Abstract
Thermal shift assay (TSA) is an increasingly popular technique used for identifying protein stabilizing conditions or interacting ligands in X-ray crystallography and drug discovery applications. Although the setting up and running of TSA reactions is a relatively simple process, the subsequent analysis of TSA data, especially in high-throughput format, requires substantial amount of effort if conducted manually. We therefore developed the Thermal Shift Assay–Curve Rapid and Automatic Fitting Tool (TSA-CRAFT), a freely available software that enable automatic analysis of TSA data of any throughput. TSA-CRAFT directly reads real-time PCR instrument data files and displays the analyzed results in a web browser. This software features streamlined data processing and Boltzmann equation fitting, which is demonstrated in this study to provide more accurate data analysis than the commonly used first-derivative method. TSA-CRAFT is freely available as a cross-operating system-compatible standalone tool (https://sourceforge.net/projects/tsa-craft/) and also as a freely accessible web server (http://tbtlab.org/tsacraft.html).
Keywords: thermal shift assay, thermofluor, differential scanning fluorimetry, automatic Tm analysis, high-throughput ligand screening
Introduction
Thermal shift assay (TSA), also known as thermofluor assay or differential scanning fluorimetry, is a technique that probes the thermal stability of proteins in various aqueous chemical environments.1–3 The assay uses a fluorescent dye, such as Sypro Orange, that does not fluoresce in the aqueous environment but fluoresces strongly when bound to hydrophobic sites of proteins exposed during thermal denaturation. As such, plotting the increase in fluorescence signal with increasing temperature generates a melting curve ( Fig. 1A ). The inflection point of the melting curve, which corresponds to the midpoint of the protein unfolding process, is known as the melting temperature (Tm). Tm reflects on the thermal stability of a protein in that particular experimental condition and can provide useful indicators for various applications. In protein X-ray crystallography, TSA is often employed to screen varying buffer conditions, such as pH and salt concentrations, as well as the presence of additives for chemical conditions that enhance a protein’s stability to facilitate its crystallization. TSA is also increasingly used for the identification of interacting ligands in drug discovery. The binding of a small molecule confers some extent of stability to the protein, resulting in a positive shift in the Tm. Thus, differences in Tm (ΔTm) in the presence of various small molecules compared with the apo protein are indicative of protein-ligand interactions.
Figure 1.
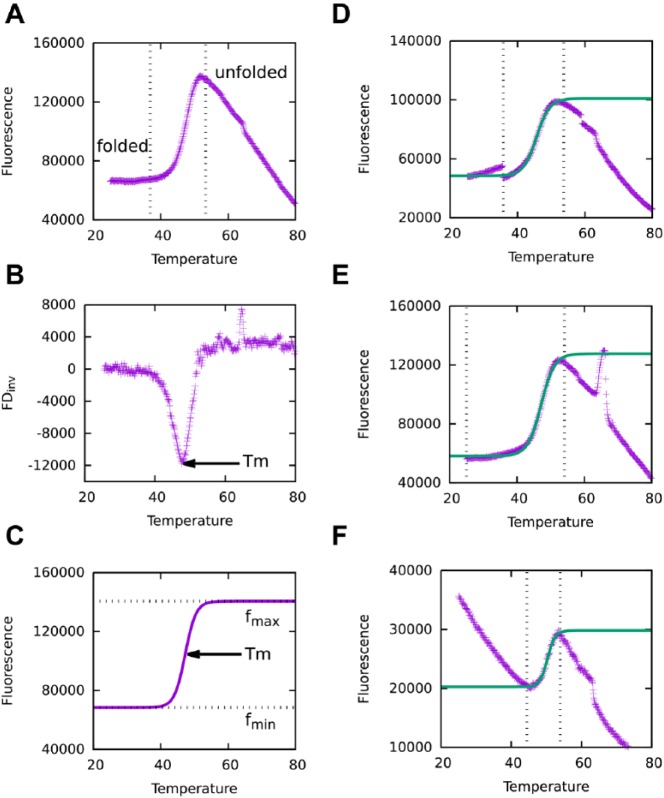
Thermal denaturation curves and Tm identification. (A) A typical melting curve obtained from monitoring protein thermal denaturation with increasing temperature. Vertical dotted lines mark the protein transition from folded to unfolded states. (B) Plot of the inverse first derivatives of the melting curve, calculated according to Eq. 1. Tm is determined by the peak of the curve. (C) Fitting of the transition part of the melting curve to the Boltzmann equation (Eq. 2). Tm is determined by the inflection point of the curve. (D) Large gaps and (E) spikes are examples of noises observed in thermal shift assay (TSA) data, which may confound automatic TSA analysis tools and lead to false or inaccurate Tm determination. TSA-CRAFT is designed to identify and filter out such noises before performing curve fitting. (F) Data with high initial fluorescence does not hinder TSA-CRAFT from determining the Tm.
TSA is widely used, as it is relatively cost-effective, easy to set up, and can be performed with a commonly available real-time PCR (qPCR) machine. As long as supported by the instrument, high-throughput experiments in 96- or 384-well formats can be conducted for efficient screening of interacting compounds or stabilizing conditions. Theoretically, Tm can be obtained simply by plotting the melting curve and determining the midpoint of the transition. However, Tm analysis can be time-consuming when performed manually for high-throughput experiments. Therefore, a number of commercial and free software tools have been developed to facilitate TSA data analysis.4–7 These existing tools, however, have limitations of operating system compatibility, restrictions of free usage, and algorithms with tendencies to pick up false Tm when the data contain substantial noise. To overcome these limitations, we developed TSA-CRAFT, a free, robust, and cross-platform tool for fast TSA data analysis of any throughput. TSA-CRAFT is an open-source software under GNU general public license. Users can download the standalone command line tool package from https://sourceforge.net/projects/tsa-craft/. Alternatively, users who prefer a quick start, graphic interface with no command line operations can access TSA-CRAFT from our web server (http://tbtlab.org/tsacraft.html) with no registration required. Instructions, input template files, and sample data are available from both package and web server options. TSA-CRAFT is designed with user-friendliness and robustness in mind. In this report, we further demonstrate the superior robustness of TSA-CRAFT compared with the existing tools.
Materials and Methods
Tm Determination Methods
First derivative and curve fitting are the two main methods used to obtain Tm from TSA data. The Tm values determined may differ between these two methods. A typical protein thermal denaturation curve follows a sigmoidal transition ( Fig. 1A ). The midpoint of the transition, which is also the inflection point of the sigmoidal curve, defines the Tm of the denaturation process. As the tangent at the inflection point of the sigmoidal curve has the steepest slope, the first-derivative method determines this point by searching for the peak of the inverse first-derivative curve ( Fig. 1B ). In this method, the first derivative of each data point is calculated according to Eq. 1, whereby f(Tn) is taken as the moving average of several neighboring points to reduce the noise of the raw data. In the second approach, which is the curve-fitting method, a sigmoidal function (usually the Boltzmann equation, Eq. 2) is used to fit the protein thermal denaturation curve such that Tm can be determined by the inflection point ( Fig. 1C ). Curve-fitting methods are known to be more robust and less sensitive to random noise than the first-derivative method. Thus, in TSA-CRAFT, we implemented curve fitting through the Boltzmann equation to identify the Tm of TSA data.
| (1) |
| (2) |
Data Processing in TSA-CRAFT
To determine the most representative fragment of TSA data for curve fitting, the most straightforward way is to search for the two points with minimal and maximal fluorescence readings, respectively, and select the data fragment in between. However, many recorded raw TSA data curves are not perfectly sigmoidal. The raw transition curves may have substantial noise, such as large gaps and spikes ( Fig. 1D, E ), as well as minor random fluctuations. In TSA-CRAFT, we designed several filters to target different types of noise before performing curve fitting. A large gap is defined as the Δf (difference of fluorescence readings) of the two adjacent points larger than six folds of standard deviations from averaged Δf. If a point is at the tip of a sharp pulse (i.e., Δf is larger than 15% of maximal transition [fmax – fmin in Eq. 2] within 5% of ΔT extended by maximal transition), it is treated as a spike in TSA-CRAFT. After filtering out large gaps and spikes, the program searches on a continuous fragment of data for the points harboring local maximal and minimal fluorescence readings. Finally, the boundaries of the most representative transition curve will be identified by searching for the pair of a local minimum and a local maximum with the largest positive Δf. The pair should not cross another local optimum unless it has a first derivative (by Eq. 1) larger than 80% of the slope obtained from the selected pair. Another nonperfect situation is when the initial fluorescence is higher than the maximum of the transition ( Fig. 1F ). In such a situation, TSA-CRAFT was able to identify the sigmoidal part of curve with maximal positive Δf for curve fitting without being affected by the high initial signal. There are situations, however, such as multiphasic denaturation, in which manual inspection and intervention are required. Multiphasic curves may be due to conditions that promote multistep denaturation or the use of proteins with multiple domains. These cases will be detected and flagged by TSA-CRAFT, and users are advised to deal with them manually (Suppl. Fig. S1). If required, users can determine each Tm by adjusting the window to fit each sigmoidal portion.
Software Design
The framework of TSA-CRAFT combines the powerful features of PERL script and Gnuplot. PERL scripts manage the entire workflow and data-processing procedures. Gnuplot is applied in graphics and curve fitting with the Boltzmann equation. The fitting results are evaluated by the coefficient of determination (R2) to indicate how closely the data are fitted to the curve. The output of the Tm analysis is coded in HTML format that can be opened by any web browser.
Experimental Setup
Here, we demonstrate the use of TSA in the detection of protein-ligand interactions involving data analysis by TSA-CRAFT. In this experiment, 36 retinoid compounds were tested for their interactions with retinoic acid receptor alpha (RARα), a well-known drug target. His-tagged RARα ligand-binding domain (aa176-421) was expressed in Escherichia coli and purified using standard nickel affinity chromatography8 followed by size exclusion chromatography in buffer containing 25 mM Tris, 300 mM NaCl, 1 mM EDTA, and 1 mM DTT. TSA reactions were set up in duplicate points in a 96-well optical PCR plate. Reactions contained 10 µM of purified protein, 50 µM of the indicated compound, and Protein Thermal Shift Dye (Life Technologies, Carlsbad, CA) in 1X reaction buffer provided by the kit. After setting up the reactions, the plate was sealed and loaded onto QuantStudio 3 (Applied Biosystems, Foster City, CA). Melting reactions were run from 25 °C to 99 °C at the rate of 0.05 °C/s. After the run was complete, the result file (.eds) was used directly as input in TSA-CRAFT. In addition, an annotation file was provided that specified well contents and indicated the control (DMSO) well to be used as a reference point for automatic ΔTm calculations.
Results and Discussion
Overview of the Process
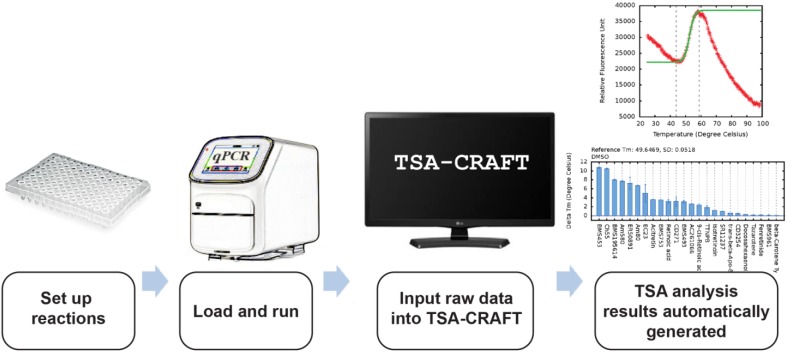
TSA-CRAFT is designed to provide users with a quick, easy, and robust method of TSA data analysis. The entire TSA is performed in three steps ( Fig. 2 ): (1) set up TSA reactions, (2) run melting reactions in a suitable machine, and (3) input data directly into TSA-CRAFT. TSA-CRAFT automatically determines the Tm of each reaction as well as ΔTm, if the appropriate annotation is supplied.
Figure 2.
Overview of the process. TSA-CRAFT is designed to provide a highly user-friendly thermal shift assay analysis tool such that users can input qPCR result files directly into the tool to obtain fast, automatic, and accurate Tm or ΔTm results with minimal effort.
Experimental Setup
A TSA reaction typically consists of a protein and a dye (e.g., SYPRO Orange) in an aqueous environment containing pH buffer and salt. Depending on the purpose of the experiment, additional components are often added, such as ligands and additives. Here, we provide an example of a ligand-binding experiment to screen recombinant RARα ligand-binding domain protein with a panel of 36 retinoid compounds. Alternatively, common reaction setups for identifying protein-stabilizing conditions include screening of chemical additives or varying of pH buffers and salts.9,10 Reactions are set up on ice in qPCR-grade tubes or multiwell plates and loaded into a qPCR machine that measures fluorescence signals over increasing temperatures.
Input Into TSA-CRAFT
Basically, TSA-CRAFT requires only one input file that contains values of fluorescence readings at each temperature. Compatible input file formats are Experiment Document Single files (.eds) generated directly by various ABI systems (e.g., 7500, StepOne, StepOnePlus, ViiA, and QuantStudio series), .cfx files from BioRad CFX systems, and text files (.txt) exported from Roche LightCycler systems. We welcome users to send us samples of exported text files (i.e., .txt or .csv) that are not currently supported so that we can extend the software’s ability to read input files from various instruments.
As an optional feature, users can provide an annotation file for labeling purposes as well as to indicate reference points for automatic ΔTm calculations. Details and examples of input data and annotation files can be found in the TSA-CRAFT user manual.
Results Presentation
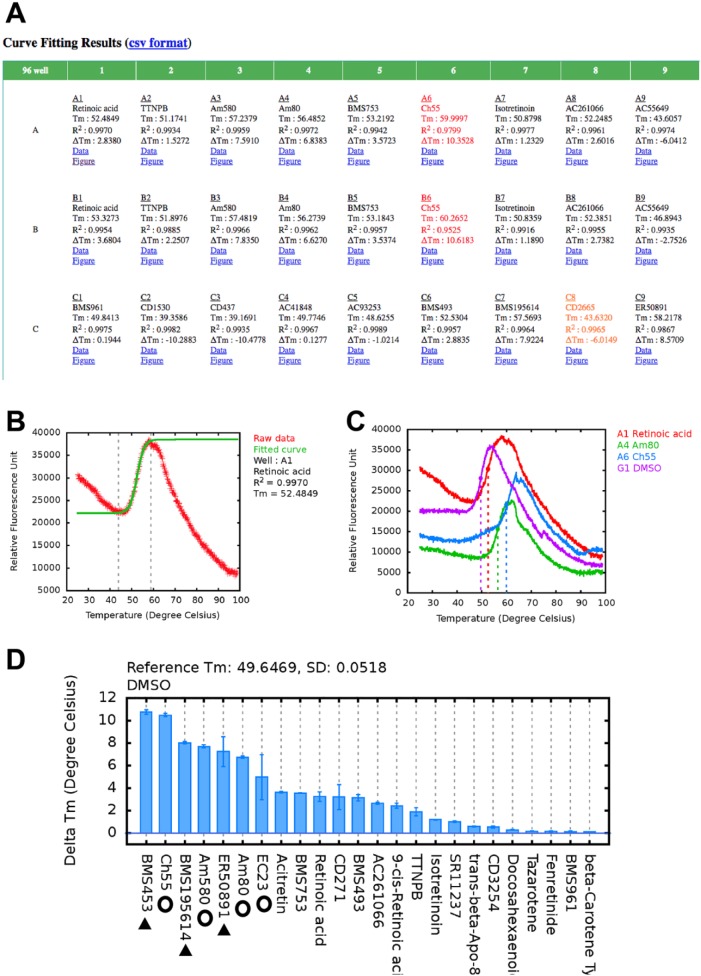
TSA-CRAFT result is presented in HTML format to provide an overview with links to display details of each curve-fitting result individually. Tm values and R2 of curve fitting of each reaction are displayed in a multiwell format ( Fig. 3A ) corresponding to the experimental layout. To flag problematic cases, fonts are colored in red if the R2 values are lower than 0.98 and in orange when the maximal Δf from the transition of the raw data deviate significantly from that of the fitted curve. Graphical data plots are accessible through the “Figure” hyperlink ( Fig. 3B ). For warning cases, users can monitor the curve-fitting quality visually and modify the fitting results by indicating the data range that better reflects the protein thermal transition. There is an option to overlay two or more (up to eight) graphs for visual comparisons ( Fig. 3C ). When annotations are provided, TSA-CRAFT integrates the well annotations and ΔTm results into the overview ( Fig. 3A ) and automatically plots a ΔTm chart ( Fig. 3D ) for easy visual analysis.
Figure 3.
TSA-CRAFT results. (A) Results are displayed in a comprehensive table according to the plate layout. (B) Graphical result and manual curve-fitting option allow monitoring, fine-tuning, and modification of the presented result if necessary. (C) TSA-CRAFT allows overlay of denaturation curves for visual presentation of the Tm shift. (D) ΔTm is calculated according to the given annotation file. Open circles and solid triangles indicate the known RARα agonists and antagonists, respectively, which expectedly generated the highest positive Tm shifts.
Demonstration of Application and Quality of Data Analysis
In this study, we screened a set of retinoid compounds for their binding to RARα ligand-binding domain protein using TSA-CRAFT for automated Tm and ΔTm calculations. RARα is a nuclear receptor that regulates critical physiological and developmental processes. Its dysregulation is linked to developmental defects and diseases such as cancers. Nuclear receptors are ligand-controlled transcription factors that are often exploited as drug targets for targeted therapies; thus, we use RARα ligand screening as a demonstration of TSA application in drug development. We tested 36 compounds and vehicle (DMSO) control in duplicates in a 96-well format for their binding to RARα ligand-binding domain. Melting reactions were performed using a QuantStudio 3 (Applied Biosystems). The output file (.eds file) generated by the instrument was then used directly as input for TSA-CRAFT. In addition, an annotation file was provided that specified well labels and reference points (DMSO control well). TSA-CRAFT automatically generated Tm and ΔTm values for each well. Expectedly, our results showed positive ΔTm with the known agonists ( Fig. 3C , open circles) and antagonists ( Fig. 3C , solid triangles) of RARα, while most of the other retinoid compounds tested, which comprised RARβ/γ- or RXR-specific compounds and carotenoids (RA precursors), showed little or negative Tm shifts.
We compared the performance and the underlying data processing and Tm determination approaches of TSA-CRAFT with three other freely available TSA analysis tools, JTSA,6 DMAN,7 and Meltdown.5 JTSA uses the curve-fitting method, whereas DMAN and Meltdown are based on first-derivative methods. For the comparison with first-derivative methods, we also included the conventional first-derivative method by applying Eq. 1. To establish a standard for the comparison, we performed an exhaustive search to define the standard Tm for each reaction. In this exhaustive search, all possibilities of data fragments were subjected to curve fitting by Eq. 2. The best-fitting results were selected based on Eq. 3, which takes into account how well the raw data are fitted to the curve, as indicated by the coefficient of determination (R2), and the magnitude and integrity of the transition.
| (3) |
Δffrag stands for Δf within the selected fragment. The magnitude of transition for the fitted curve can be evaluated by the ratio between maximal Δffrag and maximal Δf of the entire raw curve (Δfall), whereas the integrity of the transition can be measured by the ratio between Δffrag and maximal Δf of the fitted equation (i.e., fmax – fmin in Eq. 2).
Of the two curve-fitting and three first–derivative–based methods, TSA-CRAFT showed the lowest root-mean-square deviation (RMSD; Table 1 ; Suppl. Table S1). Both DMAN and TSA-CRAFT had no cases with deviation higher than 10 °C, whereas JTSA, the conventional first-derivative method, and Meltdown had 2, 8, and 16 cases of large deviations, respectively. Even after the exclusion of the large deviation cases from the analyses, TSA-CRAFT still showed the lowest RMSD. This comparison shows that Tm analyses using curve-fitting methods tend to provide more accurate Tm determination and fewer deviation cases than first-derivative methods, because regressions by curve-fitting algorithms are less sensitive to local fluctuations in the raw data.
Table 1.
Performance of Thermal Shift Assay Data Analysis in Ligand Screening for RARα.
| Method |
Curve Fitting |
First Derivative |
|||
|---|---|---|---|---|---|
| Tool | TSA-CRAFT | JTSA (MTSA6) | DMAN7 (v.5.1) | Conventional (Eq. 1) | Meltdown5 (v.2.2.0) |
| RMSD (°C)a | 0.26 (74) | 3.54 (67) | 0.33 (74) | 10.30 (74) | 14.80(39) |
| Large deviation (>10 °C) cases | 0 | 2 | 0 | 8 | 16 |
| RMSD (°C)a (without large deviation cases) | 0.26 (74) | 0.66 (65) | 0.33 (74) | 1.68 (66) | 3.41(23) |
Numbers in parentheses attached to root-mean-square deviation (RMSD) values are number of cases used for computing RMSD.
JTSA is a web server application of MTSA that provides the options of either the Boltzmann equation or Sigmoid-5 equation for curve fitting. To be comparable with TSA-CRAFT, the Boltzmann equation option was chosen for this analysis. TSA-CRAFT stood out in the comparison between the curve-fitting methods because of our elaborate design in noise reduction. As such, TSA-CRAFT successfully filtered out the noisy data in the nine challenging cases that confounded the analysis by JTSA (seven misidentified and two large deviation cases).
Of the three first-derivative methods, DMAN stood out with the lowest RMSD and no large deviation cases owing to its noise reduction capabilities and well-designed data processing. In contrast, without such additional designs, the conventional first-derivative method produced eight large deviation cases due to the spike noises in the high-temperature region of the melting curves. After eliminating these large deviation cases, the RMSD was greatly improved from 10.3 °C to 1.68 °C. As for Meltdown, the software has been specially designed for stringent quality control of TSA data. Thus, in this case study, 35 of 74 data curves were considered as monotonic, saturated, or noisy data and were discarded before Tm identification. Meltdown is more appropriate for experiments in which such stringency is required and denaturation curves tend to be relatively perfect, for instance, the screening of varying buffer conditions in experiments that involve a larger number of replicates.
TSA-CRAFT was designed to deal with substantial noise, such as spikes and large gaps ( Fig. 1D, E ), that usually affects automatic Tm analyses. Furthermore, by integrating an elaborate data-processing and curve-fitting algorithm, TSA-CRAFT achieved a promising RMSD of 0.26 °C. As there are many possibilities to how data may deviate from a typical smooth denaturation curve, some situations may still require visual inspection of raw data and manual selection of the most representative protein denaturation curve. In these cases, software such as TSA-CRAFT, JTSA, and DMAN provide a useful graphical interface to perform visual checks and modifications.
In conclusion, we have developed TSA-CRAFT, a useful tool for automatic TSA data analysis that features hassle-free preparation of input file, comprehensive display of Tm results, and easily manual modification. The robustness of TSA-CRAFT is demonstrated in this study by testing compounds for their binding to RARα. The availability of a free, user-friendly, efficient, and reliable data analysis tool will make TSA a more powerful screening platform and benefit the growing drug discovery and protein crystallography communities.
Supplemental Material
Supplemental material, Supplemental_Material_for_TSA_CRAFT_by_Lee_et_al_(2) for TSA-CRAFT: A Free Software for Automatic and Robust Thermal Shift Assay Data Analysis by Po-Hsien Lee, Xi Xiao Huang, Bin Tean Teh and Ley-Moy Ng in SLAS Discovery
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work is funded by National Medical Research Council (NMRC OFIRG16may055) and supported by the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative.
Supplemental material is available online with this article.
References
- 1. Niesen F. H., Berglund H., Vedadi M. The Use of Differential Scanning Fluorimetry to Detect Ligand Interactions That Promote Protein Stability. Nat. Protoc. 2007, 2, 2212–2221. [DOI] [PubMed] [Google Scholar]
- 2. Semisotnov G. V., Rodionova N. A., Razgulyaev O. I., et al. Study of the “Molten Globule” Intermediate State in Protein Folding by a Hydrophobic Fluorescent Probe. Biopolymers 1991, 31, 119–128. [DOI] [PubMed] [Google Scholar]
- 3. Simeonov A. Recent Developments in the Use of Differential Scanning Fluorometry in Protein and Small Molecule Discovery and Characterization. Expert Opin. Drug Discov. 2013, 8, 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Phillips K., de la, Pena A. H. The Combined Use of the Thermofluor Assay and ThermoQ Analytical Software for the Determination of Protein Stability and Buffer Optimization as an Aid in Protein Crystallization. Curr. Protoc. Mol. Biol. 2011, 10, 28. [DOI] [PubMed] [Google Scholar]
- 5. Rosa N., Ristic M., Seabrook S. A., et al. Meltdown: A Tool to Help in the Interpretation of Thermal Melt Curves Acquired by Differential Scanning Fluorimetry. J. Biomol. Screen. 2015, 20, 898–905. [DOI] [PubMed] [Google Scholar]
- 6. Schulz M. N., Landstrom J., Hubbard R. E. MTSA—A Matlab Program to Fit Thermal Shift Data. Anal. Biochem. 2013, 433, 43–47. [DOI] [PubMed] [Google Scholar]
- 7. Wang C. K., Weeratunga S. K., Pacheco C. M., et al. DMAN: A Java Tool for Analysis of Multi-Well Differential Scanning Fluorimetry Experiments. Bioinformatics 2012, 28, 439–440. [DOI] [PubMed] [Google Scholar]
- 8. Nahoum V., Perez E., Germain P., et al. Modulators of the Structural Dynamics of the Retinoid X Receptor to Reveal Receptor Function. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 17323–17328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huynh K., Partch C. L. Analysis of Protein Stability and Ligand Interactions by Thermal Shift Assay. Curr. Protoc. Protein Sci. 2015, 79, 28.9.1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reinhard L., Mayerhofer H., Geerlof A., et al. Optimization of Protein Buffer Cocktails Using Thermofluor. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_Material_for_TSA_CRAFT_by_Lee_et_al_(2) for TSA-CRAFT: A Free Software for Automatic and Robust Thermal Shift Assay Data Analysis by Po-Hsien Lee, Xi Xiao Huang, Bin Tean Teh and Ley-Moy Ng in SLAS Discovery