Abstract
Muscle damage elicits a sterile immune response that facilitates complete regeneration. Here, we utilized mass spectrometry-based lipidomics to map the mediator lipidome during the transition from inflammation to resolution and regeneration in skeletal muscle injury. We observed the temporal regulation of glycerophospholipids and the production of pro-inflammatory (e.g., leukotrienes, prostaglandins) and specialized pro-resolving (e.g., resolvins, lipoxins) lipid mediators, which were modulated by ibuprofen. These time-dependent profiles were recapitulated in sorted neutrophils and Ly6Chi and Ly6Clo muscle-infiltrating macrophages, with a distinct pro-resolving signature observed in Ly6Clo macrophages. RNA-seq of macrophages stimulated with resolvin D2 (RvD2) showed similarities to transcriptional changes found during the temporal Ly6Chi to Ly6Clo macrophage transition. In vivo, RvD2 increased Ly6Clo macrophages and functional improvement of the regenerating muscle. These results reveal dynamic lipid mediator signatures of innate immune cells and provide a proof-of-concept for their exploitable effector roles in muscle regeneration.
Cells of the innate immune system undergo dynamic changes in population size, differentiation state, and function in response to diverse stimuli to maintain organismal integrity1. However, dysregulated innate immune responses can contribute to disease2,3. Due to their roles in the initiation, maintenance and resolution during wound-healing and regeneration in different organ systems, myeloid cells, including macrophages, are therapeutic targets4,5. Our knowledge is fragmented as to how macrophages change their phenotype, employ sensory and regulatory mechanisms, and use effector functions to serve such roles. This is important because proper signaling between macrophages and other cells can ensure precisely timed repair while avoiding asynchrony, leading to regeneration delay, fibrosis and/or chronic inflammation2,3.
Injury in skeletal muscle is characterized by production of pro-inflammatory mediators including cytokines, followed by myeloid cell recruitment. Termination of pro-inflammatory mediator production and subsequent apoptotic cell clearance defines the resolution phase, which couples to tissue repair and regeneration6. Macrophages are involved in each of these phases, including clearance of necrotic debris, apoptotic cells, and tissue repair7. Pharmacologic or genetic depletion of macrophages impairs muscle regeneration. This process is characterized by an in situ transition of infiltrating monocytes to inflammatory (Ly6Chi) and to repair (Ly6Clo) macrophages8, indispensable for proper regeneration9,10. Cues from the microenvironment drive temporal gene expression changes characteristic of the switch from an inflammatory to a repair phenotype8. The signals regulating these phenotypes remain of interest.
Lipid mediators play key roles in the initiation and resolution of inflammation11. They are generated by immune cells (e.g., neutrophils and macrophages) and have direct receptor-mediated actions on them. Remodeling of membrane phospholipids occurs upon cellular activation, as esterified polyunsaturated fatty acids (PUFA) are liberated for conversion to lipid mediators12. They are enzymatically generated from PUFA, including arachidonic acid (AA), docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), through the action of cyclooxygenases (COX), lipoxygenases (LOX) and cytochrome P450 monooxygenases. These include pro-inflammatory eicosanoids such as leukotrienes (e.g., LTB4, LTC4), which regulate microvascular permeability and leukocyte recruitment, and prostaglandins (e.g., PGE2) that regulate blood vessel tone, pro-inflammatory signaling and thrombosis13. In contrast, specialized pro-resolving lipid mediators (SPM), including resolvins (e.g., RvD1, RvD2, RvE1), are produced during resolution and actively signal the termination of pro-inflammatory mediator production, promote macrophage efferocytosis, and enhance host-defense11,14,15. SPM are generated in humans in inflammatory exudates at bioactive levels and when added back to human blisters, reduce neutrophil numbers16–18. SPM can improve pathological manifestations of chronic inflammation19,20. Moreover, some SPM actively promote tissue repair in the skin and the eye, and accelerate tissue regeneration in planaria21–23. Here we mapped the global structural lipid and lipid mediator profiles during muscle injury and regeneration, investigated specific lipid mediator signatures of innate immune cell subsets, and integrated this information with transcriptomics for a systems-level comprehensive view of lipid mediator networks and immune cell phenotype.
Results
Cardiotoxin injury mobilizes PUFA from phospholipid pools
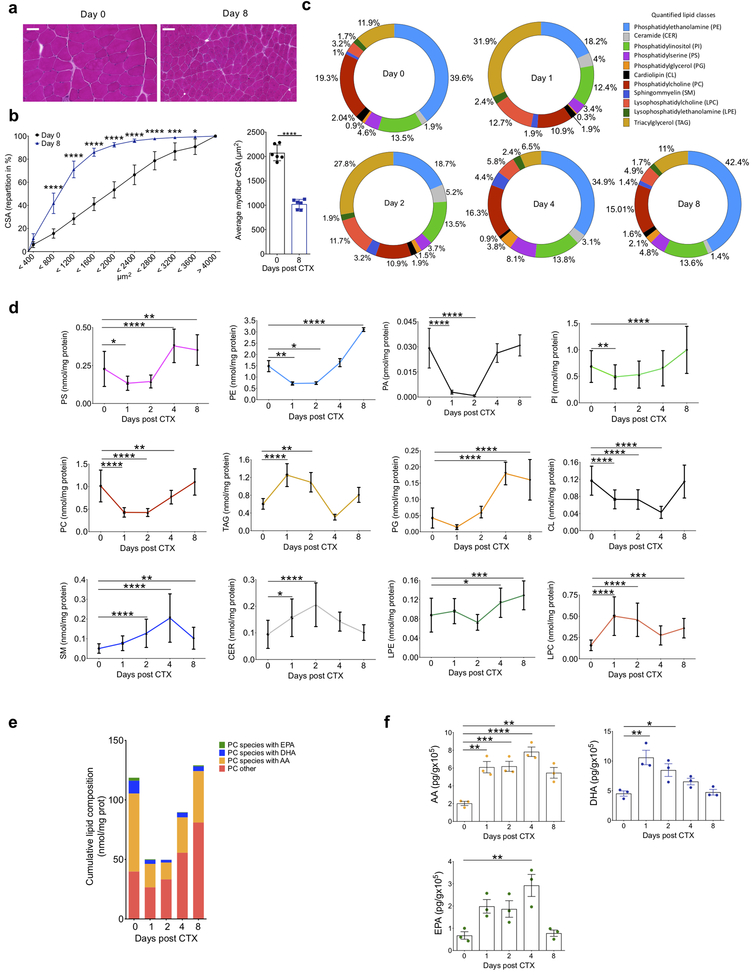
In a synchronous model of cardiotoxin (CTX)-induced injury that features coordinated inflammatory and regeneration response24, we used unbiased shotgun-lipidomics (described in Methods) to determine changes in structural lipids in the tibialis anterior (TA) muscle. We used histological analysis to document regeneration by hematoxylin and eosin (H&E) staining and morphometry. Analysis at day 0 before the CTX-induced injury and day 8 post-CTX-induced injury revealed the complete restoration of the muscle architecture by day 8. The regenerated myofibers were smaller and displayed central myonuclei (Fig. 1a), indicating myocyte differentiation. Myofiber cross-sectional area (CSA) repartition for the tissue at day 0 and day 8 showed that the cumulative CSA distribution shifted to the left and the average CSA repartition was decreased in the regenerated tissue compared to the uninjured tissue (Fig. 1b), indicating smaller myofibers.
Figure 1. Temporal regulation of structural and signaling lipids during tissue regeneration after sterile muscle injury.
(a) Representative images of H&E-stained TA muscles at days 0 and 8 after CTX-injury (at least 5 experiments were repeated independently with similar results). Scale bars are 100 μm. (b) Fiber size repartition of uninjured and regenerating muscle at day 8 post CTX injury (left panel). Data indicated mean ± SEM and p<0.05=*, p<0.001=***, p<0.0001=**** by Sidak’s multiple comparison test in two-way ANOVA. Mean (± SEM) fiber CSA is shown on the right panel. p<0.0001=**** by two-tailed unpaired Student’s t-test and n=at least 5 biologically independent muscle samples for both panels. (c) Relative distribution of lipid classes at selected time points after CTX, expressed as percentages. Color coding, names and abbreviations are shown as inset. (d) Quantification of each lipid class at indicated timepoints after CTX-injury. p<0.05=*, p<0.01=**, p<0.001=***, p<0.0001=**** by Dunnett’s multiple comparison test in two-way ANOVA. Data are shown as mean ± SEM and n=3 biologically independent muscle samples for each time point. (e) Cumulative lipid composition of four distinct groups of phosphatidyl choline (PC) species at indicated timepoints post CTX-injury. PC species groups contain Arachidonic Acid (AA), Docosahexaenoic Acid (DHA), Eicosapentaenoic Acid (EPA) or other PUFA chains at the sn-2 position of PC. (f) Levels of AA, EPA, and DHA at indicated timepoints after CTX-injury. p<0.05=*, p<0.01=**, p<0.001=***, p<0.0001=**** by Dunnett’s multiple comparison test in one-way ANOVA. Data are shown as mean ± SEM and n=3 biologically independent muscle samples for each time point.
We quantified the total lipid content of the TA in its uninjured state and at days 1, 2, 4 and 8 post injury by multi-dimensional mass spectrometry (MS)-based shotgun lipidomics (Methods and Supplementary Table 1a). We identified glycerophospholipids, sphingolipids, ceramides and glycerolipids (Fig. 1c). Phosphatidyl ethanolamine (PE, 39.6%), phosphatidyl choline (PC, 19.3%) and phosphatidyl inositol (PI, 13.5%) were the most abundant species at day 0, while lysophosphatidyl species constituted a small fraction (<5%) of the identified lipidome in normal muscle (Fig. 1c). At day 1 after CTX, triacylglycerols (TAG) and lysophosphatidylcholine (LPC) species increased compared to unijnured muscle (2.6 and 3.9 fold, respectively) and this was coupled with a simultaneous decrease in PC (1.7 fold) and PE (2.1 fold) (Fig. 1d). The amounts of PC and PE rebounded at day 4, and returned to baseline by day 8 (Fig. 1c, d). Sphingomyelin (SM) and ceramide (CER) peaked at days 2 and 4, before returning to baseline at day 8 (Fig. 1c, d). Because of the inverse relationship between PC and LPC, we assessed the composition of PUFA esterified to the sn-2 position of PC. Most of the PC contained AA, but also DHA and EPA (Fig. 1e). Their cumulative levels decreased by nearly 50% at day 1 compared to the baseline and rebounded by day 8 (Fig. 1e). During cellular activation, PUFA esterified to phospholipids are released by phospholipase A2 (PLA2) enzymes12. The amounts of free AA, EPA and DHA increased inversely to PC during the time course of muscle injury and nearly returned to baseline by day 8 (Fig. 1f). Thus, phospholipid remodeling and liberation of PUFA from phospholipids was dynamically regulated during injury and repair.
Lipid mediator class switching occurs during muscle regeneration
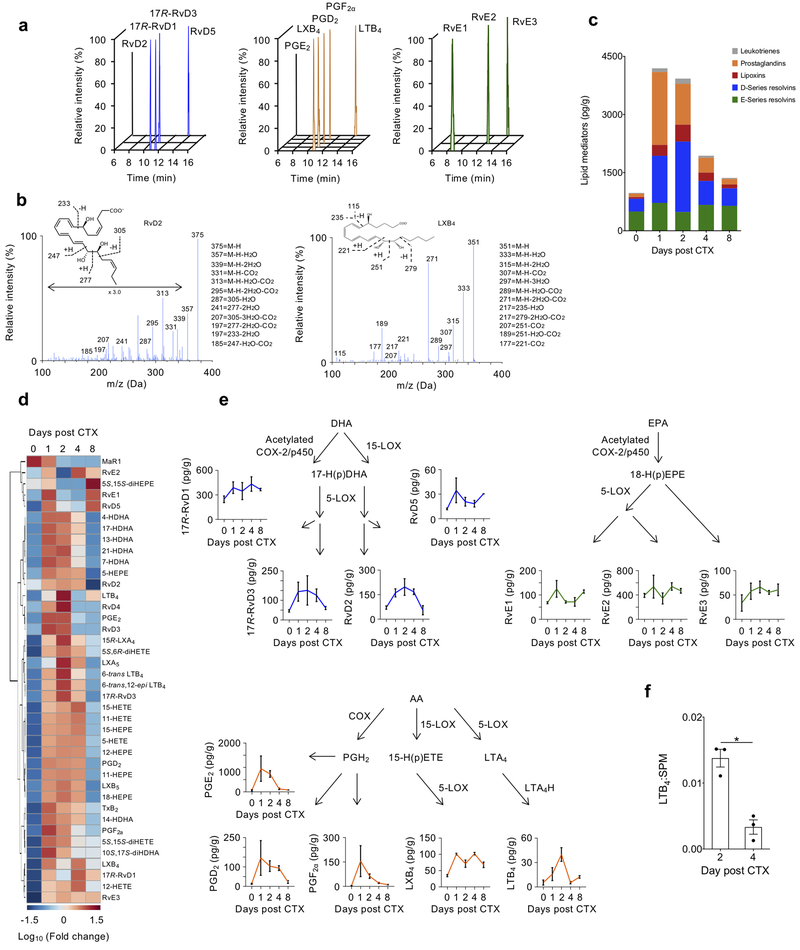
PUFA-derived lipid mediators have important roles in the initiation and resolution of inflammation. To investigate the production of lipid mediators during muscle injury, we conducted targeted liquid chromatography-tandem mass spectrometry (LC-MS/MS) at days 1, 2, 4, and 8 following injury. We assessed major families of pro-inflammatory lipid mediators (e.g., leukotrienes, prostaglandins) and SPM (e.g., resolvins, lipoxins, protectins, maresins), as well as their biosynthetic pathway markers. The abundance of each mediator was quantitated using extraction recovery of deuterated internal standards and calibration curves of external standards for each lipid mediator, identified based on retention time and diagnostic fragmentation ion assignments (see Methods). Lipid mediators and their biosynthetic pathway markers were identified across the AA, EPA and DHA bioactive metabolomes (Fig. 2a–c).
Figure 2. PUFA-derived lipid mediator profiles of TA muscle are temporally regulated following CTX-muscle injury.
(a) Representative multiple reaction monitoring (MRM) chromatograms of bioactive lipid mediators derived from DHA (blue), AA (orange) and EPA (green). 3 experiments were repeated independently with similar results. (b) Representative MS/MS spectra of RvD2 (left panel) and LXB4 (right panel) with their diagnostic ion assignments and structures shown as inset (3 experiments were repeated independently with similar results). (c) Stacked histogram showing the cumulative levels of each specific class of lipid mediators at the indicated day after CTX injury. Constituent members for each group are: Leukotrienes - LTB4, Δ6-trans-LTB4, Δ6-trans,12-epi-LTB4; Prostaglandins - PGD2, PGE2, PGF2α, TxB2; Lipoxins - 15R-LXA4, LXB4, LXA5, LXB5; D-series resolvins - 17R-RvD1, RvD2, 17R-RvD3, RvD4, RvD5; E-series resolvins - RvE1, RvE2, RvE3. (d) Heatmap displaying the relative abundance of individual lipid mediators at the indicated day post CTX. Each column represents the average of n=3 biologically independent TA muscle samples for each indicated time point. (e) Levels of selected AA (orange)-, DHA (blue)-, and EPA (green)-derived lipid mediators displayed in their biosynthetic pathways. Data shows mean ± SEM and n=3 biologically independent muscle samples for each indicated time point. (f) Ratio of LTB4 to total SPM (15R-LXA4, LXB4, LXA5, LXB5, 17R-RvD1, RvD2, 17R-RvD3, RvD4, RvD5, 10S,17S-diHDHA, MaR1, RvE1, RvE2, RvE3) at indicated days post-CTX. *p<0.05 (two-tailed unpaired Student’s t-test). Data show mean ± SEM and n=3 biologically independent muscle samples per time point.
We subdivided each of the lipid classes and compared the contribution of each to the entire lipid mediator pool at day 0, 1, 2, 4, and 8 post-injury. We observed a shift from greater levels of pro-inflammatory mediators (prostaglandins and leukotrienes) at day 1 and 2 to SPM (lipoxins and resolvins) at day 2–8 (Fig. 2c). The proportions of each class to the whole pool of lipid mediators at day 8 were similar to day 0 (Fig. 2c). Next, we examined the relative changes of each individual lipid mediator over the regeneration time course. Levels of nearly all lipid mediators increased at day 1 and 2 compared with baseline; many of the pro-inflammatory lipid mediators (LTB4, PGE2, PGF2α) then decreased by day 4, while most SPM (RvE2, RvE1, RvD5, RvD2, RvE3, LXB4) and their pathway markers remained elevated at day 4 compared with their baseline levels (Fig. 2d, e; Supplementary Table 1b). Comparison of the ratio of LTB4 to the total amount of SPM at day 2 and 4 also indicated a switch from pro-inflammatory to pro-resolving mediators (Fig. 2f). Collectively, these results indicate the dynamic class switching of lipid mediators during injury and regeneration in skeletal muscle.
Lipid mediator class switching occurs during exercise-injury
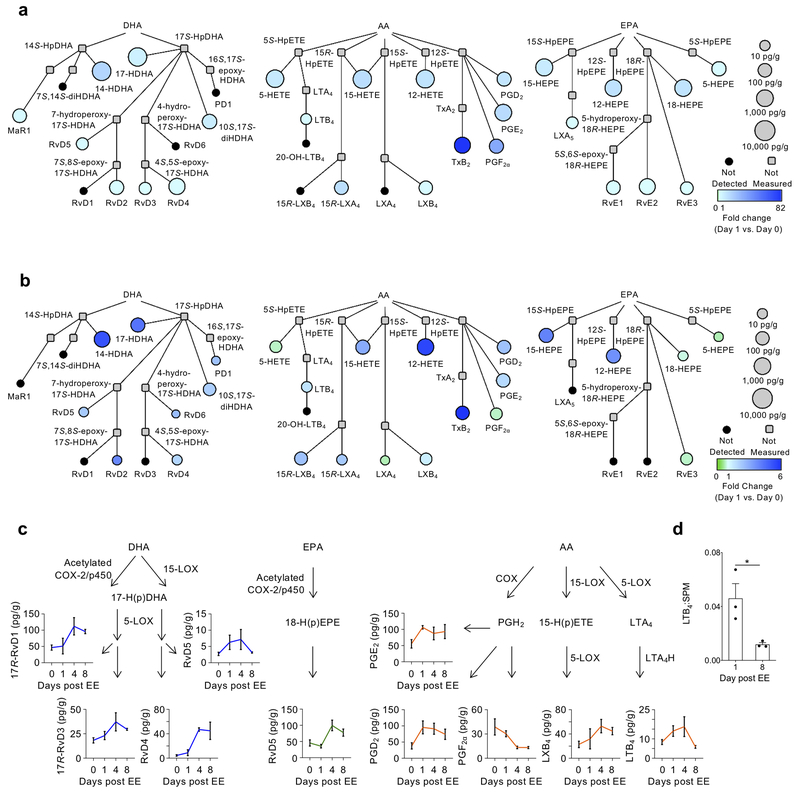
Next, we used targeted LC-MS/MS to assess the lipidomic changes that occur in eccentric exercise (EE)-induced injury (model described in Methods)25,26 which is more pathophysiologically relevant than the CTX-induced injury. We used interaction network pathway analyses to visualize changes in lipid mediator metabolomes from AA, EPA and DHA at day 1 post-injury compared to day 0 in both the CTX- and EE-induced injury (Fig. 3a,b). This analysis integrates pathway relationships with lipid mediator abundance and fold-change versus baseline (day 0). The mediators identified and their direction of change were largely consistent between the CTX- and the EE-induced injury models, although the magnitude of change of some mediators (indicated by the fold-change scale) was greater in the CTX model (Fig. 3a, b). We observed an increase in AA-derived LTB4, prostaglandins (PGE2, PGD2) and lipoxins (LXB4 and 15R-LXA4) in both models at day 1 post-injury (Fig. 3a, b). Similarly, SPM derived from EPA and DHA, including RvD2, RvD5 and RvE3, were elevated above baseline in both models at day 1 (Fig. 3a, b). Amounts of selected lipid mediators showed a similar inverse relationship between pro-inflammatory AA-derived mediators (LTB4, PGF2α) and SPM such as LXB4, 17R-RvD1, RvD4 and RvE3, which remained elevated at day 8 post-injury (Fig. 3c; Supplementary Fig. 1; Supplementary Table 1c). We also compared the amount of LTB4 to that of the total amount of SPM in the tissue at day 1 and day 8 post EE-induced injury to determine the dynamics of the switch in lipid mediator class, which occurred at day 8 compared to day 4 in CTX-induced injury (Fig. 3d). These results demonstrate that lipid mediator class switching occurs during muscle injury regardless of the stimuli, but that the less severe EE-induced injury delays the switch compared to CTX-induced injury.
Figure 3. Changes in PUFA metabolomes are conserved across pharmacological and physiological models of skeletal muscle injury.
(a, b) Interaction network pathway analyses of the docosahexaenoic (DHA), arachidonic (AA) and eicospentaenoic (EPA) acid bioactive metabolomes in TA muscle after CTX (a) or eccentric exercise-induced injury (b). The networks depict both the relative changes of each lipid mediator at day 1 compared to day 0 (color of circle) and the absolute abundance of the mediators at day 1 (size of circle). Compounds that were not detected in the analysis are shown in black, while those that were not included in the analysis are shown in grey. n=3 biologically independent muscle samples per group per time point. (c) Quantification of selected lipid mediators from the AA, DHA and EPA bioactive metabolomes depicted in their biosynthetic pathways after EE. Data show mean ± SEM. and n=3 biologically independent animals per time point (d) Ratio of LTB4 to total SPM (LXA4, 15R-LXA4, LXB4, 15R-LXB4, 17R-RvD1, RvD2, 17R-RvD3, RvD4, RvD5, RvD6, PD1, 17R-PD1, 10S,17S-diHDHA, MaR2, RvE3) at days 1 and 8 post eccentric exercise injury. *p<0.05 (two-tailed unpaired Student’s t-test). Data show mean ± SEM and n=3 biologically independent muscles per time point.
Lipid mediator signatures are distinct in immune cell subsets
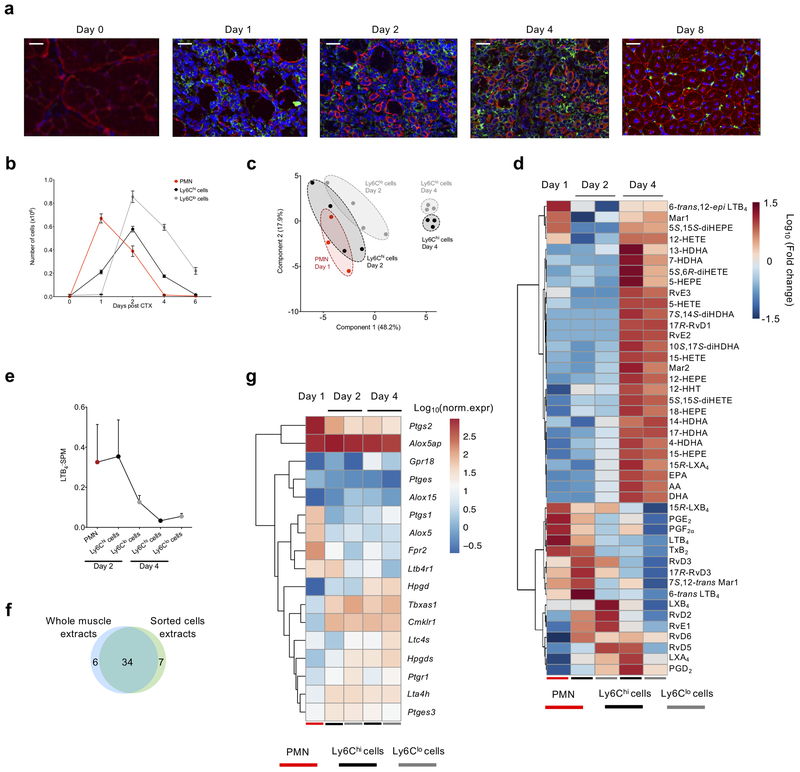
Leukocytes are sources and targets of lipid mediators, although the individual lipid mediator profile of distinct immune cell subsets during injury has not been extensively investigated. Immunohistochemistry analysis of injured muscle at days 0, 1, 2, 4 and 8 post-CTX indicated the complete destruction of the muscle architecture, as indicated by the loss of desmin+ myocytes by day 1, and the dynamic accumulation of F4/80+ macrophages starting at days 1 and 2, and increasing by day 4 (Fig. 4a). At day 8, the normal skeletal muscle architecture was restored, and the F4/80+ macrophages largely disappeared (Fig. 4a). As indicated by flow cytometry, and in line with previous observations27, Ly6G+ neutrophils reached maximum accumulation at day 1 post-injury and were rapidly cleared from the tissue by day 4 (Fig. 4b and Supplementary Fig. 2a–b), characteristic of the resolution phase. F4/80+Ly6Chi macrophages accumulated in the tissue starting at day 1 and their numbers peaked at day 2 post-injury, while F4/80+Ly6Clo macrophages appeared on day 2 and their numbers were higher than that of Ly6Chi macrophages on days 4–6 (Fig. 4b and Supplementary Fig. 2b). Similar kinetics of leukocyte accumulation were observed in the EE-induced injury model, where Ly6G+ neutrophils and Ly6Chi macrophages peaked at day 1 post-injury, and Ly6Clo macrophages were higher than Ly6Chi macrophages at day 4 post-injury (Supplementary Fig. 2c–f). Using fluorescence activated cell sorting, we isolated Ly6G+ neutrophils and both Ly6Chi and Ly6Clo macrophages from muscle at days 1–4 post-CTX in order to assess their lipid mediator profiles by LC-MS/MS (gating strategy in Supplementary Fig. 2a–b). Using partial least squares-discriminant analysis (PLS-DA) to compare global lipid mediator profiles between each sorted leukocyte population, we observed a partial overlap between the lipid mediator profile of Ly6G+ neutrophils (day 1) and that of Ly6Chi and Ly6Clo macrophages (day 2) (Fig. 4c). In contrast, the lipid mediator profile of Ly6Chi and Ly6Clo macrophages at day 4 were distinct from each other and were clearly separated from Ly6G+ neutrophils (day 1) and Ly6Chi and Ly6Clo macrophages (day 2).
Figure 4. Lipid mediator profiles and gene expression of lipid mediator receptors and enzymes are characteristic of specific immune cells during skeletal muscle regeneration.
(a) Immunofluorescence analysis of desmin (red), F4/80 (green), and nuclei (blue) in TA muscles of WT mice at the indicated day post-CTX. Scale bars are 100 μm for days 1–4 and 50 μm for day 0 and 8. Images represent 5 independent experiments with similar results. (b) Number of infiltrating neutrophils (CD45+ Ly6Ghi Ly6Cint F4/80−), inflammatory macrophages (CD45+ Ly6Chi Ly6Glo F4/80lo), and repair macrophages (CD45+ Ly6Clo Ly6G− F4/80hi) in regenerating muscle from mice at the indicated day post-CTX. Data show mean ± SEM and n=3 biologically independent experiments per group. (c) Partial least squares-discriminant analysis (PLS-DA) two-dimensional scores plot showing clustering of samples in distinct populations based on their global lipid mediator profile and specific immune cell subset at each day post-CTX (n=3 biologically independent samples per group). (d) Heatmap showing the relative abundance of individual lipid mediators of sorted cell (neutrophils, Ly6Chi and Ly6Clo macrophages) populations at the indicated day post-CTX. Each column represents the mean of at least 3 samples for each cell population per time point. (e) The ratio of LTB4 to total SPM, as described previously, in each cell population at the indicated day post CTX. Data show mean ± SEM and n=at least 3 per population per time point. (f) Venn diagram presenting the number of identified lipid mediators from whole muscle tissue and from the sorted immune cell populations (g) Normalized mRNA levels; visualized as log10(norm.expr.), of selected genes involved in lipid metabolism and signaling from Ly6Chi and Ly6Clo macrophages of days 2 and 4 post CTX and PMNs RNA-seq analysis (Ward’s clustering algorithm and Euclidean measure distance; n=3 independent samples for each cell population at the indicated time points).
To determine the changes in individual lipid mediators driving the separation or overlap among Ly6G+ neutrophils, Ly6Chi macrophages, and Ly6Clo macrophages at each day, the lipid mediator profiles were subjected to hierarchical clustering. Ly6G+ neutrophils (day 1) contained higher amounts of AA-derived pro-inflammatory mediators (e.g. LTB4, PGE2, PDF2α, TxB2) relative to Ly6Chi and Ly6Clo macrophages at day 4 (Fig. 4d; Supplementary Table 1d). While Ly6G+ neutrophils contained SPM, macrophages produced higher amounts, with a distinct SPM cluster (e.g., MaR1, 15R-LXA4, RvE3, RvE2, MaR2, RvD5, RvD6) observed at day 4 in both Ly6Chi macrophages and Ly6Clo macrophages (Fig. 4d). The Ly6Chi macrophages (day 2) had increased production of pro-inflammatory mediators (e.g. LTB4, PGE2), similar to Ly6G+ neutrophils, relative to Ly6Clo macrophages (Fig. 4d). The amount of RvD2, LXB4, LXA4 and RvD5 was higher in Ly6Clo macrophages compared to Ly6Chi macrophages at day 2 (Fig. 4d).
We next calculated the LTB4 to SPM ratio in each population, as an index of inflammation-resolution. The ratio was highest in Ly6G+ neutrophils and Ly6Chi macrophages at day 2 and was reduced in both Ly6Chi macrophages and Ly6Clo macrophages at day 4 (Fig. 4e). When we integrated the data from the whole muscle with the data from sorted cells, the majority of the lipid mediators (34 in total) were commonly identified between the two datasets (Fig. 4f). Given the dynamic signatures of lipid mediators between the leukocyte subsets, we next investigated the gene expression of enzymes and cellular receptors that regulate the production and downstream actions of lipid mediators. We performed RNA-seq analysis of Ly6G+ neutrophils, Ly6Chi macrophages and Ly6Clo macrophages sorted from muscle at day 1, 2 and 4 following CTX-injury. Several members of the phospholipases responsible for the release of PUFA from membrane lipids (e.g., Pla2g4a, Pla2g7, Pla2g15) were identified in Ly6G+ neutrophils, Ly6Chi macrophages and Ly6Clo macrophages (Supplementary Fig. 3). Expression of Pla2g4a, coding for cPLA2, was modestly higher in Ly6Chi macrophages and Ly6Clo macrophages than Ly6G+ neutrophils (Supplementary Fig. 3). Ptgs1 and Ptgs2 (encoding cyclooxygenase 1 and 2) and Ptges (prostaglandin E-synthase) were highly upregulated in Ly6G+ neutrophils at day 1compared to the Ly6Chi and Ly6Clo macrophages at days 2 and 4 (Fig. 4g), consistent with the relatively higher amounts of prostanoids (e.g., PGE2) in sorted Ly6G+ neutrophils (Fig. 4d). Hpgds (encoding hematopoietic prostaglandin-D synthase) was relatively higher in Ly6Clo macrophages compared to Ly6G+ neutrophils at day 1 (Fig. 4g), consistent with the identification of PGD2 in days 2 and day 4 Ly6Clo macrophages. Similarly, the leukotriene biosynthetic enzymes Alox5 and Alox5ap, as well as the leukotriene receptor Ltb4r1 were expressed highly in Ly6G+ neutrophils, while expression of Alox5ap and Lta4h remained constant in Ly6Chi and Ly6Clo macrophages at days 2 and 4 (Fig. 4g). The SPM identified require co-operation of both Alox5 and Alox15 for their biosynthesis11; expression of Alox15 was higher in day 2 and 4 Ly6Chi and Ly6Clo macrophages compared to Ly6G+ neutrophils (Fig. 4g). Fpr2 (encoding FPR2), the receptor for LXA4 and RvD1, was expressed in Ly6Chi and Ly6Clo macrophages at days 2 and 4, but was higher in Ly6G+ neutrophils (Fig. 4g), consistent with the role of SPM as stop signals for neutrophil trafficking8. In contrast, Gpr18, the receptor for RvD228, peaked in day 4 Ly6Chi macrophages, was expressed in day 4 Ly6Clo macrophages, and had low expression in Ly6G+ neutrophils (day 1) (Fig. 4g).
To determine whether interference with the dynamic production sequence of the bioactive lipids affected immune cell trafficking and macrophage subtype specification, mice undergoing CTX-injury were treated with the non-steroidal anti-inflammatory drug (NSAID) ibuprofen (IBP) every 24h and lipid mediator production and leukocyte dynamics were assessed at day 1, 2 and 4 post-injury. Acute treatment (24h) with IBP strongly inhibited the appearance of COX products (e.g., PGE2, TXB2) when assessed 4h after its administration (Supplementary Fig. 4a,b; Supplementary Table 1e). Daily administration of IBP resulted in dysregulated lipid mediator profiles at day 2 and 4, with the amounts of the most major lipid mediator pathways being increased when assessed 24h post-IBP administration compared to mice without IBP treatment (Supplementary Fig. 4c and Supplementary Table 1f). The number of CD45+ cells was not different between mice treated without or with IBP at either day 2 or day 4 (Supplementary Fig. 4d). IBP treatment increased the percentage of Ly6Chi macrophages and decreased the percentage of Ly6Clo macrophages at day 2, while the opposite pattern was observed at day 4 (Supplementary Fig. 4e–f). These data indicate that interfering with lipid mediator production alters the kinetics of Ly6Chi and Ly6Clo macrophage accumulation.
RvD2 induces specific macrophage gene expression changes
We next investigated the potential mediators of macrophage-dependent tissue repair. We focused on RvD2 because it’s a potent regulator of resolution of inflammation11,14; it was produced in a dynamic fashion primarily by Ly6Clo macrophages; the gene encoding its receptor, Gpr18, was expressed in Ly6Chi macrophages; and it was detected in both CTX- and EE-induced muscle injury. RvD2 potently enhances macrophage functions, such as efferocytosis14,33. However, how RvD2 affects gene expression in naïve macrophages remains incompletely characterized. We performed RNA-seq on bone marrow-derived macrophages (BMDMs) treated with RvD2 for 4 hours and compared to untreated (control-vehicle) BMDMs. 751 genes were differentially expressed (fold change>1.5; p<0.05; Supplementary Table 2a). Among the top 120 differentially expressed genes, based on the largest FC differences (absolute value), genes most highly induced by RvD2 included those involved in arginine metabolism (Arg2), transcriptional regulators (Nr4a1, Nr4a2), G-protein and cAMP signaling (Gpr35, Akap3, Adcy1, Arrdc3, Pde4b) and host defense (Rsad2, Tlr5, Gbp5, Gbp10, Masp2) (Fig. 5a). Functional classification of the differentially expressed genes with gene ontology (GO) analysis revealed multiple categories belonging to macrophage activation, function and immune system regulation (Fig. 5b; Supplementary Table 2b). Specifically, we observed an enrichment in pathways such as those associated with chemotaxis (up: Cxcl3, Cxcr4, Cxcl10, Nr4a3, Vegfa, Gab1; down: Kif5a, Plxnb3, Robo1), movement of a subcellular component (up: Enpep, Ccdc40, Wnt11; down: Elmo3, P2ry1, Kif22, Kif23, Celsr2, Cdk5r1) and response to a molecule of bacterial origin (up: Gbp6, Tlr2, Ticam2, Cmpk2, Stap1, Cd14), suggesting that RvD2 induced a unique genetic signature in BMDMs.
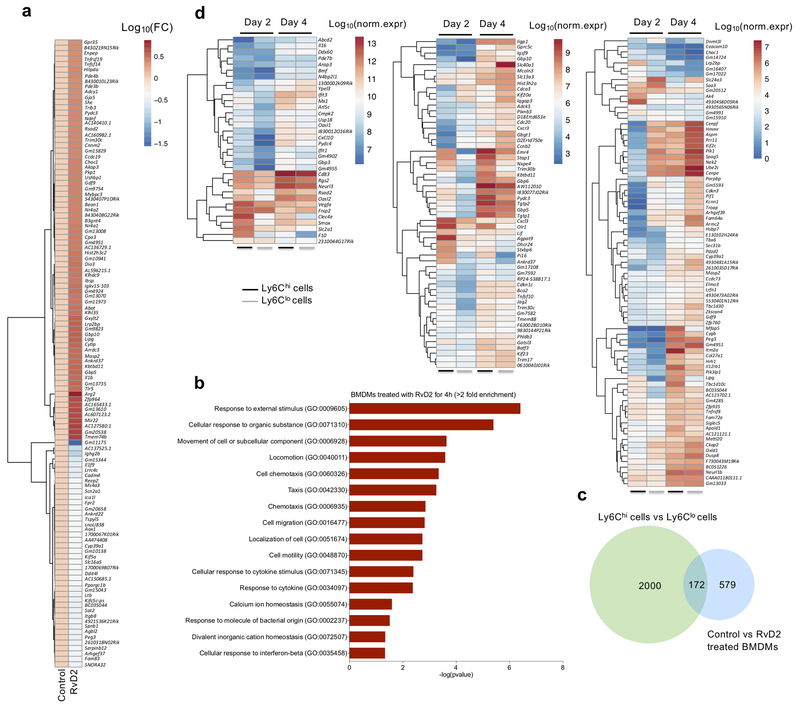
Figure 5. RvD2 induces a unique transcriptional signature in macrophages that has similarities to macrophages during the transition from inflammation to resolution and regeneration.
(a) Heat map showing the relative expression pattern of the top 120 differentially expressed genes in control and RvD2 treated (4 hours) naïve bone marrow-derived macrophages (BMDMs). Fold change values are visualized as log10(FC). (b) Gene ontology (GO) analysis of the genes that are differentially expressed in control and RvD2 treated BMDMs. Fold enrichment threshold was set at >2 and Bonferroni-corrected for p value < 0.05. (c) Venn Diagram showing the differentially expressed genes from control versus RvD2 treated BMDMs, integrated with the common differentially expressed (DE) genes in Ly6C macrophage subsets (see inset). In total, 172 genes are common between the two datasets, while 579 genes were DE only in RvD2 treated BMDMs. (d) Heatmaps showing normalized expression values of genes in Ly6Chi and Ly6Clo macrophages of days 2 and 4 post CTX that are common and DE between the two datasets (i.e., control vs RvD2 and Ly6Chi vs Ly6Clo at days 2/4). Hierarchical clustering analysis was performed for the transcript levels of the differentially expressed genes (Ward’s clustering algorithm and Euclidean measure distance and n=3 biologically independent samples for each cell populations at indicated time points).
To integrate the RvD2-induced changes in gene expression with the transcriptome of Ly6Chi and Ly6Clo macrophages during regeneration, we performed RNA-seq analysis on sorted muscle-infiltrating Ly6Chi and Ly6Clo macrophages at day 2 and 4 post-CTX. We compared the transcriptomic profile of day 2 Ly6Chi macrophages and day 4 Ly6Chi macrophages, and also the transcriptome changes between days 2 Ly6Clo macrophages and 4 Ly6Clo macrophages. We found 4219 differentially expressed genes in day 2 compared to day 4 Ly6Chi macrophages, and 3378 differentially expressed genes in day 2 compared to day 4 Ly6Clo macrophages (Supplementary Table 3a). GO analysis of the differentially expressed genes (fold change>2, p<0.05) from the two comparisons indicated these genes were involved in processes such as extracellular matrix organization, cell killing, locomotion, migration and muscle tissue development (Supplementary Fig. 5a; Supplementary Table 3b). Genes differentially expressed in Ly6Clo macrophages from day 2 to 4, included genes involved in actin filament and sarcomere organization, muscle cell differentiation and development, as well as cell proliferation and cell-cell adhesion (Supplementary Fig. 5b; Supplementary Table 3c). We next compared the genes modulated by RvD2 treatment in BMDMs, with the genes identified during the inflammatory (day 2) and repair (day 4) phase using GeneVenn software29. This analysis indicated that 172 genes were common between the differentially expressed genes of the three groups comparison: day 2 Ly6Chi versus day 4 Ly6Chi, day 2 Ly6Clo versus day 4 Ly6Clo and control-vehicle versus RvD2 treatment in BMDMs (Fig. 5c–d; Supplementary Table 3d). Several of these genes, such as Rsad2, Cd83, Rgs2, Abcd2, Il16, Arap3, Usp18, Cxcl10, Pydc4, Gbp3 and Mx1, were amongst the most highly-expressed genes in the RvD2 treatment group, and their expression was higher in day 4 Ly6Clo macrophages than in day 2 Ly6Chi macrophages (Fig. 5d). Similar gene cluster relationships were observed in those with more moderate overall expression, but which were highly induced selectively in both day 4 Ly6Chi and day 4 Ly6Clo macrophages, such as Elmo3, Gdf9, Slc13a3, Hist3h2a, Kbtbd11, Gbp6, Gbp5 and Pydc3 (Fig. 5d). GO analysis of the common-identified differentially expressed genes revealed that they were involved in functions related to mitotic cell cycle and cell division, and processes that included defense responses and response to cytokine stimulus and virus (Supplementary Fig. 5c; Supplementary Table 3e). We validated selected individual genes (i.e., Cxcl10, Gpr30, Nr4a1, Gpr35) by qRT-PCR and found that RvD2 induced their expression in a time and concentration-dependent manner (Supplementary Fig. 5d). These results indicate that RvD2 induced a unique macrophage phenotype, which had some similarities to that detected in in vivo isolated day 4 Ly6Chi and Ly6Clo macrophages, during the initial phase of repair.
RvD2 is an effector of macrophage subtype specification
We next asked whether exogenous delivery of RvD2 in mice undergoing CTX-injury can modulate the accumulation of Ly6Clo repair macrophages, a process that is widely documented to facilitate repair30. Since muscle regeneration in healthy mice is tightly coordinated and evolutionary conserved, with a very small window for intervention, we used a model of impaired muscle regeneration to test the effects of RvD2 in aiding the recovery from the injury. For this, we used a CTX injury model in which BoyJ congenic mice are irradiated to deplete satellite muscle cells, followed by bone marrow transplantation from healthy C57Bl/6J donor mice (described in Methods)31. In this model, the in situ transition from Ly6Chi to Ly6Clo macrophages is delayed due to the altered cellular composition caused by the radiation and is associated with severe impairment in tissue regeneration31. Mice were left to recover for 8 weeks following bone marrow transplantation to exclude short-term radiotoxicity effects. At this time point, circulating and muscle infiltrating myeloid cells are almost exclusively of CD45.2+ donor origin31. The chimeric mice were injured with CTX, followed by local intramuscular administration of RvD2 or saline as control either at day 2 or 3 post-CTX. The dose used (4μg/kg) achieved acute intramuscular amounts of RvD2 (~200pg at 2h post-injection), that were commensurate with the endogenous amount of RvD2 quantified during CTX injury at day 2 (Fig. 2e), as assessed using intramuscular injection of deuterium-labelled d5-RvD2 into healthy mice to differentiate injected RvD2 from endogenous RvD2 (Supplementary Fig. 6a,b). Four days after muscle injury, CD45+ immune cells (Fig. 6a) and Ly6ChiF4/80lo and Ly6CloF4/80hi cells (Fig. 6b) were detected in skeletal muscle of the RvD2-treated chimeric mice. In the saline-treated chimeric mice, Ly6Clo macrophages constituted only a small fraction (~10%) of the CD45+ cells, while in day 2 RvD2-treated mice, the frequency of Ly6Clo macrophages increased to 16% (Fig. 6b,c; Supplementary Fig. 6c), shifting closer to that observed in healthy mice undergoing regeneration at this timepoint (~45%; Supplementary Fig. 2b). Compared to saline treatment, administration of RvD2 at day 3 post-injury compared to administration at day 2 post-injury, resulted in an even higher increase in the frequency of Ly6Clo macrophages (~20%), while the frequency of Ly6Chi macrophages was reduced by 10% compared to vehicle treated animals (Fig. 6c; Supplementary Fig. 6c). More specifically, the ratio of Ly6Clo/Ly6Chi macrophages at day 4 increased from 0.22 in the saline-treated mice, to 0.36 and 0.41 upon RvD2 treatment at days 2 and 3 post-CTX injury, respectively (Supplementary Fig. 6c). These results indicated that RvD2 regulated the population transition from inflammatory Ly6Chi macrophages to repair Ly6Clo macrophages.
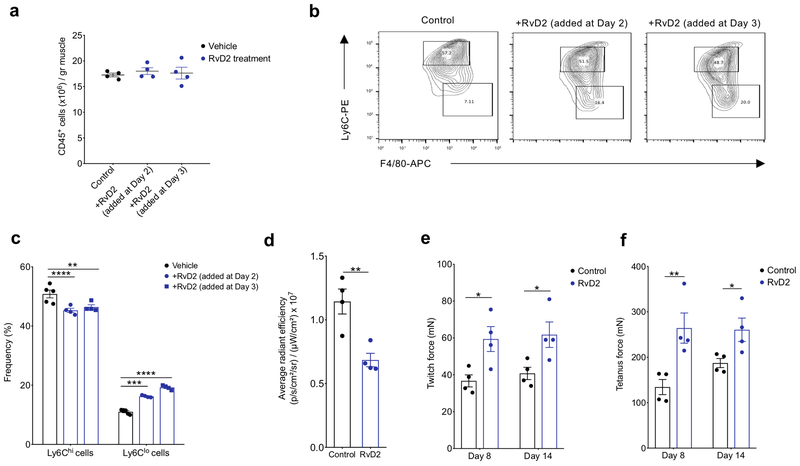
Figure 6. RvD2 administration increases the proportion of Ly6Clo macrophages and improves in vivo force generation in a model of delayed muscle regeneration.
(a) Absolute number of infiltrating CD45+ cells in TA muscle of chimeric mice administered with saline (control) or RvD2 intramuscularly on day 2 or day 3 post CTX injury. Data show mean ± SEM and n=4 biologically independent samples per treatment group. (b) Representative flow cytometry contour plots of Ly6Chi and Ly6Clo macrophages from saline (control) and RvD2 treated animals at day 4 post CTX injury. Images represent 4 independent experiments with similar results. (c) Frequency of inflammatory (Ly6Chi F4/80lo) and repair (Ly6Clo F4/80hi) macrophages from saline (control) and RvD2 treated animals at day 4 post CTX-injury. p<0.05=*, p<0.01=**, p<0.001=***, p<0.0001=**** by Sidak’s multiple comparisons test in two-way ANOVA. Data are shown as mean ± SEM and n= at least 4 mice per group. (d). 2-DG uptake FLI signals, expressed as average radiance efficiency ([p·s−1·cm−2·sr−1]/[μW·cm−2]). Data are shown as mean ± SEM (n = 4 mice per group). (e) and (f) Quantification of in vivo muscle twitch (e) and tetanus forces (f). Data are shown as mean ± SEM (n= 4 mice per group) in saline and RvD2 treated chimeric mice at day 8 and day 14 post CTX. p<0.05=*, p<0.01=**, by Sidak’s multiple comparison test in two-way ANOVA.
To further test whether RvD2 treatment affected inflammation and repair of injured muscle, we assessed the uptake of fluorescently-labeled 2-deoxy-D-glucose (2-DG), using in vivo imaging31. 2-DG is taken up by cells with high metabolic activity, such as invading macrophages, and thus is widely used as a non-invasive method to monitor inflammation31–33. Chimeric mice were injured with CTX, treated with RvD2 or saline control intramuscularly at day 3, and injected with 10 nmole 2-DG intravenously 3 hours prior to the imaging on day 4 post CTX (see Methods). The 2-DG uptake and labeling intensity at day 4 post-CTX injury, expressed as average radiant efficiency, decreased 40% following RvD2 administration at day 3, compared to saline treated controls (Fig. 6d; Supplementary Fig. 6d), supporting a pro-resolution role of RvD2. To test the functional consequence of RvD2 administration on muscle recovery, we assessed TA muscle mass at day 8, and TA twitch and tetanus force at days 8 and 14 after CTX-induced injury in vivo. RvD2 improved muscle force recovery by approximately 50% at both timepoints, compared to saline-treated chimeric mice, and increased muscle mass by 17% (Fig. 6e,f; Supplementary Fig. 6e), suggesting accelerated muscle regeneration following RvD2 treatment. Collectively, these results suggest that RvD2 can improve muscle recovery following injury, potentially by increasing the ratio of Ly6Clo/Ly6Chi macrophages.
Discussion
Here we described the dynamic regulation of the mediator lipidome during skeletal muscle inflammation and regeneration. Sorted neutrophils and Ly6Chi and Ly6Clo macrophages recapitulated the temporal profiles obtained from whole muscle and revealed a distinct pro-resolving signature expressed by Ly6Clo macrophages. Integration of transcriptomics and lipidomics results demonstrated that Ly6C+ macrophages are both sources and sensors of lipid mediators which likely facilitate macrophage population phenotypic transitions.
We observed a marked remodeling of structural lipids in the injured muscle. The liberation of PUFA from phospholipids is classically defined as the rate limiting step in lipid mediator biosynthesis and occurs by calcium-dependent PLA2 in response to agonists (e.g., pattern recognition receptors, purinergic receptors), although secreted PLA2, can also play a role12. Direct delivery of PUFA to inflammatory exudates can also facilitate lipid mediator production34. The increase in lipid mediators mirrored the appearance of PUFA, with distinct temporal regulation observed among pro-inflammatory and pro-resolving mediators, coincident with the differential expression downstream biosynthetic enzymes. These results are similar to those reported in other models of acute sterile and infectious inflammation, such as peritonitis35–37. As an index of this response, the ratio of LTB4 to the total amount of SPM decreased from day 2 to day 4 post-injury. This ratio has been associated with chronic inflammation in diseases such as atherosclerosis in humans38,39.
We used two models of injury to validate the robustness of the data. CTX causes severe muscle damage and synchronous regeneration9, while the EE-induced injury results in a less severe muscle damage and is more pathophysiologically relevant30. We identified a similar profile of lipid mediators in both models, but with a higher magnitude of change in lipid mediator amounts in the CTX model. Resistance exercise in humans also increases levels of leukotrienes, prostanoids and SPM, the production of which is negatively impacted by ibuprofen40. We observed that ibuprofen inhibited prostanoids acutely, but that prostanoids and other lipid mediators rebound after each dose, further demonstrating that lipid mediator pathways are highly connected36.
The integration of the muscle lipid mediator profiles induced by CTX injury with sorted neutrophils and macrophages showed high overlap, indicating that leukocytes were likely sources of these lipid mediators in the tissue. The contribution of macrophages to SPM production is consistent with a study demonstrating that macrophage depletion decreases SPM levels in vivo41. Levels of several SPM were higher in Ly6Clo macrophages at day 2, while a SPM cluster was observed in both macrophage subsets at day 4, coincident with expression of Alox15. These results are consistent with a report showing that Alox15 is highly expressed in resolution-phase macrophages42. Similarly, IL-4 stimulation of human macrophages increases SPM relative to macrophages polarized with IFN-γ or LPS in vitro43. SPM biosynthesis can also be regulated through heterotypic leukocyte interactions (e.g., PMN-macrophage) or leukocyte-epithelial or endothelial interactions11. Given the relatively high amounts of SPM precursors in macrophages isolated at day 4 post-injury (e.g., 17-HDHA, 15-HETE, 18-HEPE), it is possible that these intermediates were transferred to other cells for conversion to SPM.
We observed the production of RvD2 by Ly6Clo macrophages at day 2 post-injury and expression of its receptor, Gpr18, in Ly6Chi macrophages. Treatment with RvD2 caused a shift in the frequency of Ly6Chi macrophages and Ly6Clo macrophages in vivo and also lead to improvement in muscle function. Future studies using focused approaches are required to assess if macrophage phenotype is modulated by RvD2 in vivo. Transcriptomics indicated that RvD2 induced the expression of genes in BMDMs that overlapped with genes expressed in both Ly6Chi and Ly6Clo macrophages. These included genes important in host defense, chemotaxis and proliferation, the latter of which are characteristic of resolving macrophages42. Some of the genes induced by RvD2 in BMDMs would be expected to enhance host defense to both viral and bacterial pathogens, and RvD2 has been reported to enhance host defense and improve survival in sepsis14,44. RvD2 also promotes muscle regeneration and revascularization after ischemia; genes induced by RvD2 in BMDM, such as Vegfa, could underlie these effect45. In macrophages, the RvD2-GPR18 axis is also important for enhancing phagocytosis and activating STAT344, which is important for efferocytosis and M2 polarization46. In contrast, RvD2 reduces necrosis and stimulates re-epithelialization of wounds in part through direct actions on keratinocytes, indicating that RvD2 might engage multiple cellular targets during tissue repair21,47. Along these lines, PGE2 targets muscle-specific stem cells to promote regeneration48 while RvE1 regulates signaling in skeletal muscle myotubes49. Several SPM target fibroblasts, endothelial cells and epithelial cells as well11. Thus, the resources generated here could inform new roles of lipid mediators and their receptors in muscle inflammation, fibrosis, angiogenesis and regeneration. Integration of transcriptomic, lipidomic, epigenomic and (phospho)-proteomic landscapes could lead to new directions in the understanding of immune cell function and could identify pathways amenable to the development of novel therapeutics.
Methods
Mice
All animal procedures were performed in accordance with ethical regulations and pre-approved by the Institutional Animal Care and Use Committee at Johns Hopkins University (MO18C251) and University of Debrecen, School of Medicine following Hungarian (license no.: 21/2011/DEMÁB) and European regulations or Brigham and Women’s Hospital (2016N000131). Wild-type BoyJ (B6.SJL-Ptprca Pepcb /BoyJ, stock number 002014) and C57BL/6J male mice were obtained from the Jackson Laboratories and bred under specific-pathogen free (SPF) conditions.
Acute cardiotoxin (CTX) muscle injury
Mice were anaesthetized with isoflurane (adjusted flow rate or concentration to 1.5%) and 50 μl of cardiotoxin (12×10−6 M in PBS) (Latoxan) was injected in the tibialis anterior (TA) muscle.54 Muscles were recovered for flow cytometry analysis, histology, immunohistochemical analysis, and lipidomics at days 0, 1, 2, 4 and 8 post-injury. For ibuprofen treatments, mice were treated intraperitoneally with either 100mg/kg ibuprofen (Cayman Chemical; item #: 15687-27-1) or vehicle (DMSO) at day 1, 2 and 3 post CTX injury (see scheme in Supplementary Figure 4).
Histological analysis of muscle regeneration
Muscles were removed and snap frozen in nitrogen-chilled isopentane (–160°C). 8 μm thick cryosections were cut and stained with hematoxylin-eosin (H&E). For each histological analysis, at least 5 slides (per condition) were selected where the total regenerative region within the CTX injured TA muscle was at least 70%. For each TA, myofibers in the entire injured area were counted and measured. H&E muscle sections were scanned with Mirax digital slide scanner and the Cross Sectional Area (CSA) was measured with HALO software (Indica Labs). CSAs for these samples are reported in μm2.
Eccentric exercise-induced muscle injury model
Animals were maintained in a surgical plane of anesthesia (3–5% isoflurane delivered in 100% O2) for the duration of procedure. Anesthetized mice were placed on a warm heating pad and hair was removed from hind limbs using fine electric hair clippers. The hind limb was restrained at the knee firmly with a clamp to ensure that the animal’s knee did not move, and the foot strapped to a footplate/force transducer with a dual motor-arm attached (Aurora Scientific). Two 30g needle platinum electrodes were inserted intramuscularly into the thigh of the right leg, then the sciatic nerve was stimulated by an electric pulse of 100Hz at 200usec pulse width at 2.5mA and 25V for 0.4 seconds. While the muscle is under contraction, the motorized footplate applies an eccentric rotational torque, causing micro-trauma and a resultant force deficit to muscle fibers. This protocol was repeated for forty repetitions to create a significant and physiologically relevant injury to the muscle. The experimental mice were then allowed to recover for a period of anywhere from 1–14 days, and followed for muscle contraction force recovery after injury. Mice were sacrificed, and muscle tissue was collected for lipidomics and flow cytometry analysis (see below).
Injury model of delayed muscle regeneration
Recipient congenic BoyJ mice (7 weeks old) were irradiated (total body irradiation) with 11 Gy using an X-rad 320 (Precision X-ray Irradiation systems) x-ray unit for the ablation of the recipient bone marrow as described previously31. Following the irradiation, isolated bone marrow cells (in sterile RPMI-1640 medium) flushed out the femur; tibia and humerus from donor C57Bl/6J mice were transplanted into the recipient mice by retro-orbital injection (20×106 bone marrow cells per mice). This experimental bone marrow transplantation (BMT) CD45 congenic model allows the discrimination of donor, competitor and host contribution in hematopoiesis and repopulation efficiency of donor cells (congenic mice with CD45.1 versus CD45.2 that are of donor origin). The efficiency of CD45.2 repopulation was then detected by flow cytometry, 8 weeks following the BMT. In short, a cut at the tail tip of the mice provided a drop of blood that was placed into 0.5 ml PBS + 1% FBS + 10 U/ml Heparin buffer (samples kept on ice). The cells were directly stained by mouse anti-mouse CD45.2-FITC (clone 104 – dilution 1/50) and rat anti-mouse GR1-PE (clone RB6–8C5 – dilution 1/50) antibodies (BD Pharmingen) and incubated on ice for 30 min. After 2 washes with ice-cold PBS/FBS/Heparin buffer, cells were resuspended in 0.5–1 ml FACS Lysing solution (BD Cat #349202). Cells were incubated for 5 min at RT then centrifuged the cells (400g, 5 min, 4°C). We ran the double stained samples on FACS (BD FACS Aria III or MoFlo Astrios) and determined the ratio of donor cells. Blood chimerism was usually around 99% gated on the granulocyte fraction31. In parallel, chimeric mice from the same transplantation cohorts were used to validate the donor origin, and chimerism of infiltrating myeloid cells in the injured muscle at day 1 post CTX (similar ratio of donor cells; as we described previously31). Two months following BMT, the animals were injured with CTX, as described above. For in vivo RvD2 treatments, mice where treated intramuscularly with either 4 μg/kg RvD2 (Cayman Chemical; item #: 10007279) or saline at day 2 or 3 post CTX injury.
In vivo muscle force measurement.
To determine the effect of RvD2 on muscle function, we performed force measurements in the murine model of delayed regeneration, as described above. In these experiments, mice were treated with RvD2 at day 3 post-injury. For this, mice were first anesthetized with 3% vaporized isoflurane mixed with O2. Mice were then positioned under a heat lamp to maintain the body temperature at 37 °C. Knees of the animals were secured to a fixed steel post, and their feet were taped to the platform to prevent movement from the contraction of other muscle groups. Electrical stimulations were applied across two needle electrodes placed through the skin just above the knee and beneath the TA muscle to stimulate the tibial nerve. In all measurements, we used 0.1-ms pulses at a predetermined supramaximal stimulation voltage. TA muscles were stimulated with a single 0.1-ms pulse for twitch force measurements and a train of 150 Hz for 0.3-s pulses for tetanic force measurements. A two-minute rest was given to the animal while under anesthesia to allow muscles to return to normal function after tetanus. We performed five twitch and then five tetanic measurements on each muscle, with 2–3 min of recovery between each measurement. For recording the muscle force measurements, we used the 610A Dynamic Muscle Control (DMC) software from Aurora Scientific.
In vivo small animal FLI imaging using 2-deoxy-D-glucose
To determine whether RvD2 modulates inflammation in vivo, we performed imaging of 2-deoxy glucose uptake in the delayed model of regeneration (see above). For this, mice were treated with RvD2 at day 3 post-CTX injury. During the light cycle to minimize muscle activity, BoyJ chimeric mice were intravenously injected with 10 nmole of XenoLight RediJect 2-DeoxyGlucosone (DG) 750 probe (Perkin Elmer) or control 750 dye (in 0.2 mL volume) and imaged at 3 hours with IVIS Spectrum (Ex745 nm/Em820 nm). Specifically, 3 hours after 2-DG injection animals were anaesthetized by 3% isoflurane with a dedicated small animal anesthesia device and whole body FLI scans were acquired using the IVIS Spectrum system (Caliper LifeSciences). To prevent movement, the animals were fixed to the mouse chamber and positioned in the center of field of view (FOV). We acquired FLI using a 3-s exposure with field of view = 6.5. Digital images were recorded and analyzed using Living Image 4.7.2 software (IVIS Imaging Systems). We analyzed images with a consistent region of interest (ROI) placed over each hindlimb to calculate a fluorescence signal. We calculated the fluorescence signal in average radiance efficiency ([p·s−1·cm−2·sr−1]/[μW·cm−2]).
Immunohistochemistry
Tissue sections were fixed and permeabilized in ice cold acetone for 5 min and blocked for 30 minutes at 20 °C (room temperature) in PBS containing 2 % bovine serum albumin (BSA). Tissues were stained for 1 h at room temperature using a primary antibody diluted in 2 % BSA. The primary antibodies used for immunofluorescence were rabbit anti-Desmin (Abcam 32362) at a dilution 1/200 and rat anti-F4/80 (Abcam 6640) at a dilution 1/200. In all cases, the primary antibody was detected using secondary antibodies (dilution 1/200) conjugated to FITC (JIR 703-095-155) or Cy3 (JIR 711-165-152). The nuclei were counter stained with 0.1–1 μg/ml Hoechst. Fluorescent microscopy was performed using Carl Zeiss Axio Imager Z2 microscope equipped with lasers at 488, 568 and 633 nm. Figures were assembled using Fiji and Illustrator CS5 (Adobe).
Isolation of macrophages from CTX injured skeletal muscle
TA muscles from CTX-injured animals were carefully isolated with fine scissors and fascia was removed29. Despite alterations in the integrity of the muscle tissue during injury (e.g., swelling), the tissue remains intact during collection. Muscles were weighed and then placed directly into ice-cold PBS. Muscles were then dissociated in RPMI containing 0.2% collagenase B (Roche Diagnostics GmbH) at 37°C for 1 hour and filtered through a 100 μm and a 40 μm filter. CD45+ cells were isolated using magnetic sorting (Miltenyi Biotec). For FACS, macrophages were incubated with Fcγ receptor blocking antibodies and with 10% normal rat serum: normal mouse serum 1:1 mix, then stained with a combination of PE-conjugated anti-Ly6C antibody (HK1.4, eBioscience), APC-conjugated F4/80 antibody (BM8, eBioscience) and FITC-conjugated Ly6G antibody (1A8, Biolegend). Ly6Chi F4/80lo Ly6Gneg macrophages, Ly6Clo F4/80hi Ly6Gneg macrophages and Ly6Ghi Ly6Cint F4/80neg neutrophils were quantified. In each experiment, samples were processed in parallel to minimize experimental variation. Cells were analyzed on a BD FACSAria III or MoFlo Astrios sorter (Summit v6 software) and data analysis was performed using BD FACSDIVA and FlowJo V10 software. Gating strategy is shown in Supplementary Fig. 2a–b.
Shotgun lipidomics
Muscle samples were collected from mice and pulverized into a fine powder using a stainless steel biopulverizer at the temperature of liquid nitrogen. The tissue powders of 10 to 20 mg were weighed and homogenized in 0.5 mL 10x diluted PBS in 2.0 ml cryogenic vials (Corning Life Sciences, Tewksbury, MA) by using a digital sonifier (Branson 450, Danbury, CT). Protein assay on the homogenates was performed using a bicinchoninic acid protein assay kit (Thermo Scientific, Rockford, IL) with bovine serum albumin as standards. The rest of homogenate was accurately transferred into a disposable glass culture test tube, and a mixture of lipid internal standards was added prior to lipid extraction for quantification of all reported lipid species. Lipid extraction was performed by using a modified Bligh and Dyer procedure as described previously51. Individual lipid extracts were resuspended into a volume of 100 μL of chloroform/methanol (1:1, v/v) per mg of protein and flushed with nitrogen, capped, and stored at −20 °C for lipid analysis. For ESI direct infusion analysis, lipid extracts were further diluted to a final concentration of ~500 fmol/μL, and analysis was performed on a QqQ mass spectrometer (Thermo TSQ QUANTIVA, San Jose, CA) equipped with an automated nanospray device (TriVersa NanoMate, Advion Bioscience Ltd., Ithaca, NY). Identification and quantification of lipid molecular species were performed using an automated software program50–52. Data were normalized per mg of protein.
Lipid mediator identification and quantitation by liquid chromatography-tandem mass spectrometry
Samples of muscle tissue (TA) or flow-sorted leukocyte populations isolated from injured muscle were subjected to solid phase extraction (SPE) followed by a targeted liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. For muscle samples, tissue was minced with scissors in ice-cold methanol containing deuterated internal standards and stored at −80°C. For leukocyte samples, cells were isolated from CTX-treated muscle then flow sorted and plated in phenol red-free and serum-free medium. The cells and medium were then combined with two volumes of ice-cold methanol and deuterated internal standards were added. Samples were then stored at −80°C. Deuterated internal standards (d5-RvD2, d4-LTB4, d8-5-HETE, d4-PGE2 and d5-LXA4; Cayman Chemical) were used to assess extraction recovery and quantification. Samples were centrifuged (3,000 rpm) and supernatants were then subjected to SPE and LC-MS/MS analysis, as described previously53. In short, acidified water (pH 3.5 with HCl) was added to samples immediately prior to SPE using C18 column chromatography. Lipid mediators were eluted in the methyl formate fractions, the solvent was evaporated under a gentle stream of N2 gas and the samples were then resuspended in methanol:water (50:50). Samples were then injected using a high-performance liquid chromatograph (HPLC, Shimadzu) coupled to a QTrap5500 mass spectrometer (AB Sciex) operating in negative ionization mode. Individual lipid mediators were identified using specific multiple reaction monitoring (MRM) transitions, information-dependent acquisition and enhanced product ion scanning55. Identification was based on matching retention time with authentic standards, MRM transitions and diagnostic MS/MS fragmentation ions. Quantitation of mediators was then carried out based on the extraction recovery of the deuterated internal standards and calibration curves of external standards for each individual mediator.
Statistical analysis of PUFA-derived lipid mediators
Once each individual lipid mediator was quantitated, profiles for each sample set were constructed. These profiles were then subjected to further analyses using two statistical software platforms. Using the Metaboanalyst tool56 (www.metaboanalyst.ca), the data first underwent a missing value imputation in which half the minimum positive value was inserted for compounds that were not detected in all samples. A generalized log transformation was then performed and the data were autoscaled. Once these preliminary functions were completed, hierarchical cluster analysis using Euclidean distance measure and Ward’s clustering algorithm was performed to create heatmaps. Partial least squares-discriminant analysis was also conducted for the isolated leukocyte samples. In some cases, lipid mediator profiles were also subjected to interaction network pathway analysis using Cytoscape57 (http://Cytoscape.org/).
Quantification of deuterium labeled RvD2 in TA muscle
Mice were anesthetized with 2% isoflurane (with 2 L/min O2) and placed on a temperature-controlled water blanket to maintain a body temperature of 37°C. Once anesthetized, 50 μl of 4 μg/kg deuterium labeled (d5)-RvD2 (Cayman Chemical) in saline was injected into the tibialis anterior (TA) muscle. One, two or three hours later the muscles were removed and immediately placed in ice cold methanol containing d8-5-HETE and d5-LXA4. The tissue was minced with surgical scissors then subjected to LC-MS/MS targeted lipidomic analysis as described above.
Isolation and in vitro differentiation of macrophages from murine bone marrow
Bone marrow was flushed from the tibiae and femurs of 8–10-week-old male C57BL/6J mice using a 25-gauge needle and DMEM media (ATCC) containing 10% FBS, 20 mM HEPES and 1% penicillin/streptomycin. Once isolated, the bone marrow was plated in this medium supplemented with 10 ng/ml M-CSF (macrophage colony-stimulating factor; Sigma). On day four of differentiation, the media was replenished with fresh media (containing M-CSF). After seven days, the cells were liberated using a cell scraper centrifuged and re-plated in media lacking M-CSF. After 24 hours, cells were exposed to 10 nM RvD2 (Cayman Chemical) for four hours then collected in Buffer RLT (Qiagen) with 10 μl/ml 2-mercaptoethanol.
RNA isolation
Total RNA was isolated with TRIZOL reagent according to the manufacturer’s recommendation. 20ug glycogen (Ambion) was added as carrier for RNA ethanol precipitation.
qRT-PCR
Bone marrow derived macrophages (BMDMs) were treated with 0.1, 1.0 or 10 nM RvD2 for three or four hours. The cells were then dissociated and collected in RLT lysis buffer (Qiagen) and subjected to RNA extraction using the RNeasy Mini Kit with on-column DNase treatment (Qiagen) according to the manufacturer’s instructions. Reverse transcription and cDNA synthesis were performed with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR was carried out using the SsoAdvanced Universal SYBR Green Supermix on a CFX Real Time PCR Detection System with a C1000 Thermal Cycler (Bio-Rad) with verified RT2 qPCR primers for Cxcl10, Gpr30, Nr4a1, Gpr35 and Hprt (SA Biosciences). Data are presented as relative expression determined by the 2−ΔΔCT method after internal normalization to expression of Hprt.
RNA sequencing (RNA-Seq) library preparation
cDNA library for RNA-Seq was generated from 100ng-400ng total RNA using TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. Briefly poly-A tailed RNA molecules were pulled down with poly-T oligo attached magnetic beads. Following purification, mRNA was fragmented with divalent cations at 85C, and then cDNA was generated by random primers and SuperScript II enzyme (Life Technologies). Second strand synthesis was performed followed by end repair, single `A` base addition and ligation of barcode indexed adaptors to the DNA fragments. Adapter specific PCRs were performed to generate sequencing libraries. Libraries were size selected with E-Gel EX 2% agarose gels (Life Technologies) and purified by QIAquick Gel Extraction Kit (Qiagen). Libraries were sequenced on HiSeq 2500 instrument. Three independent experiments were performed to isolate and sequence each population.
RNA-seq analysis
Tophat258 was used to align the reads to the mm10 mouse assembly. Further downstream analysis of the aligned reads was performed using the StrandNGS software (version 2.8, Build 230243) from Strand Life Sciences. There, normalization of the raw read counts was performed using the DeSeq method59. 1-way ANOVA and Tukey’s post hoc test was performed for the normalized counts of BMDMs untreated/ RvD2 treated for 4 hours, and also of the sorted and isolated neutrophils of Day 1, Ly6Chi and Ly6Clo macrophages of days 2 and 4, post CTX. Changes in genes with Fold Change > 1.5 and p-value < 0.05, were considered to be statistically significant and the genes differentially expressed. Heatmaps were drawn using the R package pheatmap. The accession numbers for the RNA-seq reported in this paper are SRA: SRP145076 and GEO: GSE114291.
Gene ontology analysis
Lists of differentially expressed genes (p value<0.05 and FC>2) were analyzed using Panther tool (http://www.geneontology.org/) and the GO Enrichment Analysis to create a gene ontology (GO). GOs with p values <0.05 were selected and results were presented according to their - log10 p value.
Statistical analysis
In scatter dot plots, mean and SEM are shown in addition to individual data points. In bar graphs, bars show the mean of the indicated number of samples, error bars represent SEM. All experiments were performed using at least three independent experiments from distinct samples. No repeated measures were performed. Student’s t-tests and ANOVA analyses were performed in GraphPad Prism 6 (GraphPad Software) with 95% confidence intervals, and P<0.05 was considered significant (P<0.05=*, P<0.01=**, P<0.001=***, P<0.0001=****).
Supplementary Material
Acknowledgements
The authors are grateful for the outstanding technical contribution of M. Peloquin and acknowledge the discussions and comments on the manuscript from members of the Nagy and Spite laboratories. The authors also thank L. Thomas for excellent administrative assistance. N.G. and L.N. are supported by “Chromatin3D” ITN funded by the European Union under the Horizon2020 Framework Programme (Grant Agreement 622934). A.P. and L.N. are supported by “NR-NET” ITN PITN-GA-2013–606806 from the EU-FP7 PEOPLE-2013 program, by grants from the Hungarian Scientific Research Fund (K124298, K126885, K116855 to L.N.) and by the NIH (R01DK115924). M.S. acknowledges the support of NIH grants GM095467 (Project 3 and Core B) and HL106173. B.E.S. is supported by an NRSA from the NIH (HL136044).
Footnotes
Data availability
Shotgun lipidomics and targeted lipidomics raw data are provided in Supplementary Table 1.
These datasets are relevant to Figs. 1, 2, 3, 4 and Supplementary Figs. 1, 4. The accession numbers for the RNA-seq reported in this paper are SRA: SRP145076 and GEO: GSE114291. They have been assigned to Bioproject PRJNA466152 and are relevant to Figs. 4, 5 and Supplementary Figs. 3, 5. The RNA-seq analysis data (including complete lists with GO analysis terms) for BMDMs and muscle infiltrating macrophages are provided in Supplementary Tables 2 and 3.
Reporting Summary
Further information on research design is available in the Nature Life Sciences Reporting Summary of this article.
Competing interests
None of the authors has any conflicts of interest to disclose.
References
- 1.Kelly B & O’Neill LAJ Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Research 25, 771–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dadgar S et al. Asynchronous remodeling is a driver of failed regeneration in Duchenne muscular dystrophy. J. Cell Biol 207, 139–158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tidball JG & Villalta SA Regulatory interactions between muscle and the immune system during muscle regeneration. Am J Physiol Regul Integr Comp Physiol 298, R1173–87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chazaud B Macrophages: Supportive cells for tissue repair and regeneration. Immunobiology 219, 172–178 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Yona S et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity 38, 79–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckley CD, Gilroy DW & Serhan CN Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity 40, 315–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tidball JG Regulation of muscle growth and regeneration by the immune system. Nat. Rev. Immunol 17, 165–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varga T et al. Tissue LyC6- Macrophages Are Generated in the Absence of Circulating LyC6- Monocytes and Nur77 in a Model of Muscle Regeneration. J. Immunol 191, 5695–5701 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Arnold L et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med 204, 1057–69 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H et al. Altered macrophage phenotype transition impairs skeletal muscle regeneration. Am. J. Pathol 184, 1167–1184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serhan CN Pro-resolving lipid mediators are leads for resolution physiology. Nature 510, 92–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis EA & Norris PC Eicosanoid storm in infection and inflammation. Nature Reviews Immunology 15, 511–523 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samuelsson B Role of basic science in the development of new medicines: Examples from the eicosanoid field. J. Biol. Chem 287, 10070–10080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spite M et al. Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis. Nature 461, 1287–91 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang N et al. Infection regulates pro-resolving mediators that lower antibiotic requirements. Nature 484, 524–528 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serhan CN Discovery of specialized pro-resolving mediators marks the dawn of resolution physiology and pharmacology. Molecular Aspects of Medicine 58, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motwani MP et al. Pro-resolving mediators promote resolution in a human skin model of UV-killed Escherichia coli-driven acute inflammation. JCI Insight 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathod KS et al. Accelerated resolution of inflammation underlies sex differences in inflammatory responses in humans. J. Clin. Invest 127, 169–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spite M, Clària J & Serhan CN Resolvins, specialized proresolving lipid mediators, and their potential roles in metabolic diseases. Cell Metabolism 19, 21–36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalli J & Serhan CN Identification and structure elucidation of the proresolving mediators provides novel leads for resolution pharmacology. Br J Pharmacol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellmann J et al. Biosynthesis of D-series resolvins in skin provides insights into their role in tissue repair. J Invest Dermatol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serhan CN et al. Macrophage proresolving mediator maresin 1 stimulates tissue regeneration and controls pain. FASEB J. 26, 1755–65(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gronert K et al. A role for the mouse 12/15-lipoxygenase pathway in promoting epithelial wound healing and host defense. J. Biol. Chem 280, 15267–78 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Hardy D et al. Comparative Study of Injury Models for Studying Muscle Regeneration in Mice. PLoS One 11, e0147198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Childers MK, Grange RW & Kornegay JN In vivo canine muscle function assay. J Vis Exp 50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornegay J, Bogan J & Bogan D Canine models of Duchenne muscular dystrophy and their use in therapeutic strategies. Mamm. Genome 23, 85–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varga T et al. Highly Dynamic Transcriptional Signature of Distinct Macrophage Subsets during Sterile Inflammation, Resolution, and Tissue Repair. J. Immunol 196, 4771–82 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Chiang N, Dalli J, Colas RA & Serhan CN Identification of resolvin D2 receptor mediating resolution of infections and organ protection. J. Exp. Med 212, 1203–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirooznia M, Nagarajan V & Deng Y GeneVenn - A web application for comparing gene lists using Venn diagrams. Bioinformation 1, 420–22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sager HB, Kessler T & Schunkert H Monocytes and macrophages in cardiac injury and repair. Journal of Thoracic Disease 9(Suppl 1), S30–S35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patsalos A et al. In situ macrophage phenotypic transition is affected by altered cellular composition prior to acute sterile muscle injury. J. Physiol 595, 5815–5842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glaudemans AW, de Vries EF, Galli F, Dierckx RA, Slart RH, Signore A. The use of (18)F-FDG-PET/CT for diagnosis and treatment monitoring of inflammatory and infectious diseases. Clin Dev Immunol. 2013;2013:623036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaidyanathan S, Patel CN, Scarsbrook AF, Chowdhury FU. FDG PET/CT in infection and inflammation--current and emerging clinical applications. Clin Radiol. 2015. July;70(7):787–800. [DOI] [PubMed] [Google Scholar]
- 34.Kasuga K et al. Rapid Appearance of Resolvin Precursors in Inflammatory Exudates: Novel Mechanisms in Resolution J. Immunol 181, 8677–8687 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy BD, Clish CB, Schmidt B, Gronert K & Serhan CN Lipid mediator class switching during acute inflammation: Signals in resolution. Nat. Immunol 2, 612–19 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Dalli J et al. Resolvin D3 and aspirin-triggered resolvin D3 are potent immunoresolvents. Chem. Biol 20, 188–201 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang N et al. Infection regulates pro-resolving mediators that lower antibiotic requirements. Nature 484, 524–528 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fredman G et al. An imbalance between specialized pro-resolving lipid mediators and pro-inflammatory leukotrienes promotes instability of atherosclerotic plaques. Nat. Commun 7, 12859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thul S, Labat C, Temmar M, Benetos A & Back M Low salivary resolvin D1 to leukotriene B4 ratio predicts carotid intima media thickness: A novel biomarker of non-resolving vascular inflammation. Eur J Prev Cardiol 24, 903–906 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Markworth JF et al. Human inflammatory and resolving lipid mediator responses to resistance exercise and ibuprofen treatment. AJP Regul. Integr. Comp. Physiol 305, R1281–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halade GV, Norris PC, Kain V, Serhan CN & Ingle KA Splenic leukocytes define the resolution of inflammation in heart failure. Sci. Signal 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stables MJ et al. Transcriptomic analyses of murine resolution-phase macrophages. Blood 118, e192–208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dalli J & Serhan CN Specific lipid mediator signatures of human phagocytes: Microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood 120, e60–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiang N, de la Rosa X, Libreros S & Serhan CN Novel Resolvin D2 Receptor Axis in Infectious Inflammation. J Immunol 198, 842–851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang MJ et al. Resolvin D2 Enhances Postischemic Revascularization While Resolving Inflammation. Circulation 134, 666–680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soki FN et al. Polarization of prostate cancer-associated macrophages is induced by milk fat globule-EGF factor 8 (MFG-E8)-mediated efferocytosis. J. Biol. Chem 289, 24560–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoue Y et al. Resolvin D2 Limits Secondary Tissue Necrosis After Burn Wounds in Rats. J. Burn Care Res 39, 423–432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho ATV et al. Prostaglandin E2 is essential for efficacious skeletal muscle stem-cell function, augmenting regeneration and strength. Proc Natl Acad Sci U S A 114, 6675–6684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker LA et al. Resolvin E1 (RvE1) attenuates LPS induced inflammation and subsequent atrophy in C2C12 myotubes. J Cell Biochem (2018). [DOI] [PubMed] [Google Scholar]
- 50.Yang K, Cheng H, Gross RW & Han X Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal. Chem 81, 4356–4368 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang M, Han X. Multi-dimensional Mass Spectrometry-based Shotgun Lipidomics. Methods in molecular biology (Clifton, NJ) 1198, 203–220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang M, Wang C, Han RH, Han X. Novel Advances in Shotgun Lipidomics for Biology and Medicine. Progress in lipid research 61, 83–108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dalli J, Colas RA, Walker ME & Serhan CN in Methods in Molecular Biology 1730, 59–72 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Guardiola O et al. Induction of Acute Skeletal Muscle Regeneration by Cardiotoxin Injection. J. Vis. Exp 119, e54515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.English JT, Norris PC, Hodges RR, Dartt DA & Serhan CN Identification and Profiling of Specialized Pro-Resolving Mediators in Human Tears by Lipid Mediator Metabolomics. Prostaglandins, Leukot. Essent. Fat. Acids 117, 17–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xia J, Sinelnikov IV, Han B & Wishart DS MetaboAnalyst 3 . 0 — making metabolomics more meaningful. Nucleic Acid Research 43, 251–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shannon P et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S & Huber W Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.