [PubMed] [Google Scholar]
- Stewart L, Ford A, Sangal V, Jeukens J, Boyle B, Kukavica-Ibrulj I, et al. (2014). Draft genomes of 12 host-adapted and environmental isolates of Pseudomonas aeruginosa and their positions in the core genome phylogeny. Pathogens and Disease, 71(1), 20–25. 10.1111/2049-632X.12107. [DOI] [PubMed] [Google Scholar]
- Tranchida PQ, Maimone M, Purcaro G, Dugo P, & Mondello L (2015). The penetration of green sample-preparation techniques in comprehensive two-dimensional gas chromatography. TrAC Trends in Analytical Chemistry, 71, 74–84. 10.1016/j.trac.2015.03.011. [DOI] [Google Scholar]
- Zhu J, Bean HD, Jimenez-Diaz J, & Hill JE (2013a). Secondary electrospray ionization-mass spectrometry (SESI-MS) breath-printing of multiple bacterial lung pathogens, a mouse model study. Journal of Applied Physiology, 114(11), 1544–1549. 10.1152/japplphysiol.00099.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Bean HD, Wargo MJ, Leclair LW, & Hill JE (2014). Detecting bacterial lung infections: In vivo evaluation of in vitro volatile fingerprints. Journal of Breath Research, 7(1), 016003. 10.1088/1752-7155/7/1/016003.Detecting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Jiménez-Díaz J, Bean HD, Daphtary NA, Aliyeva MI, Lundblad LKA, & Hill JE (2013b). Robust detection of P. aeruginosa and S. aureus acute lung infections by secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting: From initial infection to clearance. Journal of Breath Research, 7(3), 037106. 10.1088/1752-7155/7/3/037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


