Abstract
Type I CRISPR-Cas system features a sequential target-searching and degradation process on double-stranded DNA, by the RNA-guided Cascade complex and the nuclease-helicase fusion enzyme Cas3, respectively. Here we present a 3.7 Å resolution cryo-EM structure of the Type I-E Cascade/R-loop/Cas3 complex, poised to initiate DNA degradation. Cas3 distinguishes Cascade conformations and only captures the R-loop forming Cascade, to avoid cleaving partially complementary targets. Its nuclease domain recruits the non-target strand (NTS) DNA at a bulged region for the nicking of single-stranded DNA. An additional 4.7 Å resolution cryo-EM structure captures the post-nicking state, in which the severed NTS retracts to the helicase entrance, to be threaded for ATP-dependent processive degradation. These snapshots form the basis for understanding RNA-guided DNA degradation in Type I-E CRISPR-Cas systems.
Introduction
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and the nearby cas (CRISPR-associated) operon establish an RNA-based adaptive immunity system in prokaryotes (1–6). They are classified into two major classes: Class 1 systems utilize a multi-subunit effector complex to search and destroy nucleic acid targets, whereas Class 2 systems use a single effector complex (7, 8). Type I CRISPR-Cas, the most prevalent CRISPR system, belongs to Class 1 and can be further categorized into six subtypes (7). It features a sequential target searching and degradation process. First, a multi-subunit surveillance complex called Cascade (CRISPR associated complex for antiviral defense) recognizes the matching dsDNA target flanked by an optimal protospacer adjacent motif (PAM), promotes the heteroduplex formation between CRISPR RNA (crRNA) and the target strand (TS) DNA and displaces the non-target strand (NTS) DNA, resulting in R-loop formation at the target site (Fig. 1A) (9–14). The helicase-nuclease fusion enzyme Cas3 is specifically recruited to the Cascade/R-loop complex, nicks the NTS (Fig. 1A), and switches to a processive DNA degradation mode (11, 15–17). Type I-E CRISPR-Cas systems from E. coli and Thermobifida fusca have been extensively studied (9–25). In the Thermobifida fusca system (Fig. 1A), high-resolution structure snapshots elucidated the PAM recognition mechanism and revealed a concerted set of conformational changes as TfuCascade traverses from the intermediate (the seed-sequence bubble) to the full R-loop state; TfuCas3 selectively binds to the latter state (25). Previous efforts to define the Cascade-Cas3 interaction was not of sufficient resolution to resolve the molecular mechanisms concerning Cas3 recruitment and DNA degradation (17). A ssDNA-bound TfuCas3 crystal structure led to the speculation that the initial R-loop nicking mechanism by Cas3 is likely different from the subsequent DNA degradation mechanism (24). To resolve these mechanistic ambiguities, we reconstituted the TfuCascade/R-loop/Cas3 complex and obtained two cryo-EM structures representing the pre- and post-R-loop-nicking states, at 3.7 Å and 4.7 Å resolutions, respectively. The former snapshot explains how Cas3 specifically captures the full R-loop forming Cascade as a mechanism to avoid mis-targeting. The latter snapshot is particularly informative in revealing how Cas3 switches from the initial ssDNA nicking mode to the later processive DNA degradation mode. Together these results provide the structural basis for us to understand crRNA-guided DNA degradation in Type I CRISPR-Cas systems.
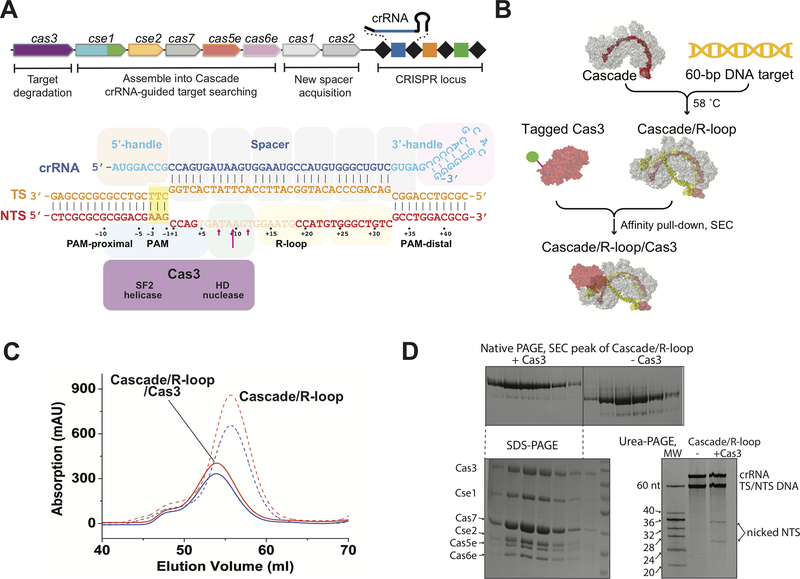
Figure 1. TfuCascade/R-loop/Cas3 reconstitution.
A. Schematic diagram of the T. fusca Type I-E CRISPR-cas operon, TfuCascadeorganization, R-loop formation, TfuCas3 binding site, and NTS nicking site. Disordered NTS nucleotides are semi-transparent, TfuCas3 nicking sites are marked by red arrows. This coloring scheme is preserved in other figures. B. Reconstitute and purification scheme of the TfuCascade/R-loop/Cas3 ternary complex. C, D. SEC, native-PAGE, SDS-PAGE, and urea-PAGE analyses of the ternary complex.
Results
Reconstitution of Cascade/R-loop/Cas3 ternary complex from Thermobifida fusca
As previously shown (25), the T. fusca Type I-E system displays a steep temperature-dependent R-loop formation behavior, allowing us to program TfuCascade into either dsDNA-binding, seed-bubble formation, or full R-loop state by changing the incubation temperatures. TfuCas3 only interacts with TfuCascade in the full R-loop state, but not the seed-bubble state, presumably by probing the conformational differences between them. In this study, we scaled up the reconstruction of the TfuCascade/R-loop/Cas3 ternary complex and purified it from individual components using a combination of affinity and size-exclusion chromatography methods (Fig. 1B–C). In the ternary complex, TfuCas3 preferentially nicked after the 7th-, 9th- (strongly preferred) and 11th-nt of the non-target strand DNA in the R-loop, counting from the PAM-proximal side. When ATP was introduced, TfuCas3 switched to a processive degradation mode, cleaving both strands of DNA to pieces (25). These behaviors are similar to the reported behaviors of Cas3 from E. coli and S. thermophilus Type I-E systems (15–17). Previously, the TfuCas3/ssDNA structure revealed the presence of two ferric ions at the HD nuclease center (24). We later found that their presence strongly inhibits the nuclease activity, whereas Co2+ supports ssDNA cleavage (25). Nonetheless, the combination of using iron-incorporated TfuCas3 and omitting ATP (or introducing non-hydrolyzable ATP analog AMPPNP) allowed us to program the TfuCascade/R-loop/Cas3 complex to a predominantly pre-nicking state; at least 74% of the NTS was intact based on ImageJ quantification, and processive degradation was not observed (Fig. 1D).
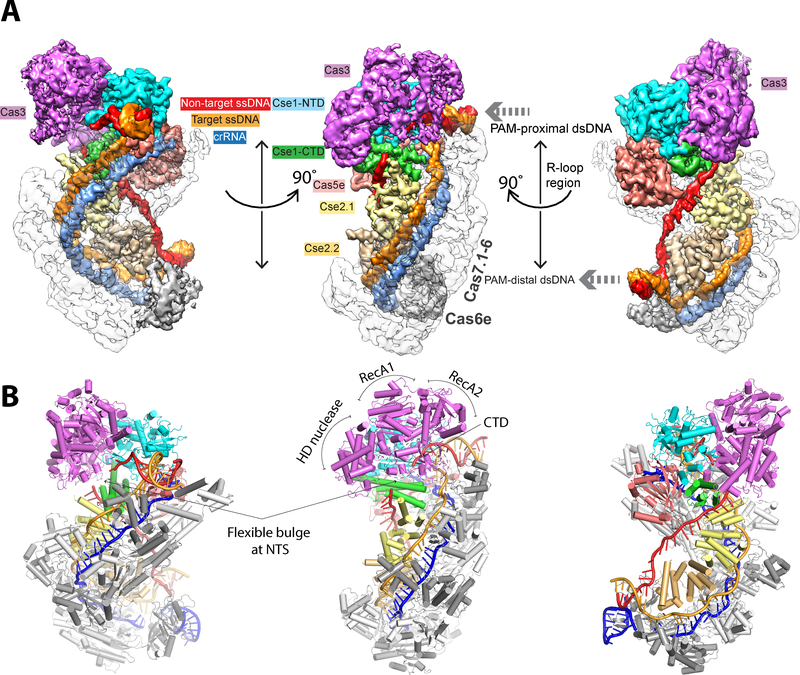
Overall architecture of the Cascade/R-loop/Cas3 complex in pre-nicking state
Single-particle cryo-EM analysis showed that the ternary complex embedded in ice was monodisperse and homogeneous, with distinct structural features clearly visible in two-dimensional (2D) averages (Fig. S1A–B). We determined a cryo-EM structure of the TfuCascade/R-loop/Cas3 interference complex at an overall resolution of 3.7 Å (Fig. S1C–D). The body of TfuCascade/R-loop is resolved at 3.5 Å or better. In contrast, finger domains of Cas7, Cas6e/crRNA, and outer shell of the dsDNAs are resolved at 4.7 Å or worse due to their conformational flexibilities (Fig. S1D, S2). The inner shell of Cas3 is resolved at better than 4.1 Å, resolving most of the interface side chains in contact with Cascade; some outer shell regions are at worse than 4.7 Å. Of the four domains, the HD nuclease and RecA1 domains of Cas3 are better resolved, RecA2 has weaker density due to hinge motion; CTD is not well resolved due to elevated thermo-motion and is modeled with a rigid-body docked crystal structure (Figs. S1D, S2).
Binding of Cas3 does not introduce further conformational changes to the R-loop forming Cascade; the overall conformation of TfuCascade in the ternary complex agrees well with that in the binary complex (PDB: 5U0A) (Fig 2B, S3) (25). These observations point to the possibility that the Cas3-Cascade interaction may utilize conformation-capture rather than induced-fit binding mechanism, although this requires conformational dynamics analysis to confirm. The Cas3 conformation inside the ternary complex is in architectural agreement with that in the crystalline state (PDB: 4QQW) (24), with the exception that the RecA2 and CTD domains undergo rigid body movements in the range of 2 and 10 Å, respectively (Fig. S3). Cas3 binding is mediated exclusively by the Cse1 subunit of Cascade. The location and domain orientation of Cas3 are consistent with the biochemistry data (15–17, 25, 26). However, the nuclease center of Cas3 is ~20 Å away from the path of NTS. Therefore, a special mechanism must be in place to hand over the DNA substrate from Cascade to Cas3.
Figure 2. Overall architecture of TfuCascade/R-loop/Cas3 in the pre-nicking state.
Cryo-EM map (A) and cartoon model (B) of the pre-nicking ternary complex in three different orientations, revealing the Cas3 protein binding to the Cse1 subunit of Cascade. The cryo-EM maps are displayed without applying B-factor sharpening.
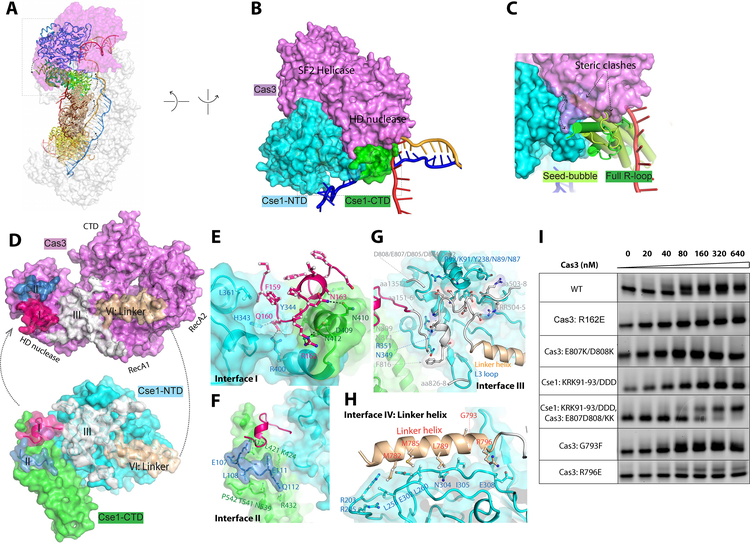
Cas3-Cascade interface: how does Cas3 capture the R-loop forming Cascade?
The Cas3-Cascade interaction is exclusively mediated by the Cse1 subunit in Cascade. Previously we showed that Cse1_CTD pivots 15° when Cascade transitions from the seed-bubble to the full R-loop state (Fig. 3A) (25). This hinge motion alters the orientation and distances between Cse1_NTD and CTD. Therefore, any long-range contact from Cas3 that spans the two Cse1 domains would be sensitive to the conformation of Cse1. The conformational changes also altered the surface features at the Cse1 NTD-CTD interface. Any Cas3 contacts recognizing the newly exposed Cse1 NTD-CTD interface would also help distinguish the Cascade conformation.
Figure 3. Selective binding of Cas3 to the R-loop forming Cascade.
A. Conformational differences between seed-bubble and full R-loop Cascade. Cas3 contacts the Cse1 subunit of Cascade exclusively. B. A hind view of Cas3-Cse1 contact. Cas3 inserts a wedge into the groove between Cse1-NTD and CTD. C. This groove is absent in the seed-bubble state, and Cas3-binding would lead to steric clash. D. The four interfaces on Cas3 and Cse1, colored in pink, blue, white, and wheat, respectively. E-H. Detailed contacts at Interface I through IV, respectively. I. Native EMSA to evaluate the interface residues. The lower and upper bands represent the Cascade/R-loop and Cascade/R-loop/Cas3 complexes, respectively.
Both principles are explored by Cas3 to sense the Cascade conformation (Fig. 3B, C). The Cascade-Cas3 interface buries a large surface area of 2,520 Å2 and spans an entire side of Cas3. It can be roughly divided into four sectors (Interfaces I-IV) (Fig. 3D). At Interface I, Cas3 inserts a protruding loop from its HD nuclease domain (aa157–171) into a groove between the Cse1 NTD and CTD (Fig. 3E) which is only exposed when the Cascade is in the full R-loop conformation (Fig. 3B, C). Severe clashing would result if Cas3 were to bind Cascade in the seed-bubble conformation (Fig. 3C). Contacts at Interface I feature shape complementarity; favorable hydrogen bonding contacts mediated by Q160, R161, and N163 from Cas3 further strengthen the interaction. Charge-swapping mutagenesis, R162E, completely disrupted the Cascade-Cas3 contact (Fig. 3I), underlining the functional importance of these polar contacts.
Interface II and III flank interface I and serve as a molecular ruler to sense angular and distance changes between the two Cse1 domains. This serves as a proxy to readout whether Cascade has opened an R-loop on the target site. At the Cse1_CTD side, a surface loop from the Cas3 HD domain (aa105–114) inserts into a groove perpendicular to the four-helical bundle in Cse1_CTD (Interface II; Fig. 3F). While the specific contacts are weak, the two surfaces would misalign and clash if Cascade were not in the full R-loop state. On the Cse1_NTD side, the entire Cas3 linker region, composed of an α-helix (aa779–798) and a flexible loop (aa799–817), is buried at the interface (Fig. 3G). The long linker region is unique to the Cas3 branch of Super Family II helicases and has been implicated in our previous study to mediate interactions with Cascade; replacing this region with a Gly/Ser linker completely abolished Cas3 binding (24). Here we define the area contacted by the flexible linker loop and the nearby Cas3 surface loops (aa 151–156, 503–508, and 826–828) as Interface III (Fig. 3D, G). In addition to mediating shape complementarity, contacts at this interface emphasize charge complementarity. In particular, an acidic patch in the linker loop (D802, D804, D805, E807, D808, N809, and N811) complements a basic patch in Cse1_NTD (N87 N89, K91, R92, and K93; Interface III). Charge-swapping mutagenesis from either the Cas3 (E807K/D808K) or Cse1 (KRK91–93/DDD) side of the interface led to a complete loss-of-binding phenotype. Strikingly, combining these two charge-swapping mutations rescued the binding defect (Fig. 3I), underlining the importance of charge complementarity at Interface III.
The entire linker helix (aa779–798) is buried at the Cas3-Cascade interface (Interface IV), packing at a tilted angle against two parallel loops in Cse1_NTD (aa258–269 and aa301–309) (Fig. 3H). A set of hydrophobic residues at the N-terminal half of the linker helix interdigitates with hydrophobic residues on the Cse1 surface. However, alanine-scanning mutagenesis targeting the hydrophobic residues along the helix had little effect on Cascade-Cas3 interaction (data not shown). The C-terminal half of the linker helix in Cas3 mediates a few salt-bridge interactions to Cse1. An R796E charge-swapping mutation in Cas3 significantly weakened its affinity for Cascade/R-loop (Fig. 3I). G793F mutation in Cas3 had a mild effect, presumably by perturbing the shape complementarity at the interface (Fig. 3I).
Taken together, our structural observations explain why Cas3 is only capable of binding to Cascades that have completed the target validation process. This serves as a mechanism to avoid the degradation of a target DNA with partial complementarity. Interestingly, despite strong sequence and structural homology, we failed to assemble a hetero-complex from T. fusca Cas3 and E. coli Cascade/R-loop (Fig. S4A). Docking of these two structures resulted in reasonable surface complementarity at the putative interface (Fig. S4B–C). However, non-ideal charge complementarity can be found in several major interfaces and steric clashes are present at Interface III (Fig. S4D). Sequence alignment further revealed that the interface residues in Cas3 are not conserved (Fig. S5). These observations rationalized why T. fusca Cas3 and E. coli Cascade/R-loop failed to hetero-assemble. Given that multiple Type I systems frequently coexist in one bacterium – T. fusca itself maintains two Type I-E systems – this may be a mechanism to prevent cross-operon Cascade-Cas3 interaction, which may lead to mis-targeting or mis-incorporation of new spacers during primed adaptation.
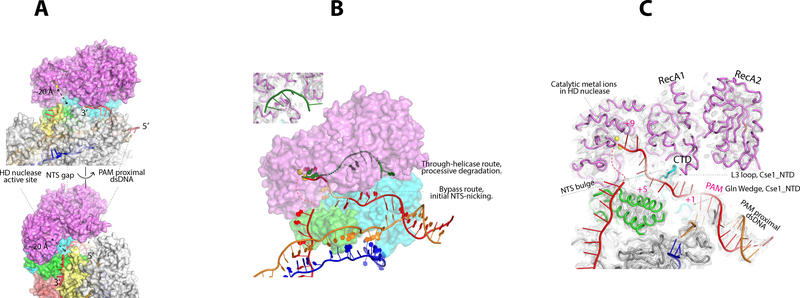
NTS handover and nicking mechanism
Available biochemical data suggest Cas3 nicks the NTS after binding to Cascade and then ATP hydrolysis mobilizes the helicase. These concerted actions lead to processive DNA degradation. In the TfuCascade/R-loop/Cas3 complex structure, the proximity of Cas3 to the PAM-side NTS is in architectural agreement with the observed DNA-nicking pattern. The Cas3 orientation relative to the NTS is consistent with the 3’-to-5’ directionality of its helicase (Fig. 4A). However, the ternary structure clearly reveals that the initial DNA-nicking mechanism is different from the processive degradation mechanism, which is depicted in the TfuCas3/ssDNA structure (24). The NTS in the ternary complex bypasses the helicase completely prior to NTS-nicking; nucleic acid densities are absent from the helicase core as seen in the TfuCas3/ssDNA structure (Fig. 4B). Rather, the first four nucleotides 3’-to-PAM travel underneath the L3 loop of Cascade. The bases are not resolved in the EM map due to the lack of specific contacts, but the backbone can be traced with good confidence (Fig 4C). The NTS density then disappears from the EM map at regions between the L3 loop and Cse1_CTD, before reappearing at the rim of Cse1_CTD and heading towards the backside of Cse2.1. A similar scenario was observed in the Cascade/R-loop binary structure and we demonstrated that this was because the NTS assumed a flexible 8–10-nt bulge in this region (25). This was interpreted as a mechanism to handover the NTS substrate to Cas3 for strand-nicking (25). Here the ternary structure provides direct evidence that a handover mechanism is indeed essential for Type I-E CRISPR interference. In the cryo-EM structure, the HD nuclease center in Cas3 is ~20 Å away from the NTS on the surface of Cascade (Fig 4A). Without a protruding ssDNA bulge, it would be impossible for Cas3 to cleave the NTS. Importantly, extra densities corresponding to the scissile phosphate and the backbone of the two preceding nucleotides are found in the HD nuclease center (Fig. 4C). These nucleotides are accommodated in a similar fashion by the di-metal HD active site as observed in the TfuCas3/ssDNA structure (24). It takes a maximum of three additional nucleotides to model the entire 9-nt PAM-side NTS into the nuclease center of Cas3 (Fig. 4C), which rationalizes the strong preference of Tfu- and EcoCas3 to nick after PAM+9 (25). Taken together, the structural observations converge with previous biochemistry to suggest that the Cas3 nuclease recruits the NTS ssDNA directly for strand-nicking. Substrate capture relies on the presence of a flexible bulge in the NTS, and the location of the nicking reaction is predetermined by the length of the recruitment pathway.
Figure 4. NTS-nicking bypasses Cas3 helicase, and HD nuclease captures NTS at a flexible bulge.
A. The nuclease center of Cas3 is ~20 Å away from the NTS traveling on the Cascade surface. B. The cryo-EM densities suggest NTS (in red) bypasses the Cas3 helicase and accesses its HD nuclease center directly. This route is different from the through-helicase route (in green) observed in the TfuCas3/ssDNA structure. Inset: cryo-EM density showing that no nucleic acid densities are found inside the Cas3 helicase. C. Path of NTS inside the TfuCascade/R-loop/Cas3 complex. A flexible bulge is present in NTS between the L3 loop and CTD of Cse1, where the EM density is absent. The bulge allows the Cas3 nuclease to capture and nick the NTS. Structure building and modeling explains why Cas3 preferentially nicks after PAM+9 in NTS. The backbone of nucleotides PAM+1–4 and PAM+8–9 can be traced from the cryo-EM map; PAM+5–7 are modeled. The minor nicking sites (PAM+7 and +11) can be rationalized based on the observed NTS flexibility.
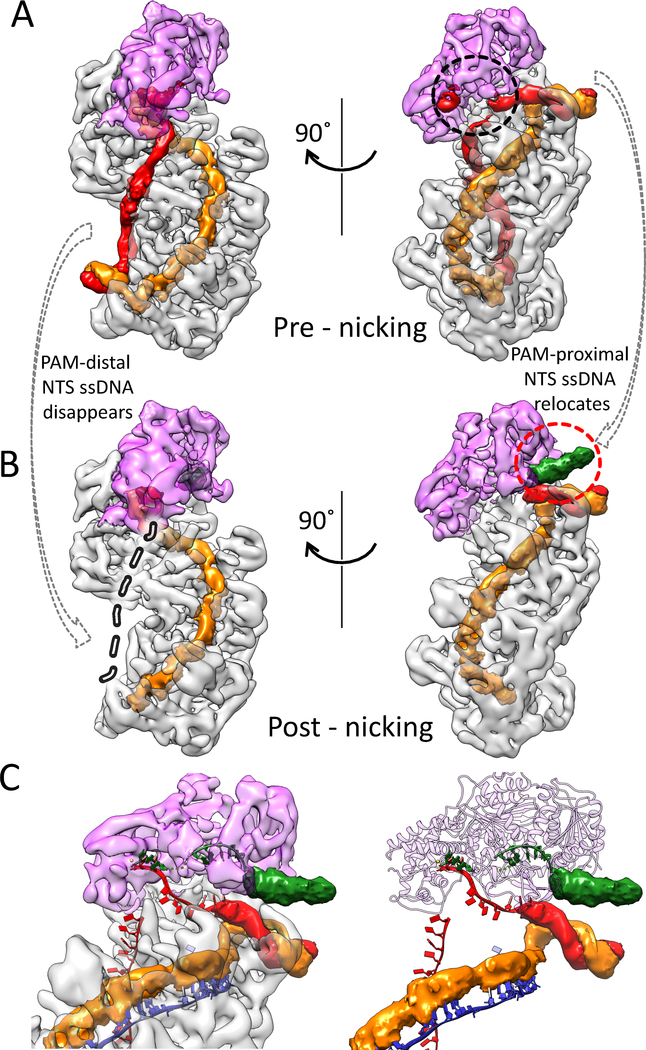
Post-nicking snapshot: nicked NTS retracts to Cas3 helicase, awaiting re-threading
While most ternary complexes were trapped at the pre-nicking stage, a fraction of the complexes did undergo NTS nicking (Fig. 1D). These particles were partitioned computationally during 3D classification. They converge to give a cryo-EM structure at 4.7 Å resolution; variations in local resolution follows the same trend as in the pre-picking structure (Fig. S2). The following structural differences led us to conclude that this snapshot represents the post-nicking ternary complex (Fig. 5A). First, the entire NTS DNA disappears from its original path in the R-loop region (Fig. 5B). The scenario seems analogous to when a rubber band is severed - the two halves snap back and become flexible. The PAM-distal dsDNA also disappears from the density, presumably due to increased flexibility, while the PAM-proximal dsDNA is held in its original location (Fig. 5B). Importantly, as the PAM-side NTS disappears from its original path, a new blob of density appears by the side of the PAM-proximal dsDNA, pointing towards the opening of the Cas3 helicase moiety (Fig. 5B, C). The density lacks recognizable structural feature, but its volume can accommodate ~6–7 nt ssDNA. The most logical explanation is that the density corresponds to the relocated NTS after strand-nicking. It follows then that upon ATP hydrolysis, the Cas3 helicase would thread the ssDNA through itself and further into the HD nuclease, as depicted in our previous published TfuCas3/ssDNA structure (24). This switches Cas3 into a processive DNA degradation mode. We note that the Cascade-Cas3 contact at the helicase region is insulated by the linker region of Cas3. Therefore, the anticipated conformational changes in the Cas3 helicase in each ATP hydrolysis cycle would not disrupt the Cas3-Cascade complex. This further rationalizes the observation that Cas3 is capable of degrading dsDNA while on top of Cascade before eventually breaking free (26, 27).
Figure 5. NTS rearrangement observed in the post-nicking state.
A, B. Density differences between the pre- and post-NTS-nicking states. Both maps are filtered to 6 Å without applying a B-factor. Extra densities in the post-nicking 3D reconstruction are colored in green, in the red dotted circle. Disappeared densities are highlighted in grey dashed line and in black dotted circles. C. Zoom-in of the post-nicking state highlighting the fate of NTS after strand-nicking. Densities previously leading to the HD nuclease center is now missing, while extra densities accumulate at the opening of the Cas3 helicase.
Discussion
With this study, we complete the structure-function characterization of the key molecular events leading to the onset of CRISPR interference in Type I-E system. The theme prior to DNA target degradation appears to emphasize stringency, by establishing a sequence of causal events. To reduce off-targeting, two separate effector macromolecules are required, and their interaction is controlled by an R-loop dependent conformation-capture mechanism. This ensures that the DNA substrate is not prematurely cleaved before the entire targeted sequence is validated. At the onset of CRISPR interference, upon NTS nicking, the mechanism then switches to favor efficiency. Rather than stopping at generating a double strand break, as Cas9, Cas12a, and other single-effector CRISPR systems do, the helicase-nuclease Cas3 elicits more severe damage through processive DNA degradation. These characteristics may explain why Type I evolved to be the most prevalent CRISPR-Cas system found in nature. It would be interesting to see if Type I system could be reprogrammed to function as a genome editing tool.
Following Cas3 recruitment and NTS nicking, Cas3 enters into a processive DNA degradation mode. Single molecule studies revealed that initially Cas3 remains bound to Cascade and reels dsDNA towards itself, trapping a DNA loop and creating a ssDNA region within (26, 27). As tension accumulates, Cas3 dissociates from Cascade in a stochastic fashion and travels alone for kilo-base distances (26, 27). Surprisingly, no obvious double-stranded DNA break was observed in these studies. This does not agree with the interference efficiency observed in vivo. The inconsistency between bulk and single molecule biochemistry leaves an empty spot in our molecular understanding of Type I CRISPR system.
Phages can evade CRISPR surveillance by acquiring mutations at PAM or the protospacer region. A mechanism called primed adaptation is in place in Type I CRISPR-Cas system where Cascade and Cas3 drive the preferential acquisition of new spacers by Cas1-Cas2 from evader phages that acquired mutations in the protospacer region. The molecular details remain murky, however, recent single-molecule imaging directly visualized Cas1-Cas2 enabling Cascade to specifically recruit Cas3 to protospacers adjacent to a mutated PAM (26, 27). It is intriguing to speculate that the priming PAM sequences may trigger the Cascade to adopt a currently undefined conformation to recruit Cas3 and Cas1-Cas2, thereby initiating the primed spacer acquisition process. Additional studies are required to address precisely how Cascade/Cas3/Cas1-Cas2 selects protospacers during primed adaptation.
Materials and methods
See supplemental materials for a detailed description of materials and methods. Briefly, TfuCascade/R-loop and TfuCas3 were prepared as described (24, 25). The ternary complex was assembled by incubating TfuCascade/R-loop and Twin-Strep-HRV-TfuCas3 at a 1:2 molar ratio at 4°C for 60 min, followed by Strep-tag pull-down to remove the unbound TfuCascade/R-loop, PreScission Protease cleavage to remove the Twin-Strep tag, and size-exclusion chromatography to remove the excess Cas3. Cryo-EM data acquisition, image acquisition, and structure reconstruction followed a similar procedure as described (25). Data processing and refinement statistics for the two cryo-EM structures is summarized in Table S1.
Supplementary Material
Acknowledgements
We thank R. Battaglia, A. Dolan, and S. Ng for critical reading of the manuscript, and I. Finkelstein for discussions.
Funding: Research in the Ke lab is supported by the National Institutes of Health (GM118174 and GM102543).
Footnotes
Competing interests: The authors have no competing interest.
Data and materials availability: The 3D cryo-EM density maps of the TfuCascade/R-loop/Cas3 complex have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-7347 (pre-nicking state) and EMD-7346 (post-nicking state). The atomic coordinate for the pre-nicking state has been deposited in the Protein Data Bank under the accession number 6C66.
References
- 1.Barrangou R et al. , CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD, Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Mojica FJ, García-Martínez J, Soria E, Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of molecular evolution 60, 174–182 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Pourcel C, Salvignol G, Vergnaud G, CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Marraffini LA, Sontheimer EJ, CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV, A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1, 7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova KS, Koonin EV, Annotation and Classification of CRISPR-Cas Systems. Methods Mol Biol 1311, 47–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmakov S et al. , Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell 60, 385–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouns SJ et al. , Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiedenheft B et al. , Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477, 486–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westra ER et al. , CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46, 595–605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rutkauskas M et al. , Directional R-Loop Formation by the CRISPR-Cas Surveillance Complex Cascade Provides Efficient Off-Target Site Rejection. Cell reports, (2015). [DOI] [PubMed] [Google Scholar]
- 13.Blosser TR et al. , Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex. Mol Cell 58, 60–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jore MM et al. , Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nature structural & molecular biology 18, 529–536 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Sinkunas T et al. , In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. The EMBO journal 32, 385–394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulepati S, Bailey S, In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. The Journal of biological chemistry 288, 22184–22192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochstrasser ML et al. , CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proceedings of the National Academy of Sciences of the United States of America 111, 6618–6623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA, Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329, 1355–1358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sashital DG, Wiedenheft B, Doudna JA, Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell 46, 606–615 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson RN et al. , Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345, 1473–1479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao H et al. , Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature 515, 147–150 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Mulepati S, Heroux A, Bailey S, Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science 345, 1479–1484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes RP et al. , Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature 530, 499–503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huo Y et al. , Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nature structural & molecular biology 21, 771–777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Y et al. , Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System. Cell 170, 48–60 e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redding S et al. , Surveillance and Processing of Foreign DNA by the Escherichia coli CRISPR-Cas System. Cell 163, 854–865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown MW et al. , Assembly and translocation of a CRISPR-Cas primed acquisition complex. bioRxiv, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ru H et al. , Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell 163, 1138–1152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant T, Grigorieff N, Measuring the optimal exposure for single particle cryo-EM using a 2.6 angstrom reconstruction of rotavirus VP6. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mindell JA, Grigorieff N, Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142, 334–347 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Scheres SH, RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S et al. , High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kucukelbir A, Sigworth FJ, Tagare HD, Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.